Abstract
Here we present a technique to label the trajectories of small groups of DRG neurons into the embryonic spinal cord by diffusive staining using the lipophilic tracer 1,1'-dioctadecyl-3,3,3',3'-tetramethylindocarbocyanine perchlorate (DiI)1. The comparison of axonal pathways of wild-type with those of mouse lines in which genes are mutated allows testing for a functional role of candidate proteins in the control of axonal branching which is an essential mechanism in the wiring of the nervous system. Axonal branching enables an individual neuron to connect with multiple targets, thereby providing the physical basis for the parallel processing of information. Ramifications at intermediate target regions of axonal growth may be distinguished from terminal arborization. Furthermore, different modes of axonal branch formation may be classified depending on whether branching results from the activities of the growth cone (splitting or delayed branching) or from the budding of collaterals from the axon shaft in a process called interstitial branching2 (Fig. 1).
The central projections of neurons from the DRG offer a useful experimental system to study both types of axonal branching: when their afferent axons reach the dorsal root entry zone (DREZ) of the spinal cord between embryonic days 10 to 13 (E10 - E13) they display a stereotyped pattern of T- or Y-shaped bifurcation. The two resulting daughter axons then proceed in rostral or caudal directions, respectively, at the dorsolateral margin of the cord and only after a waiting period collaterals sprout from these stem axons to penetrate the gray matter (interstitial branching) and project to relay neurons in specific laminae of the spinal cord where they further arborize (terminal branching)3. DiI tracings have revealed growth cones at the dorsal root entry zone of the spinal cord that appeared to be in the process of splitting suggesting that bifurcation is caused by splitting of the growth cone itself4 (Fig. 2), however, other options have been discussed as well5.
This video demonstrates first how to dissect the spinal cord of E12.5 mice leaving the DRG attached. Following fixation of the specimen tiny amounts of DiI are applied to DRG using glass needles pulled from capillary tubes. After an incubation step, the labeled spinal cord is mounted as an inverted open-book preparation to analyze individual axons using fluorescence microscopy.
Keywords: Neuroscience, Issue 58, neurons, axonal branching, DRG, Spinal cord, DiI labeling, cGMP signaling
Protocol
1. Dissection procedure
Note: Experimental use of mice should follow officially approved guidelines for the care and use of laboratory animals.
Before preparation, set up your dissecting microscope and lay out the surgical instruments needed for dissection including large and small scissors, large toothed forceps, curved forceps and four sets of Dumont No.5 dissecting forceps (two of which have inside-polished tips) (for details see the table of specific reagents and equipment). Place a sheet of filter paper in a 100-mm Sylgard-coated Petri dish. Pour cold PBS in the wells of a 12-well plate, a 100-mm petri dish and a 12-ml tube and leave on ice. Pipette 2ml of fixation buffer (4% paraformaldehyde in PBS, pH 7.4) in each well of a second 12-well plate and place on ice.
After anesthetization, sacrifice the timed pregnant dam at E12.5 (detection of vaginal plug was designated as E0.5), place the mouse with its ventral side up on three sheets of paper towels and soak the abdominal area with 70% ethanol.
Open the abdominopelvic cavity, isolate the bilateral uterine horns and transfer them into one of the 100-mm dishes with ice-cold PBS.
Under microscopic control, make a longitudinal incision of the uterine wall with small, straight scissors and cut away each amniotic sac from the placenta.
Peel away the amniotic sac from each embryo and cut the umbilical cord.
Using small scissors, decapitate the embryos and put the heads or a part of them aside for the isolation of genomic DNA if genotyping should be required. Subsequently, transfer the torsi to the respective wells of the 12-well plate with cold PBS.
Wet the filter paper in the Sylgard dish with a few drops of PBS and position an embryonic torso with its dorsal side up on the paper. For better stability, straighten its tail and extremities away from the body.
Working under a dissecting microscope with a magnification of approximately 16x, carefully pinch the skin of the embryo above the spinal cord with two pairs of fine tipped forceps and gently tear it apart. Beginning in the middle, proceed first towards the tail and then resuming from the middle towards the anterior side.
Wet the embryo from time to time with two or three drops of PBS to prevent it from drying.
To prevent that DRG are teared off from the spinal cord when it is removed from the embryo detach the DRG and the spinal cord from the surrounding cartilaginous vertebral column by horizontal sliding movements with the blade of a fine forceps with inside-polished tips. Starting in the middle of the embryo's right side, work your way towards the tail and then to the anterior end. Afterwards, repeat the procedure on the other side. Be careful not to rip off the associated DRG.
To detach completely the loosened spinal cord from the embryo, pick up its cervical part with fine forceps and pull it out in one piece towards its caudal end.
Submerge the isolated spinal cord with attached DRG in a well of the 12-well plate filled with fixation buffer and proceed with the preparation of spinal cords from the remaining embryos.
