Abstract
Gene expression patterns are specified by cis-regulatory element (CRE) sequences, which are also called enhancers or cis-regulatory modules. A typical CRE possesses an arrangement of binding sites for several transcription factor proteins that confer a regulatory logic specifying when, where, and at what level the regulated gene(s) is expressed. The full set of CREs within an animal genome encodes the organism′s program for development1, and empirical as well as theoretical studies indicate that mutations in CREs played a prominent role in morphological evolution2-4. Moreover, human genome wide association studies indicate that genetic variation in CREs contribute substantially to phenotypic variation5,6. Thus, understanding regulatory logic and how mutations affect such logic is a central goal of genetics.
Reporter transgenes provide a powerful method to study the in vivo function of CREs. Here a known or suspected CRE sequence is coupled to heterologous promoter and coding sequences for a reporter gene encoding an easily observable protein product. When a reporter transgene is inserted into a host organism, the CRE′s activity becomes visible in the form of the encoded reporter protein. P-element mediated transgenesis in the fruit fly species Drosophila (D.) melanogaster7 has been used for decades to introduce reporter transgenes into this model organism, though the genomic placement of transgenes is random. Hence, reporter gene activity is strongly influenced by the local chromatin and gene environment, limiting CRE comparisons to being qualitative. In recent years, the phiC31 based integration system was adapted for use in D. melanogaster to insert transgenes into specific genome landing sites8-10. This capability has made the quantitative measurement of gene and, relevant here, CRE activity11-13 feasible. The production of transgenic fruit flies can be outsourced, including phiC31-based integration, eliminating the need to purchase expensive equipment and/or have proficiency at specialized transgene injection protocols.
Here, we present a general protocol to quantitatively evaluate a CRE′s activity, and show how this approach can be used to measure the effects of an introduced mutation on a CRE′s activity and to compare the activities of orthologous CREs. Although the examples given are for a CRE active during fruit fly metamorphosis, the approach can be applied to other developmental stages, fruit fly species, or model organisms. Ultimately, a more widespread use of this approach to study CREs should advance an understanding of regulatory logic and how logic can vary and evolve.
Keywords: Developmental Biology, Issue 58, Cis-regulatory element, CRE, cis-regulatory module, enhancer, site-specific integration, reporter transgenes, confocal microscopy, regulatory logic, transcription factors, binding sites, Drosophila melanogaster, Drosophila
Protocol
Overview:
This video demonstrates a protocol capable of quantitatively measuring the gene regulatory activities for cis-regulatory element (CRE) sequences in Drosophila (D.) melanogaster. This protocol can be used to compare the regulatory activities possessed by: wild type and mutant CRE forms, naturally-occurring CRE alleles found within a species, or orthologous CREs between diverged species.
1. Site-specific integration of reporter transgenes into the Drosophila melanogaster genome
A. Reporter transgene construction
As a first step, any CRE evaluated must be separately cloned into a reporter vector that contains an (1) attB bacterial attachment site sequence used for site-specific transgene integration8-11, (2) multiple cloning site upstream of a (3) heterologous promoter that is followed by the (4) coding sequence for a fluorescent protein (such as EGFP or DsRed).
While any vector can be made compatible for phiC31 mediated site-specific integration by introducing an attB sequence into the vector backbone, the vector pS3aG12-14 can be obtained from the addgene plasmid repository (http://www.addgene.org/pgvec1) pS3aG is a customized version of the P-based transformation vector15 in which one of the two gypsy insulators was replaced by a SfIb insulator, it contains an enhanced multiple cloning site, and possesses a 250 base pair sequence containing an attB attachment site. CREs of interest are inserted into the multiple cloning site upstream of an hsp70 minimal promoter and the EGFP gene that encodes an enhanced version of Green Fluorescent protein that localizes to the nucleus.
B. Site-specific integration of reporter transgenes
For a set of CREs whose activities are to be compared as a part of reporter transgene vectors, the created vectors are separately integrated into the same genomic landing site. Here the phiC31 phage integrase protein catalyzes the unidirectional recombination between the bacterial (attB) attachment site in the transgene vector and a genomically located phage (attP) attachment site9. Several groups8-11 have made a variety of transgenic lines that include an attP site (so-called landing sites) and contain a genomic source8 of phiC31 that expresses this protein in the germ cells. These fly stocks can be readily obtained from the Bloomington Drosophila stock center (http://flystocks.bio.indiana.edu/).
