Abstract
Hepatitis C virus (HCV) is a pandemic disease affecting an estimated 180 million individuals worldwide and infecting each year another ~3-4 million people making HCV a global public health issue. HCV is the main cause for chronic hepatitis, cirrhosis, and hepatocellular carcinoma. In the United States, HCV-related chronic liver disease is a leading cause of liver transplantation. Despite significant improvements in antiviral drugs, only ~50% of treated patients with HCV have viral clearance after treatment. Showing unique species specificity, HCV has a narrow range of potential hosts infecting only chimpanzees and humans. For decades, the chimpanzee model has been the only and instrumental primate for studying HCV infection; however, availability, economic, and ethical issues make the chimpanzee an unsuitable animal model today. Thus, significant research has been devoted to explore different models that are suitable in studying the biology of the virus and application in the clinical research for developing efficient and tolerable treatments for patients. This review focuses on experimental models that have been developed to date and their findings related to HCV.
1. Introduction
Hepatitis C virus (HCV) is a small positive sense single-stranded RNA virus that causes acute and chronic hepatitis C in humans [1, 2]. HCV is one of the major causative agents of liver disease worldwide, with more than 180 million people infected [2, 3]. It is estimated that 3-4 million people are newly infected each year [2]. In the United States, hepatitis-C-related chronic liver disease is a leading cause of liver transplantation and causes thousands of deaths annually [4]. Although therapeutic options are improving, viral clearance fails in about 80% of infected patients, resulting in a chronic viral disease [5]. In 4–20% of patients with chronic hepatitis C, liver cirrhosis develops within 20 years, with 1–5% of these patients developing hepatocellular carcinoma (HCC). Persistent HCV infections are facilitated by the ability of virus to incorporate adaptive mutations in the host and exist as genetically distinct quasispecies. Moreover, the persistent infection may also result from the ability of the virus to disrupt host defense by blocking phosphorylation and function of interferon regulatory factor-3 (IRF-3), an antiviral signaling molecule [6]. Unlike for hepatitis A and B, there is no vaccine to prevent HCV infection; therefore, current treatment is a combination therapy of pegylated interferon-alpha (IFN-α) and ribavirin, which results in sustained clearance of serum HCV-RNA. However, this treatment is only efficacious in approximately 50% of patients [7–9]. Several host factors, such as age, stage of liver fibrosis, body mass index (BMI), liver steatosis, insulin resistance, ethnicity, and IL28B single nucleotide polymorphisms, as well as viral genotype, can potentially influence the treatment outcome [10, 11]. For instance, patients with HCV genotypes 2 and 3 respond more favorably to treatment than patients with genotype 1 and 4 [12]. Therefore, new antiviral compounds that are more efficacious and better-tolerated need to be developed.
One of the biggest challenges in developing and implementing therapy for HCV infection is finding the appropriate models to examine the translational capability. The focus of this review emphasizes the biological importance of the virus and discusses a number of relevant in vitro and in vivo small animal models that are used for preclinical evaluations prior to translating to clinical trials in humans.
2. HCV and Its Life Cycle
HCV was originally referred to as non-A non-B Hepatitis (NANB). In 1989, a major breakthrough in HCV research was discovered in which the complete sequence of the viral genome was identified and cloned by Choo and collaborators [13]. HCV is the only member of the Hepacivirus genus that belongs to the Flaviviridae family [14, 15]. Structural analysis of the virus revealed that the genetic material is surrounded by a protective nucleocapside, composed mainly of the protein core (C), and further protected by a lipid envelope [16]. The lipid envelope contains two major glycoproteins, envelope protein 1 (E1) and E2, that are embedded in the envelope [17]. The genome consists of a single open-reading frame (ORF), that is, ~9,600 nucleotides long, which is made into a single polyprotein (3,010 or 3,033 amino acids) product (Figure 1) [18, 19]. The HCV genome is flanked by two nontranslated regions (NTRs), which are essential in the replication and synthesis of viral proteins. Viral and cellular proteases mediate the processing of the polyprotein into structural (core, E1, E2, and p7) and nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) as illustrated in Figure 1 [20–22]. The HCV life cycle is entirely cytoplasmic and replication occurs mainly in hepatocytes, but the virus may also replicate in peripheral blood mononuclear cells (PBMCs). The virus enters the host cells through a complex interaction between virions and cell surface molecules CD81, LDL receptor, scavenger receptor class B type 1 (SR-B1), Claudin-1, and Occludin [23–26]. Recent studies by Ray et al. and Sainz et al. have identified Niemann-Pick C1-like 1 (NPC1L1) cholesterol absorption receptor as a new HCV entry factor [27, 28]. Once inside the cell, the virus takes over the intracellular machinery to replicate [29]. Due to its high-mutation rate caused by the virus' RNA-dependent RNA polymerase (NS5B), which lacks 3′-5′ exonuclease activity [30], HCV is considered a quasispecies composed of 6 genotypes with several subtypes [12]. The eleven genotypes have differences in geographic distribution, disease progression, and response to therapy. Genotypes 1, 2, and 3 are distributed worldwide, with genotypes 1a and 1b accounting for 60% of global infections. In the United States, genotypes 1a and 2b are more commonly encountered.
