Abstract
This study provides with a first insight on Mycobacterium tuberculosis complex epidemiology and genetic diversity in the Cross River State, Nigeria. Starting with 137 smear positive patients recruited over a period of 12 months (June 2008 to May 2009), we obtained 97 pure mycobacterial isolates out of which 81 (83.5%) were identified as M. tuberculosis complex. Genotyping revealed a total of 27 spoligotypes patterns with 10 clusters (n = 64% or 79% of clustered isolates, 2–32 isolates/cluster), with patients in the age group range 25–34 years being significantly associated with shared-type pattern SIT61 (p = 0.019). Comparison with SITVIT2 database showed that with the exception of a single cluster (SIT727/H1), all other clusters observed were representative of West Africa; the two main lineages involved were LAM10-CAM (n = 42/81% or 51.8%) of M. tuberculosis and AFRI_2 sublineage of Mycobacterium africanum (n = 27/81% or 33.3%). Subsequent 12-loci MIRU typing resulted in a total of 13 SIT/MIT clusters (n = 52 isolates, 2–9 isolates per cluster), with a resulting recent n − 1 transmission rate of 48.1%. Available drug-susceptibility testing (DST) results for 58/81 clinical isolates revealed 6/58% or 10.4% cases of multiple drug-resistance (MDR); 5/6 MDR cases were caused by strains belonging to LAM10-CAM lineage (a specific cluster SIT61/MIT266 in 4/6 cases, and an orphan spoligotype pattern in 1/6 case). Additionally, MIT266 was associated with streptomycin resistance (p = 0.016). All the six MDRTB isolates were concomitantly resistance to streptomycin and ethambutol; however, 4/6 MDR strains with identical MIRU patterns were characterized by consecutive strain numbers hence the possibility of laboratory cross contamination could not be excluded in 3/4 serial cases. The present preliminary study underlines the usefulness of spoligotyping and 12-loci MIRU–VNTRs to establish a baseline of circulating genotypic lineages of M. tuberculosis complex in Nigeria.
Keywords: Mycobacterium tuberculosis, Epidemiology, Spoligotyping, MIRU–VNTRs, Cross River State, Nigeria
1. Introduction
Mycobacterium tuberculosis is one of the most successful bacterial pathogens in the history of mankind and M. tuberculosis continues to exert an enormous toll on world health. The resurgence of tuberculosis (TB) around the world has renewed interest in understanding the epidemiology and pathogenesis of this disease (Rastogi and Sola, 2007). One important advance in the field of TB research has been the development of molecular techniques that allow the identification and tracking of individual strains of M. tuberculosis, making it possible to differentiate between recent active transmission versus endogenous reactivation (García de Viedma et al., 2011). The contribution of molecular epidemiological methods on the prevention and control of TB is made possible through their ability to differentiate between infecting strains, assessment of the overall diversity of circulating M. tuberculosis complex strains, pinpointing differences by regions and subpopulations, and measurement of the prevalence of endemic strains (van Soolingen, 2001; van Soolingen et al., 1995; Rastogi and Sola, 2007; García de Viedma et al., 2011). Unfortunately, this powerful tool has yet to be applied extensively in many of the 22 high TB burden countries which account for 80% of the world’s total TB burden; Nigeria being one of them with the world’s fourth largest TB burden (http://www.stoptb.org/countries/tbdata.asp). The country adopted the Directly Observed Treatment Scheme (DOTS) strategy for TB control since 2003; unfortunately, with a treatment success rate for new smear positive cases in 2008 at 78%, and a case detection rate comprised between 19% and 24% in 2009, Nigeria still rates among the lowest of high-TB burden countries (WHO, 2010). In this context, we hereby provide with a first insight on M. tuberculosis complex molecular epidemiology and genetic diversity in the Cross River State, Nigeria.