A published protocol exists detailing how to create transgenic Drosophila site-specifically using phiC31 integrase16. The equipment and technical expertise to make transgenic Drosophila lines is no longer required as several vendors exist (Table 1) to whom this service can be outsourced.
Transgene behavior can vary greatly from one tissue to the next11. Thus a single attP landing site is not optimal for the analysis of all CREs, developmental stages, and/or tissues of study. It is advisable to assess the activity of a CRE of interest in several different landing sites in order to find a site where the CRE's activity is representative of the endogenous target gene(s) expression pattern.
For obtained transgenic lines it is advisable to make them homozygous for the transgene. CRE activity is generally more robust in individuals with two copies of the transgene and for quantitative measurements it is imperative that comparisons are made between individuals with the same number of transgene copies. Homozygous individuals can be obtained through the use of balancer chromosomes. For well characterized landing sites though, it is often simpler to collect virgin males and females, and cross those with the more intense eye color phenotype conferred by the mini-white gene (Fig. 1), as the white mutant rescue provided by the integrated transgene vector is more dramatic in individuals with two vector copies.
2. Obtaining specimens or tissues of the appropriate developmental stage
Obtain specimens for analysis at the time in development when the gene expression pattern(s) of interest occur(s) endogenously, hence the time when the CRE under investigation should activate reporter gene expression. For Drosophila, this can be either embryonic, larval, pupal or adult stages. Below we describe a method to stage adult and metamorphic flies; the latter of which takes place inside the puparium, a hard larval skin that contains the immobile specimen until it ecloses as an adult.
Expand transgenic lines to ensure specimens of the proper stage are available when ready to analyze reporter gene expression by confocal microscopy. Set up two vials each with several (˜5- 10) male and female flies. On alternating days, bump the flies from one of the vials to an unused vial. Repeat this for a week. By the end of two weeks, transgenic fruit flies will be available that range from larval to adult developmental stages.
Staging adult flies: The duration of time spent inside a puparium is 98 hours for females and 102 hours for males when reared at 25°C. Adult flies can be easily collected when they emerge from the puparium (called eclosion). This timepoint can be considered 0 hours post eclosion. Adult flies can be reared until the required timepoint for analysis. For example, a CRE called mel-oe1 that controls expression of the desatF gene in the adult oenocytes of D. melanogaster females is not active immediately after eclosion but CRE activity switches on after one day14. When the correct stage is obtained, anesthetize flies and situate in a drop of Halocarbon oil on a slide and proceed to confocal evaluation.
Staging specimens during metamorphosis: At the end of the third-instar larval stage the puparium is formed. Though at this stage the animal technically is still a third instar larva, this developmental stage is easily identifiable as the specimen is non-mobile, the puparium is soft and white, and the spiracles have everted (Fig. 2A). This timepoint can be considered 0 hours After Puparium Formation (hAPF). At this stage, males can be distinguished from females by the presence of bilaterally situated gonads near the midpoint of the body that appear as translucent circles (boxed in Fig. 2A and indicated by black arrow in 2A").
1. Staging based upon length of metamorphosis:
At 0 hAPF, specimens can be transferred to a fresh vial or Petri dish using a moistened paint brush. To keep specimens from drying out, add a piece of Kimwipe and moisten with water. Transfer vial or dish to a 25°C incubator until the desired hAPF.
2. Staging by visible morphological features:
It is often inconvenient to analyze specimens for CRE activity at a specific timepoint following the collection of 0 hAPF specimens. Alternatively, one can identify correctly staged specimens at a time convenient for analysis based upon the presence and position of several morphological markers (detailed below) that are visible through the puparium or after puparium removal (Fig. 2).
2a. Morphological criteria for approximating metamorphic stage:
0 hAPF: White prepupa stage where larva stops moving; anterior spiracles everted (arrow in Fig. 2A); lateral trachea visible; soft white puparium (Fig. 2A).
˜14 hAPF: Puparium tanned and hardened; head sac everted; lateral trachea and gonads less distinct; legs and wings fully extended along abdomen; Malpighian tubules not yet prominent and green (dashed boxed region in Fig. 2B); eyes are unpigmented (Fig. 2B').
