Abstract
Proper cell fate choice in myelopoiesis is essential for generating correct numbers of distinct myeloid subsets manifesting a wide spectrum of subset-specific activities during development and adulthood. Studies have suggested that myeloid fate choice is primarily regulated by transcription factors; however, new intrinsic regulators and their underlying mechanisms remain to be elucidated. Zebrafish embryonic myelopoiesis gives rise to neutrophils and macrophages and represents a promising system to derive new regulatory mechanisms for myeloid fate decision in vertebrates. Here we present an in vivo study of cell fate specification during zebrafish embryonic myelopoiesis through characterization of the embryos with altered Pu.1, Runx1 activity alone, or their combinations. Genetic analysis shows that low and high Pu.1 activities determine embryonic neutrophilic granulocyte and macrophage fate, respectively. Inactivation and overexpression of Runx1 in zebrafish uncover Runx1 as a key embryonic myeloid fate determinant that favors neutrophil over macrophage fate. Runx1 is induced by high Pu.1 level and in turn transrepresses pu.1 expression, thus constituting a negative feedback loop that fashions a favorable Pu.1 level required for balanced fate commitment to neutrophils versus macrophages. Our findings define a Pu.1-Runx1 regulatory loop that governs the equilibrium between distinct myeloid fates by assuring an appropriate Pu.1 dosage.
Introduction
Myeloid cells are a collection of morphologically, phenotypically, and functionally distinct blood cells that conventionally consist of 2 separated lineages, granulocytes (neutrophils, basophils, and eosinophils), and monocytes/macrophages. The importance of this class of cells is exemplified by their wide engagement in diverse physiologic processes, including organogenesis, tissue homeostasis, and immune defense. Perturbation in the formation, differentiation, and function of these cells will incur devastating consequences, such as leukemia. Hence, a comprehensive understanding of how functional myeloid system is established will render novel therapeutic opportunities for curing myeloid malformation-associated diseases. During ontogeny, there exist multiple waves of myeloid cell production, termed myelopoiesis. In the adult myelopoiesis, all granulocytes and monocytes/macrophages are derived from the hierarchical transformation of multipotential hematopoietic stem cells (HSCs) into lineage-restricted progenitors and subsequent maturation of unilineage-restricted progenitors.1 In the developing embryos where HSCs are not formed, myeloid cells are transitorily supplied by unipotent or bipotent progenitors, which appear to arise directly from mesoderm without transiting through HSC-like multipotent progenitors.
A key indispensable step shared by different waves of myelopoiesis is the specification of granulocyte or monocyte/macrophage fate from initially equivalent pool of myeloid progenitors. Resolving granulocyte versus monocyte/macrophage fate is thought to be primarily dictated by the interplay among various transcription factors and their abundances at appropriate developmental time points.2 Although a number of transcription factors have been shown to be required for the general or certain specific aspect of myeloid development, few transcription factors are demonstrated to have a primary role in regulating granulocyte versus monocyte/macrophage fate choices.2 PU.1, also known as Spi-1, is a member of Ets-family transcription factors. Apart from the earlier obligatory role of PU.1 in generating myeloid progenitors,3,4 PU.1 activity level appears to be critical for fate decision of myeloid progenitors. In vitro reconstitution of PU.1 expression in PU.1−/− progenitors showed that granulocytes developed from progenitors supplied with low PU.1 expression whereas macrophages developed from progenitors supplied with high PU.1 expression.5,6 However, there is still controversy over whether different PU.1 dosages cause distinct myeloid fate7 partly because the consequence of altering PU.1 expression on myeloid fate choice has not been directly tested in vivo. The transcriptional regulation of Pu.1 expression has been studied over years, which leads to the identification of an upstream cis-regulatory element important for proper expression of Pu.1 in mice.8 However, such regulation mediated by this cis-regulatory element has not been causatively linked to the control of PU.1 dosage in the context of myeloid fate decision. Thus, the mechanism by which appropriate PU.1 level is attained in vivo to ensure balanced commitment toward different myeloid subsets remains elusive.
Zebrafish, as a genetically tractable system, is increasingly used to dissect vertebrate specific developmental mechanisms. Embryonic myelopoiesis in zebrafish occurs in anteriorly located rostral blood island (RBI)9–11 where cephalic mesoderm-derived myeloid progenitors differentiate to produce either macrophages or neutrophils.10,12 Embryonic myeloid cells might also arise from a transient population of erythromyeloid progenitors (EMPs),13 yet the relative contribution of this source to the total embryonic myeloid pool is not clear. Nevertheless, the derivation of zebrafish embryonic myeloid cells without transiting through multipotential HSCs allows for bypassing the potential epistatic role of transcription factors in HSCs and directly accessing their roles in the myeloid development per se.14 The utility of zebrafish embryonic myelopoiesis as a model to study the common mechanisms of vertebrate myelopoiesis is supported by numerous reports showing the conserved expression and function of genes for both myeloid fate determination and terminal differentiation.14,15 Like in the adult mouse myelopoiesis, the early undifferentiated embryonic myeloid cells in the RBI express transcription factor pu.1, and maximal knockdown of pu.1 disrupted the formation of both macrophages and neutrophils.16,17 However, how embryonic macrophages and neutrophils in zebrafish establish their respective fates is still unclear.
Here we describe a previously unidentified role of transcription factor, Runx1, in regulating zebrafish embryonic myeloid fate choices through directly suppressing pu.1 expression in a negative feedback loop. Loss- and gain-of-function studies established a differential requirement of Pu.1 activity in vivo for the formation of RBI-originated neutrophil versus macrophage fates with low Pu.1 activity permissive for neutrophil fate while high Pu.1 activity instructive for macrophage fate. Using a candidate gene approach along with loss- and gain-of-function study, Runx1 is shown to regulate embryonic myeloid fate decision by favoring neutrophil over macrophage fates. Expression analyses followed by the compound mutant studies indicate that Runx1 allows for neutrophil commitment by feedback repressing pu.1 expression. Further biochemical analyses suggest that Runx1 binds to pu.1 promoter and directly suppresses pu.1 expression. Together, we propose that Runx1, acting as a direct feedback repressor to constrain pu.1 expression, is an important mechanism that determines fate choice between different embryonic subsets.
Methods
Zebrafish husbandry
Zebrafish were raised, bred, and staged according to standard protocols.18 The following strains were used: AB, Tg(mpx:eGFP)i114,19 Tg(lyz:Dsred2)nz50,20 Tg(lyz:eGFP)nz117,20 Tg(−5.3pu.1:eGFP),21 csf1raj4e1,22 runx1w84x,23,24 pu.1G242D, Tg(hsp70:myc-runx1)hkz02t, and Tg(hsp70:myc-pu.1)hkz03t.
Lineage tracing
Lineage tracking using 4,5-dimethoxy-2-nitrobenzyl caged fluorescein was performed essentially as described previously.25
Targeted induced local lesions in genomes and isolation of pu.1G242D
Tilling isolation of pu.1G242D was performed essentially as described previously.26 The mutation was identified by sequencing the genomic DNA of pu.1 locus.
In vitro synthesis of antisense RNA probe and mRNA
Antisense RNA probes were synthesized by in vitro transcription reaction according to standard protocols.18 The probes used are listed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Histology
Single color whole-mount in situ hybridization using nitroblue tetrazolium/5-bromo-4-chloro-3-inodolyl phosphate as a staining substrate was performed based on standard protocol.18 Single-color antibody staining and 2-color FISH were carried out as previously described.27 Conventional Sudan Black (SB) staining and combined detection of SB staining and protein targets were essentially performed according to previous report.12 The antibodies used are listed in supplemental Methods.
Scoring myeloid cells expressing specific lineage markers
The number of positive cells in embryos having been stained for a particular lineage marker was manually counted under Nikon AZ100 microscope. Each embryo was scored twice for all the identifiable positive cells, and the average count was calculated. After scoring, individual embryo was transferred into 96-well plates for genotyping. Individual counts were eventually sorted based on genotype for comparison among different genotype groups.
MO injection
pu.1 sp3 morpholino oligonucleotide (MO; 5′-AATAACTGATACAAACTCACCGTTC-3′) targeting exon2 intron2 boundary was designed and synthesized by Gene Tools.28 Control MO used was a standard control morpholino purchased from Gene Tools. MO was administrated at 16 ng per embryo.