Fix spinal cords at least two hours on ice.
2. DiI-labeling of DRG neurons
For each spinal cord to be labeled, prepare two glass needles with a micropipette puller. We use a Sutter model P-97 electrode puller (programme setting: 1. HEAT=625, VEL= 35, TIME=175; 2. HEAT=600, VEL= 25, TIME=175; 3. HEAT=630, VEL= 30, TIME=150).
Under microscopic control dip the tip of each glass needle three to five times in a 5 % (w/v) solution of DiI in ethanol. Evaporation of the ethanol creates a thin layer of DiI crystals on the needle.
Working under a dissecting microscope with a magnification of approximately 40x, place a spinal cord with its ventral side up on a slide. Using spring scissors, open the cord on the ventral side at its whole length by cutting through the floorplate to allow the later mounting of the preparation in an inverted open-book mode (see section C).
Position the spinal cord with the dorsal side up on a slide. Then manually approach the cord with the DiI-covered tip of the glass needle under microscopic visualization and carefully pierce every second DRG on one side of the spinal cord. Almost no traces of DiI should be visible in the pierced DRG to ensure the labeling of only a small number of neurons. Take a second needle for the other side of the cord. Aiming only every second DRG avoids an overlap of labeled axonal projections from neighboring DRG which might complicate the differentiation of individual axons.
Return the spinal cord to the fixation buffer and incubate in the dark for at least six hours at ambient temperature or overnight at 4 °C to give time for the dye to diffuse along the axon within the plasma membrane. Allow for a longer incubation period (up to two days at 4 °C) if the analysis of collateral growth into the spinal cord is desired.
3. Mounting and microscopic analysis
Following dye diffusion, position a single spinal cord with its dorsal side down on a coverslip in a drop of PBS. Take care that the attached DRG are correctly oriented laterally and do not intermingle. Aspirate excess fluid and flatten out the cord in an inverted open-book mode using forceps.
Mount the preparation on a microscope slide using PBS.
To prevent the increase of unwanted background staining fluorescence microscopic analysis of labeled axonal projections should be carried out the day of mounting. Until then keep the slides in the dark at 4 °C.
4. Representative Results:
The spinal cord of the mouse receives afferent projections from 8 pairs of cervical, 13 pairs of thoracic, 5 pairs of lumbar and 4 pairs of sacral DRG totaling to 60 spinal ganglia. After some training a spinal cord with most DRG still attached can be isolated from an embryo in under five minutes. This procedure is suited for the isolation of spinal cords with attached DRG from E11.5 to E13.5 mouse embryos. However, best results are achieved from E12.5. Exemplary outcomes of the labeling procedure described here are shown in Figure 3. The labeling of DRG over the entire length of the spinal cord may then be used to quantify axonal branching behaviour at different vertebral levels (Fig. 4).
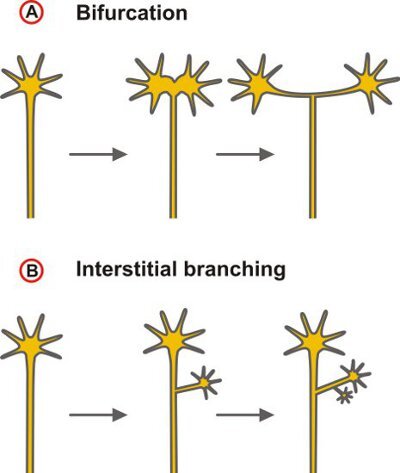 Figure 1. Scheme depicting the two principal modes of axonal branching. (A) Branching by the activities of the growth cone that could lead to a bifurcation - as indicated here - as well as complex terminal arbors or (B) collateral formation at the axon shaft (interstitial branching). The latter mode is the dominating branching type of projecting cortical and thalamocortical axons6,7.
Figure 1. Scheme depicting the two principal modes of axonal branching. (A) Branching by the activities of the growth cone that could lead to a bifurcation - as indicated here - as well as complex terminal arbors or (B) collateral formation at the axon shaft (interstitial branching). The latter mode is the dominating branching type of projecting cortical and thalamocortical axons6,7.
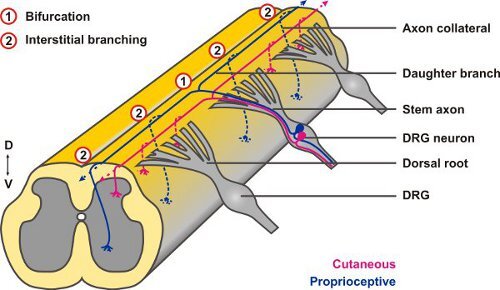 Figure 2. Afferent projections of DRG neurons into the embryonic spinal cord display both types of axonal branching: axons first branch at the DREZ by bifurcation (1) and from the resulting daughter branches collaterals form after a waiting period by interstitial branching (2).