˜25 hAPF: Two parallel Malpighian tubules prominent and green in color (dashed boxed region in Fig. 2C).
˜40 hAPF: Dark green yellow body positioned between the anterior ends of the Malpighian tubules (red arrowhead in Fig. 2D).
˜50 hAPF: Yellow body positioned at the mid-point of the Malpighian tubules (boxed region in Fig. 2E and enlarged in 2E'') and the eyes are amber in color if wild type for white gene (needed for red eye color). Eye color is lacking at this stage when the white mutant phenotype is rescued by the mini-white gene that is part of the integrated reporter transgene vector (Fig. 2E').
˜70 hAPF: Dorsal thoracic microchaetae and macrochaetae visible (Fig. 2F, arrow); yellow body positioned at the posterior of the Malpighian tubules (boxed region in Fig. 2F boxed and enlarged in 2F''); eye color is pale pink when rescued by the mini-white gene (Fig. 2F').
˜80 hAPF: Wing tips gray; Malpighian tubules located between anterior to the midpoint of the abdomen (boxed region in Fig. 2G and enlarged in 2G"); folds between abdominal segments and bristles on the abdominal tergites are visible (Fig. 2G and 2G"); and rescued eye color is bright red (Fig. 2G').
˜90 hAPF: Wings darkened to black; Malpighian tubules and yellow body obscured by tanning of tergites; thoracic (arrow) and abdominal bristles are mature and darkened (Fig. 2H); and green meconium appears at the dorsal posterior tip of the abdomen (Fig. 2H'').
Notes:
At late stages of metamorphosis, the sex of the specimens can be determined by genital morphology (arrowheads in Fig. 2I and 2J) or the presence of darkened sex combs on the first set of male legs.
Further precision in staging can be achieved by consideration of additional morphological markers as described previously17.
D. Steps to remove specimen from puparium:
Using lab tape adhere a piece of packing tape to a dissection board with the sticky surface facing upwards.
Wet a paint brush and apply moisture to pupariated specimens. Using a paint brush, transfer specimens to a Kimwipe.
Using forceps or a paintbrush, transfer specimens to the packing tape and adhere with the dorsal surface facing up (curved side of puparium).
Allow specimens to dry for ˜15 minutes. Dry puparium are much easier to open up than those that are moist. At this point specimens can be coarsely staged by morphological markers visible through the puparium.
Using forceps, progressively open up the puparium beginning at the anterior operculum and proceed towards the posterior end. If a more fine-scale dissection is needed to isolate a specific tissue this can be done in a watch glass with PBS solution. Otherwise, transfer pupae to a drop of viscous HC700 Halocarbon oil (Sigma-Aldrich) on a microscope slide.
Situate specimens on a slide, using a stereomicroscope, and using the above described criteria (section 2a) a specimen's stage can be determined..
3. Confocal microscopic evaluation of reporter gene expression in a whole mount sample
For whole mount specimens, use either the 4X or 10X objectives. Using fluorescence stimulated by mercury lamp exposure, or alternatively by viewing in bright field, adjust the microscope stage to position specimen in the optical field then bring specimen into focus.
Using the confocal microscope software, adjust the excitation wavelength for EGFP (or a suitable wavelength when using another fluorescent protein).
To prevent photobleaching a specimen, begin with a laser intensity of 5-10%. If signal is underwhelming, then increase the intensity as needed.
In order to improve the signal to noise ratio for confocal images, enable software settings that average pixel measurements from replicate scans; such as Kalman averaging or line and stack averaging. In our experience Kalman averaging for three scans improves the signal to noise ratio without making the time for image collection too long.
For quantitative comparisons of CRE activity it is essential that fluorescence intensity for a specimen has not saturated many pixels. Using a z-section where fluorescence appears most intense, adjust settings so that few pixels are saturated. This can be done by switching to a saturation-warning lookup table and adjusting the channel voltage, gain, and offset to settings where few pixels are saturated. Lastly, in order to collect sufficient signal from a specimen it is often necessary to adjust the confocal aperture.
Once the optimal settings are determined, run the z-scan.
When the scan is complete convert the image stack into a projection and save this image in the Tagged Image File Format or "TIFF".
Note:
It is essential to use the same confocal settings for all replicate specimens and reporter transgene lines that will be included in a quantitative comparison of CRE activities.