Quantitative and semiquantitative RT-PCR
The RNA preparation, cDNA synthesis, and quantitative RT-PCR were performed as described (supplemental Methods).29
Pu.1 promoter reporter assay
The activities of various pu.1 promoter fragment in wild-type (WT) or runx1w84x backgrounds were analyzed by either injecting DNA constructs (−9.0pu.1:eGFP; −9.0pu.1ΔR:eGFP) into one-cell stage embryos or using a stable transgenic line Tg(−5.3pu.1:eGFP). The expression levels of GFP were determined by living monitoring and quantified by quantitative RT-PCR at 17.5 hpf (supplemental Methods).
ChIP
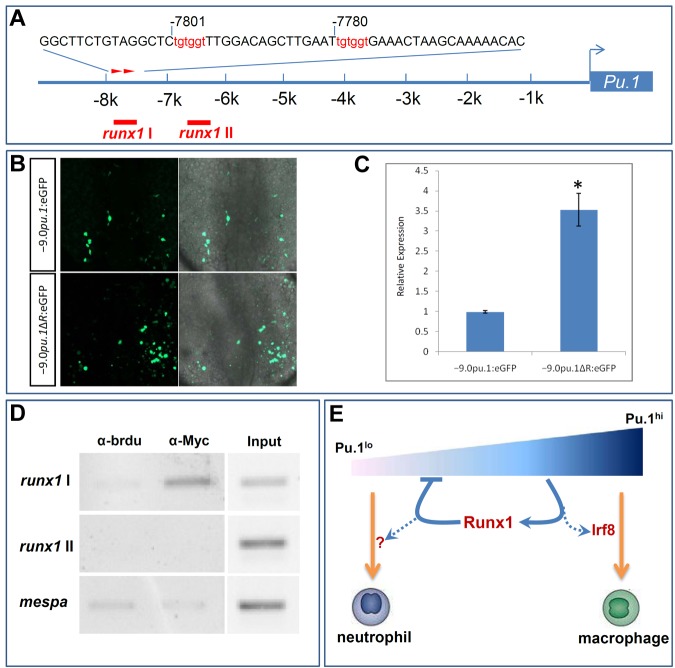
For ChIP analysis, 450 embryos derived from Tg(hsp70:myc-runx1)hkz02t+/− crossed with AB WT were heat shocked at 14 hpf stage for 45 minutes and harvested at 20 hpf for brief fixation. Cross-linked chromatin was immunoprecipitated with anti-myc antibody or anti-BrdU antibody (negative control) according to the procedure described.30 The resultant immunoprecipitated samples were subjected to semiquantitative PCR using primer pairs: runx1 I, 5′-cagcgtgtattaatataaaga-3′/5′-acatttatttatcgattcatt-3′; runx1 II, 5′-agtaacacagtgacacacatt-3′/5′-atgaatgtcactttactgtca-3′; and mespa, 5′-aagagtaagctggtggagaaaaact-3′/5′-ctcttctcaagtcctgactgaaatc-3′.
Results
RBI gives rise to 2 distinct embryonic myeloid lineages, neutrophils and macrophages, which are distinguishable by distinct markers
During zebrafish embryonic myelopoiesis, 2 alternative myeloid lineages, neutrophils and macrophages, have been reported to arise.9,10,17,31,32 It has been suggested that both embryonic neutrophils and macrophages were originated from the RBI in zebrafish.10,12,33 To validate this observation, we used a more rigorous assay in which we photo-labeled the RBI myeloid cells with fluorescein (flu+) at 14 to 16 hours postfertilization (hpf) and subsequently identified their fates based on the cellular and morphologic characteristics of flu+ progeny in 36 to 48 hpf living embryos (supplemental Figure 1A). Very often in the same embryo, certain portion of the flu+ progeny were found to be macrophages because these cells were loaded with engulfed material (supplemental Figure 1B), such as apoptotic cells, whereas other flu+ progeny were determined as neutrophils as they bore segmented nucleus and actively moving granules (supplemental Figure 1B). Thus, it confirms the notion that both embryonic neutrophils and macrophages derive from the RBI. We then sought to define the segregation of these 2 embryonic myeloid lineages in the RBI and their subsequent differentiation with molecular markers. CCAAT/enhancer binding protein 1 (cebp1) is a presumptive functional ortholog of C/ebpϵ, a transcription factor indispensable for neutrophil maturation in mammals,34–36 and lysozyme C (lyz) encodes a primary and secondary granule protein in granulocytes.37,38 We found the expression of cebp1 and lyz extensively overlapped with that of myeloperoxidase (mpx),9,31 a canonical neutrophil specific marker in zebrafish but not with that of macrophage specific markers, such as interferon regulatory factor 8 (irf8)28 and macrophage colony-stimulating factor receptor (csf1ra)33 (supplemental Figure 2A-B, data not shown).17,32 In addition, most of lyz:Dsred+ cells20 coexpressed mpx:GFP19 in the double transgenic line Tg(lyz:Dsred; mpx:eGFP) and lyz:GFP+ cells exhibited defining neutrophil characteristics,12 such as motile SB+ granules and poor phagocytotic activity (supplemental Figure 2C-D). Thus, embryonic neutrophils and macrophages preferentially express cebp1/mpx/lyz/SB and irf8/csf1ra, respectively. Such discrimination of embryonic myeloid subsets is supported by the expanded expression of cebp1/mpx/lyz/SB, but the diminished expression of csf1ra in irf8 knocked down embryos, which have a skewed output toward neutrophils.28
We next used these lineage specific markers to define the developmental sequence of embryonic neutrophils and macrophages. Consistent with the transcriptional activation of lyz by cebp1,39 we found that the expression of cebp1 preceded that of lyz and mpx with cebp1 expression detectable as early as 18 hpf when that of lyz and mpx was barely seen (supplemental Figure 2E). The early cebp1+ population was presumably early neutrophil progenitors that later transited to differentiated neutrophils because by 24 hpf, a subset of cebp1+ cells turned on mpx/lyz expression, but none of them expressed csf1ra (supplemental Figure 2A-B). Mpx+/lyz+ cells proceeded to be weak SB+ at approximately 32 hpf and strong SB+ from 36 hpf onwards (supplemental Figure 2D, data not shown). Hence, we postulate a defined sequence for embryonic neutrophil development through which cebp1+ early neutrophilic progenitors progressively mature into cebp1+/mpx+/lyz+ intermediate progenitors and SB+ granule containing maturing neutrophils. On the other hand, irf8 expression was detected at 16 hpf preceding csf1ra expression, which initiated at 21.5 hpf, whereas phagocytic macrophages and apoeb+ microglia emerged at a later stage, 24 hpf and 55 hpf, respectively (data not shown).10,28,33 Correlated with the temporal appearance of these cell populations, suppression of irf8 abolished the development of csf1ra+ macrophages, phagocytic macrophages, and apoeb+ microglia,28 whereas only phagocytic macrophages and microglia were affected by csf1ra mutation (supplemental Figure 3, data not shown). Thus, parallel to neutrophil development, embryonic macrophages differentiate along the pathway whereby irf8+ macrophage progenitors, csf1ra+ young macrophages, and phagocytic and apoeb+ resident macrophages sequentially arise.