2. Materials and methods
2.1. Study setting, population, and bacteria
The study was carried out in Cross River State, located in the southernmost region of Nigeria. A total of 137 smear-positive TB patients (age range, <15 years to >64 years) recruited over a period of 12 months (June 2008 to May 2009) from major hospitals and TB care facilities were included. According to all smear-positive TB cases notified in 2009 (n = 44863) in Nigeria with a population of 155 million out of which 2.89 million or 1.86% lived in the Cross-River State, the expected number of smear-positive TB cases in our setting for the 12 months period studied may be estimated at 835 persons (https://extranet.who.int/sree/Reports?op=Replet&name=%2FWHO_HQ_Reports%2FG2%2FPROD%2FEXT%2FTBCountry-Profile&ISO2=NG&outtype=pdf).Thus the 137 smear-positive patients represented 16.4% of all smear-positive TB cases. Relevant demographic and clinical information was obtained through a standard questionnaire. The specimens collected were preserved using sodium carbonate (75 mg), and/or refrigerated until transported for processing at the TB research laboratory, Zankli Medical Centre, Abuja, Nigeria. Each specimen was decontaminated using modified Petroff method and cultured using MGIT960 Mycobacterial Detection System (Becton Dickinson, Franklin Lakes, NJ07417, USA) using standard procedure (Siddiqi and Rüsch-Gerdes, 2006). Each positive liquid culture was subcultured into two Lowenstein–Jensen (L–J) slants, one of which contained sodium pyruvate. The cultures were examined twice a week and their rate of growth and colonial morphologies recorded. Contaminated slants were further re-decontaminated and re-cultured. Primary identification was performed using Capilia TB-Neo (TAUNS Laboratories, Inc., Japan) according to manufacturer’s instructions (http://capilia.jp/english/capilia_tb_neo.html). Drug susceptibility testing (DST) was done using the proportion method on L–J medium against isoniazid (INH, 0.2 µg/ml), rifampicin (RIF, 40 µg/ml), streptomycin (SM, 8 µg/ml), and ethambutol (EMB, 2 µg/ml).
2.2. Molecular typing, database comparison, and phylogenetical analysis
The DNA from heat-killed mycobacterial cells suspensions (80 °C for 30 min), was extracted using the cetyltrimethylammonium bromide (CTAB) method (van Embden et al., 1993), and subjected to spoligotyping (Kamerbeek et al., 1997) and classical 12-loci MIRU–VNTR typing (Supply et al., 2001); genotypes were compared using the SITVIT2 proprietary database of the Institut Pasteur de la Guadeloupe as reported recently (Vanhomwegen et al., 2011). In this database, Spoligotype International Type (SIT) and MIRU International Type (MIT) designate spoligotypes and MIRU patterns shared by two or more patient isolates, respectively, whereas “orphan” designates patterns reported for a single isolate. Major phylogenetic clades were assigned according to signatures provided in SpolDB4 (Brudey et al., 2006). The phylogenetical relationships between the spoligotypes and 12-loci MIRUs were studied by building a combined minimum spanning tree (MST) using categorial coefficient with the Bionumerics software Version 3.5 (Applied Maths NV, Sint–Martens–Latem, Belgium) with the parameters “priority rules”, maximum cross-link distance”, and “maximum neighbor distance” set at the default values; MSTs were drawn both with and without hypothetical types or missing links. Lastly, the recent transmission rate based on combined spoligotyping and MIRU–VNTRs data was calculated using the n − 1 method as described earlier (Small et al, 1994).
3. Results
3.1. Study population and bacteria
Starting from a total of 137 smear positive TB patients for MGIT960-based culture, we initially obtained 125 (91.2%) positive vials, among which 122 (97.6%) were AFB positive by Ziehl Neelsen staining. However six vials were mislabeled (possibly due to excess workload) prior to subculture in the L–J medium, another 15 were heavily contaminated after subculture and discarded, and four were negative. This reduced our working cultures to 97 pure isolates. Of the 97 isolates, 81 (83.5%) were identified as members of the M. tuberculosis complex.
3.2. Cluster analysis
The cluster analysis is summarized in Table 1 (for detailed typing results using spoligotyping and MIRU typing, the readers are referred to the Supplemental Table S1). A total of 27 spoligotype patterns were obtained from the 81 isolates, out of which 10 patterns shown in Table 2 corresponded to clustered isolates (10 clusters containing 64% or 79% of clustered isolates, 2–32 isolates/cluster). Seventeen SITs containing 64 isolates matched a preexisting shared type in the SITVIT2 database, whereas four SITs (containing 11 Isolates) were newly-created either within this study or after a match with an orphan from the database. Six were orphan strains. Sixteen MIRUs shared types (MITs) were observed; 12 MITs containing 56 isolates matched a preexisting pattern in the database, whereas four MITs (containing nine isolates) were newly-created either within this study or after a match with an orphan from the database. Thirteen MIRU patterns corresponded to orphans while DNA from three strains did not amplify (strains classified as MIRU patterns being unknown). The comparison of the characteristics such as sex, HIV status and drug resistance of the patients to clustered vs. non clustered strains in a univariate analysis showed no statistically significant differences; however, the age group range of 25–34 years was significantly associated with SIT61 (p = 0.019). In fact, 18/32 (56.3%) of the patients harboring SIT61/LAM10-CAM lineage belonged to this age group.