Figure 1.
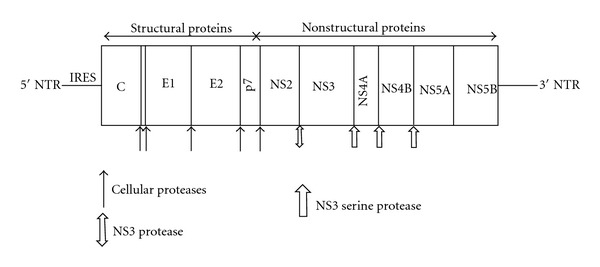
HCV genome and polyprotein cleavage products. A schematic representation of the HCV genome indicating the structural and nonstructural regions, including the 5′ and 3′ NTRs. The polyprotein cleavage products are drawn within. The cleavage site and the corresponding protease are indicated (arrows).
3. Development of Animal Models Used to Study HCV
HCV infects only humans and chimpanzees. Although the virus was discovered more than 20 years ago using molecular biological methods and the entire genome of the virus sequenced, acquiring further knowledge of the virus has been hampered by the lack of a small animal model. Much of the understanding of HCV replication has been based on subgenomic and genomic replicon systems developed by Bartenschlager and colleagues, described below [31]. Viruses are obligate intracellular parasites that require a permissive host cell in order to study replication. Therefore, development of a suitable small animal is critical in understanding the pathogenesis of the virus, establishing a relevant translational platform for therapeutic methods, and developing effective vaccines. The following describes various models used to study the pathogenicity of HCV and its applicable progression into in vivo models, which are summarized in Table 1.
Table 1.
In vitro and in vivo models to study HCV.
| In Vitro | In Vivo |
|---|---|
| Human fetal Hepatocytes | Chimpanzee |
| Chimpanzee Hepatocytes | Tree Shrew (Tupaias sp.) |
| Human Hepatocytes | Chimeric mouse model |
| Peripheral Blood Mononuclear Cells (PBMCs) | Humanized mouse model |
| HepG2 | |
| HuH-7 | |
| Li23 | |
| HEp3B | |
| PH5CH | |
| MOLT-4 | |
| MT-2 | |
| B-Cell Daudi |
4. Cell-Based or In Vitro Models
Viral infection and propagation requires specific host factors that are mainly expressed in highly differentiated cells. To mimic the host factors in an in vitro system, development of a cell-based model is essential. A number of cell-based models have been established; however, most of them have yielded limited success. Poor reproducibility and low levels of HCV replication mainly contribute to the shortfall of these models. Furthermore, highly sensitive techniques are needed for transcript and protein detection. Strand-specific real-time-polymerase chain reaction (rt-PCR) was used to detect minus-strand RNA intermediates during HCV replication; however, due to false priming, this technique is not reliable. As a result, several other genetic and biological indicators are refined and employed to show viral replication such as detection of plus-strand RNA, inhibition of viral replication using IFN-α or antisense oligonucleotides, transmission of cell culture grown HCV to naïve cells, detection of viral antigens by immunofluorescences, and the long-term propagation of HCV [22].