The development of embryonic macrophages but not neutrophils requires full Pu.1 activity
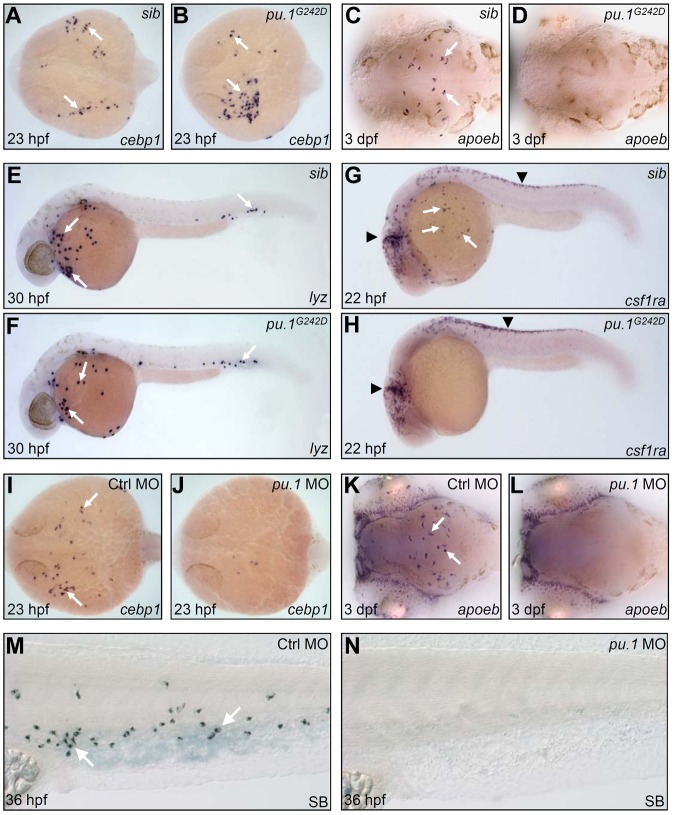
To test whether the formation of embryonic macrophage and neutrophil fates requires different Pu.1 levels, we generated a partial loss of function or hypomorphic pu.1 zebrafish allele pu.1G242D through targeted induced local lesion in genome approach.26 The G242D allele harbored a mis-sense mutation, which resulted in a conserved residue Gly (Gly 242) replaced by Asp (supplemental Figure 4). We observed that, although pu.1 transcript expressed comparably between pu.1G242D mutants and siblings (supplemental Figure 5A-B), Pu.1 protein expression was markedly reduced (supplemental Figure 5C-E), indicating that pu.1G242D mutation destabilized Pu.1 protein in vivo and thereby weakened overall Pu.1 activity. Assessment of embryonic myeloid development revealed that the initiation of the RBI myelopoietic program and subsequent commitment toward neutrophils in pu.1G242D mutants were essentially maintained as evidenced by normal expression of the markers of early undifferentiated myeloid progenitors (cebpα and pu.1) at 14 hpf (supplemental Figure 5A-B, data not shown) and neutrophils of various differentiated stages (cebp1, lyz, mpx, and SB; Figure 1A-B,E-F, and data not shown). In contrast, the development of embryonic macrophage lineage was completely interrupted in pu.1G242D mutants as shown by the loss of the macrophage markers at various differentiation stages (Figure 1C-D,G-H, and data not shown). In accordance with previous studies,16,17 only maximal knockdown of Pu.1 expression by a high dose of pu.1 antisense MOs abolished the initiation of embryonic myelopoietic program in the RBI and subsequent neutrophil development (Figure 1K-L, n = 29 of 29; Figure 1I-J, n = 35 of 38; Figure 1M-N, n = 45 of 48). Based on these data, we conclude that, similar to studies in mice,5,6,8,40 full Pu.1 activity is required for embryonic macrophage specification, whereas low Pu.1 activity suffices for embryonic myeloid initiation and neutrophil specification in zebrafish.
Figure 1.
High Pu.1 level supports the production of embryonic macrophages, whereas low Pu.1 level supports embryonic neutrophil production. (A-B) WISH of cebp1 expression in 23 hpf siblings (sib; panel A arrows) and pu.1G242D mutants (panel B arrows). (C-D) WISH of apoeb expression in 3 dpf sib (panel C arrows) and pu.1G242D mutants (D). (A-D) Embryos are viewed dorsally with the anterior to the left. (E-F) WISH of lyz expression in 30 hpf sib (panel E arrows) and pu.1G242D mutants (panel F arrows). (G-H) WISH of csf1ra expression in 22 hpf sib (G) and pu.1G242D mutants (H). White arrows indicate csf1ra expression in macrophage; and black arrowheads, its expression by neural crest cells. (I-J) WISH of cebp1 expression in 23 hpf control (panel I arrows) and pu.1 morphants (panel J, n = 35 of 38). (K-L) WISH of apoeb expression in 3 dpf control (panel K arrows) and pu.1 morphants (panel L, n = 29 of 29). (I-L) Embryos are viewed dorsally with the anterior to the left. (M-N) SB staining of 36 hpf control (panel M arrows) and pu.1 morphants (N, n = 45 of 48).
Overexpression of Pu.1 promotes embryonic macrophage fate at the expense of neutrophil fate
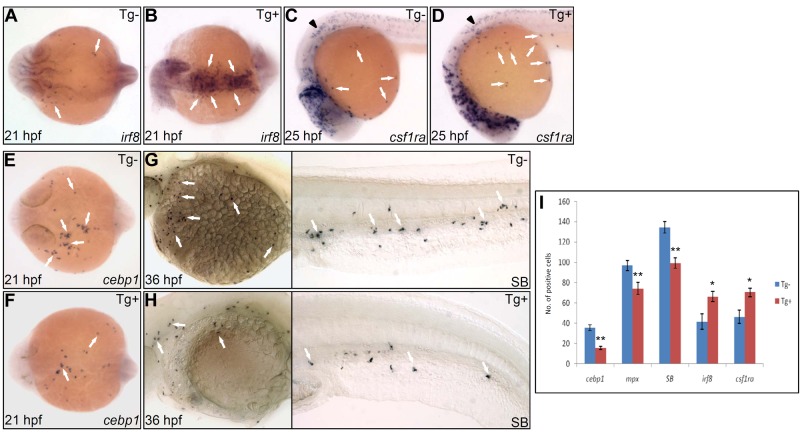
To investigate whether high Pu.1 activity is permissive to macrophage development or instructively influences macrophage versus neutrophil fate choice, we elevated Pu.1 level using an inducible pu.1 overexpression line Tg(hsp70:myc-pu.1)hkz03t in which pu.1 expression was controlled by the heat shock protein (hsp) 70 promoter.41 When raised at 28°C, Tg(hsp70:myc-pu.1)hkz03t embryos did not produce detectable exogenous Myc-Pu.1 expression and had a normal composition of embryonic macrophages and neutrophils (supplemental Figure 6A). Heat-shock treatment of Tg(hsp70:myc-pu.1)hkz03t embryos triggered high level of Myc-Pu.1 expression (supplemental Figure 6A-B). As a result, heat-shock–treated Tg(hsp70:myc-pu.1)hkz03t embryos had 50% increase in both irf8+ macrophage progenitors (Figure 2A-B,I) and csf1ra+ young macrophages (Figure 2C-D,I) but 50% reduction in cebp1+ neutrophil progenitors (Figure 2E-F,I) and 30% reduction in mpx+ (Figure 2I) and SB+ maturing neutrophils (Figure 2G-I). Thus, in the presence of uniformly high Pu.1 activity, myeloid commitment shifted toward macrophage lineage, suggesting an instructive role of high Pu.1 activity in promoting macrophage over neutrophil fate. Besides, it can also be inferred from these loss-of-function and gain-of-function studies that, to ensure balanced commitment toward macrophage versus neutrophil lineage, endogenous Pu.1 level must be appropriately tuned to a proper range.
Figure 2.
Enforced Pu.1 expression promotes the formation of embryonic macrophages at the expense of embryonic neutrophils. (A-B) WISH of irf8 expression at 21 hpf in heat-shocked Tg(hsp70:myc-pu.1)hkz03t (Tg+, panel B arrows) and nontransgenic sibling (Tg-, panel A arrows). (C-D) WISH of csf1ra expression at 25 hpf in heat-shocked Tg(hsp70:myc-pu.1)hkz03t (Tg+, panel D arrows) and nontransgenic sibling (Tg-, panel C arrows). Arrowheads indicate csf1ra expression in neural crest cells. (E-F) WISH of cebp1 expression at 21 hpf in heat-shocked Tg(hsp70:myc-pu.1)hkz03t (Tg+, panel F arrows) and nontransgenic sibling (Tg-, panel E arrows). (G-H) SB staining at 36 hpf in heat-shocked Tg(hsp70:myc-pu.1)hkz03t (Tg+, panel H arrows) and nontransgenic sibling (Tg-, panel G arrows). (I) Summary of effects of transient Pu.1 overexpression on the development of cebp1+, mpx+, SB+ neutrophils and irf8+, csf1ra+ macrophages. Embryos from Tg(hsp70:myc-pu.1)hkz03t+/− crossed with AB WT are heat shocked at 11 hpf and fixed at 21 hpf, 33 hpf, 36 hpf, 21 hpf, 25 hpf for WISH detection of cebp1 (E-F), mpx, SB (G-H), irf8 (A-B), csf1ra (C-D), respectively. Number (No.) of cells positive for these markers were counted and compared between Tg(hsp70:myc-pu.1)hkz03t (Tg+) and nontransgenic sibling (Tg-) embryos having undergone the same heat-shock and staining protocol. The asterisks indicate statistical difference (t test, cebp1Tg-(mean/SE/n) = 35.6/2.9/12, cebp1Tg+ = 15.6/1.7/11; mpxTg- = 96.9/4.8/13; mpxTg+ = 74.4/5.8/10; SBTg- = 134.4/5.6/18, SBTg+ = 99.3/5.0/16; irf8Tg- = 41.8/7.6/4; irf8Tg+ = 66.3/5.2/12; csf1raTg- = 46.4/6.6/12; csf1raTg+ = 70.5/4.3/12; *P < .05, **P < .001). (A-B,E-F) Embryos are viewed dorsally with the anterior to the left.