Table 1.
Spoligotypes and MIRU12 patterns of M. tuberculosis complex clinical isolates from the Cross River State, Nigeria.
| Parameters | Spoligotyping | MIRU12 | ||
|---|---|---|---|---|
| Patterns, n | Strains, n (%) | Patterns, n | Strains, n (%) | |
| Already described | ||||
| Clustered | 6 | 53 (65.4) | 8 | 52 (64.2) |
| Unique | 11 | 11 (13.6) | 4 | 4 (5) |
| Newly created | ||||
| Clustered | 4 | 11 (13.6) | 4 | 9 (11.1) |
| Unique | ||||
| Orphan | 6 | 6 (7.4) | 13 | 13 (16) |
| Unknown | 3 | 3 (3.7) | ||
| Total | 27 | 81 | 32 | 81 |
Table 2.
Description of the clustered M. tuberculosis complex spoligotypes observed in Cross River State, Nigeria and their worldwide distribution in the SITVIT2 database, with additional information on the subclustering of spoligotyping based clusters by 12-loci MIRUs.
| SIT (Clade) a Spoligotype Description Octal Code |
Strains, n (%) in study |
% in study vs. SITVIT2 |
Distribution in Regions with ≥5% of a given SITs b |
Distribution in countries with ≥5% of a given SIT c |
MIRU-based subclustering (MIT designation and number of strains) |
|---|---|---|---|---|---|
| 32 (39.51) | 4.75 | AFRI-W 28.78, AFRI-M 26.11, AMER-N 11.87, EURO-W 10.39, ASIA-W 6.53 | NGA 26.11, CMR 25.07, USA 11.87, SAU 6.53, FXX 5.93 | MIT12 (n=9), MIT263 (n=9), MIT266 (n=7), Orphan (n=3), MIT264 (n=2), MIT265 (n=1), MIT784 (n=1) | |
| 2 (2.47) | 22.22 | AFRI-W 44.44, AFRI-M 22.22, AMER-N 22.22, EURO-S 11.11 | NGA 44.44, CMR 22.22, USA 22.22, ESP 11.11 | MIT 348 (n=2) | |
| 10 (12.35) | 23.81 | AFRI-W 64.29, AMER-N 23.81, AFRI-M 7.14 | NGA 54.76, USA 23.81, CMR 7.14, CIV 7.14 | MIT1228 (n=3), MIT934 (n=2), MIT1266 (n=2), Orphan (n=2), Unknown (n=1) | |
| 2 (2.47) | 5.71 | AMER-S 65.71, AMER-N 20, AFRI-W 5.71 | COL 62.86, USA 20, NGA 5.71 | MIT43 (n=1), Orphan (n=1) | |
| 5 (6.17) | 38.46 | AFRI-W 46.15, AFRI-M 30.77, EURO-W 7.69, AFRI-N 7.69, ASIA-W 7.69 | NGA 46.15, CMR 30.77, SDN 7.69, SAU 7.69, BEL 7.69 | MIT266 (n=5) | |
| 2 (2.47) | 33.33 | AFRI-W 33.33, AMER-N 33.33, AFRI-N 16.67, ASIA-W 16.67 | NGA 33.33, USA 33.33, LBY 16.67, SAU 16.67 | MIT1225 (n=2) | |
| 2 (2.47) | 100.00 | AFRI-W 100 | NGA 100 | MIT956 (n=2) | |
| 3 (3.70) | 100.00 | AFRI-W 100 | NGA 100 | MIT934 (n=3) | |
| 2 (2.47) | 100.00 | AFRI-W 100 | NGA 100 | MIT1138 (n=2) | |
| 4 (4.94) | 80.00 | AFRI-W 80, EURO-S 20 | NGA 80.00, ITA 20 | MIT348 (n=4) |
SIT followed by an asterisk indicates “newly created shared-type”.