4. Quantifying cis-regulatory element activity
While some confocal acquisition software allows for further processing of projection images, we prefer to use the freely available Image J software program to evaluate confocal images18. Using Image J 18 recorded GFP expression patterns can be quantified as pixel value statistics within a specified area. Although images do not need to be in gray scale we prefer to do so.
With the Image J program started, open a TIFF image to be evaluated.
Click on the freehand selection button and outline the area of the specimen where quantitation is desired; for example the region in Figure 3 within the dashed yellow border. (see Note a below regarding the selection of an area to quantitate.)
To get a pixel value statistic, click on the Analyze tab and then select measure. A results box will appear showing the area, and the mean, minimum and maximum pixel value scores. Record the mean pixel value score and repeat to generate pixel value statistics for replicate specimens (see Note b regarding replicate number).
We recommend collecting a second mean pixel value from a control region where the CRE is not active; for example the region in Figure 3 within the dashed red border. This latter value can be subtracted from the former value to remove background effects from the measurement to get an adjusted mean pixel value. In our experience this further reduces variation between replicate specimens (see Note c regarding size selection for control region).
Use the adjusted mean pixel values from replicate specimens to calculate the average regulatory activity for a CRE and the standard error of the mean. This regulatory activity value and measurement of error can be compared to other reporter transgenes where the CRE's sequence differs (for example Figure 3).
Notes:
Image J allows for the user to select a defined shape and area from which a mean pixel value score will be determined. However, as specimen size often varies we prefer to use the Freehand selection tool to specify the area to be evaluated for each specimen.
A larger number of replicates (N) results in a better estimation of the mean regulatory activity for a given CRE. We find that an N between 4 and 8 is generally sufficient.
In our experience the size of the control area selected has little affect on the measured mean pixel value for the control region. In general, we select an area proportional in size to the area of interest.
5. Representative Results:
A wild type version of a D. melanogaster CRE, called the Dimorphic Element13, drives a high level of EGFP reporter expression in the A6 abdominal segment of female pupae (Fig. 3A). A 13 base pair sequence within this element was found to be a binding site for the DSX transcription factor, and the in vivo importance of this sequence can be demonstrated by quantifying the regulatory activity for this CRE when this binding site is mutated. Considering the regulatory activity of the wild type Dimorphic Element to be 100±5%, mutation of this DSX site reduced the regulatory activity to 28±3%. Similar comparisons can be done of regulatory activities for intraspecific CRE alleles or between orthologous CREs from separate species. For example, considering the levels of EGFP in female segment A6 driven by the D. melanogaster Canton S strain as a regulatory activity of 100±4%, orthologous CREs for the species D. simulans and D. willistoni were found to respectively possess regulatory activities equal to 75±4% and 0±0% (Fig. 3B).
 Figure 1. Using the intensity of the white-rescue eye phenotype to make transgenic lines homozygous. In order to quantitatively evaluate reporter transgenes it is important that each specimen have the same reporter transgene genotype. (A) A white gene mutant genetic background with an attP landing site sequence is utilized for site-specific transgene integration. Transgenic individuals (B) hemizygous and (C) homozygous for the integrated vector can typically be distinguished by the intensity of the rescued eye color phenotype by the number of copies of the integrated mini white gene. Homozygous lines can be established by crossing (C) male and female flies with the darkest eye color phenotype.
Figure 1. Using the intensity of the white-rescue eye phenotype to make transgenic lines homozygous. In order to quantitatively evaluate reporter transgenes it is important that each specimen have the same reporter transgene genotype. (A) A white gene mutant genetic background with an attP landing site sequence is utilized for site-specific transgene integration. Transgenic individuals (B) hemizygous and (C) homozygous for the integrated vector can typically be distinguished by the intensity of the rescued eye color phenotype by the number of copies of the integrated mini white gene. Homozygous lines can be established by crossing (C) male and female flies with the darkest eye color phenotype.