Figure 2. Afferent projections of DRG neurons into the embryonic spinal cord display both types of axonal branching: axons first branch at the DREZ by bifurcation (1) and from the resulting daughter branches collaterals form after a waiting period by interstitial branching (2).
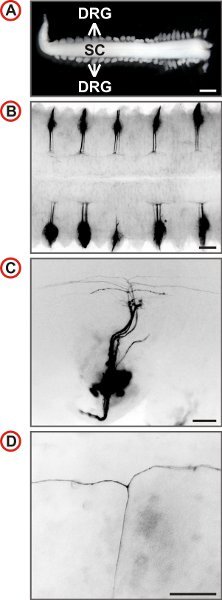 Figure 3. Visualization of single axonal trajectories of embryonic mouse DRG neurons. (A) Spinal cord with attached DRG prepared from an E12.5 mouse embryo. (Scale bar, 1 mm.) (B-D) Dorsal views of DiI-labeled DRG of wild-type mice at increasing magnification are shown. In B every second DRG is labeled by DiI. Fluorescence images are inverted, caudal is at the left and in C and D, lateral is at the bottom. In C a small number of axons is labeled and at higher magnification the presence of T-like branches can be identified in the DREZ of the spinal cord. (Scale bars, 250 μm (B), 100 μm (C) and 50 μm (D).)
Figure 3. Visualization of single axonal trajectories of embryonic mouse DRG neurons. (A) Spinal cord with attached DRG prepared from an E12.5 mouse embryo. (Scale bar, 1 mm.) (B-D) Dorsal views of DiI-labeled DRG of wild-type mice at increasing magnification are shown. In B every second DRG is labeled by DiI. Fluorescence images are inverted, caudal is at the left and in C and D, lateral is at the bottom. In C a small number of axons is labeled and at higher magnification the presence of T-like branches can be identified in the DREZ of the spinal cord. (Scale bars, 250 μm (B), 100 μm (C) and 50 μm (D).)
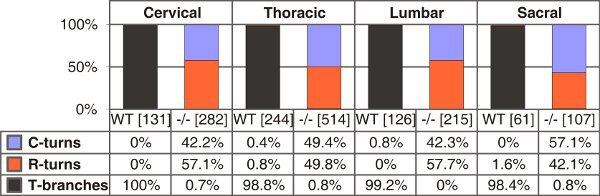 Figure 4. Quantification of T-shaped branches, single rostral or caudal turns in wild-type and C-type natriuretic peptide (CNP)-deficient mice at E13.5. The numbers of single axons counted are given in brackets for different trunk levels for each genotype. C- or R-turns - growth only in caudal or rostral direction, respectively.
Figure 4. Quantification of T-shaped branches, single rostral or caudal turns in wild-type and C-type natriuretic peptide (CNP)-deficient mice at E13.5. The numbers of single axons counted are given in brackets for different trunk levels for each genotype. C- or R-turns - growth only in caudal or rostral direction, respectively.
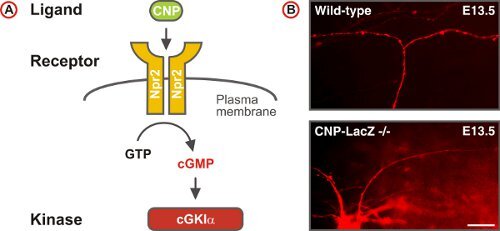 Figure 5. A cGMP signaling pathway triggers sensory axon bifurcation at the DREZ of the spinal cord. (A) Scheme of the cGMP signaling pathway composed of the ligand CNP, the receptor guanylyl cyclase Npr2 and the serine/threonine kinase cGKIα in embryonic DRG neurons. Npr2 generates cGMP from GTP upon stimulation by CNP. (B) DiI tracing of single axons of DRG neurons in wild-type and CNP-deficient mice. (Scale bar, 25 μm.)
Figure 5. A cGMP signaling pathway triggers sensory axon bifurcation at the DREZ of the spinal cord. (A) Scheme of the cGMP signaling pathway composed of the ligand CNP, the receptor guanylyl cyclase Npr2 and the serine/threonine kinase cGKIα in embryonic DRG neurons. Npr2 generates cGMP from GTP upon stimulation by CNP. (B) DiI tracing of single axons of DRG neurons in wild-type and CNP-deficient mice. (Scale bar, 25 μm.)
Discussion
Stereotyped projection patterns comprising both types of axonal branch formation together with ease of preparation in combination with the use of fixed tissue for DiI labeling makes the embryonic spinal cord with attached DRG a favourable model to study axonal branching. The application of minute amounts of DiI using coated glass needles allows - in contrast to the bulk labeling of DRG - the visualization of small groups of DRG neurons and thereby the analysis of individual axons and their branching patterns. The described method could be further improved by iontophoretic injection of the lipophilic tracer which further decreases the number of labeled neurons8. However, this would be more time-consuming.