Worldwide distribution is reported for regions with ≥5% of a given SITs as compared to their total number in the SITVIT2 database. The definition of macro-geographical regions and sub-regions is according to the United Nations (http://unstats.un.org/unsd/methods/m49/m49regin.htm); Regions: AFRI (Africa), AMER (Americas), ASIA (Asia), EURO (Europe), and OCE (Oceania), subdivided in: E (Eastern), M (Middle), C (Central), N (Northern), S (Southern), SE (South-Eastern), and W (Western). Furthermore, CARIB (Caribbean) belongs to Americas, while Oceania is subdivided in 4 sub-regions, AUST (Australasia), MEL (Melanesia), MIC (Micronesia), and POLY (Polynesia). Note that in our classification scheme, Russia has been attributed a new sub-region by itself (Northern Asia) instead of including it among rest of the Eastern Europe. It reflects its geographical localization as well as due to the similarity of specific TB genotypes circulating in Russia (a majority of Beijing genotypes) with those prevalent in Central, Eastern and South-Eastern Asia.
The 3 letter country codes are according to http://en.wikipedia.org/wiki/ISO_3166-1_alpha-3; countrywide distribution is only shown for SITs with ≥5% of a given SITs as compared to their total number in the SITVIT2 database.
As summarized in Table 2, the most predominant cluster SIT61/LAM10-CAM (n = 32% or 39.51% of isolates) was subdivided into seven MIRUs genotypes: MIT12 (n = 9), MIT263 (n = 9), MIT266 (n = 7), orphan (n = 3), MIT264 (n = 2), MIT265 (n = 1), MIT784 (n = 1). It was followed by SIT331/AFRI_2 cluster (n = 10 isolates) split into five MIRU patterns: MIT1228 (n = 3), MIT934 (n = 2), MIT1226 (n = 2), orphan (n = 2), and unknown (partial MIRU results, n = 1). The only other spoligotyping cluster that were split upon MIRU typing was SIT727/H1 (n = 2), resulting in two individual MIRU patterns (MIT43 n = 1, and orphan n = 1). The remaining seven clusters did not subdivide by MIRU–VNTRs, and included following patterns: SIT320/AFRI_2 (n = 2), SIT838/LAM10-CAM (n = 5), SIT1204/Unknown (n = 2), SIT3068/LAM10-CAM (n = 2), SIT3069/AFRI_2 (n = 3), SIT3070/H3 (n = 2), and SIT3071/AFRI_2 (n = 4). Among the seven unsplit clusters, four spoligotype patterns (SIT3068, n = 2; SIT3069, n = 3; SIT3070, n = 2; SIT3071, n = 4) were not reported elsewhere in the world, and constitute newly created shared-types in our study. Comparison with SITVIT2 database (Table 2) showed that with the exception of a single cluster (SIT727/H1), all other clusters observed were representative of West Africa and more specifically of Nigeria (Brudey et al., 2006); the two main lineages involved were LAM10-CAM (n = 42/81% or 51.8%) of M. tuberculosis and AFRI_2 sublineage of M. africanum (n = 27/81% or 33.3%). One may therefore raise the question if these 2 SIT727/H1 isolates (subdivided by MIRU typing to SIT727/MIT43 strain resistant to INH and and SM, isolated from a HIV-negative male, age group 25–34 years; and the pansusceptible SIT727/orphan strain from a HIV-positive female, age group 15–24 years) did not represent imported cases of disease. Unfortunately, available information did not allow to conclude on this aspect. Furthermore, as summarized in Table 2 as well as by the color codes used in the phylogenetical analysis below (Fig. 1), LAM10–CAM lineage conserved significantly bigger clusters than M. africanum. In conclusion, a total of 13 SIT/MIT clusters containing 52 isolates (2–9 isolates per cluster) were observed in this study which corresponded to a recent n − 1 transmission rate of 48.1%.