4.1. Primary Cell Lines
Primary cell lines obtained from humans and chimpanzees have been used to study HCV infection. Cultivation of HCV in tissue culture was achieved by Iacovacci et al. in which primary fetal human hepatocytes were injected with sera isolated from patients with HCV. Although, these studies demonstrated an increase in copy number of the minus-strand RNA [32, 33], the total efficiency after 24 days was low, expressing a maximum of 20,000 copies of RNA in 106 cells. Following a similar strategy as used by Iacovacci, Lanford et al. demonstrated a rapid increase in positive-strand RNA from days 1 to 4 and sustained constant levels of transcripts using primary hepatocytes from chimpanzees [34]. Using strand-specific rt-PCR, the authors detected minus-strand RNA replication intermediates, which indicate that the virus is undergoing replication within the hepatocytes. In addition, they showed that primary liver cells obtained from baboons could not be used to cultivate the virus. This observation supports the concept that HCV is quite species selective and has a narrow range of hosts. In 1999, Rumin et al. developed specific tissue culture conditions that could support the culturing of primary human hepatocytes for 4 months, without any morphogenic changes [35]. Although they were able to detect increasing levels of RNA during the 3 months of culturing, the efficiency had many uncontrollable parameters such as the infectivity of the sera and the quality of the hepatocytes. In addition to the potential to infect hepatocytes, HCV has also been shown to replicate in PBMCs, indicating its ability to replicate in extrahepatic cells [35]. Consistent with this observation, HCV has been reported to replicate within PBMCs isolated from chronically infected patients. Cribier et al. reported detection of viral RNA 28 days after infecting a mixture of white blood cells (obtained from 10 donors) that were infected in vitro with high-titer serum [36]. However, the levels and quality of RNA were similar to those reported in hepatocytes.
4.2. Nonprimary Cell Lines
The most critical shortfalls in culturing primary cell lines have been the availability and the technical challenges associated with culturing these cells in vitro. As a result, tremendous efforts have been made in developing a nonprimary cell line that is able to mimic physiological hepatocytes. Although a number of cell lines have been tested to show persistent replication, the most extensive studies have been conducted with the non-neoplastic cell line PH5CH, which was obtained from a human hepatocyte immortalized with the simian virus 40 large T antigen. Two important studies by Ikeda et al. and Kato et al. have shown that HCV plus-strand RNA can be detected more than 100 days after infection with HCV [37, 38]. In addition, they determined that only certain variants, variants in the hypervariable region 1 (HVR1) of the E2 protein, could infect and replicate in PH5CH cells. More importantly, these two groups demonstrated that infected PH5CH cells treated with IFN-α showed significant loss of plus-strand RNA of the virus; thereby, it can serve as an ideal in vitro platform to examine potential therapeutic molecules.
In addition to nonneoplastic cell lines, human B- and T-cell lines have been used as in vitro model to study HCV infection. Mizutani et al. using the T-cell line MT2 isolated a clone containing HCV RNA after 200 days postinfection [39]. Moreover, T- and B-cell lines; HPB-Ma and Daudi, respectively, have been shown to sustain virus propagation lasting for more than one year [40]. A study by Shimizu et al. has demonstrated that supernatant from HCV infected Daudi cells exhibited remarkable infectious capacity in chimpanzee [41]. HCV RNA was detected in the chimpanzee serum after 5 weeks postinfection, however, the levels of HCV replication in the infected animal were low and gradually disappeared after 25 weeks postinfection.
5. Transfected Cell Lines
5.1. Cloned HCV Genomes
Generation of clones from the HCV genome has permitted the genetic analysis of a variety of different aspects in the HCV life cycle. Introduction of cloned virus genome is superior to infection using HCV infected patient serum because the clone is well defined and can be generated in high quantities. Studies by Dash et al. and Yoo et al. have shown that transfecting HepG2 and HuH-7, respectively, with truncated HCV genomes that lacked the 3′ NTR were able to maintain extended culture infected with HCV [42, 43]. However, the usage of this truncated genome contradicts the finding that the 3′ NTR is essential for replicating in vivo [44, 45]. In addition, this method is not useful in infecting chimpanzee with HCV producing cells, unlike other transfected cell methods.
5.2. Subgenomic Replicons
In 1999, Bartenschlager et al. developed a system that consisted of subgenomic replicons of HCV that could replicate autonomously in hepatic cell cultures [31, 46]. To obtain the subgenomic replicons, they isolated sera from patients infected with HCV and purified the viral genome. Complementary DNA was synthesized and amplified; therefore, the final clones selected contained the complete genome including the 5′ and 3′ NTR, which was stably expressed in the pCR2.1 expression vector containing a T7 promoter. The final replicon contained a gene resistant to neomycin and the region encoding the structural proteins was eliminated. The structural proteins were removed from this replicon because it was previously observed that for several plus-strand RNA viruses did not require the structural proteins for RNA replication [47–49]. This replicon was able to replicate itself within the cell; however, it was not capable of producing infectious viruses. In addition, this replicon was able to reproduce in HuH-7 cells with high efficiency and for an extended period of time [50]. Studies have also been conducted to determine if certain mutations improve replication. It was determined that mutations in the NS5a region and in the NS4b region increase replication more than 1,000 times [51, 52].