DiI labeling of single axonal trajectories in whole mount preparations of the embryonic spinal cord was instrumental in the identification of a cGMP signaling cascade -comprising the ligand CNP, its receptor Npr2, and the cGMP-dependent kinase cGKIα - that regulates one type of axonal branching: the bifurcation of DRG neurons at the DREZ 4,5,9-12. In contrast to the wild-type, axons of DRG neurons in cGMP signaling mutants turn only in one direction, instead of bifurcating at the DREZ (Fig. 5). These studies also revealed that collateral formation was not affected in cGMP signaling mutants indicating that this second type of branch formation observed in DRG neurons is differently regulated4.
Alternatively, the branching behaviour of single DRG axons may also be studied in transgenic mouse lines expressing either a fluorescent protein in only a small subset of DRG neurons13 or containing a combination of fluorescent markers which allows one to distinguish between individual neurons14. However, the effort to cross-breed the mutant mouse model to be analyzed with the fluorescent mouse lines has to be considered. Furthermore, due to the promotor (Thy1) used expression of the fluorescent proteins in the transgenic mouse lines starts only at postnatal stages making it difficult to find out whether an observed branching phenotype is the result of impaired branch formation during embryonic development or a later degeneration process instead. In contrast, DiI labeling of embryonic DRG has the advantage to study the process of axonal branching and its disruption in appropriate mouse mutants at the time when axonal branching occurs.
Preparations of embryonic spinal cords with attached DRG might be further used for other labeling procedures (whole mount in-situ hybridization, LacZ- or AP-staining of transgenic mice), collection of spinal cord and DRG tissue for RNA-isolation, or when adapted to sterile conditions for tissue culture experiments and downstream applications like calcium imaging or electrophysiological measurements.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors wish to thank Dr. Alistair Garratt (Max Delbrück Center, Berlin) for helpful comments. This work was supported by the collaborative research center (SFB665) of the German Research Council (DFG).
References
- Honig MG, Hume RI. Dil and diO: versatile fluorescent dyes for neuronal labelling and pathway tracing. Trends. Neurosci. 1989;12(9):333–333. [PubMed] [Google Scholar]
- Acebes A, Ferrus A. Cellular and molecular features of axon collaterals and dendrites. Trends. Neurosci. 2000;23(11):557–557. doi: 10.1016/s0166-2236(00)01646-5. [DOI] [PubMed] [Google Scholar]
- Ozaki S, Snider WD. Initial trajectories of sensory axons toward laminar targets in the developing mouse spinal cord. J. Comp. Neurol. 1997;380(2):215–215. [PubMed] [Google Scholar]
- Schmidt H. The receptor guanylyl cyclase Npr2 is essential for sensory axon bifurcation within the spinal cord. J. Cell Biol. 2007;179(2):331–331. doi: 10.1083/jcb.200707176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DA, Ma L. Developmental regulation of axon branching in the vertebrate nervous system. Development. 2011;138(2):183–183. doi: 10.1242/dev.046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary DD, Terashima T. Cortical axons branch to multiple subcortical targets by interstitial axon budding: implications for target recognition and "waiting periods" Neuron. 1988;1(10):901–901. doi: 10.1016/0896-6273(88)90147-x. [DOI] [PubMed] [Google Scholar]
- Portera-Cailliau C. Diverse modes of axon elaboration in the developing neocortex. PLoS. Biol. 2005;3(8):e272–e272. doi: 10.1371/journal.pbio.0030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan WB. Vital imaging and ultrastructural analysis of individual axon terminals labeled by iontophoretic application of lipophilic dye. J. Neurosci. Methods. 1999;93(1):13–13. doi: 10.1016/s0165-0270(99)00096-5. [DOI] [PubMed] [Google Scholar]
- Schmidt H. C-type natriuretic peptide (CNP) is a bifurcation factor for sensory neurons. Proc. Natl. Acad. Sci. U. S. A. 2009;106(39):16847–16847. doi: 10.1073/pnas.0906571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. Regulate axon branching by the cyclic GMP pathway via inhibition of glycogen synthase kinase 3 in dorsal root ganglion sensory neurons. Journal of Neuroscience. 2009;29(5):1350–1350. doi: 10.1523/JNEUROSCI.3770-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Ma L. Regulation of axonal development by natriuretic peptide hormones. Proc. Natl. Acad. Sci. U. S. A. 2009;106(42):18016–18016. doi: 10.1073/pnas.0906880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Rathjen FG. Signalling mechanisms regulating axonal branching in vivo. Bioessays. 2010. [DOI] [PubMed]
- Feng G. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28(1):41–41. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Livet J. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56–56. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]


