Abstract
Identification of emerging bacterial pathogens generally results from a chain of events involving microscopy, serology, molecular tools, and culture. Because of the spectacular molecular techniques developed in the last decades, some authors think that these techniques will shortly supplant culture. The key steps that led to the discovery of emerging bacteria have been reviewed to determine the real contribution of each technique. Historically, microscopy has played a major role. Serology provided indirect evidence for causality. Isolation and culture were crucial, as all emerging bacteria have been grown on artificial media or cell lines or at least propagated in animals. With the use of broad-range polymerase chain reaction, some bacteria have been identified or detected in new clinical syndromes. Culture has irreplaceable advantages for studying emerging bacterial diseases, as it allows antigenic studies, antibiotic susceptibility testing, experimental models, and genetic studies to be carried out, and remains the ultimate goal of pathogen identification.
Keywords: emerging bacteria, unrecognized pathogens, bacterial detection, bacterial identification, histologic stain, broad-range PCR, 16S rDNA, culture, animal model, complete nucleotide sequence
In the last 20 years, advances in knowledge have resulted in a broad expansion of the spectrum of microorganisms regarded as human pathogens. Most advances have evolved in a series of small steps based on several techniques that have been used successively by different investigators who faced clinically suspect diseases. These include the traditional techniques of microscopy, serology, and culture, as well as more recent molecular tools (Figure 1). In addition to aiding in discovering new pathogens, these techniques also contributed to studies of the epidemiology, pathophysiology, and treatment response of the newly recognized diseases, providing further evidence for causal relationships between disease and organism (1). As a diagnostic and research laboratory specializing in fastidious, intracellular bacteria, we have been particularly interested in assessing the specific role played by culture in identifying emerging pathogens. Historical examples, such as Lyme or Legionnaires’ diseases, and recent successes, such as culture of the Whipple bacillus, support the effectiveness of this technique (2). Moreover, culture provided the basis of other supplemental tools to elucidate the causes of microbial disease and to study the clinical and biological features of emerging bacterial diseases. These tools are not only antigenic and serologic assays but also in vitro and in vivo disease models for pathophysiologic studies and antimicrobial susceptibility testing, plus extensive genetic sequencing. The isolation of emerging pathogens serves, therefore, not only as a means for diagnosis but also as a route to enhance understanding of the diversity and epidemiology of emerging bacteria and the infections they cause.
Figure 1.
Diagram describing the respective places of culture-, polymerase chain reaction-, serology- and histology-based approaches for the diagnosis of acute bacterial infections, according to the natural course of the disease. Isolation and culture are possible as long as live bacteria are present in tissues, i.e., from the colonization to the treatment or to the end of the clinical manifestations (or shortly earlier). Bacterial DNA can be detected during the same period and also as far as dead microorganisms remain in tissues. Specific antibodies appear during the clinical course of the disease and persist generally for months or years. Pathologic changes can be observed soon after the contamination and, in an acute infection, will decline rapidly after elimination of the bacteria.
Despite these unique advantages, however, culture has been challenged by the recent development of genotype-based methods such as broad-range polymerase chain reaction (PCR) (3). Because culture as a tool is still threatened by the possible existence of uncultivatable organisms, several authors have emphasized the critical role that molecular, culture-independent techniques could play in further investigations of emerging infectious diseases, affirming that a reassessment of Koch’s postulates for disease causation was required (4). What actually are the respective roles of these two techniques? Should we consider that broad-range PCR has made culture and traditional techniques obsolete, or is it only a step among others in the sequence of events leading to isolation of a new microorganism? To answer these questions, we examined the key steps that led to identification of most bacterial diseases that have been discovered during the last 20 years. Table 1 presents the main biological evidence that allowed merging bacteria to be recognized and disease causation to be demonstrated. We examined the contribution of traditional and molecular techniques to understand their respective roles, and we emphasize the specific advantages of culture.
Table 1. Key steps that led to identification and demonstration of disease causation for emerging bacteriaa.
| Group | Species | Clinical picture | Histologic detection | Serology | Molecular Detection (gene) | Culture system | Year of culture | Ref. |
|---|---|---|---|---|---|---|---|---|
| Alpha1 Proteobacteria | ||||||||
| Ehrlichia chaffeensis | Fever, cytopenia | Smear | Antibodies to Ehrlichia canis] | 16S rRNA | Cell line (DH82) | 1991 | 5 | |
| E. ewingii | Fever, cytopenia | Smear | Western blot | 16S rRNA | Cell line | 1971 | 6 | |
| Human granulocytic Ehrlichia | Fever, cytopenia | Smear | Antibodies to E. phagocytophila, E. equis | 16S rRNA | Cell line (HeLa) | 1996 | 7 | |
| Rickettsia felis | Fever | gltA | Cell line (XTC-2) | 2000 | 8 | |||
| R. japonica ] | Spotted fever | Antibodies to Spotted fever group rickettsiae | Cell line (Vero) | 1989 | 9 | |||
| R. mongolotimonae | Febrile rash | Antibodies to Spotted fever group rickettsiae | rOmpA | Embryonated egg, guinea pig | 1991 | 10 | ||
| R. slovaca | Fever, eschar, lymphadenitis | Specific antibodies | rOmpA | Cell line | 1968 | 11 | ||
| Alpha2 Proteobacteriae | ||||||||
| Afipia broomae | Wrist abscess | Axenic (specific) | 1981 | 12 | ||||
| A. clevelandensis | Osteitis | Axenic (specific) | 1988 | 12 | ||||
| Bartonella elizabethae | Endocarditis | Axenic (nonspecific) | 1993 | 13 | ||||
| B. grahamii | Neuro-retinitis | Antibodies to B. henselae | 16S rRNA | Axenic (nonspecific) | 1995 | 14 | ||