Subgenomes of other genotypes of HCV have been shown to be efficient in maintaining high infectious potential and long-term in vitro culture, for example, genotype 2a clone isolated from a Japanese patient with a rare case of fulminant hepatitis C, designated as JFH-1 (Japanese fulminant hepatitis C). The data have indicated the isolate could replicate in HuH-7 cells without the requirements for adaptive mutations that were required for previous isolates [53]. HuH-7 cells infected with cloned JFH-1 genomes produced viruses (designated as HCVcc for cell culture derived HCV) that were capable of infecting naïve HuH-7 cells [54]. In addition, the virus particles could be neutralized with a monoclonal antibody against the viral glycoprotein E2 [55]. The study was the first in vitro experiment that showed the complete lifecycle of HCV. More importantly, virus obtained from the cell culture was highly infectious in vivo by readily infecting chimpanzees [54, 56] as well as immunodeficient mice with partial human livers (chimeric mice) described later [56].
In addition to HuH-7 cells supporting propagation of HCVcc, additional cell lines have been developed. Human hepatoma Li23-derived cells were found to possess the necessary components required for HCV RNA replication and persistent production of infectious HCV [57]. Similar expression levels of HCV entry factors were observed between Li23- and HuH-7-derived cells, suggesting that certain factors are necessary for infectivity and propagation of HCVcc. Recent studies have shown that expression of a liver-specific microRNA, miR-122, in HEp3B cells can propagate HCVcc [58]. A lentiviral vector expressing miR-122 was placed in HEp3B cells at comparable levels with HuH-7 cells, which lead to the production of infectious particles. The levels were comparable to those observed in HuH-7-infected cells. Shimakami et al. have shown that miR-122 forms a complex with Ago2 to protect and stabilize HCV RNA from 5′ exonuclease activity of the host mRNA decay machinery [59]. This finding may explain why expression of miR-122 in HEp3B cells can support HCV propagation.
6. Animal Model
6.1. Chimpanzees
The chimpanzee animal model (Pan troglodytes) is currently the only established animal or primate model for HCV infection. The chimpanzee model for HCV infection was instrumental in the initial studies of non-A, non-B hepatitis, including observations on the clinical course of infection, determination of the physical properties of the virus, and eventual cloning of the HCV nucleic acid [60]. The chimpanzee model has been invaluable in demonstrating that the cDNA clones of HCV developed from HCV strains of genotypes 1a, 1b, and 2a were infectious. In addition, using this primate model that is evolutionarily close and associated with humans has provided important insight regarding the etiology of liver disease caused by HCV. The chimpanzee has provided evidence that infection with HCV did not provide complete protective immunity when challenged with homologous or heterologous viral strains [61, 62]. As a result, it has been difficult to design effective vaccines against the virus, even though chimpanzees have allowed for the identification of important viral genetic elements. Several studies have identified the importance of the active sites of various enzymatic functions as well as the role of the p7 protein. Thus, chimpanzees are certainly one of the ideal models to study the pathogenesis of the HCV and serve as a great model for translation research; however, it is quite a challenge to work with this model due to limited availability, the costs associated with acquiring and maintaining the animal for scientific research, and public resistance.
6.2. Tupaias
The tree shrew, Tupaias (T. belangeri chinensis), was previously shown to be susceptible to the hepatitis B virus [63, 64]. Xie et al. have demonstrated that Tupaia were susceptible to HCV infection [65]. However, persistent HCV infection could not be established and only 25% of infected animals developed transient or intermittent viremia. In addition, Tupaia must undergo severe immunosuppression before they can be infected with HCV. However, sera or plasmas obtained from patients with HCV were able to infect primary Tupaia hepatocyte cultures [66]. The authors demonstrated that the hepatocytes could produce infectious viruses that were capable of infecting naïve hepatocytes. Sequence analysis of cloned Tupaia cDNA revealed a high degree of homology between Tupaia and human CD81 large extracellular loops (LELs), suggesting CD81 aids in viral entry. In addition, the study indicated that viral entry can also occur through receptors other than CD81 since cellular binding of E2 and anti-CD81 antibodies or soluble CD81-LEL could not inhibit HCV infection.