Runx1 represses pu.1 expression in a negative feedback loop
To interrogate how appropriate Pu.1 level is attained in vivo, we hypothesized that it could be achieved by transcription factors tuning pu.1 expression at the transcription level; therefore, analysis of pu.1 promoter might provide a handle. Prompted by this hypothesis, we performed in silico analysis to search for transcription factor binding sites within the pu.1 upstream regulatory region. Among some other putative transcription factor binding sites, 8 putative runx1 recognition motifs were identified in the 9-kb pu.1 promoter region. In light of the presence of putative runx1 motifs in the pu.1 promoter and the expression of runx1 in embryonic myeloid cells in the RBI (Figure 3E,G), we set out to address the potential regulation of pu.1 expression by runx1 in subsequent study.
Figure 3.
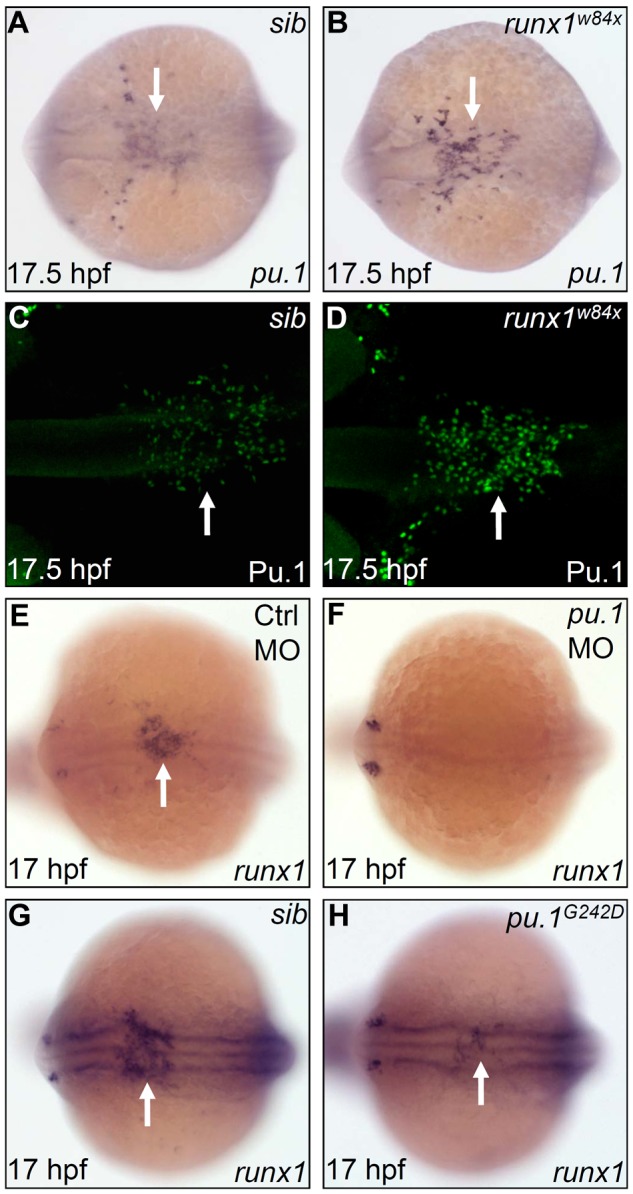
Pu.1 initiates the expression of runx1, whereas Runx1 suppresses late-phase expression of pu.1. (A-B) WISH of pu.1 expression in 17.5 hpf sib (A) and runx1w84x (B) embryos. Arrows indicate WISH signal of pu.1. (C-D) Whole-mount antibody staining of Pu.1 protein in 17.5 hpf sib (C) and runx1w84x (D) embryos. Arrows indicate fluorescent antibody staining signal for Pu.1 protein. (E-F) WISH of runx1 expression in 17 hpf control (E) and pu.1 morphants (F, n = 47 of 48). The arrow indicates WISH signal of runx1 in control morphants (E), which is absent in pu.1 morphants (F). (G-H) WISH of runx1 expression in 17 hpf sib (G) and pu.1G242D mutants (H). Arrows indicate WISH signal of runx1, which is markedly reduced in pu.1G242D mutants (H) compared with sib (G). (A-H) Embryos are viewed dorsally with the anterior to the left.
To test the role of runx1 in controlling pu.1 expression, we investigated the expression of pu.1 in the runx1 loss-of-function mutants (runx1w84x), which harbor a premature stop codon in the essential functional domain, the runt domain.23,24 The initial pu.1 RNA expression at 12 hpf was found to be unaffected by runx1w84x mutation (data not shown). However, per-cell pu.1 expression at 17.5 hpf was markedly intensified in the runx1w84x mutants, especially in the midline population, which normally expressed less pu.1 than those emigrating to the yolk sac (Figure 3A-B). A similarly marked enhancement (per-cell basis) was detected at protein level using an anti-Pu.1 antibody (Figure 3C-D). These expression analyses indicate that runx1 limits the pu.1 expression after the onset of embryonic myelopoiesis in the RBI. To further explore the hierarchical regulation between pu.1 and runx1, we evaluated the effect of Pu.1 deficiency on runx1 expression. We found that runx1 expression was nearly absent when Pu.1 was maximally depleted with a pu.1 MO and was markedly reduced when Pu.1 activity was partially removed by the pu.1G242D allele (Figure 3E-F, n = 47 of 48; Figure 3G-H). As the count of pu.1+ cells in pu.1G242D mutants was comparable with that in WT siblings (supplemental Figure 5A-B) and runx1+ cells coexpressed pu.1 in the RBI, the decreased expression of runx1 in pu.1G242D mutants thus indicated that the expression of runx1 rather than the frequency of runx1+ cells was regulated by high Pu.1 level. Taken together, the observations that runx1 expression is suppressed on Pu.1 deficiency and only late phase of pu.1 expression is elevated in runx1w84x mutants suggest a negative feedback loop whereby pu.1 initiates runx1 expression and runx1 in turn represses pu.1 expression.
