Abstract
Glycosylation, the addition of covalently linked sugars, is a major post-translational modification of proteins that can significantly affect processes such as cell adhesion, molecular trafficking, clearance, and signal transduction1-4. In eukaryotes, the most common glycosylation modifications in the secretory pathway are additions at consensus asparagine residues (N-linked); or at serine or threonine residues (O-linked) (Figure 1). Initiation of N-glycan synthesis is highly conserved in eukaryotes, while the end products can vary greatly among different species, tissues, or proteins. Some glycans remain unmodified ("high mannose N-glycans") or are further processed in the Golgi ("complex N-glycans"). Greater diversity is found for O-glycans, which start with a common N-Acetylgalactosamine (GalNAc) residue in animal cells but differ in lower organisms1.
The detailed analysis of the glycosylation of proteins is a field unto itself and requires extensive resources and expertise to execute properly. However a variety of available enzymes that remove sugars (glycosidases) makes possible to have a general idea of the glycosylation status of a protein in a standard laboratory setting. Here we illustrate the use of glycosidases for the analysis of a model glycoprotein: recombinant human chorionic gonadotropin beta (hCGβ), which carries two N-glycans and four O-glycans 5. The technique requires only simple instrumentation and typical consumables, and it can be readily adapted to the analysis of multiple glycoprotein samples.
Several enzymes can be used in parallel to study a glycoprotein. PNGase F is able to remove almost all types of N-linked glycans6,7. For O-glycans, there is no available enzyme that can cleave an intact oligosaccharide from the protein backbone. Instead, O-glycans are trimmed by exoglycosidases to a short core, which is then easily removed by O-Glycosidase. The Protein Deglycosylation Mix contains PNGase F, O-Glycosidase, Neuraminidase (sialidase), β1-4 Galactosidase, and β-N-Acetylglucosaminidase. It is used to simultaneously remove N-glycans and some O-glycans8 . Finally, the Deglycosylation Mix was supplemented with a mixture of other exoglycosidases (α-N-Acetylgalactosaminidase, α1-2 Fucosidase, α1-3,6 Galactosidase, and β1-3 Galactosidase ), which help remove otherwise resistant monosaccharides that could be present in certain O-glycans.
SDS-PAGE/Coomasie blue is used to visualize differences in protein migration before and after glycosidase treatment. In addition, a sugar-specific staining method, ProQ Emerald-300, shows diminished signal as glycans are successively removed. This protocol is designed for the analysis of small amounts of glycoprotein (0.5 to 2 μg), although enzymatic deglycosylation can be scaled up to accommodate larger quantities of protein as needed.
Keywords: Molecular Biology , Issue 58, Glycoprotein, N-glycan, O-glycan, PNGase F, O-glycosidase, deglycosylation, glycosidase
Protocol
1. Enzymatic deglycosylation
Use PCR tubes to minimize water loss due to evaporation. Label one set of tubes 1 to 7.
Thaw the 10X G7 buffer, the 10X glycoprotein denaturing buffer, and the 10% NP-40 and gently tap the tubes to mix the contents. Keep at room temperature.
Place the enzyme-containing vials on ice. Try to minimize thaw/freeze cycles.
Dissolve the contents of the hCGβ vial (150 μg) in 600 μl of dH2O and keep on ice.
Prepare 1 ml of 1X G7 buffer by diluting the 10X stock in dH2O.
Dilute 0.5 μl of PNGase F in 25 μl 1X G7 buffer and keep on ice.
Prepare the exoglycosidase mix (EG mix) by combining 2 μl each of α-N-Acetylgalactosaminidase, α1-2 Fucosidase, β1-3 Galactosidase, and α1-3, 6 Galactosidase.
Set up PCR tubes as indicated:
| Tube/sample# | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| hCGβ (0.25mg/ml) | 9μl | 9μl | 9μl | 9μl | -- | -- | -- |
| 10X Glycoprotein Denaturing Buffer | 1μl | 1μl | 1μl | 1μl | 1μl | 1μl | 1μl |
| dH2O | -- | -- | -- | -- | 9μl | 9μl | 9μl |
Cap the tubes, mix gently and place in the thermocycler, close the lid and denature the proteins by incubating 10 minutes at 94°C, followed by a 4°C hold (using PCR tubes in a thermocycler greatly prevents evaporation in small volume samples).
Remove the tubes from thermocycler and centrifuge to remove any visible condensation.