6.3. Chimeric Human Liver Mouse Model
By genetically manipulating the uPA transgenic mouse, Mercer et al. have generated a chimeric mouse model with human hepatocytes. This was done by transplanting normal human hepatocytes into severe combined immunodeficiency (SCID) mice carrying a plasminogen activator transgene [67]. The human hepatocytes transplanted were able to integrate into the parenchyma and repopulate the diseased mouse liver without losing their metabolic functions. Successfully generated chimeric mice exhibited prolonged infection with high viral titers following inoculation with HCV and HBV isolated from human serum. In addition, the authors have demonstrated that the model was able to exhibit horizontal transmission in which HCV can be transmitted from one infected animal to another. Since the mice were immunodeficient, they were not appropriate models to study HCV pathogenesis, although they were useful in assessing the activity of antiviral compounds, specifically the effect of IFN-α, HCV protease inhibitors and cyclophilin inhibitor DEBIO-025 on virus propagation and infectivity [68]. Moreover, this chimeric mouse model has been used to show that neutralizing antibodies can prevent an HCV infection in vivo [69].
6.4. Genetically Humanized Mouse Model
Studies by Ploss et al. have previously shown that CD81 and occludin (OCLN) were the minimum human factors required for HCV uptake by rodent cells [70]. To determine these human factors for efficient HCV infection, the authors constructed recombinant adenovirus expressing human CD81, scavenger receptor type B class 1 (SCARB1), claudin 1 (CLDN1), and OCLN. Inoculating mice with these human factors expressed in adenoviral vector was sufficient for HCV infection [71]. Furthermore, the authors employed a bicistronic HCV genome expressing CRE recombinase (Bi-nlsCre-Jc1FLAG2, abbreviated HCV-CRE), which activates a loxP-flanked luciferase reporter in Rosa26-Fluc mice. The mice were infected with the HCV-CRE and the mice that were expressing all four transgenes had luciferase signal that peaked at 72 hours postinfection. Interestingly, only animals that expressed both human CD81 and OCLN displayed 6–10 fold increase in virus infection. In addition, they were also able to show that SCARB1 is a legitimate HCV entry factor. This system allows for the studies of HCV coreceptor biology in vivo and evaluation of passive immunization strategies. This, therefore, represents the first immunocompetent small animal model for HCV study.
7. Conclusions
Although, great strides in small animal models for the study of HCV have been made in the past decades, HCV remains a global public health issue. In past years, advancements have been made in vitro and in vivo models for the study of hepatitis C, in particular, the chimeric and genetically humanized mouse models, which have provided a platform to study new antiviral treatments and evaluate immunization strategies. With the advent of autoantibodies recognizing structural proteins found in HCV patients, both in vitro and in vivo models could provide an important foundation for the discovery of novel biomarkers for prognosis and treatment [72, 73]. With new insights on HCV biology obtained from in vitro models and the ability to infect animals with active immune systems, it should be possible to develop new therapies and possibly a vaccine for HCV.
Acknowledgments
This work was supported by the Sjögren's Syndrome Foundation, PHS Grant K99/R00 DE018958, grant from the NIDCR (CQN). W. C. Carcamo is supported by Bridges to Doctorate, University of Florida Graduate School and University of Florida Alumni Graduate.