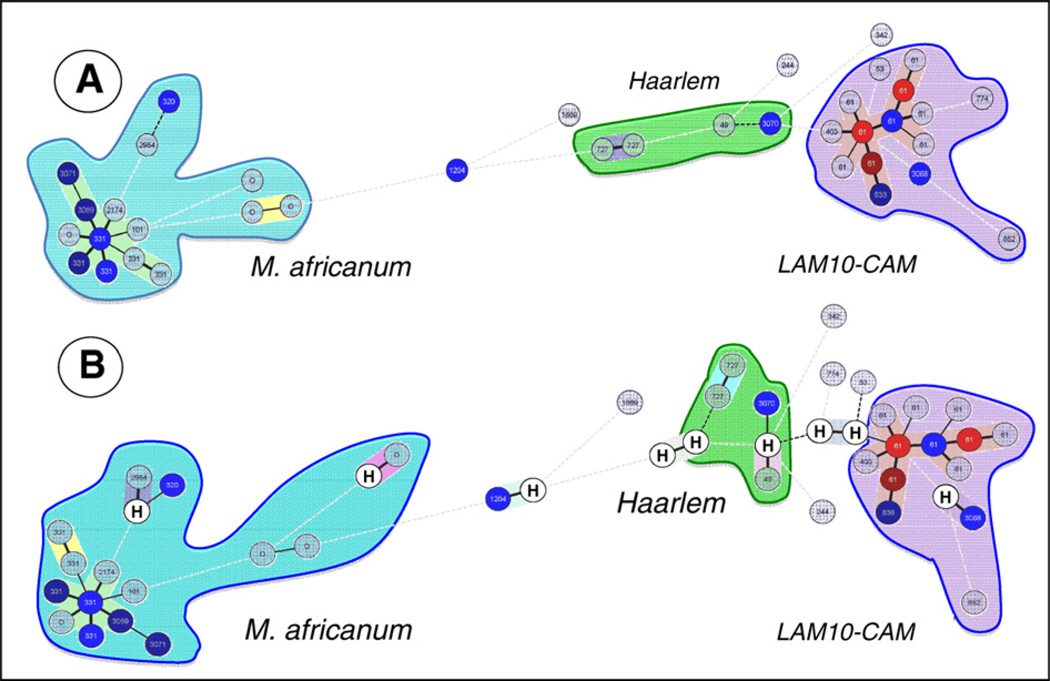
Fig. 1.
Unrooted minimum spanning tree (MST) showing evolutionary relationships between the spoligotypes and 12-loci MIRUs (1A), and the same tree drawn after allowing hypothetical types or missing links (shown by letter “H” within a circle) created using Bionumerics software (1B). In this figure, the length and form of the branches indicates the level of changes (continuous line, 1 change; black dotted line, 2 changes; dotted gray line, 3 or more changes), while the color of circles is proportional to the number of clinical isolates within the sample studied (gray = 1, blue = 2, dark blue = 5, brown = 8, red = 9 or more strains).
3.3. DST results
Available results for 58/81 clinical isolates revealed 6/58% or 10.4% MDR cases (Table 3); 5/6 MDR cases were traced to LAM10–CAM lineage (a specific cluster SIT61/MIT266 in 4/6 cases, and an orphan spoligotype pattern in 1/6 case). The cluster SIT61/LAM10-CAM/MIT266 was found among relatively younger patients, underlining active ongoing transmission of this MDR clone in our setting. Additionally, MIT266 was associated with streptomycin resistance (p = 0.016). All the six MDRTB isolates were concomitantly resistance to streptomycin and ethambutol; however, 4/6 MDR strains with identical MIRU patterns were characterized by consecutive strain numbers hence the possibility of laboratory cross contamination could not be excluded in 3/4 serial cases.
Table 3.
Description of the MDR M. tuberculosis complex strains among smear-positive tuberculosis patients in Cross River State, Nigeria*.
| ID strain |
Year of isolation |
SIT (Clade) Spoligotype Description |
12-loci MIRU Pattern (MIT)** |
Sex (Age group) |
HIV serology, Drug resistance*** |
|---|---|---|---|---|---|
| 123TH | 2009 | 223315153321 (266) | F (25–34) | HIV-negative, SIRE | |
| 124TH | 2009 | 223315153321 (266) | M (25–34) | HIV Unk, SIRE | |
| 125TH | 2009 | 223315153321 (266) | M (15–24) | HIV-negative, SIRE | |
| 126TH | 2009 | 223315153321 (266) | M (35–44) | HIV Unk, SIRE | |
| 127TH | 2009 | 22222514332X (orphan) | F (25–34) | HIV-positive, SIRE | |
| 64IDH | 2008 | No result | M (25–34) | HIV Unk, SIRE |
Drug-susceptibility testing (DST) was done for 58/81 clinical isolates; MDR strains correspond to 10.4% (n = 6/58) of strains with known DST.
Among MIRU patterns shown, the letter “X” denotes loci that could not be amplified.