Suppressed embryonic neutrophil but enhanced macrophage development in runx1w84x mutants
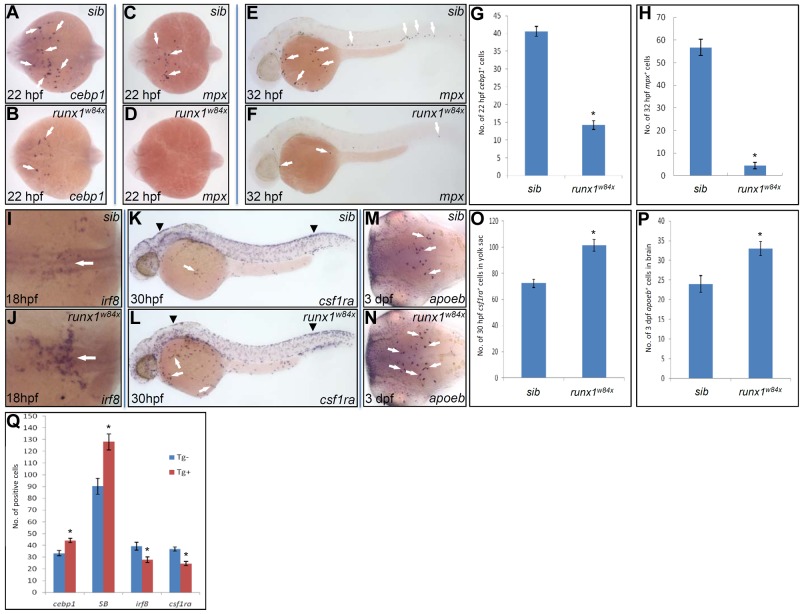
As Pu.1 overexpression alone was able to shift the fate of embryonic neutrophil to that of macrophage, we thus speculated that similar fate switching might also occur in runx1w84x mutants, given the up-regulated expression of pu.1 in runx1w84x mutants. To test this possibility, we first scrutinized embryonic myeloid development in runx1w84x mutants with a panel of markers that discern embryonic myeloid subsets at different development stages. We observed that the expression of pu.1 at 12 hpf was essentially unaffected in runx1w84x mutants, indicating the uncommitted embryonic myeloid progenitors was normally formed in runx1w84x mutants (data not shown). By contrast, the expression of cebp1, a marker of early neutrophil progenitor, was markedly decreased in runx1w84x mutants from the time when this marker became first detectable in the RBI of the embryos, and quantification showed that the number of cebp1+ cells at 22 hpf in runx1w84x mutants was only approximately one-third of that of siblings (Figure 4A-B,G). Accordingly, the expression of more differentiated neutrophil markers, including mpx at 22 hpf (Figure 4C-D), mpx/lyz at 32 hpf (Figure 4E-F; supplemental Figure 7A-B), and SB at 36 hpf (supplemental Figure 7C-D), was also significantly decreased in runx1w84x mutants compared with siblings. Quantification showed that the total number of mpx+ cells at 32 hpf in runx1w84x mutants was approximately one-tenth of that of siblings (Figure 4H). Neutrophil scoring with in vivo video enhanced DIC microscopy further confirmed that living runx1w84x mutants at 36 to 48 hpf indeed contained far less number of mature neutrophils (supplemental Figure 7E-F). BrdU pulse-chase experiments and Hoechst DNA content analysis indicated that decreased embryonic neutrophil development was not the result of the inhibited cell proliferation (supplemental Figure 8). Altogether, diminished neutrophil progenitors in the RBI but intact uncommitted progenitors and unchanged cell division suggest that the diminution of neutrophil number in runx1w84x mutants results from a blockage of neutrophil specification in the RBI. As embryonic neutrophils were also implicated to be derived from EMPs residing in the posterior blood island (PBI),13 we thus inferred the impact of runx1w84x mutation on EMP-derived neutrophils by comparing the number of neutrophils in the PBI with the total neutrophil number. It showed that, in the runx1w84x mutants, the extent of decrease in the neutrophil number in the PBI and in the total neutrophil number was comparable (data not shown), indicating that neutrophils arising from EMPs might also be affected by runx1w84x in a way similar to their RBI counterparts.
Figure 4.
Runx1 regulates embryonic neutrophil versus macrophage fate choice. (A-B) WISH of cebp1 expression in 22 hpf siblings (sib; A arrows) and runx1w84x mutants (B arrows). (C-D) WISH of mpx expression in 22 hpf sib (panel C arrows) and runx1w84x mutants (D). (A-D) Embryos are viewed dorsally with the anterior to the left. (E-F) WISH of mpx expression in 32 hpf sib (E arrows) and runx1w84x mutants (F arrows). (G) Quantification of the number of cebp1+ cells in 22 hpf sib and runx1w84x mutants. *P < .001 (t test, cebp1sib(mean/SE/n) = 40.6/1.4/54, cebp1runx1w84x = 14.2/1.3/17). (H) Quantification of the number of mpx+ cells in 32 hpf sib and runx1w84x mutants. *P < .001 (t test, mpxsib(mean/SE/n) = 56.7/3.6/30, mpxrunx1w84x = 4.5/1.4/9). (I-J) WISH of irf8 expression in 18 hpf sib (I, arrow) and runx1w84x mutants (J, arrow). Embryos are viewed dorsally with the anterior to the left. (K-L) WISH of csf1ra expression in 30 hpf sib (K) and runx1w84x mutants (L). Arrows indicate csf1ra expressed by macrophage; and arrowheads, csf1ra expressed by neural crest cells. (M-N) WISH of apoeb expression in 3 dpf sib (M arrows) and runx1w84x mutants (N arrows). Embryos are viewed dorsally with the anterior to the left. (O) Quantification of the number of csf1ra+ cells in the yolk sac of 30 hpf sib and runx1w84x mutants. *P < .001 (t test, csf1rasib(mean/SE/n) = 72.4/3.2/7, csf1rarunx1w84x = 101.5/4.5/11). (P) Quantification of the number of apoeb+ cells in the brain of 3 dpf sib and runx1w84x mutants. *P < .01 (t test, apoebsib(mean/SE/n) = 24.0/2.1/24, apoebrunx1w84x = 33.0/1.7/33). (Q) Effects of transient Runx1 overexpression on the development of cebp1+, SB+ neutrophils and irf8+, csf1ra+ macrophages. Embryos from Tg(hsp70:myc-runx1)hkz02t+/− crossed with AB WT are heat shocked at 11 hpf and fixed at 22, 36, 20, and 24 hpf for WISH detection of cebp1,SB, irf8, and csf1ra, respectively. The number of cells positive for these markers were counted and compared between Tg(hsp70:myc-runx.1)hkz02t (Tg+) and nontransgenic sibling (Tg−) embryos having undergone the same heat-shock and staining protocol. *P < .01 (t test, cebp1Tg-(mean/SE/n) = 33.4/2.4/27, cebp1Tg+ = 44.1/1.9/32; SBTg− = 90.3/6.8/17, SBTg+ = 128/6.9/15; irf8Tg− = 39.4/3.3/20; irf8Tg+ = 27.9/2.4/18; csf1raTg− = 37/1.8/22, csf1raTg+ = 24.4/1.9/22).
We next examined whether reduced specification of embryonic neutrophils in runx1w84x mutants was coupled with a skew toward the alternative lineage, macrophage. Contrary to the profound decrease of neutrophils, we noted an expanded macrophage compartment, including irf8+ macrophage progenitors, csf1ra+ young macrophages, and apoeb+ microglia (Figure 4I-N). Cell counting revealed that both csf1ra+ macrophages and apoeb+ microglia in runx1w84x mutants increased by 40% compared with siblings (Figure 4O-P). The enhanced macrophage but concomitantly reduced neutrophil formation in runx1w84x mutants indicates that, akin to Pu.1 overexpression, Runx1 deficiency biases myeloid output to macrophage fate.
Overexpression of Runx1 promotes embryonic neutrophil fate at the expense of macrophage fate
The opposing effect of runx1 loss-of-function mutation (runx1w84x) on neutrophil versus macrophage formation implies that runx1 might regulate embryonic myeloid fate choices by favoring neutrophil fate over macrophage fate. To further support this notion, we examined the consequence of Runx1 overexpression by creating a heat-inducible runx1 overexpression line Tg(hsp70:myc-runx1)hkz02t. Tg(hsp70:myc-runx1)hkz02t embryos did not yield detectable exogenous Myc-Runx1 and displayed normal embryonic myelopoiesis if grown at normal temperature 28°C (supplemental Figure 6C). On heat-shock treatment (at 39.5°C), a high level of Myc-Runx1 expression was induced in Tg(hsp70:myc-runx1)hkz02t embryos (supplemental Figure 6C-D). Heat-shock induction of Myc-Runx1 expression resulted in an enhanced neutrophil development, indicated by 30% increase of cebp1+ early neutrophil progenitors and 40% increase of SB+ neutrophils, and a suppressed macrophage development, as evidenced by 30% decrease of both irf8+ macrophage progenitors and csf1ra+ macrophages (Figure 4Q). Thus overexpression of runx1 promotes neutrophil formation but inhibits that of macrophages. Collectively, our loss-of-function and gain-of-function studies demonstrate that runx1 critically regulates embryonic myeloid cell fate choices through promoting neutrophil fate over that of macrophage.