| B. henselae | Fever, cat scratch disease, bacillary angiomatosis | Tissue section | Specific antibodies | 16S rRNA | Axenic (nonspecific) | 1990 | 15 , 16 | |
| Beta Proteobacteriae | ||||||||
| Bordetella trematum | Chronic otitis | Axenic (nonspecific) | 1996 | 17 | ||||
| Neisseria weaveri | Infected wound | Axenic (nonspecific) | 1993 | 18 | ||||
| Spirochetae | ||||||||
| Borrelia burgdorferi sensu stricto, B. afzelii, B. garinii | Erythema chronicum migrans, acrodermatitis chronica atrophicans, Lyme arthritis, neuro-borreliosis | Specific antibodies | Axenic (specific) | 1981 | 19 | |||
| B. duttonii | Relapsing fever | Smear | Specific antibodies | Axenic (specific), animal model | 1999 | 20 | ||
| B. recurrentis | Relapsing fever | Smear | Axenic (specific) | 1994 | 21 | |||
| Delta-Xi Proteobacteriae | ||||||||
| Campylobacter coli, C. jejuni | Febrile diarrhea | Specific antibodies | Axenic (nonspecific) | 1977 | 22 | |||
| Helicobacter cinaedi, H. fennelliae | Rectitis | Axenic (nonspecific) | 1984 | 23 | ||||
| H. heilmanii | Chronic gastritis | Tissue section | Mouse | 1989 | 24 | |||
| H. pylori | Gastritis, gastroduodenal ulcer | Tissue section | Specific antibodies | Axenic (nonspecific) | 1982 | 25 | ||
| Gamma Proteobacteriae | ||||||||
| Escherichia coli O48:H21, O103:H2, O157:H7 | Bloody diarrhea, HUS | slt | Axenic (nonspecific) | 1982-1996 | 26 | |||
| Haemophilus influenzae biogroup aegyptius | Brazilian purpuric fever | Axenic (nonspecific) | 1986 | 27 | ||||
| Legionella anisa | Pneumonia, Pontiac fever | Specific antibodies | Axenic (specific) | 1989 | 28 | |||
| L. bozemanii | Pneumonia | Smear | Specific antibodies | Axenic (specific) | 1983 | 29 | ||
| L. dumoffii | Pneumonia | Smear | Specific antibodies | Axenic (specific) | 1978 | 29 | ||
| L. feeleii | Pneumonia, Pontiac fever | Specific antibodies | Axenic (specific) | 1986 | 30 | |||
| L. micdadei | Pneumonia | Specific antibodies | Embryonated egg, guinea pig | 1979 | 29 | |||
| L. oakridgensis | Pneumonia | Smear | Specific antibodies | Axenic (specific) | 1987 | 29 | ||
| L. pneumophila | Pneumonia | Tissue section | Specific antibodies | Embryonated egg, Guinea pig | 1947 | 31 | ||
| Legionella like amoebal pathogen | Pneumonia | Specific antibodies | Amoeba | 1991 | 32 | |||
| Vibrio alginolyticus | Conjunctivitis, wound infection | Axenic (nonspecific) | 1977 | 33 | ||||
| V. cholerae O:139 | Diarrhea | Axenic (nonspecific) | 1992 | 33 | ||||
| V. fluvialis | Diarrhea | Axenic (nonspecific) | 1980 | 33 | ||||
| V. furnissii | Diarrhea | Axenic (nonspecific) | 1983 | 33 | ||||
| V. metschnikovii | Cholecystitis | Axenic (nonspecific) | 1981 | 33 | ||||
| V. mimicus | Diarrhea, otitis | Axenic (nonspecific) | 1981 | 33 | ||||
| Mycobacteria | ||||||||
| Mycobacterium asiaticum | Pneumopathy | Axenic (specific) | 1983 | 34 | ||||
| M. celatum | Pneumopathy | Axenic (specific) | 1992 | 34 | ||||
| M. genavense | Disseminated infection, lymphadenitis | Tissue section | Axenic (specific) | 1992 | 34 | |||
| M. malmoense | Pneumopathy, lymphadenitis | Axenic (specific) | 1977 | 34 | ||||
| M. simiae | Pneumopathy, osteitis, kidney infection | Axenic (specific) | 1984 | 34 | ||||
| Mycoplasmas | ||||||||
| M. fermentans | Pneumopathy, nephritis | Tissue section | Insertion sequence-like | Axenic (specific) | 1993 | 35 | ||
| M. genitalium | Urethritis | Smear | Adhesion protein | Axenic (specific), Animal model | 1981 | 35 | ||
| Gram-positive bacteria | ||||||||
| Tropheryma whipplei | Whipple disease | Tissue section | Specific antibodies | 16S rRNA | Cell line (HEL) | 2000 | 2 , 36 | |
| Corynebacterium auris | Acute otitis | Axenic (nonspecific) | 1995 | 37 | ||||
| Staphylococcus lugdunensis, S. schleiferi | Skin abscess, osteoarthritis | Axenic (nonspecific) | 1988 | 38 | ||||
| Streptococcus iniae | Meningitis, endocarditis, cellulitis | Axenic (nonspecific) | 1995 | 39 | ||||
aHistologic detection can be performed with morphologic techniques, in blood or tissue smears, or in tissue sections. Serologic assays can detect specific antibodies to the suspected agent or to a related organism in tissues or in biological fluids. The year of the first isolation and the culture system used are indicated. HUS = hemolytic uremic syndrome. HLE=human embryonic lung fibroblasts; ref = reference.
Traditional Techniques Other Than Culture: Microscopy and Serology
Optic Microscopy
Direct Detection in Smears
Historically, morphologic methods have played an important role in detecting new microorganisms, and they are still crucial for diagnosing infections caused by agents not routinely cultured, such as Mycobacterium leprae (40). Because microscopic examination of stained smears from biologic fluids or tissue imprints is usually rapid and easy, it has often been performed in patients who have an unexplained disease, although its interpretation is subjective and its sensitivity and specificity are generally low. The first evidence for the responsibility of Ehrlichia species in humans with an acute febrile illness was provided by examining blood smears stained with a Romanowsky stain, in which these as-yet-uncultivated organisms could be observed forming intracytoplasmic morulae within leukocytes (6,7). Borrelia burgdorferi were first observed in Giemsa-stained smears from midgut diverticula of ticks (19). Examination of smears can also be helpful when multiple organisms are cultured from a nonsterile site, as microbial culture alone, as well as molecular detection, cannot distinguish between colonization or asymptomatic shedding and tissue invasion: in such a situation, the morphology of the predominant organism visualized in the tissue sections can suggest the true causative agent (40).
Detection in Tissue Sections
Although individual bacteria generally are not detected in hematoxylin and eosin (H&E)-stained tissue sections, exceptions do exist. Clumps of finely particulate basophil material were seen in H&E-stained sections of bacillary angiomatosis and subsequently identified as Bartonella (41). In H&E-stained sections of gastric biopsy specimens that show acute gastritis, curved bacteria consistent with Helicobacter pylori may be seen in the layer of mucus on the crypt epithelium (25). Moreover, as histopathologic damage and causal microorganisms usually have a long-established association, microscopic examination of H&E-stained tissue sections during the course of an unexplained disease may lead to hypotheses about the nature of the etiologic agent (40).
