Abstract
Fluorescence in situ hybridization (FISH) is a technique that allows specific DNA sequences to be detected on metaphase or interphase chromosomes in cell nuclei1. The technique uses DNA probes with unique sequences that hybridize to whole chromosomes or specific chromosomal regions, and serves as a powerful adjunct to classic cytogenetics. For instance, many earlier studies reported the frequent detection of increased chromosome aberrations in leukemia patients related with benzene exposure, benzene-poisoning patients, and healthy workers exposed to benzene, using classic cytogenetic analysis2. Using FISH, leukemia-specific chromosomal alterations have been observed to be elevated in apparently healthy workers exposed to benzene3-6, indicating the critical roles of cytogentic changes in benzene-induced leukemogenesis.
Generally, a single FISH assay examines only one or a few whole chromosomes or specific loci per slide, so multiple hybridizations need to be conducted on multiple slides to cover all of the human chromosomes. Spectral karyotyping (SKY) allows visualization of the whole genome simultaneously, but the requirement for special software and equipment limits its application7. Here, we describe a novel FISH assay, OctoChrome-FISH, which can be applied for Chromosomics, which we define here as the simultaneous analysis of all 24 human chromosomes on one slide in human studies, such as chromosome-wide aneuploidy study (CWAS)8. The basis of the method, marketed by Cytocell as the Chromoprobe Multiprobe System, is an OctoChrome device that is divided into 8 squares, each of which carries three different whole chromosome painting probes (Figure 1). Each of the three probes is directly labeled with a different colored fluorophore, green (FITC), red (Texas Red), and blue (Coumarin). The arrangement of chromosome combinations on the OctoChrome device has been designed to facilitate the identification of the non-random structural chromosome alterations (translocations) found in the most common leukemias and lymphomas, for instance t(9;22), t(15;17), t(8;21), t(14;18)9. Moreover, numerical changes (aneuploidy) in chromosomes can be detected concurrently. The corresponding template slide is also divided into 8 squares onto which metaphase spreads are bound (Figure 2), and is positioned over the OctoChrome device. The probes and target DNA are denatured at high-temperature and hybridized in a humid chamber, and then all 24 human chromosomes can be visualized simultaneously.
OctoChrome FISH is a promising technique for the clinical diagnosis of leukemia and lymphoma and for detection of aneuploidies in all chromosomes. We have applied this new Chromosomic approach in a CWAS study of benzene-exposed Chinese workers8,10.
Keywords: Genetics, Issue 60, Chromosomics, OctoChrome-FISH, fluorescence in situ hybridization (FISH), Chromosome-wide aneuploidy study (CWAS), aneuploidy, chromosomal translocations, leukemia, lymphoma
Protocol
1. Sample slide preparation
OctoChrome FISH is designed to examine chromosomal changes in metaphase spreads of human cells, including but not limited to cultured peripheral blood cells, stem/progenitor cells, and cell lines. Samples should be prepared following the standard procedure for harvesting metaphases3,11-13: by employing colcemid treatment to arrest cells in metaphase, hypotonic treatment to swell cells, and Carnoy's fixative to fix cells in suspension at approximately 0.5-1 x 106 cells per ml.
Clean the 8-square slide (provided in the OctoChrome Kit, Catalogue #: PMP 803, Cytocell Ltd, Cambridge, UK, Figure 2) by dipping into pure ethanol for 2 min.
Before making the sample slide, drop a small amount of cells onto a slide to check the cell density. If the cell density is too high (too many overlapping cells), dilute the suspension with fresh fixative. If the cell density is too low (too few cells on slide), spin down the cells and re-suspend in a smaller volume of fresh fixative. Pipette 5-10 μl of cell suspension onto each of 8 areas of the template slide in a sequence of alternating squares 1/7 - 2/8 - 3/5 - 4/6. Once the first two squares has air-dried, spot the remaining squares in the same manner. This will prevent the cell spreads from interfering with each other.
The dried slide can be stored at -20°C in nitrogen atmosphere for greater than 5 years before hybridization.
2. Localization of metaphases using an automated system
Apply 20 μl of 4',6-diamidino-2-phenylindole (0.2 μg/ml) to the center of each half of the slide and then cover with a coverslip.
Perform an automated scan of the sample slide to localize the metaphase cells using Metafer software (MetaSystems, Altlussheim, Germany). This facilitates location of all the metaphases and future re-evaluation of all cell images.