Add the following reagents as indicated:
| Sample# | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 10% NP-40 | 2.5μl | 2.5μl | 2.5μl | 2.5μl | 2.5μl | 2.5μl | 2.5μl |
| 10X G7 Buffer | 2.5μl | 2.5μl | 2.5μl | 2.5μl | 2.5μl | 2.5μl | 2.5μl |
| PNGase F 1:50 dil. | -- | 2μl | -- | -- | 2μl | -- | -- |
| Deglycosylation Mix | -- | -- | 2μl | 2μl | -- | 2μl | 2μl |
| EG mix | -- | -- | -- | 2μl | -- | -- | 2μl |
| dH2O | 10μl | 8μl | 8μl | 6μl | 8μl | 8μl | 6μl |
| Total reaction vol. | 25μl | 25μl | 25μl | 25μl | 25μl | 25μl | 25μl |
Close the PCR tubes using new caps (discard the used ones since they do not fit properly after one incubation cycle).
Mix the tubes by gently tapping 4 times and then spin the contents down.
Place the tubes in the thermocycler and incubate at 37 °C for 4 hours then cool the samples to 4 °C.
2. SDS-PAGE of deglycosylated samples
Prepare 130 μl fresh 3X reducing SDS loading buffer by adding 4 μl of 1.25M DTT.
Add 12.5 μl of the prepared 3X reducing SDS loading buffer to each sample.
Close the tubes with new caps and gently tap the tubes to mix.
Incubate the tubes in a thermocycler at 94°C for 5 minutes and then cool to 4 °C.
Load 30 μl of each sample and 10 μl of the protein marker on a 10-20% Tris-Glycine gel. Save the remainder of the sample for part 3.1. Load 10μl of ColorPlus Prestained Protein Marker.
Electrophorese the gel at 130 volts at room temperature until the dye front is near the bottom of the gel.
When the gel has finished running, remove the gel from the cast and place it in a small plastic box with enough Coomasie blue stain to cover the gel.
Stain the gel for 1 hour with gentle agitation.
Wash the gel three times for 30 minutes in 50 ml of destain solution.
Record the images using a white light transilluminator or scanner. Alternatively, the gel can be dried between sheets of cellophane in a frame.
3. Pro-Q Emerald 300 for detection of glycosylated proteins in SDS-PAGE gels
In parallel with the gel in 2.1), load the remainder of the samples on a 10-20% Tris-Glycine gel. Load 10μl of ColorPlus Prestained Protein Marker.
While the gel is running dissolve the Pro-Q Emerald reagent with DMF and prepare the stock Fix, Wash and Oxidizing solutions for the Pro-Q Emerald 300 stain following the product manual provided with the kit.
When the electrophoresis is complete, remove the gel from cast and place it in a plastic box.
Fix the gel by adding 100 ml of the Fix solution and leaving it overnight at room temperature with gentle agitation.
Wash the gel with 100 ml of the Wash solution for 10 to 20 minutes at room temperature with gentile agitation. Repeat the wash with fresh Wash solution.
Oxidize the carbohydrates by incubating the gel with gentle agitation for 30 minutes in 25 ml of Oxidizing solution.
Wash the gel as described in step 3.5.
While the gel is washing prepare fresh Pro-Q Emerald 300 stain by adding 500 μl of the Pro-Q Emerald 300 reagent solution dissolved in step 3.2 to 25 ml of staining buffer provided in the kit.
Stain the gel by adding 25 ml of the stain prepared in step 3.8 and incubating in the dark with gentle agitation for 90 to 120 minutes.
Repeat the two wash steps described in step 3.5.
Record the images with an UV transilluminator at 300nm. Use the 80 kDa marker, which is labeled with Pro-Q Emerald Green, to overlap the UV image to a white light picture of the gel showing the prestained ladder.
Compare the images of the Coomasie stained gel in step 2.10 with the Pro-Q Emerald stained gel from step 3.11.
4. Representative results
The changes in protein migration after enzymatic deglycosylation are shown in Figure 2. Compare the control sample (panel A, lane 1) with the PNGase F treatment (removal of N-glycans, lane 2), and Deglycosylation Mix (PNGase F; plus endo-and exoglycosidases to remove O-glycans, lane 3). No further reduction in size is seen after digesting with additional glycosidases (lane 4). Besides a change in mass, bands become sharper as glycans are removed. A band running under the 17 kDa marker (asterisk) represents the fully deglycosylated hCGβ polypeptide (MW: 16 kDa). Other bands might derive from incomplete deglycosylation or from the multiple unidentified proteins present in the hCGβ sample (see lane 1). Lanes 5 to 7 (controls) show the bands corresponding to the glycosidases.