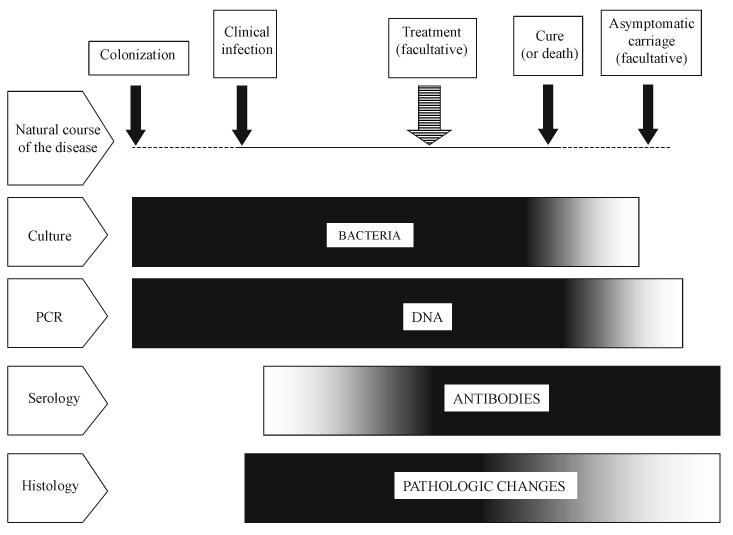
Gram stain has also proven useful to routinely diagnose H. pylori and H. heilmanii in the gastric mucosa of patients with gastritis, as well as that of B. henselae in cardiac valves (10,24,25). Silver impregnation is among the most useful methods for detecting bacteria, especially for that stained weakly with a tissue Gram stain. Thus, bacillary angiomatosis lesions were found to contain clusters of bacilli on Warthin-Starry staining 2 years before the etiologic role of B. henselae was elucidated. With the same stain, this bacterium was also detected in cardiac valves of patients with endocarditis (Figure 2) (41). The first observation of Whipple agent was reported in 1907 by George Whipple in silver-stained sections of a lymph node, although the author did not link this observation with the cause of the disease (2).
Figure 2.
Demonstration of Bartonella henselae in cardiac valve of a patient with blood culture-negative endocarditis. The bacilli appear as black granulations (Warthin Starry, original magnification X250).
Special stains have also played a role in establishing the etiologic role of new bacteria. Gimenez’ and Pinkerton’s stains allowed the detection of rickettsial organisms in tissue sections from patients with acute febrile disease (40).New mycobacteria were initially detected by using Ziehl-Nielsen, Kinyoun, or auramine O stains. For example, in an HIV-infected boy, examination of a retroperitoneal lymph node showed granuloma with large numbers of intracellular acid-fast bacilli that were later characterized as a new Mycobacterium species, M. genavense (34). Morphologic techniques, indeed, do not allow specific identification of the detected organisms. Despite this limitation, the approach consisting of detecting infectious lesions and agents by using cytologic and histologic examination appeared to be sometimes more valuable than the cultural or molecular techniques (40).
Electron Microscopy
Among morphologic techniques, transmission and scanning electron microscopy (EM) has substantial advantages resulting from its high flexibility and sensitivity (42). Negative staining is a rapid EM method that can be useful in patients with persisting or unexplained disease. Further, its specificity and sensitivity can be enhanced by using immuno-capture assay. Thus, in patients with chronic gastritis, EM provided the first detection of H. pylori in the gastric mucosa (25). EM can resolve details many hundreds of time smaller than can be seen through light microscopes, and resolution of major taxonomic features can help to characterize new microorganisms (42). Thus, the agent of Whipple disease was recognized as a bacillus through ultrastructural examination of the bacilli (42). Nevertheless, limitations of EM include its availability, cost, and need for experienced staff. EM requires knowledge of histology and ultrastructure of the tissue being examined and organisms likely to be encountered and is very time-consuming, since every specimen must be examined individually (42).
Serology and Antigenic Detection
Serology
By showing rising antibody titers or seroconversion, serology can provide indirect evidence for causal relationships between a disease and a newly identified bacterium. Conversely, in the absence of serologic evidence, the role of a cultured organism should be interpreted cautiously, as shown by the example of Afipia felis, which was first thought to be the cause of cat-scratch disease, but was finally identified as a water contaminant (12,43). Serology is also useful to assess the involvement in human diseases of microorganisms that had been initially recovered from the environment, such as novel Legionella species, or from animal hosts, as for the tick-associated bacteria Borellia burgdorferi or Rickettsia slovaca (11,19,29). Further, serology is a valuable tool for exploring the disease spectrum of a bacterium. Thus, serologic testing contributed to the recognition of B. henselae as the main agent of cat-scratch disease (16), as well as implicating Campylobacter jejuni as a possible cause of Guillain-Barré syndrome (44).
Moreover, the contribution of serologic studies to the identification of new bacterial pathogens should not be underrated. Serologic cross-reactions are common between members of the same bacterial genus, and antibodies specific to a bacterial species can suggest the role of a closely related, still unidentified organism. Thus, specific antibodies to Ehrlichia canis, E. phagocytophila, and E. equis, then known only as veterinary pathogens, were detected in patients and led to description of the agents of human ehrlichioses (E. chaffeensis, E. ewingii, and human granulocytic ehrlichiosis) (5–7). Involvement of Bartonella grahamii in neuroretinitis was first suggested by detection of specific antibodies to B. henselae in the patient’s blood (14). Reliable interpretation of such serologic cross-reactions, however, would not have been possible without considering other evidence, such as intraleukocytic morulae for ehrlichioses.
Antigenic Detection
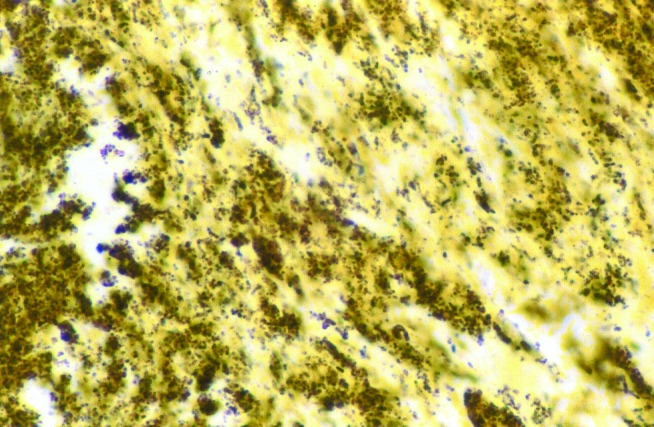
Production of specific antibodies in experimental animal studies allowed immunochemical detection techniques to be developed. Direct immunofluorescence staining can be performed in smears in respiratory fluids of patients with pneumonia (29). Immunohistochemistry is useful for demonstrating disease causation, as it provides evidence for in situ association between microorganisms and histologic structures. With this technique, Tropheryma whipplei was detected in a patient’s mitral valve and later in intestinal mucosae (Figure 3) (2). Immunohistochemistry also suggested the role of M. fermentans in pulmonary infections (35). Immunologic techniques are dependent, however, on the availability of specific antibodies or antigens, which in most cases requires previous isolation of the agent; therefore, such techniques indirectly contribute to culture.