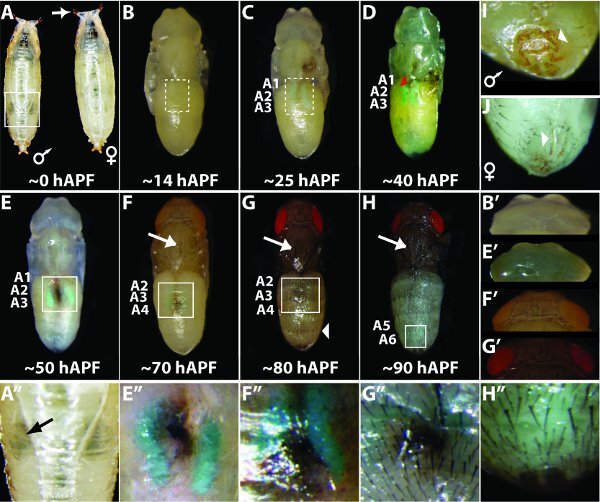 Figure 2. Using morphological markers to determine the metamorphic stage for Drosophila. During metamorphosis from a larva to an adult fruit fly, the specimen transitions through a series of stereotypic morphologies that can be used to determine the sex and approximate the development stage. Specimens in B-H have been removed from their puparium. Boxes in panels A, E, F, G and H indicate the zoomed in regions respectively for panels A", E", F", G", and H".
Figure 2. Using morphological markers to determine the metamorphic stage for Drosophila. During metamorphosis from a larva to an adult fruit fly, the specimen transitions through a series of stereotypic morphologies that can be used to determine the sex and approximate the development stage. Specimens in B-H have been removed from their puparium. Boxes in panels A, E, F, G and H indicate the zoomed in regions respectively for panels A", E", F", G", and H".
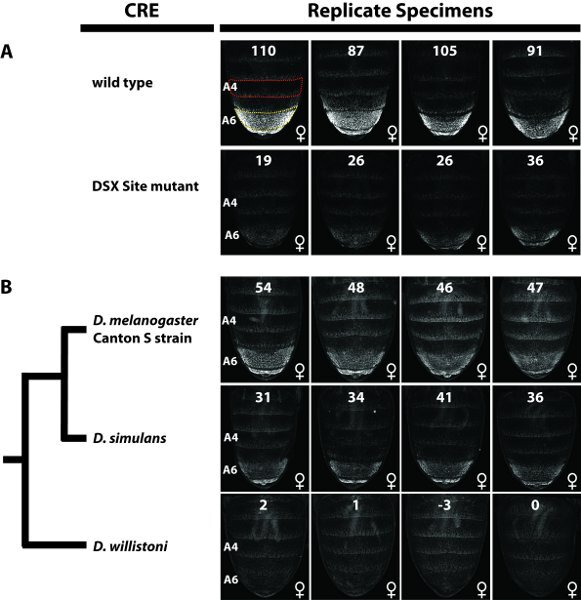 Figure 3. Quantitative comparison of cis-regulatory element activities. For all samples EGFP-reporter gene expression mediated by a particular Dimorphic Element was assessed in females at 80 hours after puparium formation. The number at the top of each image is the EGFP-expression level for the specimen that is calculated as the mean pixel value for the A6 abdominal segment (indicated for the upper left most image as the region within the dashed yellow border) minus the mean pixel value for the A4 segment (background correction; indicated for the upper left most image as the region within the dashed red border). For each reporter transgene four replicates were assessed to calculate a mean segment A6 pixel value. Regulatory activity is reported as the % of the (A) wild type or (B) Canton S strain female mean pixel value ±SEM. (A) GFP expression for females possessing a wild type allele of the Dimorphic Element and a mutant version where a single binding site for the DSX transcription factor was ablated. This mutant binding site reduced Dimorphic Element activity to 28±3% of the wild type sequence. (B) Comparison of the regulatory activities for sequences orthologous to the D. melanogaster (Canton S strain) Dimorphic Element. Considering the GFP expression mediated by a D. melanogaster Dimorphic Element allele as 100%, the orthologous sequences from the related species D. simulans and D. willistoni respectively possess activities of 75±4% and 0±0%.