Reducing pu.1 level in the runx1w84x mutant rescues its phenotype
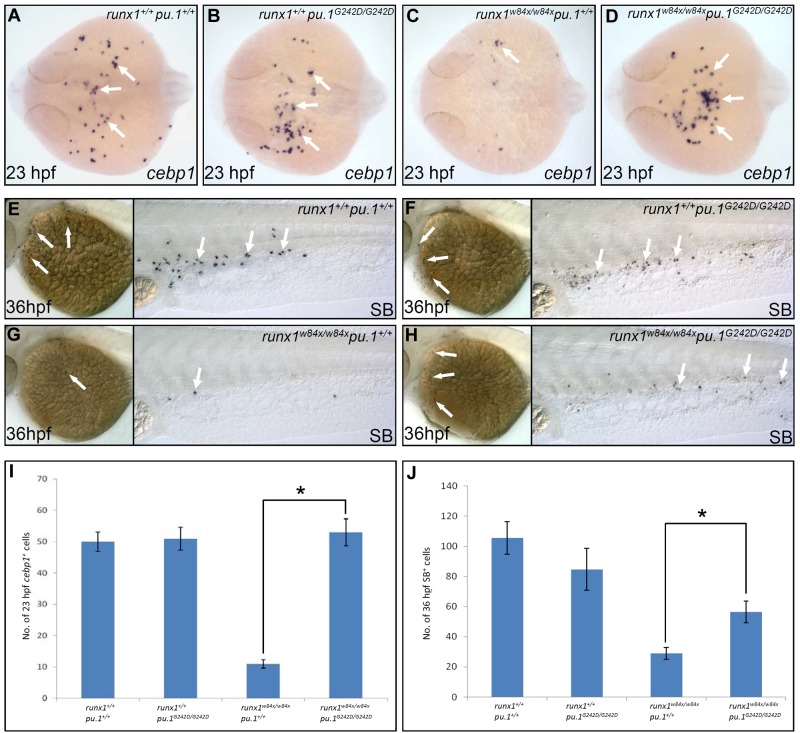
To directly demonstrate that lineage skewing and the resultant diminished neutrophil lineage development in the runx1w84x mutants were indeed the result of unconstrained Pu.1 activity, we tested whether reducing pu.1 level in runx1w84x mutants could reverse the runx1w84x phenotype by crossing hypomorphic pu.1G242D line and runx1w84x fish together. Introduction of one allele of pu.1G242D mutation into runx1w84x/w84x mutants partially restored the number of cebp1+ neutrophil progenitors (Table 1; supplemental Figure 9A-D,O; comparing runx1w84x/w84xpu.1G242D/+ with runx1w84x/w84xpu.1+/+). Biallelic pu.1G242D mutation in the runx1w84x/w84x background fully reverted the number of cebp1+ neutrophil progenitors to the level comparable with those in WT and pu.1 single homozygous mutants (Figure 5A-D,I; Table 1; comparing runx1w84x/w84xpu.1G242D/G242D with runx1w84x/w84xpu.1+/+ and with runx1+/+pu.1+/+ and runx1+/+pu.1G242D/G242D). Furthermore, both the numbers of lyz+ intermediate cells and SB+ mature cells in runx1w84x/w84x mutants were also significantly increased by combining runx1w84x/w84x with 1 or 2 alleles of pu.1G242D (Figure 5E-H,J; Table 1; supplemental Figure 9E-N,P-Q; comparing runx1w84x/w84xpu.1G242D/+ and runx1w84x/w84xpu.1G242D/G242D with runx1w84x/w84xpu.1+/+). Thus, reducing Pu.1 activity fully corrected embryonic neutrophil specification defects and partially rescued subsequent neutrophil differentiation in runx1w84x/w84x mutants. For macrophage development, akin to pu.1 single homozygous mutants, pu.1 and runx1 double homozygous mutants completely lacked macrophage cells as shown by WISH assay of multiple macrophage specific markers, underscoring the indispensable requirement for high Pu.1 level in driving macrophage fate (supplemental Figure 10). Collectively, the rescue of runx1w84x/w84x mutants by pu.1G242D, together with the up-regulated pu.1 expression in runx1w84x/w84x mutants, demonstrates that runx1 regulates bifurcate fate choice between neutrophils and macrophages via confining endogenous pu.1 expression.
Table 1.
Counts of cebp1+, lyz+, and SB+ neutrophil cells scored in various genetic combinations of runxw84x and pu.1G242D
| Genotype |
runx1+/+ |
runx1+/+ |
runx1+/+ |
runx1w84x/w84x |
runx1w84x/w84x |
runx1w84x/w84x |
|---|---|---|---|---|---|---|
| Marker | pu.1+/+ | pu.1G242D/+ | pu.1G242D/G242D | pu.1+/+ | pu.1G242D/+ | pu.1G242D/G242D |
| cebp1* (mean/SE/embryo no.) | 50.3/3.1/13 | 54.2/3.2/23 | 51.2/3.7/12 | 11/1.3/10 | 24.0/2/20 | 53.2/4.3/15 |
| lyz* (mean/SE/embryo no.) | 79.5/6.0/8 | 63.0/2.9/21 | 72.3/12.8/6 | 23.0/2.0/13 | 32.7/3.5/18 | 34.0/3.9/9 |
| SB* (mean/SE/embryo no.) | 105.6/10.7/7 | 100.8/4.9/18 | 84.7/13.8/7 | 28.9/3.9/11 | 52.3/6.2/15 | 56.5/7.3/11 |
The stage of embryos scored for cebp1+, lyz+, and SB+ cells was 23, 30, and 36 hpf, respectively.
Figure 5.
Reducing Pu.1 level rescues neutrophil deficit phenotype in runx1w84x mutants. (A-D) WISH of cebp1 expression in 23 hpf runx1+/+pu.1+/+ (A arrows), runx1+/+pu.1G242D/G242D (B arrows), runx1w84x/w84xpu.1+/+ (C arrow), runx1w84x/w84xpu.1G242D/G242D (D arrows). Embryos are viewed dorsally with the anterior to the left. (E-H) SB staining in 36 hpf runx1+/+pu.1+/+ (E arrows), runx1+/+pu.1G242D/G242D (F arrows), runx1w84x/w84xpu.1+/+ (G arrows), and runx1w84x/w84xpu.1G242D/G242D (H arrows). (I) Quantification of cebp1+ cell numbers in 23 hpf runx1+/+pu.1+/+, runx1+/+pu.1G242D/G242D, runx1w84x/w84xpu.1+/+, and runx1w84x/w84xpu.1G242D/G242D embryos. *P < .001 (t test, cebp1runx1+/+pu.1+/+(mean/SE/n) = 50.3/3.1/13, cebp1runx1+/+pu.1G242D/G242D = 51.2/3.7/12, cebp1runx1w84x/w84x pu.1+/+ = 11/1.3/10, cebp1runx1w84x/w84x pu.1G242D/G242D = 53.2/4.3/15). (J) Quantification of SB+ cell numbers in 36 hpf runx1+/+pu.1+/+, runx1+/+pu.1G242D/G242D, runx1w84x/w84xpu.1+/+, and runx1w84x/w84xpu.1G242D/G242D embryos. *P < .01 (t test, SBrunx1+/+ pu.1+/+ (mean/SE/n) = 105.6/10.7/7, SBrunx1+/+ pu.1G242D/G242D = 84.7/13.8/7, SBrunx1w84x/w84x pu.1+/+ = 28.9/3.9/11, SBrunx1w84x/w84x pu.1G242D/G242D = 56.5/7.3/11.
Runx1 directly represses pu.1 promoter
The presence of putative Runx1 recognition motifs in the 9.0-kb pu.1 promoter raises the possibility that Runx1 might directly suppress pu.1 promoter. As these 8 putative Runx1 recognition motifs were dispersed throughout the 9.0-kb pu.1 promoter, we first conducted promoter deletion analysis to assess the contribution of identified Runx1 sites to transcription repression by runx1. We took the advantage of 2 available pu.1 reporters, Tg(−5.3pu.1:eGFP) and Tg(−9.0pu.1:eGFP),21,42 and assayed GFP expression driven by these 2 regulatory fragments in runx1w84x mutants and sibling controls. We found that the level of GFP expressed from Tg(−5.3pu.1:eGFP) was comparable between runx1w84x mutants and clutchmates (supplemental Figure 11A) measured by quantitative RT-PCR. In contrast, the level of GFP expressed from injected Tg(−9.0pu.1:eGFP) DNA construct was substantially higher in runx1w84x mutants compared with clutchmates (supplemental Figure 11B). The finding that GFP driven by 9.0-kb but not 5.3-kb pu.1 upstream regulatory region exhibited higher expression in runx1w84x mutants than in clutchmates indicates that Runx1 binding sites in the 3.7-kb distal region (−9.0 to −5.3 kb) might be candidate cis-elements mediating the suppressive effect by runx1. Two adjoining runx1 recognition motifs of high similarity score were present in the 3.7-kb distal promoter region (Figure 6A).
Figure 6.