Figure 3.
Demonstration of Tropheryma whipplei by immuno-histochemistry in the lamina propria of the villous tips. Bacilli are revealed in foamy macrophage cytoplasm as red-brown deposits (polyclonal rabbit.
Culture: A Traditional Technique of Expanding Potential
Culture Media
Axenic Media
Broad-spectrum media allowed several previously unrecognized gram-positive bacteria, such as novel corynebacteria or Staphylococcus species, as well as novel beta-Proteobacteria, to be isolated, mainly from blood or pus of patients (18,37,38). The first isolation of B. elizabethae, B. quintana, and B. henselae was also achieved on blood agar (15). Use of Campylobacter-selective medium allowed novel Campylobacter and Helicobacter species to be grown from stools and rectal swabs, respectively (23), and provided further evidence for the association between C. jejuni infection and Guillain-Barré syndrome (44). For Campylobacter spp., selective, antibiotic-containing media could be satisfactorily replaced by nonselective blood agar, provided stool specimens had been filtered with a cellulose acetate membrane (23). Newly recognized serotypes of enterohemorrhagic Escherichia coli were isolated on MacConkey-sorbitol agar from stools or urine of patients with hemolytic-uremic syndrome (26). For Vibrio cholerae O:139 and most novel Vibrio species, the most convenient, highly selective medium was thiosulfate-citrate-bile salts sucrose agar (33).
The usefulness of broad-spectrum media should not obscure the fact that some emerging bacteria would not have been isolated without specific media. Buffered charcoal-yeast extract (BCYE) agar facilitated the recovery of most novel Legionella species, as well as Afipia broomeae and A. clevelandensis, from human respiratory sources (12,29) The first cultivation of Borrelia burgdorferi was achieved in 1981 in a modified Kelly medium (19). In 1994, 20 years after the first attempts, the Kelly growth medium itself allowed first cultivation of B. recurrentis from the blood of an Ethiopian patient with louse-borne relapsing fever, and B. duttonii, agent of East African tick-borne relapsing fever, was isolated for the first time in 1999 in BSK II medium (21). Generally, combining different types of medium, using both solid and liquid media, increases the effectiveness of culture, perhaps because of a preference of the bacterium for one type of medium over another or simply from the increased sensitivity obtained by culturing a large volume of specimen. For example, B. elizabethae and B. henselae were detected in BACTEC blood culture medium before inoculation in blood agar (13,15). Isolation of most novel Mycobacterium species required both solid- and liquid-specific media (34).
Living Systems
While more expensive and less easy to use than artificial media, animal models can provide certain advantages not available with artificial media. For example, until recently, inoculation to mice was the only means available to propagate B. duttonii (21). Today, animals are still necessary for isolating organisms such as Treponema pallidum or Mycobacterium leprae. Animal inoculation can help to reduce the contaminant flora. Thus, a combination of passage in guinea pigs and subsequent transfer into embryonated eggs was the key for isolating L. pneumophila from lung autopsy specimens (31). Embryonated eggs themselves have been recognized as a standard for rickettsial isolation, allowing, for example, the first isolation of Astrakhan fever rickettsia (45).
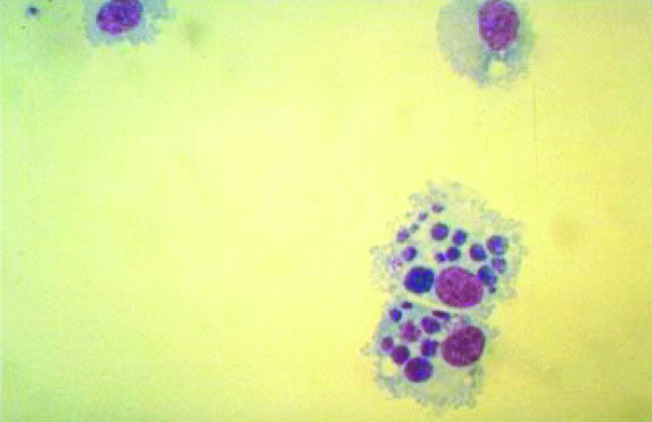
Cell culture is easy to use and may be very sensitive. Isolation of T. whipplei was obtained from valve and duodenal biopsy specimens by using human embryonic lung fibroblasts (HEL) (2,36). Ehrlichia chaffeensis and R. japonica were grown from blood samples of patients on canine macrophage cells (Figure 4) and African green monkey cells, respectively (9,46). Cultivation of facultative intracellular bacteria also was facilitated by cell culture. L. pneumophila has been isolated by using HEL cells while inoculated BCYE and agar plates remained sterile (47). With a bovine endothelial cell line, B. quintana was isolated for the first time from cutaneous biopsy material of a bacillary angiomatosis patient (48). Such enhanced sensitivity is a major advantage for an infection with low levels of bacteremia or when limited biopsy material is available (49). Indirectly, HEL cells also provided the first evidence for the role of a toxic factor in pseudomembranous colitis, which could be neutralized by clostridial antiserum. This observation led to the discovery of Clostridium difficile as the responsible agent (50).
Figure 4.
Canine monocytes (DH82) cultivated in vitro and heavily infected with Ehrlichia chaffeensis, as viewed by light microscopy after Giemsa staining. Typical ehrlichial inclusions (morulae) are observed within the cytoplasm of the infected cells (Giemsa, original magnification X600).
The search for appropriate media that could allow the growth of still uncultivatable or unrecognized bacteria has led us to try coculture with nonmammalian cells. Cell lines from toads (XTC-2) have been used in our laboratory to grow Rickettsia felis, a flea-associated Rickettsia pathogenic for humans (8). Coculture with arthropod cells will probably enhance our ability to detect intracellular, arthropod-transmitted bacteria. For example, tick cells (IDE8) have been used to grow the agent of human granulocytic ehrlichiosis (7). Cocultivation of samples with free-living amebae has allowed recovery of otherwise uncultivatable microorganisms from patients and the environment. This technique provided evidence for the role of several Legionella or Legionella-like species and of Parachlamydia acanthamoeba as etiologic agents of community-acquired pneumonia (51).