Figure 3. Quantitative comparison of cis-regulatory element activities. For all samples EGFP-reporter gene expression mediated by a particular Dimorphic Element was assessed in females at 80 hours after puparium formation. The number at the top of each image is the EGFP-expression level for the specimen that is calculated as the mean pixel value for the A6 abdominal segment (indicated for the upper left most image as the region within the dashed yellow border) minus the mean pixel value for the A4 segment (background correction; indicated for the upper left most image as the region within the dashed red border). For each reporter transgene four replicates were assessed to calculate a mean segment A6 pixel value. Regulatory activity is reported as the % of the (A) wild type or (B) Canton S strain female mean pixel value ±SEM. (A) GFP expression for females possessing a wild type allele of the Dimorphic Element and a mutant version where a single binding site for the DSX transcription factor was ablated. This mutant binding site reduced Dimorphic Element activity to 28±3% of the wild type sequence. (B) Comparison of the regulatory activities for sequences orthologous to the D. melanogaster (Canton S strain) Dimorphic Element. Considering the GFP expression mediated by a D. melanogaster Dimorphic Element allele as 100%, the orthologous sequences from the related species D. simulans and D. willistoni respectively possess activities of 75±4% and 0±0%.
6. Materials
| Material Name | Type | Company | Catalogue Number | Comment |
| Site-specific transgene integration | Service | Best Gene | Plan H | Choose a landing site, receive transformant lines |
| Halocarbon Oil | Reagent | Sigma-Aldrich | H8898 | Used to mount specimens for confocal imaging |
| Dumont #5 Forceps | Tool | Fine Science Tools | 11252-40 | Fine pointed and durable for dissection |
| SZ61 Zoom Stereo microscope | Tool | Olympus | Customizable | Excellent optics for working with fruit flies |
| FluoView Confocal Microscope | Tool | Olympus | Customizable | Provides high-resolution confocal images |
Discussion
cis-regulatory elements encode the genomic program that specifies gene expression patterns and thereby the process of development1, and are prominent locations for both mutations underlying morphological evolution2-4 and phenotypic variation for human traits5,6,19. In spite of this importance, the regulatory logic for CREs remain poorly understood. A prominent reason for this understanding deficit has been the lack of suitable methods to quantitatively compare functional sequences of CREs. Here we present a protocol that capitalizes on improved transgenesis methods for D. melanogaster to add a quantitative aspect to the study of CREs.
In our experience when quantifying a CRE’s regulatory activity, variation between replicate specimens is of 10% or less of the adjusted mean pixel value when replicates are (1) of the same developmental stage and (2) the reporter transgene is in the same genomic landing site. This amount of variation is consistent with that determined for the CREs presented in Figure 3, and places a limitation on the use of this quantitative method to situations where genetically distinct versions of a CRE differ in regulatory activity by a value greater than 10%. Importantly though, this limitation is a significant improvement over the qualitative descriptions of “reduced” or “increased” that those studying CREs previously had to use when describing varying activity.
When versions of a CRE are known or found to quantitatively differ in their regulatory activities, this protocol makes feasible determining which of the mutational differences are responsible for or contribute to the activity difference12,13. The ability to identify these functionally-relevant mutations is a necessary first step to determine the molecular mechanisms, such as the gain or loss of transcription factor binding sites, causing CRE activities to differ. Looking forward, the use of this protocol for other Drosophila CREs and the application of similar methods in protocols for other model organisms should allow for a better understanding of CRE logic to emerge. This includes an ability to discriminate functionally-relevant CRE mutations from the morass of functionally-neutral mutations.
Disclosures
We have nothing to disclose.
Acknowledgments
We thank: Nicolas Gompel and Benjamin Prud'homme for their contributions to the development of this protocol; Melissa Williams and four anonymous reviewers for comments on the manuscript; the University of Dayton Graduate School for research fellowships to WAR; and the University of Dayton Biology Department and Research Institute (UDRI) for research support for TMW. This work was supported by the American Heart Association Grant 11BGIA7280000 to TMW.
References
- Davidson EH. Gene Regulatory Networks in Development and Evolution. Academic Press; 2006. The Regulatory Genome. [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Stern DL. Evolutionary developmental biology and the problem of variation. Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24:489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N, editors. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. United States: 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science. 2009;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TM. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS biology. 2009;7:e1000168–e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques. 2000;29:726–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Fish MP, Groth AC, Calos MP, Nusse R. Creating transgenic Drosophila by microinjecting the site-specific phiC31 integrase mRNA and a transgene-containing donor plasmid. England: 2007. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS, editors. Drosophila: A laboratory handbook. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Abramoff MD, Magelhaes PS, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Musunuru K. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–721. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]