The glycoprotein staining by Emerald Green is shown in panel B. This reagent oxidizes and stains all glycans present in a protein molecule. Therefore the intensity of the signal decreases as hCGβ is enzymatically deglycosylated (lane 1 to lane 4). The residual signal in lanes 3 and 4 indicate the presence of glycan motifs, which are resistant to the enzymes used. The additional glycosidases used in lane 4 remove a few extra sugar residues: the protein migration is the same, but a slight reduction in intensity in staining can be seen. Resistant sugar moieties were not present in all protein species: some bands were not detected by Emerald Green (absent in the UV image, in brackets), indicating they were extensively deglycosylated. Additional data support the conclusion that hCGβ is heterogeneously glycosylated. The lower band on lane 2 (arrow) is faint on the Emerald Green image, while the upper band on lane 2 is bright, indicating that many glycans groups are still present. These data support the conclusion that recombinant hCGβ expressed in mouse cells contains multiple glycoforms9 . These different glycoforms are due to the inherent heterogeneity of glycosylation where some polypeptides do not receive a glycan in each consensus site and/or some glycans are extended while others even on the same protein are not.
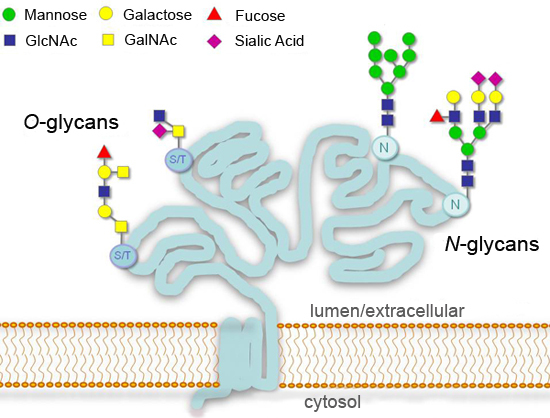 Figure 1. Typical glycosylation patterns for secreted or cell-surface glycoproteins.
Figure 1. Typical glycosylation patterns for secreted or cell-surface glycoproteins.
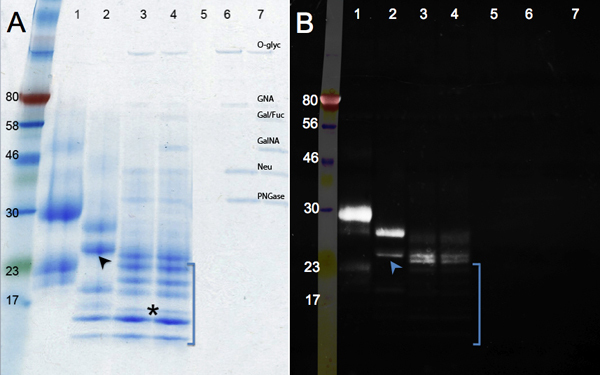 Figure 2. SDS-PAGE gels showing enzymatic deglycosylation of hCGβ. Panel A shows a Coomasie blue staining while Panel B shows the results of Pro-Q Emerald 300 for visualization of glycosylated proteins. Sample number: 1, hCGβ control; 2, PNGase F digestion; 3, Deglycosylation Mix digestion; 4, Deglycosylation Mix plus exoglycosidases digestion, samples 5 to 7 are the reagent controls (O-Glyc, O-Glycosidase; GNA, β-N-Acetylglucosaminidase; Gal/Fuc, β1-3 Galactosidase, α1-3,6 Galactosidase, and α1-2 Fucosidase; GalNA, α-N-Acetylglalactosaminidase; Neu, Neuraminidase; PNGase; PNGase F)
Figure 2. SDS-PAGE gels showing enzymatic deglycosylation of hCGβ. Panel A shows a Coomasie blue staining while Panel B shows the results of Pro-Q Emerald 300 for visualization of glycosylated proteins. Sample number: 1, hCGβ control; 2, PNGase F digestion; 3, Deglycosylation Mix digestion; 4, Deglycosylation Mix plus exoglycosidases digestion, samples 5 to 7 are the reagent controls (O-Glyc, O-Glycosidase; GNA, β-N-Acetylglucosaminidase; Gal/Fuc, β1-3 Galactosidase, α1-3,6 Galactosidase, and α1-2 Fucosidase; GalNA, α-N-Acetylglalactosaminidase; Neu, Neuraminidase; PNGase; PNGase F)
Discussion
The method described here using enzymatic deglycosylation and SDS-PAGE can provide valuable information about the glycosylation state of a protein of interest, while glycan-specific reagents facilitate the interpretation of the data. This protocol is intended for the initial studies of protein glycosylation and it is particularly suited for secretory and membrane glycoproteins from mammalian cells: the enzymes chosen in this case will specifically remove all N-glycans, and or N-glycans plus and the most common sugars extending and forming the core of O-glycans. Glycosidases have the additional advantage of being mild, compared with chemical deglycosylation methods, preserving the integrity of both sugars and protein backbone.
To elucidate the rate of occupancy (which amino acids are glycosylated), extent of glycosylation, or to determine the fine structure of glycans, more sophisticated techniques such as mass spectrometry, liquid chromatography or NMR are required.