View the scanning results manually and delete non-metaphase artifacts.
3. Hybridization
- Preparation of the sample slide and OctoChrome device
- Dip the sample slide in 2x SSC buffer at room temperature for 30 min to wash away DAPI.
- Dehydrate the sample slide through a series of ethanol washes (2 minutes each in 70%, 85% and 100% ethanol), allow to dry and place on a 37°C hotplate for 5 minutes.
- Place the Chromoprobe Multiprobe Hybridization Chamber (provided in the OctoChrome Kit) in a 37°C water bath and allow to equilibrate to 37°C (+/- 1°C).
- Mix the hybridization solution (provided in the OctoChrome Kit) by repeated pipetting and pre-warm a 25 μl aliquot per OctoChrome device to 37°C.
- Pre-warm each OctoChrome device (provided in the OctoChrome Kit, see Figure 2) to 37°C by placing the device label side down on a hotplate. DO NOT touch the embossed surfaces of the OctoChrome device as they contain the probes.
- Positioning of the sample slide over the OctoChrome device (Figure 2)
- Add 2 μl of pre-warmed hybridization solution to each of the eight areas on the pre-warmed OctoChrome device while it remains at 37°C.
- Carefully invert the template slide over the OctoChrome device such that the number 1, which is now upside down, is located over the top right hand area of the OctoChrome device. To help locate square 1, its position on the device has been marked in Orange.
- Make sure that the template slide is carefully aligned with the matching areas on the OctoChrome device. Carefully lower the slide over the OctoChrome device so that the drops of hybridization solution make contact with the slide. Apply gentle, even pressure to ensure that the hybridization solution is spread to the edges of each of the raised areas on the OctoChrome device.
- Lift the slide/OctoChrome, carefully holding the frosted end of the glass slide, and invert so that the slide is underneath the OctoChrome device. Make sure the device does not smear across the template slide as this could cause cross-contamination of the probes.
- Place on a 37°C (+/- 1°C) hotplate for 10 minutes.
- Denaturation
- Transfer the sample slide/OctoChrome device to the hotplate taking particular care to hold it level. Ensure the sample slide evenly contacts the hotplate.
- Denature on the hotplate at 75°C (+/- 1°C) for 5 minutes.
- Hybridization
- Place sample slide/OctoChrome device in the Chromoprobe Multiprobe Hybridization Chamber.
- Replace the lid and float the chamber in the 37°C (+/- 1°C) water bath (non - stirring) overnight. Please note: DO NOT seal the lid on the hybridization chamber; DO NOT seal the lid on the water bath; DO NOT hybridize in an incubator. These steps are critical to controlling humidity.
- Post-hybridization washes
- Preparation of wash solutions Solution 1: Prepare a Coplin/Hellendahl jar containing 0.4x SSC. Equilibrate to 72°C (+/- 1°C) in a water bath and adjust the pH to 7.0. Solution 2: Prepare a Coplin/Hellendahl jar containing 2x SSC and 0.05% Tween 20. Allow to stand at room temperature.
- Remove the OctoChrome device carefully from the slide and place the slide in Solution 1 for 2 minutes.
- Place the slide into Solution 2 for 30 seconds.
4. Mounting and visualization of results
Apply 20 μl of DAPI to the center of each half of the slide and apply a coverslip.
Incubate in the dark for 10 minutes at room temperature before viewing by fluorescence microscopy.
5. Representative Results
Figure 3A shows a normal metaphase cell in Square 2. (1)-(3): Chromosomes 8, 21 and 12 were painted red, green, and blue, and are visualized through a Texas Red, FITC, and DEAC filter, respectively; (4): Visualization through a DAPI/FITC/Texas Red triple filter. Generally, the chromosomes painted with blue are not clear through the triple filter and need to be viewed under the specific DEAC filter.
Figure 3B shows representative abnormal cells with leukemia-specific chromosomal translocation and aneuploidy. (1): t(8;21), a common chromosomal translocation in acute myeloid leukemia; (2): Trisomy 21, three copies of chromosome 21, a common aneuploidy in leukemia.