Other Critical Issues in Culture
In addition to the choice of an appropriate medium, the main critical issues in culturing concern inoculation of the specimen and incubation of the culture, both summarized in Table 2. Since successful culture usually results from the selection of a unique cultivatable clone, the quantity of inoculated pathogen should be as high as possible. Samples should be collected from anatomic sites that are likely to contain a high concentration of bacteria, and inoculation of a large volume of tissue sample is preferable. In patients with Bartonella endocarditis, the sensitivity of cell cultures has been shown to be higher when performed with valvular biopsy samples than with peripheral blood samples (49). These criteria, however, are not always feasible, as patients may reject, for instance, invasive explorations that are required to obtain the specimens. Arthropod-transmitted bacteria, which are often rare in infected human tissues, may be sometimes more easily recovered from samples collected from infected vectors; this was the key leading to the identification of B. burgdorferi (19). If initially such a result was insufficient in a clinical diagnostic approach, it has since led to efficient serologic and molecular tools, which would not have been available without culture. For intracellular bacteria, the use of a lysis method for eukaryotic cells before inoculation substantially enhances the ability to grow the organisms, especially when inoculation is performed in an axenic media, as for Bartonella or Mycobacterium species (15). Since low-speed centrifugation may also increase infectivity, the centrifugation-shell vial technique for isolating cytomegalovirus has been adapted to detect intracellular bacteria and used successfully to cultivate Rickettsia species from blood and skin biopsies and T. whipplei from the mitral valve of a patient with endocarditis (2).
Table 2. Key issues for isolating main emerging bacteria.
| Medium |
Conditions for incubation |
||||||
|---|---|---|---|---|---|---|---|
| Group | Axenic specific medium | Living system (embryonated egg, cell line) | Low temperature (<37°C) | O2 and CO2 conditions | Extended incubation | ||
| Alpha1 Proteo- bacteriae | Ehrlichia sp. Rickettsia sp. Chlamydia sp. | ELB agent (“Rickettsia felis”) (28°C) | Ehrlichia sp. Rickettsia sp. | ||||
| Alpha2 Proteo- bacteriae | Afipia sp. | Afipia sp. Bartonella sp. | Bartonella bacilliformis (28°C) | Bartonella sp. | |||
| Spirochetae | Borrelia sp. | Treponema pallidum | |||||
| Delta-Xi Proteo- bacteriae | Campylobacter sp. (microaerophilic) Helicobacter sp. (microaerophilic) | Helicobacter pylori | |||||
| Gamma Proteo- bacteriae | Legionella sp. | Legionella sp. | Yersinia pestis | ||||
| Mycobacteria | Mycobacterium sp. | Mycobacterium leprae | Mycobacterium malmoense (microaerophilic) | Mycobacterium sp. | |||
| Mycoplasmas | Mycoplasma sp. | Mycoplasma fermentans | |||||
| Gram-positive bacteria | Tropheryma whipplei | Clostridium difficile (anaerobic) | Tropheryma whipplei | ||||
Special attention should be accorded to the duration, temperature, and atmosphere of incubation. For some of the most important newly discovered pathogens, such as H. pylori, patience has been a key to successful cultivation (25). With T. whipplei, the first evidence of cytopathic effect and microorganisms did not occur until day 65 after inoculation (2). Isolation of Bartonella henselae from blood or tissue samples from infected patients required up to 33 days’ incubation (15,49). Although most pathogenic bacteria have been cultured at 35°C to 37°C, which is close to the physiologic temperature of the human body, several pathogens need a lower temperature. In addition to well-known examples such as M. leprae and Treponema pallidum, several arthropod-borne pathogens, including arboviruses, Yersinia pestis, B. bacilliformis, or R. felis may be more easily cultivated at <32°C (8).
A More Recent Technique: 16S rDNA Amplification and Sequencing
With the use of universal primers that recognize highly conservative loci such as the 16S rDNA encoding gene, species-specific sequences can be amplified directly from diseased host tissues and compared with a reference-sequence database to infer phylogenetic relationships (3,4). This broad-range PCR technique has expanded the ability of laboratories to partially characterize organisms that had never been cultured. Thus, in the last decade, it has enabled two unexplained illnesses to be associated with novel etiologic agents: B. henselae in bacillary angiomatosis and 1 year later T. whipplei in patients with Whipple disease (52,53). These remarkable successes of molecular techniques, however, should not obscure the fact that a bacterial origin was previously established for both diseases on the basis of histologic studies and clinical responses to antimicrobial treatment (2,48). Further, isolation and culture were achieved at the same time as molecular identification (for B. henselae) or soon after (for T. whipplei) (2,15). In both cases, successful isolation resulted from laboratory practices generally used to enhance the detection of fastidious pathogens. Although it has been suggested that specific culture conditions could be inferred from molecular phylogenetic data, such a situation has never occurred for any bacterium (3). These examples suggest, therefore, that molecular techniques are particularly useful for taxonomic studies and identification, while traditional methods remain powerful to detect pathogens.
For viruses, several species, such as the Sin Nombre virus (SNV) or the Hepatitis C virus (HCV), were detected by reverse transcriptase PCR before any morphologic, serologic, or cultural detection. Although SNV was subsequently cultured in vitro, the HCV agent has only been cultured recently in chimeric mice (54). Because of its high sensitivity, broad-range PCR also expands the ability to detect organisms present in very low quantity and those that are difficult to grow, such as intracellular bacteria. Ehrlichia ewingii, previously known as a canine parasite, was detected by this technique in circulating leukocytes of four patients with febrile illness. Note, however, that morulae had been identified in neutrophils from two of the four patients, providing strong evidence for an ehrlichial origin for the disease, and that serologic evidence was reported before the PCR assays (6). Advantages of molecular techniques seem more obvious for Bartonella grahamii and B. vinsonii subsp. berkhoffii, which have been implicated in human disease solely on the basis of 16S rDNA amplification and sequencing (14,55). Molecular tools are also particularly useful in diseases associated with dormant or latent organisms, such as chronic Lyme arthritis, and for which the sensitivity of culture from body fluids remains very low (4,). The advantages of broad-range PCR, however, are offset by the problem of microbial DNA contamination. Even after rigorous technical precautions are taken to minimize contamination of PCR reaction, false-positive reactions can occur. Another noticeable limitation of broad-range PCR is the examination of sites that are not normally sterile, such as feces or sputum; use of family-restricted primers, in situ hybridization with specific nucleic probes, or expression library screening with immune sera may help to evercome such limitations (3,4). Another potential problem is interpretation of the microheterogeneity found in microbial sequences derived directly from host tissues, especially when these sequences become the sole basis for defining the existence of an organism. For example, attempts to characterize and classify nanobacteria using 16S rDNA sequence analysis provided doubtful results, and these organisms were later considered contamination (56). Additionally, current databases contain an insufficient number of entries with which to define species and other taxon boundaries over a wide range of microorganisms (3).