References
- 1.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36(5, supplement 1):S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 2.National Institutes of Health Consensus Development Conference Statement: management of hepatitis C 2002 (June 10–12, 2002) Gastroenterology. 2002;123(6):2082–2099. doi: 10.1053/gast.2002.1232082. [DOI] [PubMed] [Google Scholar]
- 3.Alter HJ, Seeff LB. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Seminars in Liver Disease. 2000;20(1):17–35. doi: 10.1055/s-2000-9505. [DOI] [PubMed] [Google Scholar]
- 4.Charlton M. Hepatitis C infection in liver transplantation. American Journal of Transplantation. 2001;1(3):197–203. doi: 10.1034/j.1600-6143.2001.001003197.x. [DOI] [PubMed] [Google Scholar]
- 5.Seeff LB. Natural history of hepatitis C. Hepatology. 1997;26(3, supplement 1):21S–28S. doi: 10.1002/hep.510260704. [DOI] [PubMed] [Google Scholar]
- 6.Foy E, Li K, Wang C, et al. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300(5622):1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 7.Fried MW, Shiffman ML, Rajender Reddy K, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. New England Journal of Medicine. 2002;347(13):975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 8.Hadziyannis SJ, Sette H, Morgan TR, et al. Peginterferon-α2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Annals of Internal Medicine. 2004;140(5):346–I67. doi: 10.7326/0003-4819-140-5-200403020-00010. [DOI] [PubMed] [Google Scholar]
- 9.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferonalfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358(9286):958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 10.Kau A, Vermehren J, Sarrazin C. Treatment predictors of a sustained virologic response in hepatitis B and C. Journal of Hepatology. 2008;49(4):634–651. doi: 10.1016/j.jhep.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Smith KR, Suppiah V, O'Connor K, et al. Identification of improved IL28B SNPs and haplotypes for prediction of drug response in treatment of hepatitis C using massively parallel sequencing in a cross-sectional European cohort. Genome Medicine. 2011;3(8):p. 57. doi: 10.1186/gm273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42(4):962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 13.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 14.Lauer GM, Walker BD. Hepatitis C virus infection. New England Journal of Medicine. 2001;345(1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 15.Dustin LB, Rice CM. Flying under the radar: the immunobiology of hepatitis C. Annual Review of Immunology. 2007;25:71–99. doi: 10.1146/annurev.immunol.25.022106.141602. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg S. Recent advances in the molecular biology of Hepatitis C virus. Journal of Molecular Biology. 2001;313(3):451–464. doi: 10.1006/jmbi.2001.5055. [DOI] [PubMed] [Google Scholar]
- 17.De Beeck AO, Dubuisson J. Topology of hepatitis C virus envelope glycoproteins. Reviews in Medical Virology. 2003;13(4):233–241. doi: 10.1002/rmv.391. [DOI] [PubMed] [Google Scholar]
- 18.Kato N. Genome of human hepatitis C virus (HCV): gene organization, sequence diversity, and variation. Microbial and Comparative Genomics. 2000;5(3):129–151. doi: 10.1089/omi.1.2000.5.129. [DOI] [PubMed] [Google Scholar]
- 19.Ohba KI, Mizokami M, Lau JYN, Orito E, Ikeo K, Gojobori T. Evolutionary relationship of hepatitis C, pesti-, flavi-, plantviruses, and newly discovered GB hepatitis agents. FEBS Letters. 1996;378(3):232–234. doi: 10.1016/0014-5793(95)01441-1. [DOI] [PubMed] [Google Scholar]
- 20.Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39(1):5–19. doi: 10.1002/hep.20032. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann V, Koch JO, Bartenschlager R. Processing pathways of the hepatitis C virus proteins. Journal of Hepatology, Supplement. 1996;24(supplement 2):11–19. [PubMed] [Google Scholar]
- 22.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. Journal of General Virology. 2000;81(7):1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 23.Evans MJ, Von Hahn T, Tscherne DM, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446(7137):801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 24.Bartosch B, Cosset FL. Cell entry of hepatitis C virus. Virology. 2006;348(1):1–12. doi: 10.1016/j.virol.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 25.Cocquerel L, Voisset C, Dubuisson J. Hepatitis C virus entry: potential receptors and their biological functions. Journal of General Virology. 2006;87(5):1075–1084. doi: 10.1099/vir.0.81646-0. [DOI] [PubMed] [Google Scholar]
- 26.Kohaar I, Ploss A, Korol E, et al. Splicing diversity of the human OCLN gene and its biological significance for hepatitis C virus entry. Journal of Virology. 2010;84(14):6987–6994. doi: 10.1128/JVI.00196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray K. Hepatitis: NPC1L1 identified as a novel HCV entry factor. Nature Reviews Gastroenterology and Hepatology. 124;9(3) doi: 10.1038/nrgastro.2012.4. [DOI] [PubMed] [Google Scholar]