Runx1 represses pu.1 expression through acting on pu.1 promoter. (A) Schematic diagram of pu.1 promoter. Two adjoining putative runx1 binding sites (red arrowheads) in the distal portion of pu.1 promoter are predicted by promo, Version 3.0 online software. Red bars represent positions of primers designed to test for Runx1 binding. runx1-I amplifies the region containing 2 putative Runx1 sites, whereas runx1-II amplifies the region devoid of runx1 sites. (B) Representative fluorescent images (left panels) of transient eGFP expression at 17.5 hpf in the RBI of WT embryos injected with −9.0pu.1:eGFP (top panels) and −9.0pu.1ΔR:eGFP (bottom panels) constructs. Right panels: Overlays with bright field images. (C) Quantitative RT-PCR for GFP expression at 17.5 hpf in WT embryos injected with −9.0pu.1:eGFP and −9.0pu.1ΔR:eGFP. Units on y-axis represent the relative fold change of GFP expression in WT embryos injected with −9.0pu.1:eGFP and −9.0pu.1ΔR:eGFP. Expression level was normalized with elf1α expression and the amount of injected DNA. Error bars represent SE. (D) Semiquantitative PCR analysis with chromatin before (input) and after immunoprecipitation with anti-Myc antibody or anti-BrdU antibody (negative control). Sequence of mespa gene promoter serves as a negative control. (E) A model for the regulatory network in controlling embryonic neutrophil and macrophage fate segregation. In this model, a graded Pu.1 level specifies embryonic neutrophil and macrophage fates with high Pu.1 activity required for macrophage fate formation and low Pu.1 supporting neutrophil fate formation. High Pu.1 activity might switch on the expression of its binding partner, Irf8, to establish the embryonic macrophage fate. High Pu.1 activity, on the other hand, turns on the expression of Runx1, which is a direct feedback repressor of pu.1 expression. This Pu.1-Runx1 negative feedback loop thus stabilizes a favorable Pu.1 level that is essential for the formation of neutrophil fate.
To demonstrate the occupancy of these sites by Runx1, we performed a ChIP experiment with extracts from heat-shocked Tg(hsp70:myc-runx1)hkz02t embryos overexpressing a myc tagged version of Runx1. We immunoprecipitated recombinant Runx1 protein and its associated chromatin with an anti-myc antibody and then subjected the immunoprecipitated DNA to semiquantitative PCR analysis using primer pairs that tiled the 3.7-kb pu.1 distal promoter region (Figure 6D). Using this assay, we demonstrate the specific binding of Runx1 protein to the region harboring the 2 putative Runx1 sites. To further determine whether Runx1 acts through these 2 Runx1 motifs to repress pu.1 expression in vivo, we generated a reporter construct (−9.0pu.1ΔR:eGFP) harboring GFP driven by the 9.0-kb pu.1 upstream regulatory region specifically lacking the 2 Runx1-binding motifs. GFP expressed from transiently injected −9.0pu.1ΔR:eGFP construct maintained the same spatial expression pattern as that from the intact −9.0pu.1:eGFP construct, suggesting that removal of Runx1-binding motifs does not affect the tissue specificity of pu.1 promoter. However, eGFP expression in the RBI of the embryos receiving −9.0pu.1ΔR:eGFP construct was evidently increased compared with that in the embryos receiving −9.0pu.1:eGFP construct (Figure 6B). Quantitatively, deletion of Runx1-binding motifs resulted in a 3.5-fold increase of GFP when measured in a transient transgenic assay (Figure 6C). Together, these data suggest that Runx1 feedback represses pu.1 expression via directly acting on pu.1 promoter.
Discussion
In the present study, we exploited the strength of this highly tractable zebrafish system to establish a transcriptional regulatory hierarchy required for the homeostasis of embryonic macrophage and neutrophil specification. The core of this regulatory circuit is a Pu.1-Runx1 negative feedback loop wherein Pu.1 activates the expression of a transcriptional repressor, Runx1, to limit its own activity (Figure 6E). Unlike positive feedback circuit, which leads to rapid spiral change of activities, negative feedback loop attains an equilibrium state of the output concentration and renders resistance to perturbation. Given the central role of Pu.1 dosage in driving alternative myeloid fates suggested by the in vitro5,6 and our current in vivo studies, the Pu.1-Runx1 negative feedback loop uncovered here would thus ensure the stabilization of Pu.1 concentration within a range favorable for balanced macrophage and neutrophil commitment.
Several possibilities can explain how Pu.1-Runx1 loop contribute to the regulation of embryonic myeloid fates. One possibility is that this loop may actively specify neutrophil fate by facilitating the expression of unknown factors. An alternative possibility is that this loop may be used to maintain cell competence for responding to neutrophil inducing factors by preventing the dominant macrophage program from being overactive. Candidate dominant macrophage fate promoting factors emerging might include interferon regulatory factor 8 (irf8)28,43 and miR-146a44 as enforced expression of both factors were shown to drive macrophage development. In particular, irf8 expression was completely lost in pu.1G242D mutants (supplemental Figure 10B) but augmented in runx1w84x mutants (Figure 4I-J). In addition, overexpression of irf8 in WT embryos shifted fate toward macrophages,28 while knocking down of irf8 rescues the phenotype of runx1w84x mutants. Elucidating the function relationship between Pu.1-Runx1 loop and these macrophage promoting molecules will thus aid comprehension of the regulatory circuits governing macrophage versus neutrophil fate choice.
Our study shows that Runx1 plays critical roles in regulating embryonic myeloid fate choice through promoting neutrophil fate over that of macrophage. However, whether this myeloid lineage selection role of Runx1 is relevant to adult phase of myelopoiesis has not been investigated. Blood profiling of adult kidney marrow of viable runx1w84x mutants revealed that the extent of decrease in neutrophil counts significantly exceeds that in macrophage counts (S. Jiping, L.L., and W.Z., unpublished data, June 2010), thus favoring a similar role of Runx1 in adult myelopoiesis. It has occurred to us that adult ablation of RUNX1 in mice instead causes either no observable myeloid phenotype45 or mild myeloid expansion with a basis of increased granulocyte-macrophage progenitors.46 Although such discrepancy might be attributed to species difference or hematopoiesis phase difference, alternative explanation might lie in the difference of these 2 systems used to study these processes. The lineage specific role of RUNX1 in mice was inferred by classic approach in which conditional alleles of RUNX1 were removed in most adult blood cells by inducible Mx-Cre and the consequence of the deletion was assessed thereafter. In such assay, the effect of RUNX1 depletion in myeloid development per se might be confounded by the loss of Runx1 in various upstream progenitors of myeloid potential and those blood cells capable of modulating myeloid differentiation, thus potentially masking the myeloid specific role of RUNX1. Moreover, if a phenotype is discerned, it often suffers from the inadequacy to track down the exact underlying cellular mechanism. For instance, it often remains to show in many of these studies whether the alteration of a given population arising from a particular gene targeting is because of changes in upstream progenitors, or changes in cell fate, division, or migration.44 By contrast, zebrafish embryonic myeloid cells arise without transiting from HSCs and with minimal influence from other blood cells (erythropoiesis occurs in an anatomically different site),14 allowing to directly access the role of Runx1 in myelopoiesis. In addition, the sequence of myeloid development in fish embryo could be defined by temporally ordered expression of molecular markers, and many in vivo cell tracking tools, including photo-activatable dye47 and photo-switchable protein48 are now available, making it possible to definitively pinpoint the underlying cellular defect. Hence, it can be anticipated that these merits combined with the genetic tractability of fish system would accelerate the identification of new conserved players in myelopoiesis. Indeed, the conserved role of Runx1 in myeloid fate choices is supported by findings showing that lozenge, the Drosophila ortholog of Runx genes, specifies crystal fate from undifferentiated prohemocytes, which otherwise give rise to plasmatocytes, a fly equivalent of macrophage.49
Our current work does not deny any potential role of Runx1 in subsequent maturation or differentiation of neutrophils after the fate of these cells is established. Indeed, the expression of more advanced neutrophil markers, such as mpx, lyz, and SB, was more severely disrupted compared with that of early neutrophil progenitor marker, cebp1. In addition, we noted that introducing biallelic pu.1G242D mutation to runx1w84x mutants was inadequate to fully restore the expression of more differentiated neutrophil markers, such as lyz, SB to the level in pu.1G242D single mutants and WT, despite full recovery of cebp1+ neutrophil progenitors in pu.1G242Drunx1w84x double mutants. These observations are consistent with studies showing that Runx1 regulates the promoter of myeloid differentiation genes50 and point to a probable additional role of Runx1 in subsequent neutrophil maturation or differentiation apart from its early Pu.1 repressive role in fate decision. It will thus be of interest in the future to pursue how Runx1 coordinates with other myeloid factors to fulfill its differentiation role.