Advantages of Culture for the Study of Emerging Bacterial Diseases
Antibiotic Susceptibility Testing
When culture and isolation are achieved, susceptibility of emerging bacteria to a large panel of antimicrobial drugs can be easily tested, providing essential data to guide clinical treatment, particularly when resistant strains are reported and empiric therapy may be ineffective. This antimicrobial testing would have been difficult, if not impossible, with molecular techniques, as genetic determinants of antibiotic resistance have been identified in only a few situations (57). Thus, isolation of H. pylori has revolutionized the treatment of duodenal ulcers, which are now definitively healed by appropriate antimicrobial regimens. As strains resistant to either metronidazole or clarithromycin have been increasingly reported, culture of the agent is very helpful in case of proven treatment failure, to assess the antibiotic resistance pattern of local strains of H. pylori (58). Coculture of bacteria with cell lines has brought new insights about antibiotic susceptibility patterns for obligate and facultative intracellular organisms. For example, while patients with human ehrlichiosis have been treated for a long time, with variable results, with chloramphenicol, in vitro studies showed that E. chaffeensis was resistant to this antibiotic (59).
Experimental Animal Models for Pathogenicity
With viable microorganisms, disease models can often be established in animals. Rodent models are the most commonly used. For Legionella oakridgensis, originally isolated from industrial cooling towers, demonstration of its pathogenicity for guinea pigs suggested for the first time, before any clinical involvement, that it might be an unrecognized human pathogen (29). For assessing the capability of various Vibrio species to elaborate an enterotoxin, rabbit and mouse intestinal models were used (33). Human tissues can also now be maintained in immunodeficient mice (SCID-hu), which can then serve as useful models for human host-specific pathogens (56,60). Although less accessible, primate models supported, for example, the implication of Mycoplasma genitalium in genital tract infections (35). Finally, experimental animal models are useful for immunization studies, as for H. pylori in mouse and primate models. Following culture, immunodominant antigens can be cloned, expressed, and inoculated to animals to identify candidate vaccines (61).
Genetic Studies
Isolated Genes
For noncultured organisms, molecular techniques have been proposed to identify isolated bacterial genes directly from clinical specimens. These techniques, however, are quite difficult to use and can identify only a few, short genetic fragments (3). On the other hand, by providing pure microbial cell mass, culture enables genes to be identified in high numbers through recombinant chromosomal libraries built from the extracted DNA. Genes identified in this fashion can then be utilized as more refined diagnostic tools. For example, Rickettsia mongolotimonae and R. slovaca were associated with human disease on the basis of amplification of a species-specific rOmpA gene fragment from skin biopsy specimens (11,12). DNA probes developed after isolation of Chlamydia pneumoniae enabled this organism to be detected by in situ hybridization in coronary atherosclerotic plaques (62). Further, molecular subtyping of cultured strains has offered new perspectives for epidemiologic studies. Thus, comparison of nucleotide sequences of 16S rDNA, OspA, and Fla genes for different strains of B. burgdorferi provided phylogenetic data that consistently supported the division of B. burgdorferi sensu lato into three geographically distinct genotypes, which were subsequently shown to have different pathogenic potentials (63). Correlation between genotypes and biologic characters is a key to understanding the pathophysiology of bacterial diseases.
Complete Genome Sequence
Because of the importance of organisms such as H. pylori, M. genitalium, and C. pneumoniae as emerging human pathogens and the value of complete genome sequence information for drug discovery and vaccine development, the complete nucleotide sequences of these three organisms has been determined by the whole-genome random sequencing method as described initially for Haemophilus influenzae. Sequence analyses allowed identification of several predicted coding regions that included genes required for DNA replication, transcription and translation, DNA repair, cellular transport, and energy metabolism (64). With the availability of complete genome sequences, further assessment of microbial genetic diversity is possible; based on the large number of sequence-related genes encoding outer membrane proteins, H. pylori was predicted to use recombination as a mechanism for antigenic variation and adaptative evolution (65). As the genome sequences of new bacterial species or strains are determined, comparative genomics will be an increasingly useful method to provide insights into physiologic differences among microorganisms (64).
Conclusion
A comprehensive study of the histories of emerging bacterial diseases provided new insights into the respective roles played by the different identification techniques. Because of the spectacular development of molecular methods, traditional techniques have been prematurely considered obsolescent. We hope to have shown, however, that such a statement does not reflect the real contribution of these techniques. The undoubted value of novel molecular methods, especially for rapid bacterial detection and phylogenetic studies, should not hide the crucial role that traditional techniques have historically played. Moreover, these traditional techniques have never stopped evolving towards increased sensitivity and specificity. Today, these techniques appear complementary. If broad-range PCR was helpful in determining the taxonomic position of new, still uncultured organisms, most of the novel infectious diseases were finally described after culture and isolation of the responsible agents. In the current, fast-changing world of emerging infections, fulfillment of Koch’s postulates, which requires culture, remains a very necessary model of rigorous proof and scientific thinking (1). Culture is still an irreplaceable key for studying emerging bacterial diseases, even if routine diagnosis can be efficiently achieved by using other (although generally culture-derived) tools, including genetic amplification. The history of infectious diseases shows that no human bacterial pathogen is uncultivable so far: the real issue seems to be whether we are able to determine the environmental conditions required by prokaryotic agents for growth (2).
Biographies
Pierre Houpikian is a physician working on the epidemiology of infectious diseases. He is currently in a postdoctoral position.
Didier Raoult is the head of l’Unité des Rickettsies, a leading research laboratory working on rickettsial diseases and emerging pathogens.