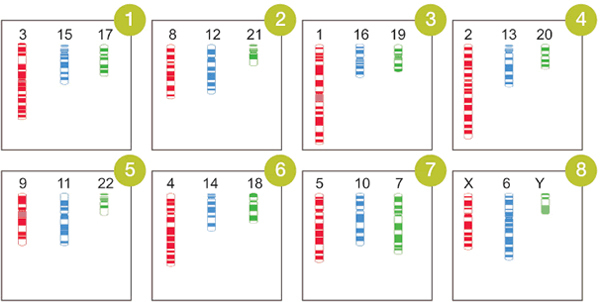
Figure 1. Arrangement of chromosome combinations on the OctoChrome device and expected chromosomal painting results.
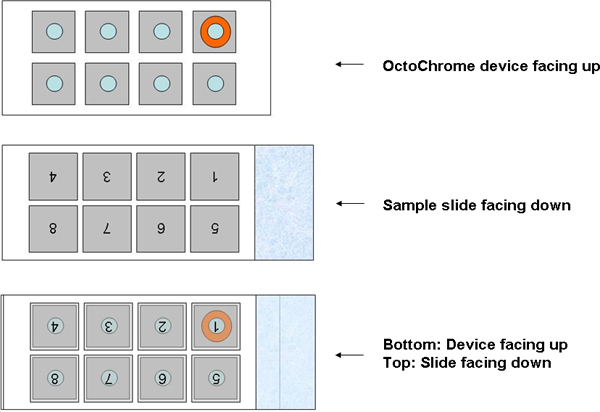
Figure 2. Positioning of sample slide over the OctoChrome device.
Figure 3. Representative results obtained from OctoChrome FISH (Square 2).
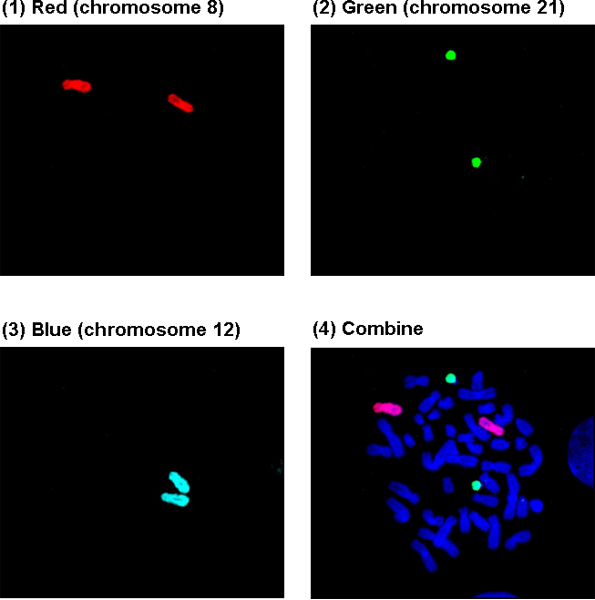
Figure 3A. A normal cell with chromosomes 8, 12, 21 painted in Square 2.
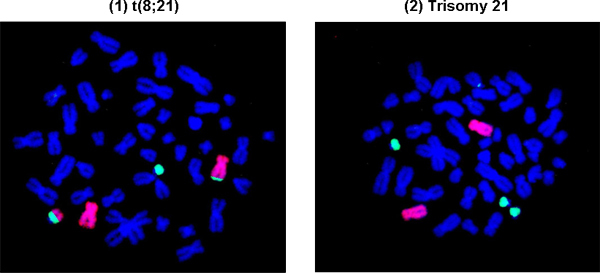
Figure 3B. Representative abnormal cells with leukemia-specific chromosomal translocation and aneuploidy in Square 2.
Discussion
This novel OctoChrome-FISH assay allows one to simultaneously examine all 24 human chromosomes in a single hybridization on one slide. The arrangement of chromosome combinations on the 8 squares has been designed to facilitate the identification of the non-random chromosome rearrangements in the most common leukemias and lymphomas. Thus, the assay can detect numerical (aneuploidy) as well as structural (translocations) chromosome alterations concurrently.
The most critical step in this assay is controlling the humidity during the overnight hybridization in the floating chamber in the water bath. The FISH probes and hybridization buffer easily dry out if the humidity is low, or become too dilute by absorbing too much moisture if the humidity is high. Either outcome could adversely impact the results. A possible modification to overcome this challenge is to seal the OctoChrome device and sample slide with tape and hybridize on a 37°C hotplate in the dark overnight.
Sample slides that have been aged too much may be unsuitable for use in this assay directly. While digesting such samples with pepsin will improve hybridization, storing prepared sample slides in a nitrogen atmosphere at -20°C is strongly recommended to minimize aging.