Abbreviations: S, Streptomycin; I, Isoniazid; R, Rifampicin; E, Ethambutol; Unk, unknown.
3.4. Phylogenetical analysis
The MST connected each single spoligotype and MIRU pattern based on degree of differences among them; the length and form of the branches indicated the level of change(s) required to go from one allele to another while the color of circles was meant to be proportional to the number of clinical isolates within the sample studied. The tree shown in Fig. 1 is represented by branches, continuous vs. dotted lines, and open vs. closed circles; the complexity of the lines (continuous, black dotted and gray dotted) denotes the number of combined level of changes required to go from one allele to another (spoligotype + MIRU) that differ between two isolates: continuous for one MIRU locus and/or one spacer change, black dotted for two loci/spacers and dotted gray line for three or more loci/spacers. The color of circles is proportional to number of clinical isolates in the study (gray = 1, blue = 2, dark blue = 5, brown = 8 strains, red = 9 or more). Note that if a same SIT was split in three dinstinct MITs, each of the subcluster is individually represented in the MST. In the Fig. 1A showing combined spoligotyping and MIRU data, one can observe the two major lineages AFRI_2 and the LAM10–CAM. As represented by the color of the circles, LAM10–CAM strains are more prevalent, and its central node is represented by its prototype SIT61 which is highly predominant in our study sample (32/81% or 39.5% of the isolates). Similarly, the central node for AFRI_2 strains is represented by its prototype SIT331 (n = 10/81% or 12.3% of the isolates). Contrary to M. africanum subpopulation which was well conserved around its central prototype node (with some cases of ongoing transmissions with genotypes that might have evolved overtime starting from its prototype SIT331; examples include newly defined shared-types SIT3069, and SIT3071), there was no central node observed for Haarlem strains (n = 5) in our sample, and none of the SITs present corresponded to any of the Haarlem sublineage prototypes in SpolDB4 (Brudey et al., 2006).
One may hypothesize that LAM10–CAM strains have successfully established themselves in the Cross River State, as evidenced through the LAM10–CAM isolates had undergone significant transmission of highly related clones since the central node SIT61/MIT12 (n = 9) evolved to give other predominant nodes made-up of SIT61/MIT263 (n = 9), and SIT61/MIT266 (n = 7); the subsequent allelic changes therefore only involved a single locus variation (SLV) at MIRU-loci 16, and/or a higher variation at MIRU-loci 40 (MIT12 = 223315153323; MIT263 = 223215153324; MIT266 = 223315153321). Discrete transmission due to Haarlem SIT727/H1 and SIT3070/H1 was also observed. The evolutionary distances between all the three major lineages observed (Fig. 1A) argue in favor of three different pockets of ongoing TB transmission, most of it due to LAM10–CAM and M. africanum (AFRI_2 sublineage) that are phylogeographically specific for West-Africa (Groenheit et al., 2011), and a much smaller fraction due to Haarlem lineage phylogeographically specific for European and North American regions, that could probably involve imported cases of the disease, or strains that initially arrived through settlers but adapted to the local population as can be presumed by the presence of a newly described clone SIT3070/MIT1138 (Table 2, Fig. 1A). The number of repeats for MIRU loci-24 in our study was equal to two for all M. africanum strains (and a single SIT342 strain belonging to EAI lineage) that corresponds to ancestral lineages (Supply et al., 2006), as opposed to one for all other evolutionary modern strains that also included LAM10–CAM strains (Fig. 1A).
The fact that four orphan patterns were observed for M. africanum as opposed to none for LAM10–CAM, supports the recent suggestion that the proportion of evolutionary ancestral M. africanum is slowly decreasing in West-Africa, and these strains are increasingly getting replaced by more successful evolutionary recent M. tuberculosis (Groenheit et al., 2011). We decided to look into the above supposition by allowing hypothetical (H) types or missing links in the MST drawn in Fig. 1A; these were created only if the total distance decreased by at least one change, and if among the neighbors at least one neighbor had no more than one change. The result obtained (Fig. 1B), showed that almost all hypothetical missing links were created for Haarlem strains as opposed to two diametrically opposed and well-conserved M. africanum, and LAM10–CAM lineages. However, even though this conclusion seems reasonable in evolutionary terms, further studies will be needed to shed light on the role of evolution versus extinction of pre-existing strains for M. africanum. Lastly, despite the creation of hypothetical links, the Haarlem strains still appeared scattered on the tree shown in Fig. 1B without any central node(s), an observation that comforts a recent introduction of these strains in Cross River State, Nigeria.