Footnotes
Suggested citation: Houpikian P and Raoult D. Traditional and Molecular Techniques for the Study of Emerging Bacterial Diseases: One Laboratory’s Perspective. Emerg Infect Dis. [serial on the Internet]. 2002 Feb [date cited]. http://dx.doi.org/10.3201/eid0802.010141
References
- 1.Krause RM. Microbes and emerging infections: the compulsion to become something new. ASM News. 2001;65:15–20. [Google Scholar]
- 2.Raoult D, Birg ML, La Scola B, Fournier PE, Enea M, Lepidi H, et al. Cultivation of the bacillus of Whipple disease. N Engl J Med. 2000;342:620–5. 10.1056/NEJM200003023420903 [DOI] [PubMed] [Google Scholar]
- 3.Relman DA. The search for unrecognized pathogens. Science. 1999;284:1308–10. 10.1126/science.284.5418.1308 [DOI] [PubMed] [Google Scholar]
- 4.Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maeda K, Markowitz N, Hawley RC, Ristic M, Cox D, McDade JE. Human infection with Ehrlichia canis, a leukocytic rickettsia. N Engl J Med. 1987;316:853–6. [DOI] [PubMed] [Google Scholar]
- 6.Buller RS, Arens M, Hmiel SP, Paddock CD, Sumner JW, Rikhisa Y, et al. Ehrlichia ewingii, a newly recognized agent of human ehrlichiosis. N Engl J Med. 1999;341:148–55. 10.1056/NEJM199907153410303 [DOI] [PubMed] [Google Scholar]
- 7.Goodman JL, Nelson C, Vitale B, Madigan JE, Dumler JS, Kurtti TJ, et al. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N Engl J Med. 1996;334:209–15. 10.1056/NEJM199601253340401 [DOI] [PubMed] [Google Scholar]
- 8.Raoult D, La Scola B, Enea M, Fournier PE, Roux V, Fenollar F, et al. Isolation and characterization of a flea-associated rickettsia pathogenic for humans. Emerg Infect Dis. 2001;7:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uchida T. Rickettsia japonica, the etiologic agent of oriental spotted fever. Microbiol Immunol. 1993;37:91–102. [DOI] [PubMed] [Google Scholar]
- 10.Raoult D, Brouqui P, Roux V. A new spotted fever group Rickettsiosis. Lancet. 1996;348:412. 10.1016/S0140-6736(05)65037-4 [DOI] [PubMed] [Google Scholar]
- 11.Raoult D, Berbis P, Roux V, Xu W, Maurin M. A new tick-transmitted disease due to Rickettsia slovaca. Lancet. 1997;350:112–3. 10.1016/S0140-6736(05)61814-4 [DOI] [PubMed] [Google Scholar]
- 12.Brenner DJ, Hollis DG, Moss CW, English CK, Hall GS, Vincent J, et al. Proposal of Afipia gen. nov., with Afipia felis sp. nov. (formerly the cat scratch disease bacillus), Afipia clevelandensis sp. nov. (formerly the Cleveland Clinic Foundation strain), Afipia broomeae sp. nov., and three unnamed genospecies. J Clin Microbiol. 1991;29:2450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daly JS, Worthington MG, Brenner DJ, Moss WC, Hollis DG, Weyant RS, et al. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerkhoff FT, Bergmans AMC, van der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol. 1999;37:4034–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slater LN, Welch DF, Hensel D, Coody DW. A newly recognized fastidious gram-negative pathogen as a cause of fever and bacteremia. N Engl J Med. 1990;323:1587–93. [DOI] [PubMed] [Google Scholar]
- 16.Regnery RL, Olson JG, Perkins BA, Bibb W. Serological response to Rochalimaea henselae antigen in suspected cat-scratch disease. Lancet. 1992;339:1443–5. 10.1016/0140-6736(92)92032-B [DOI] [PubMed] [Google Scholar]
- 17.Vandamme P, Heyndrickx M, Vancanneyt M, Hoste B, de Vos P, Falsen E, et al. Bordetella trematum sp. nov., isolated from wounds and ear infections in humans, and reassessment of Alcaligenes denitrificans Rüger and Tan 1983. Int J Syst Bacteriol. 1996;46:849–85. [DOI] [PubMed] [Google Scholar]
- 18.Homes B, Costas M, On SL, Vandamme P, Falsen E, Kersters K. Neisseria weaveri sp. nov. (formerly CDC group M-5), from dog bite wounds of humans. Int J Syst Bacteriol. 1993;43:693. [DOI] [PubMed] [Google Scholar]
- 19.Burgdorfer W. Discovery of the Lyme disease spirochete and its relation to tick vector. Yale J Biol Med. 1984;57:515–20. [PMC free article] [PubMed] [Google Scholar]
- 20.Cutler SJ, Fekade D, Hussein K, Knox KA, Melka A, Cann K, et al. Successful in-vitro cultivation of Borrelia recurrentis. Lancet. 1994;343:242. 10.1016/S0140-6736(94)91032-4 [DOI] [PubMed] [Google Scholar]
- 21.Cutler SJ, Moss J, Fukunaga M, Wright DJM, Fekade D, Warrell D. Borrelia recurrentis characterization and comparison with relapsing-fever, lyme-associated, and other Borrelia spp. Int J Syst Bacteriol. 1997;47:958–68. [DOI] [PubMed] [Google Scholar]
- 22.Taylor DN, Kiehlbauch JA, Tee W, Pitarangsi C, Echeverria P. Isolation of group 2 aerotolerant Campylobacter species from Thai children with diarrhea. J Infect Dis. 1991;163:1062–7. [DOI] [PubMed] [Google Scholar]
- 23.Totten PA, Fennell CL, Tenover FC, Wezenberg JM, Perine PL, Stamm WE, et al. Campylobacter cinaedi (sp. nov.) and Campylobacter Campylobacter (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J Infect Dis. 1985;151:131–9. [DOI] [PubMed] [Google Scholar]
- 24.Heilmann KL, Borchard F. Gastritis due to spiral shaped bacteria other than Helicobacter pylori: clinical, histological, and ultrastructural findings. Gut. 1991;32:137–40. 10.1136/gut.32.2.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall B. Unidentified curved bacillis on gastric epithélium in active chronic gastritis. Lancet. 1983;I:1273–5. [PubMed] [Google Scholar]
- 26.Tarr PI, Fouser LS, Stapleton AE, Wilson RA, Kim HH, Vary JC, et al. Hemolytic uremic syndrome in a six-year-old girl after a urinary tract infection with Shiga-toxin-producing Escherichia coli O103:H2. N Engl J Med. 1996;335:635–8. 10.1056/NEJM199608293350905 [DOI] [PubMed] [Google Scholar]
- 27.The Brazilian Purpuric Fever Study Group. Haemophilus aegyptus bacteremia in Brazilian purpuric fever. Lancet. 1987;331:761–3. [PubMed] [Google Scholar]
- 28.Fernstersheib MD, Miller M, Diggins C, Liska S, Detwiler L, Werner SB, et al. Outbreak of Pontiac fever due to Legionella anisa. Lancet. 1990;336:35–7. 10.1016/0140-6736(90)91532-F [DOI] [PubMed] [Google Scholar]
- 29.Korvick JA, Yu VL, Fang GD. Legionella species as hospital-acquired respiratory pathogens. Semin Respir Infect. 1987;2:34–47. [PubMed] [Google Scholar]
- 30.Herwaldt LA, Gorman GW, McGrath T, Toma S, Brake B, Hightower AW, et al. A new Legionella species, Legionella feeleii species nova, causes Pontiac fever in an automobile plant. Ann Intern Med. 1984;100:333–8. [DOI] [PubMed] [Google Scholar]
- 31.McDade JE, Brenner DJ, Bozeman M. Legionnaires’ disease bacterium isolated in 1947. Ann Intern Med. 1979;90:659–61. [DOI] [PubMed] [Google Scholar]