OctoChrome-FISH is a promising technique for the clinical diagnosis of leukemia and lymphoma. It is also very useful for examining most specific chromosomal rearrangements related to human leukemia and lymphoma in populations exposed to potential leukemogens and lymphomagens and for studying the aneuploidy-inducing effects of chemicals in a chromosome-wide manner. Our previous CWAS study using this technique demonstrated that certain chromosomes may be more affected than others by exposure to benzene8,10, a primary industrial chemical and a ubiquitous environmental pollutant that causes human leukemia14. This phenomenon of “selective aneuploidy” could be explored in other chemical exposures using CWAS.
Disclosures
We have nothing to disclose.
Acknowledgments
The authors would like to thank Dr. Cliona M. McHale for critically reading and editing the manuscript. This work was funded by the National Institute of Environmental Health Sciences, National Institute of Health grant R01ES017452 to L Zhang.
References
- Levsky JM, Singer RH. Fluorescence in situ hybridization: past, present and future. J. Cell. Sci. 2003;116:2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- Zhang L, Eastmond DA, Smith MT. The nature of chromosomal aberrations detected in humans exposed to benzene. Crit. Rev. Toxicol. 2002;32:1–42. doi: 10.1080/20024091064165. [DOI] [PubMed] [Google Scholar]
- Zhang L. Aberrations in chromosomes associated with lymphoma and therapy-related leukemia in benzene-exposed workers. Environ. Mol. Mutagen. 2007;48:467–474. doi: 10.1002/em.20306. [DOI] [PubMed] [Google Scholar]
- Zhang L. Benzene increases aneuploidy in the lymphocytes of exposed workers: a comparison of data obtained by fluorescence in situ hybridization in interphase and metaphase. Environ. Mol. Mutagen. 1999;34:260–268. [PubMed] [Google Scholar]
- Zhang L. Increased aneusomy and long arm deletion of chromosomes 5 and 7 in the lymphocytes of Chinese workers exposed to benzene. Carcinogenesis. 1998;19:1955–1961. doi: 10.1093/carcin/19.11.1955. [DOI] [PubMed] [Google Scholar]
- Smith MT. Increased translocations and aneusomy in chromosomes 8 and 21 among workers exposed to benzene. Cancer. Res. 1998;58:2176–2181. [PubMed] [Google Scholar]
- Bayani J, Squire JA. Advances in the detection of chromosomal aberrations using spectral karyotyping. Clin. Genet. 2001;59:65–73. doi: 10.1034/j.1399-0004.2001.590201.x. [DOI] [PubMed] [Google Scholar]
- Zhang L. Chromosome-wide aneuploidy study (CWAS) in workers exposed to an established leukemogen, benzene. Carcinogenesis. 2011;32:605–612. doi: 10.1093/carcin/bgq286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. The role of chromosome translocations in leukemogenesis. Semin. Hematol. 1999;36:59–72. [PubMed] [Google Scholar]
- Zhang L. Use of OctoChrome fluorescence in situ hybridization to detect specific aneuploidy among all 24 chromosomes in benzene-exposed workers. Chem. Biol. Interact. 2005. pp. 153–154. [DOI] [PubMed]
- Barch MJ, Lawce HJ, Arsham MS. In: In The ACT cytogenetics laboratory. Barch MJ, editor. Raven Press; 1991. pp. 17–30. [Google Scholar]
- Zhang L, Yang W, Hubbard AE, Smith MT. Nonrandom aneuploidy of chromosomes 1, 5, 6, 7, 8, 9, 11, 12, and 21 induced by the benzene metabolites hydroquinone and benzenetriol. Environ. Mol. Mutagen. 2005;45:388–396. doi: 10.1002/em.20103. [DOI] [PubMed] [Google Scholar]
- Smith MT. Hydroquinone, a benzene metabolite, increases the level of aneusomy of chromosomes 7 and 8 in human CD34-positive blood progenitor cells. Carcinogenesis. 2000;21:1485–1490. [PubMed] [Google Scholar]
- Smith MT. Advances in understanding benzene health effects and susceptibility. Annu. Rev. Public. Health. 2010;31:133–148. doi: 10.1146/annurev.publhealth.012809.103646. [DOI] [PMC free article] [PubMed] [Google Scholar]