4. Discussion
This study reported a total of 27 spoligotypes patterns from the 81 M. tuberculosis complex isolates, out of which 10 patterns corresponded to clustered isolates (n = 64 isolates, 2–32 isolates/cluster) with a very high clustering rate of 79%, with SIT61/LAM10–CAM being the main genotypic lineage observed. This is not surprising since Cross River State in Nigeria shares a common frontier with Cameroon where this family (lacking spacer 23–25) was found to be dominant (responsible for 42% of smear positive pulmonary TB; Niobe-Eyangoh et al., 2003). A similar occurrence (39.5%) was seen in our study. Republic of Chad which also shares boundary with Nigeria has reported predominance (33%) of this strain (Diguimbaye et al., 2006). Movement of populations across the boundaries of these three countries for business purposes is massive and constant, thereby supporting the observation of a stable association of specific clones with geographically localized human population (Hirsh et al., 2004). Another study carried out in Ibadan in the Western part of Nigeria (Cadmus et al., 2006) showed a higher predominance of this strain (66%), and other studies carried out in the West African region, notably in Benin (Affolabi et al., 2009), and in Burkina Faso (Godreuil et al., 2007) have also reported the “Cameroon family” as the major cause of TB (21% and 30%, respectively). From Cameroon to Burkina Faso, through Nigeria and Benin Republic, this family could be associated with recent transmission of TB, and may represent an expanding clone. This predominant spoligotype has equally been reported in other West African countries such as Senegal and Ivory Coast (Niobe-Eyangoh et al., 2003).
Although the exact reasons for Cameroon family selection and dissemination are unknown, it has been suggested that the M. bovis BCG vaccination, which is common practice in Cameroon (as well as in Nigeria) may play a role in its selection (Niobe-Eyangoh et al., 2003). The possible role of BCG vaccination in the selection of resistant strains with BCG-induced immunity was previously suggested to explain the expansion of Beijing family strains and the predominance of other families in certain geographic settings (Hermans et al., 1995; van Soolingen, 2001). This could also be a factor to explain why M. africanum, which is less virulent than M. tuberculosis in experimental models (Castets and Sarrat, 1968), dramatically diminished as a cause of TB after the generalization of BCG vaccination in Nigeria like in Cameroon. Indeed, a decreasing trend in M. africanum transmission over the last three decades was reported (Niobe-Eyangoh et al., 2003). Nonetheless, as reviewed recently, phenotypic differences exist within M. africanum lineages in mouse models (de Jong et al. 2010); it seems that virulence is more related to specific biological traits than lineage restricted, something that have been reported before for M. tuberculosis Beijing lineage in which one may find drug-susceptible and not so successful (virulent) strains. On the other hand, a recent review on possible underlying mechanisms for successful emergence of the Beijing genotype strains emphasized its selective advantage over other M. tuberculosis strains by its ability to escape from BCG vaccination, higher virulence, and interaction with the human immune system (Parwati et al. 2010). Further research therefore is needed to determine the mechanism(s) underlying the decreasing trend in M. africanum transmission vs. Cameroon family selection and dissemination in certain parts of Africa.
In our study, M. africanum (SIT331) represented 12.35% of TB cases. M. africanum is generally responsible for TB in patients living in or coming from sub-Saharan African countries (Castets and Sarrat, 1968; Frothingham et al., 1999). Recent surveys have shown highly variable prevalence of M. africanum in different regions of Africa; e.g., approximately 5% of patients with TB from the Ivory Coast, 10% from Cameroon (Niobe-Eyangoh et al., 2003), and about 60% from Guinea-Bissau (Källenius et al., 1999; Bonard et al., 2000) were found to be infected with M. africanum. However, the proportion of M. africanum in Western Africa, and more precisely in Guinea–Bissau is diminishing, e.g., a recent report showed the proportion of M. africanum in Guinea-Bissau at 47.1% (Groenheit et al., 2011). In this context, one reason for the diminished reported incidence of M. africanum in some regions of Africa as the cause of human TB could also be due to misidentification of the M. africanum. However, one of the most astonishing aspect regarding phylogeographical regional specificity of M. africanum is the fact that almost 99% M. africanum strains in Guinea-Bissau belong to the AFRI_1 sublineage (essentially SIT181, SIT187) as opposed to our study in Cross River State where it is solely imputable to AFRI_2 sublineage (essentially SIT331). In a study carried out in Ibadan, Nigeria (Cadmus et al., 2006), the authors reported that approximately 13% isolates were more likely to be M. africanum and M. bovis rather than M. tuberculosis. The authors highlighted the need for molecular characterization of clinical isolates to ensure that correct estimates are made of the true burden of infection due to M. bovis and M. africanum in developing countries; M. africanum isolates have substantial phenotypic heterogeneity with some strains resembling M. bovis and others resembling M. tuberculosis (Collins et al., 1982; Wayne et al., 1982; Frottier et al., 1990; Hoffner et al., 1993; Haas et al., 1997). This has led several authors to question the validity of the species designation as M. africanum (Wieten et al., 1983; Tsukamura et al., 1985; Wayne and Kubica, 1986).