- 32.Birtles RJ, Rowbotham TJ, Raoult D, Harrison TG. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology. 1996;142:3525–30. [DOI] [PubMed] [Google Scholar]
- 33.Janda JM, Powers C, Bryant RG, Abbott SL. Current perspectives on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin Microbiol Rev. 1988;1:245–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falkinham JO. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996;9:177–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor-Robinson D. Infections due to species of Mycoplasma and Ureaplasma: an update. [quiz 683-4]. Clin Infect Dis. 1996;23:671–82. [DOI] [PubMed] [Google Scholar]
- 36.La Scola B, Fenollar F, Fournier PE, Altwegg M, Mallet MN, Raoult D. Description of Tropheryma whipplei gen. nov. sp. nov., the Whipple disease bacillus. Int J Syst Bacteriol. 2001;51:1471–9. [DOI] [PubMed] [Google Scholar]
- 37.Funke G, Lawson PA, Collins MD. Heterogeneity within human-derived centers for disease control and prevention (CDC) coryneform group ANF-1-like bacteria and description of Corynebacterium auris sp. nov. Int J Syst Bacteriol. 1995;45:735–9. [DOI] [PubMed] [Google Scholar]
- 38.Freney J, Brun Y, Bes M, Meugnier H, Grimont F, Grimont PAD, et al. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int J Syst Bacteriol. 1988;38:168–72. [Google Scholar]
- 39.Centers for Disease Control and Prevention. Invasive infection with Streptococcus iniae--Ontario, 1995-1996. MMWR Morb Mortal Wkly Rep. 1996;45:650–3. [PubMed] [Google Scholar]
- 40.Woods GL, Walker DH. Detection of infection or infectious agents by use of cytologic and histologic stains. Clin Microbiol Rev. 1996;9:382–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurin M, Raoult D. Bartonella (Rochalimaea) quintana infections. Clin Microbiol Rev. 1996;9:273–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curry A. Electron microscopy as a tool for identifying new pathogens. J Infect. 2000;40:107–15. 10.1053/jinf.2000.0634 [DOI] [PubMed] [Google Scholar]
- 43.La Scola B, Raoult D. Afipia felis in a hospital water supply in association with free-living amoebae. Lancet. 1999;353:1330. 10.1016/S0140-6736(99)00906-X [DOI] [PubMed] [Google Scholar]
- 44.Sovilla JY, Regli F, Francioli PB. Guillain-Barré syndrome following Campylobacter jejuni enteritis: report of three cases and review of the literature. Arch Intern Med. 1988;148:739–41. 10.1001/archinte.148.3.739 [DOI] [PubMed] [Google Scholar]
- 45.Tarasevich IV, Makarova VA, Fetisova NF, Stepanov AV, Miskarova ED, Balayeva N, et al. Astrakhan fever, a spotted-fever rickettsiosis. Lancet. 1991;337:172–3. 10.1016/0140-6736(91)90833-B [DOI] [PubMed] [Google Scholar]
- 46.Dumler J, Walker D. Tick-borne ehrlichioses. Lancet Infect Dis. 2001;:21–8.11871406 [Google Scholar]
- 47.La Scola B, Michel G, Raoult D. Isolation of Legionella pneumophila by centrifugation of shell vial cell cultures from multiple liver and lung abscesses. J Clin Microbiol. 1999;37:785–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koehler JE, Quinn FD, Berger TG, Leboit PE, Tappero JW. Isolation of Rochalimaea species from cutaneous and osseous lesions of bacillary angiomatosis. N Engl J Med. 1992;327:1625–31. [DOI] [PubMed] [Google Scholar]
- 49.La Scola B, Raoult D. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998). J Clin Microbiol. 1999;37:1899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knoop FC, Owens M, Crocker IC. Clostridium difficile: clinical disease and diagnosis. Clin Microbiol Rev. 1993;6:251–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birtles RJ, Rowbotham TJ, Storey C, Marrie TJ, Raoult D. Chlamydia-like obligate parasite of free-living amoebae. Lancet. 1997;349:925–6. 10.1016/S0140-6736(05)62701-8 [DOI] [PubMed] [Google Scholar]
- 52.Relman AD, Schmidt TM, MacDermott RP, Falkow S. Identification of the uncultured bacillus of Whipple disease. N Engl J Med. 1992;327:293–301. [DOI] [PubMed] [Google Scholar]
- 53.Relman DA, Loutit JS, Schmidt TM, Falkow S, Tompkins LS. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–80. [DOI] [PubMed] [Google Scholar]
- 54.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, et al. Hepatitis C virus replication in mice with chimeric human liers. Nat Med. 2001;7:927–33. 10.1038/90968 [DOI] [PubMed] [Google Scholar]
- 55.Roux V, Eykyn SJ, Wyllie S, Raoult D. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J Clin Microbiol. 2000;38:1698–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cisar JO, Thompson J, Swain W, Hu L, Kopecko D. An alternative interpretation of nanobacteria induced biomineralization. Proc Natl Acad Sci U S A. 2000;97:11511–5. 10.1073/pnas.97.21.11511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drancourt M, Raoult D. Characterization of mutations in the rpoB gene in naturally rifampin-resistant Rickettsia species. Antimicrob Agents Chemother. 1999;43:2400–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goodwin CS, Mendall MM, Northfield TC. Helicobacter pylori infection. Lancet. 1997;349:265–9. 10.1016/S0140-6736(96)07023-7 [DOI] [PubMed] [Google Scholar]
- 59.Brouqui P, Raoult D. In vitro antibiotic susceptibility of the newly recognized agent of ehrlichiosis in humans, Ehrlichia chaffeensis. Antimicrob Agents Chemother. 1992;36:2799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.MacCune JM, Namikawa R, Kaneshima H, Shultz LD, Lieberman M, Weissman IL. The SCID-hu mouse: murine model for the analysis of human hematolymphoid differentiation and function. Science. 1988;241:1632–9. 10.1126/science.2971269 [DOI] [PubMed] [Google Scholar]
- 61.Sutton P, Lee A. Review article: Helicobacter pylori vaccines – the current status. Aliment Pharmacol Ther. 2000;14:1107–18. 10.1046/j.1365-2036.2000.00825.x [DOI] [PubMed] [Google Scholar]
- 62.Kuo CC, Jackson LA, Campbell LA, Grayston JT. Chlamydia pneumoniae (TWAR). Clin Microbiol Rev. 1995;8:451–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balmelli T, Piffaretti JC. Association between different clinical manifestations of Lyme disease and different species of Borrelia burgdorferi sensu lato. Res Microbiol. 1995;146:329–40. 10.1016/0923-2508(96)81056-4 [DOI] [PubMed] [Google Scholar]
- 64.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–406. 10.1093/nar/28.6.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleishmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–47. 10.1038/41483 [DOI] [PubMed] [Google Scholar]