Supplementary Material
Acknowledgments
The authors thank Drs Nathan Lawson and Koichi Kawakami for providing pTol vector, Dr Tingxi Liu for providing −9.0pu.1:eGFP construct, Dr Graham J. Lieschke for providing Tg(−5.3pu.1:eGFP) line, Dr Phil Crosier for providing Tg(lyz:Dsred) line, Drs Philip W. Ingham and Steve Renshaw for providing Tg(mpx:eGFP) line, and Dr Philippe Herbomel for his instrumental help in setting up the video-enhanced DIC microscopy.
This work was supported by the General Research Fund (Research Grants Council of the HKSAR, grant HKUST6/CRF/09), the National Basic Research Program of China (grant 2012CB945102), the National Natural Science Foundation of China (grants 31171403 and 30828020), and the Intramural Research Program of National Human Genome Research Institute, National Institutes of Health.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.J., L.L., J.X., F.Z., and L.Z. designed the research, performed experiments, and analyzed data; P.P.L. provided reagents; M.J.Z. analyzed data; and W.Z. and Z.W. designed the research and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Zilong Wen, State Key Laboratory of Molecular Neuroscience, Division of Life Science, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, PR China; e-mail: zilong@ust.hk; and Wenqing Zhang, Key Laboratory of Zebrafish Modeling and Drug Screening for Human Diseases of Guangdong Higher Education Institutes, Department of Cell Biology, Southern Medical University, Guangzhou 510515, PR China; e-mail: zzwwqq@smu.edu.cn.
References
- 1.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26(6):726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7(2):105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 3.McKercher SR, Torbett BE, Anderson KL, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15(20):5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 4.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 5.Dahl R, Walsh JC, Lancki D, et al. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat Immunol. 2003;4(10):1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- 6.Laslo P, Spooner CJ, Warmflash A, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126(4):755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 7.Dakic A, Wu L, Nutt SL. Is PU.1 a dosage-sensitive regulator of haemopoietic lineage commitment and leukaemogenesis? Trends Immunol. 2007;28(3):108–114. doi: 10.1016/j.it.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Rosenbauer F, Wagner K, Kutok JL, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36(6):624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 9.Bennett CM, Kanki JP, Rhodes J, et al. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98(3):643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- 10.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126(17):3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 11.Lieschke GJ, Oates AC, Paw BH, et al. SPI-1 (PU.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev Biol. 2002;246(2):274–295. doi: 10.1006/dbio.2002.0657. [DOI] [PubMed] [Google Scholar]
- 12.Le Guyader D, Redd MJ, Colucci-Guyon E, et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111(1):132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134(23):4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman JN, Kanki JP, Look AT. Zebrafish as a model for myelopoiesis during embryogenesis. Exp Hematol. 2005;33(9):997–1006. doi: 10.1016/j.exphem.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 15.de Jong JL, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- 16.Rhodes J, Hagen A, Hsu K, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8(1):97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Su F, Juarez MA, Cooke CL, et al. Differential regulation of primitive myelopoiesis in the zebrafish by Spi-1/Pu.1 and C/ebp1. Zebrafish. 2007;4(3):187–199. doi: 10.1089/zeb.2007.0505. [DOI] [PubMed] [Google Scholar]
- 18.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene, OR: University of Oregon Press; 2000. [Google Scholar]
- 19.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108(13):3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 20.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward AC, McPhee DO, Condron MM, et al. The zebrafish spi1 promoter drives myeloid-specific expression in stable transgenic fish. Blood. 2003;102(9):3238–3240. doi: 10.1182/blood-2003-03-0966. [DOI] [PubMed] [Google Scholar]
- 22.Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127(14):3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 23.Jin H, Sood R, Xu J, et al. Definitive hematopoietic stem/progenitor cells manifest distinct differentiation output in the zebrafish VDA and PBI. Development. 2009;136(4):647–654. doi: 10.1242/dev.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sood R, English MA, Belele CL, et al. Development of multilineage adult hematopoiesis in the zebrafish with a runx1 truncation mutation. Blood. 2010;115(14):2806–2809. doi: 10.1182/blood-2009-08-236729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin H, Xu J, Wen Z. Migratory path of definitive hematopoietic stem/progenitor cells during zebrafish development. Blood. 2007;109(12):5208–5214. doi: 10.1182/blood-2007-01-069005. [DOI] [PubMed] [Google Scholar]
- 26.Wienholds E, Schulte-Merker S, Walderich B, Plasterk RH. Target-selected inactivation of the zebrafish rag1 gene. Science. 2002;297(5578):99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- 27.Jin H, Xu J, Qian F, et al. The 5′ zebrafish scl promoter targets transcription to the brain, spinal cord, and hematopoietic and endothelial progenitors. Dev Dyn. 2006;235(1):60–67. doi: 10.1002/dvdy.20613. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Jin H, Xu J, Shi Y, Wen Z. Irf8 regulates macrophage versus neutrophil fate during zebrafish primitive myelopoiesis. Blood. 2011;117(4):1359–1369. doi: 10.1182/blood-2010-06-290700. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Jin H, Li L, Qin FX, Wen Z. cMyb regulates hematopoietic stem/progenitor cell mobilization during zebrafish hematopoiesis. Blood. 2011;118(15):4093–4101. doi: 10.1182/blood-2011-03-342501. [DOI] [PubMed] [Google Scholar]
- 30.Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450(7172):1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98(10):3087–3096. doi: 10.1182/blood.v98.10.3087. [DOI] [PubMed] [Google Scholar]
- 32.Meijer AH, van der Sar AM, Cunha C, et al. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev Comp Immunol. 2008;32(1):36–49. doi: 10.1016/j.dci.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238(2):274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- 34.Lekstrom-Himes J, Xanthopoulos KG. CCAAT/enhancer binding protein epsilon is critical for effective neutrophil-mediated response to inflammatory challenge. Blood. 1999;93(9):3096–3105. [PubMed] [Google Scholar]
- 35.Lyons SE, Shue BC, Oates AC, Zon LI, Liu PP. A novel myeloid-restricted zebrafish CCAAT/enhancer-binding protein with a potent transcriptional activation domain. Blood. 2001;97(9):2611–2617. doi: 10.1182/blood.v97.9.2611. [DOI] [PubMed] [Google Scholar]
- 36.Yamanaka R, Barlow C, Lekstrom-Himes J, et al. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein epsilon-deficient mice. Proc Natl Acad Sci U S A. 1997;94(24):13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cramer EM, Breton-Gorius J. Ultrastructural localization of lysozyme in human neutrophils by immunogold. J Leukoc Biol. 1987;41(3):242–247. doi: 10.1002/jlb.41.3.242. [DOI] [PubMed] [Google Scholar]
- 38.Shaft D, Shtalrid M, Berebi A, Catovsky D, Resnitzky P. Ultrastructural characteristics and lysozyme content in hypergranular and variant type of acute promyelocytic leukaemia. Br J Haematol. 1998;103(3):729–739. doi: 10.1046/j.1365-2141.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 39.Kitaguchi T, Kawakami K, Kawahara A. Transcriptional regulation of a myeloid-lineage specific gene lysozyme C during zebrafish myelopoiesis. Mech Dev. 2009;126(5):314–323. doi: 10.1016/j.mod.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 40.Dakic A, Metcalf D, Di RL, Mifsud S, Wu L, Nutt SL. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201(9):1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halloran MC, Sato-Maeda M, Warren JT, et al. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127(9):1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- 42.Hsu K, Traver D, Kutok JL, et al. The pu.1 promoter drives myeloid gene expression in zebrafish. Blood. 2004;104(5):1291–1297. doi: 10.1182/blood-2003-09-3105. [DOI] [PubMed] [Google Scholar]
- 43.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13(2):155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 44.Ghani S, Riemke P, Schonheit J, et al. Macrophage development from HSCs requires PU.1-coordinated microRNA expression. Blood. 2011;118(8):2275–2284. doi: 10.1182/blood-2011-02-335141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10(3):299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 46.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106(2):494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong TP, Childs S, Leu JP, Fishman MC. Gridlock signalling pathway fashions the first embryonic artery. Nature. 2001;414(6860):216–220. doi: 10.1038/35102599. [DOI] [PubMed] [Google Scholar]
- 48.Scott EK, Mason L, Arrenberg AB, et al. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4(4):323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- 49.Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science. 2000;288(5463):146–149. doi: 10.1126/science.288.5463.146. [DOI] [PubMed] [Google Scholar]
- 50.Lutterbach B, Hiebert SW. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene. 2000;245(2):223–235. doi: 10.1016/s0378-1119(00)00014-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.