- 28.Sainz B, Jr, Barretto N, Martin DN, et al. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nature Medicine. 2012;18(2):281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436(7053):933–938. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- 30.Pavio N, Lai MMC. The hepatitis C virus persistence: how to evade the immune system? Journal of Biosciences. 2003;28(3):287–304. doi: 10.1007/BF02970148. [DOI] [PubMed] [Google Scholar]
- 31.Bartenschlager R. Hepatitis C virus replicons: potential role for drug development. Nature Reviews Drug Discovery. 2002;1(11):911–916. doi: 10.1038/nrd942. [DOI] [PubMed] [Google Scholar]
- 32.Iacovacci S, Manzin A, Barca S, et al. Molecular characterization and dynamics of hepatitis C virus replication in human fetal hepatocytes infected in vitro. Hepatology. 1997;26(5):1328–1337. doi: 10.1002/hep.510260535. [DOI] [PubMed] [Google Scholar]
- 33.Iacovacci S, Sargiacomo M, Parolini I, Ponzetto A, Peschle C, Carloni G. Replication and multiplication of hepatitis C virus genome in human foetal liver cells. Research in Virology. 1993;144(4):275–279. doi: 10.1016/s0923-2516(06)80040-3. [DOI] [PubMed] [Google Scholar]
- 34.Lanford RE, Sureau C, Jacob JR, White R, Fuerst TR. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202(2):606–614. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 35.Rumin S, Berthillon P, Tanaka E, et al. Dynamic analysis of hepatitis C virus replication and quasispecies selection in long-term cultures of adult human hepatocytes infected in vitro. Journal of General Virology. 1999;80(11):3007–3018. doi: 10.1099/0022-1317-80-11-3007. [DOI] [PubMed] [Google Scholar]
- 36.Cribier B, Schmitt C, Bingen A, Kirn A, Keller F. In vitro infection of peripheral blood mononuclear cells by hepatitis C virus. Journal of General Virology. 1995;76(10):2485–2491. doi: 10.1099/0022-1317-76-10-2485. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda M, Sugiyama K, Mizutani T, et al. Human hepatocyte clonal cell lines that support persistent replication of hepatitis C virus. Virus Research. 1998;56(2):157–167. doi: 10.1016/s0168-1702(98)00063-x. [DOI] [PubMed] [Google Scholar]
- 38.Kato N, Ikeda M, Mizutani T, et al. Replication of hepatitis C virus in cultured non-neoplastic human hepatocytes. Japanese Journal of Cancer Research. 1996;87(8):787–792. doi: 10.1111/j.1349-7006.1996.tb02101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizutani T, Kato N, Ikeda M, Sugiyama K, Shimotohno K. Long-term human T-cell culture system supporting hepatitis C virus replication. Biochemical and Biophysical Research Communications. 1996;227(3):822–826. doi: 10.1006/bbrc.1996.1591. [DOI] [PubMed] [Google Scholar]
- 40.Nakajima N, Hijikata M, Yoshikura H, Shimizu YK. Characterization of long-term cultures of hepatitis C virus. Journal of Virology. 1996;70(5):3325–3329. doi: 10.1128/jvi.70.5.3325-3329.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu YK, Igarashi H, Kiyohara T, et al. Infection of a chimpanzee with hepatitis C virus grown in cell culture. Journal of General Virology. 1998;79(6):1383–1386. doi: 10.1099/0022-1317-79-6-1383. [DOI] [PubMed] [Google Scholar]
- 42.Yoo BJ, Selby MJ, Choe J, et al. Transfection of a differentiated human hepatoma cell line (Huh7) with in vitro-transcribed hepatitis C virus (HCV) RNA and establishment of a long- term culture persistently infected with HCV. Journal of Virology. 1995;69(1):32–38. doi: 10.1128/jvi.69.1.32-38.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dash S, Halim AB, Tsuji H, Hiramatsu N, Gerber MA. Transfection of HepG2 cells with infectious hepatitis C virus genome. American Journal of Pathology. 1997;151(2):363–373. [PMC free article] [PubMed] [Google Scholar]
- 44.Yanagi M, Claire MS, Emerson SU, Purcell RH, Bukh J. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(5):2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3’ nontranslated region are essential for virus replication in vivo. Journal of Virology. 2000;74(4):2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohmann V, Körner F, Koch JO, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285(5424):110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 47.Behrens SE, Grassmann CW, Thiel HJ, Meyers G, Tautz N. Characterization of an autonomous subgenomic pestivirus RNA replicon. Journal of Virology. 1998;72(3):2364–2372. doi: 10.1128/jvi.72.3.2364-2372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9(12):1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 49.Khromykh AA, Westaway EG. Subgenomic replicons of the flavivirus Kunjin: construction and applications. Journal of Virology. 