Because of its simplicity, several steps in this protocol can be adjusted, substituted, and/or combined to accommodate various experimental needs. However, in order to obtain results that can be clearly interpreted it is important to understand its strengths and limitations. First, the specificity and purity of the glycosidases are crucial: only well-characterized enzymes tested to be free of proteases and other contaminating activities should be used. Unfortunately, there is no standard enzyme unit definition for glycosidases; the user should determine the appropriate substitution according to the manufacturer's specifications. Second, a careful choice of detection is necessary: a) Protein staining reagents are useful only if deglycosylation results in a significant shift in molecular mass. Such a clear result as shown here is not always obtained. In other cases, we have seen abnormal migration after deglycosylation (far from the predicted molecular weight, or even slower migration). This phenomenon is not well understood, but it can be said that any change in migration is evidence that the protein has been deglycosylated. b) Sugar detection with antibodies presents unique challenges limiting their applicability. It has been very difficult to generate general anti-glycan antibodies; they are usually raised against a complex glycan target, which restricts their use. Moreover, several monoclonal anti-glycan antibodies display undesired cross reactivity10. c) Lectins (proteins with intrinsic sugar affinity) are well suited for sugar detection, plus they give insight on glycan structure. However, not all of them have a narrow specificity; and many are only partially characterized (meaning that they could have unknown affinities). As a consequence positive lectin staining provides indication, not proof, of the presence of a given sugar. d) Chemical labeling kits (based on periodate oxidation of sugars) are the method of choice to stain all glycoproteins, and thus well suited to follow deglycosylation.
When processing an unknown sample, it is a good practice to include glycoprotein controls. Fetuin is a readily available N- and O-glycoprotein; chorionic gonadotropin (both subunits) is also a good choice. Bovine serum albumin (BSA) can be used as a negative control. However, it should be noted that some non-glycosylated proteins react slightly with the Pro-Q Emerald 300, particularly when used at high concentrations. Non-glycosylated molecular weight standards, such as the prestained protein marker used in this protocol, have the advantage of displaying sharp bands. Only by chance is one of these proteins (80 kDa) reactive to Pro-Q Emerald 300. Therefore, the user might want to run a glycoprotein standard ladder instead, such as Candy-Cane from Invitrogen.
Finally, simple in-gel detection of nucleocytosolic glycoproteins (which are modified with a single O-GlcNAc) is possible with the use of a monoclonal antibody, along β-N-Acetylglucosaminidase for specificity control 11.The description of this technique is beyond the scope of this article, but it should be mentioned as glycosidases are useful tools for the study of many other glycoproteins and glycoconjugates present in cells. A comprehensive treatment of all known forms of glycosylation can be found in the second edition of the "Essentials of Glycobiology"; available free online at the NCBI Bookshelf (Bookshelf ID: NBK1908; PMID: 20301239)12.
Disclosures
The authors are employed by New England Biolabs, which produces many of the reagents used in this article.
Acknowledgments
Don Comb
References
- Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12(4):43–56. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. Glycosylation in Cellular Mechanisms of Health and Disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annual Review of Immunology. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- Mitra N, Sinha S, Ramya TNC, Surolia A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends in Biochemical Sciences. 2006;31:156–163. doi: 10.1016/j.tibs.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Carlsen RB, Bahl OP, Swaminathan N. Human chorionic gonadotropin. Linear amino acid sequence of the beta subunit. Journal of Biological Chemistry. 1973;248(19):6810–6812. [PubMed] [Google Scholar]
- Tarentino AL, Plummer TH. Deglycosylation of Asparagine-linked Glycans by PNGaseF. Trends in Glycoscience and Glycotechnology. 1993;5(23):163–170. [Google Scholar]
- Tretter V, Altmann F, Marz L. Peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached α1-3 to the asparagine-linked N-acetylglucosamine residue. European Journal of Biochemistry. 1991;199:647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- Koutsioulis D, Landry D, Guthrie EP. Novel endo-α-N-acetylgalactosaminidases with broader substrate specificity. Glycobiology. 2008;18(10):799–805. doi: 10.1093/glycob/cwn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur D. Profiling the glycoforms of the intact alpha subunit of recombinant human chorionic gonadotropin by high-resolution capillary electrophoresis-mass spectrometry. Analytical Chemistry. 2009;81(2):8900–8907. doi: 10.1021/ac901506p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Lee MR, Shin I. Carbohydrate microarrays as powerful tools in studies of carbohydrate-mediated biological processes. Chemical Communications. 2008;37:4389–4399. doi: 10.1039/b806699j. [DOI] [PubMed] [Google Scholar]
- Zachara NE, Vosseller K, Hart GW. Detection and analysis of proteins modified by O-linked N-acetylglucosamine. Current Protocols in Molecular Biology. 2011;95:17–17. doi: 10.1002/0471142727.mb1706s95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. Essentials of Glycobiology. 2nd Ed. Clod Spring Harbor Laboratory Press; 2008. [PubMed] [Google Scholar]