1997;71(2):1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. Journal of Virology. 2001;75(3):1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krieger N, Lohmann V, Bartenschlager R. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. Journal of Virology. 2001;75(10):4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lohmann V, Körner F, Dobierzewska A, Bartenschlager R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. Journal of Virology. 2001;75(3):1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato T, Date T, Miyamoto M, et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125(6):1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 54.Wakita T, Pietschmann T, Kato T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature Medicine. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindenbach BD, Evans MJ, Syder AJ, et al. Virology: complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 56.Lindenbach BD, Meuleman P, Ploss A, et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(10):3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kato N, Mori K, Abe KI, et al. Efficient replication systems for hepatitis C virus using a new human hepatoma cell line. Virus Research. 2009;146(1-2):41–50. doi: 10.1016/j.virusres.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 58.Kambara H, Fukuhara T, Shiokawa M, et al. Establishment of a novel permissive cell line for the propagation of hepatitis C virus by expression of microRNA miR122. Journal of Virology. 2012;86(3):1382–1393. doi: 10.1128/JVI.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimakami T, Yamane D, Jangra RK, et al. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(3):941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39(6):1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 61.Farci P, Alter HJ, Govindarajan S, et al. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258(5079):135–140. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 62.Prince AM, Brotman B, Huima T, Pascual D, Jaffery M, Inchauspe G. Immunity in hepatitis C infection. Journal of Infectious Diseases. 1992;165(3):438–443. doi: 10.1093/infdis/165.3.438. [DOI] [PubMed] [Google Scholar]
- 63.Yan RQ, Su JJ, Huang DR, Gan YC, Yang C, Huang GH. Human hepatitis B virus and hepatocellular carcinoma II. Experimental induction of hepatocellular carcinoma in tree shrews exposed to hepatitis B virus and aflatoxin B1. Journal of Cancer Research and Clinical Oncology. 1996;122(5):289–295. doi: 10.1007/BF01261405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walter E, Keist R, Niederöst B, Pult I, Blum HE. Hepatitis B virus infection of tupaia hepatocytes in vitro and in vivo. Hepatology. 1996;24(1):1–5. doi: 10.1002/hep.510240101. [DOI] [PubMed] [Google Scholar]
- 65.Xie ZC, Riezu-Boj JI, Lasarte JJ, et al. Transmission of hepatitis C virus infection to tree shrews. Virology. 1998;244(2):513–520. doi: 10.1006/viro.1998.9127. [DOI] [PubMed] [Google Scholar]
- 66.Zhao X, Tang ZY, Klumpp B, et al. Primary hepatocytes of Tupaia belangeri as a potential model for hepatitis C virus infection. Journal of Clinical Investigation. 2002;109(2):221–232. doi: 10.1172/JCI13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mercer DF, Schiller DE, Elliott JF, et al. Hepatitis C virus replication in mice with chimeric human livers. Nature Medicine. 2001;7(8):927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 68.Meuleman P, Leroux-Roels G. The human liver-uPA-SCID mouse: a model for the evaluation of antiviral compounds against HBV and HCV. Antiviral Research. 2008;80(3):231–238. doi: 10.1016/j.antiviral.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 69.Vanwolleghem T, Bukh J, Meuleman P, et al. Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver-chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology. 2008;47(6):1846–1855. doi: 10.1002/hep.22244. [DOI] [PubMed] [Google Scholar]
- 70.Ploss A, Evans MJ, Gaysinskaya VA, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457(7231):882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dorner M, Horwitz JA, Robbins JB, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474(7350):208–212. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carcamo WC, Satoh M, Kasahara H, et al. Induction of cytoplasmic rods and rings structures by inhibition of the CTP and GTP synthetic pathway in mammalian cells. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0029690. Article ID e29690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Covini G, Carcamo WC, Bredi E, von Muhlen CA, Colombo M, Chan EK. Cytoplasmic rods and rings autoantibodies developed during pegylated interferon and ribavirin therapy in patients with chronic hepatitis C. doi: 10.3851/IMP1993. Antiretroviral Therapy. In press. [DOI] [PubMed] [Google Scholar]