The LAM10–CAM clade (SIT61) in our study was associated with recent transmission among the age group of 25–34 years (p = 0.019), since 18/32 (56.3%) of the patients harboring this lineage belonged to this age group. Although less predominant, the M. africanum AFRI_2 clade was also present and clustered in our study suggesting that it probably was actively transmitted, although in a smaller number of patients. The comparison of the characteristics such as sex, HIV status and drug resistance of the patients to clustered vs. non clustered strains in a univariate analysis showed no statistically significant differences. Although one reason could be the small sample size as a whole and when stratified, several studies from developing countries have had difficulties to demonstrate a statistically significant link between demographic data and clustering (Affolabi et al., 2009; Godreuil et al., 2007; Niobe-Eyangoh et al., 2003); it may nonetheless be noticed that young age was associated with clustering in developed countries (Small et al., 1994). In conclusion, spoligotyping followed by MIRU typing resulted in a total of 13 SIT/MIT clusters containing 52 isolates (2–9 isolates per cluster), with a resulting recent n − 1 transmission rate of 48.1% during the 1 year recruitment period. This rate is higher than recently reported for Ndola, Zambia (Mulenga et al., 2010), comparable to Burkina Faso (Godreuil et al., 2007), but below the rate reported in Zimbabwe (Easterbrook et al., 2004).
In conclusion, the present preliminary study underlined the usefulness of spoligotyping and 12-loci MIRU–VNTRs to establish a baseline of circulating genotypic lineages of M. tuberculosis complex. Although we are unable to exclude the possibility of laboratory cross contamination for 3/6 MDR strains, available results showed a much higher proportion of MDR strains than expected, and an identical cluster of LAM10–CAM lineage (SIT61/MIT266) found among relatively younger patients. Future studies over extended period of time targeting a detailed mapping of circulating tubercle bacilli lineages in conjunction with their drug-susceptibility profiles would allow to determine the precise situation of TB transmission and drug-resistance in Nigeria.
Supplementary Material
Acknowledgements
The authors are grateful to the Fondation Mérieux, Lyon, France for funding this research. Benjamin Thumamo benefited from the fellowship award of the International Union of Microbiological Societies, IUMS-SGM. The molecular typing work done at Nalin Rastogi’s laboratory was financed by the International Network of the Pasteur Institutes. Veronique Hill was awarded a Ph.D. fellowship by the European Social Funds through the Regional Council of Guadeloupe. The SITVIT2 database project was partially financed by the Regional Council of Guadeloupe (CR/08-1612: Biodiversité et Risque Infectieux dans les modèles insulaires).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.meegid.2011.08.011.
References
- Affolabi D, Anyo G, Faïhun F, Sanoussi N, Shamputa IC, Rigouts L, Kestens L, Anagonou S, Portaels F. First molecular epidemiological study of tuberculosis in Benin. Int. J. Tuberc. Lung Dis. 2009;13:317–322. [PubMed] [Google Scholar]
- Bonard D, Msellati P, Rigouts L, Combe P, Coulibaly D, Coulibaly IM, Portaels F. What is the meaning of repeated isolation of Mycobacterium africanum? Int. J. Tuberc. Lung Dis. 2000;4:1176–1180. [PubMed] [Google Scholar]
- Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuno L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, Garcia de Viedma D, Garzelli C, Gazzola L, Gomes HM, Gutierrez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ly HM, Martin C, Mokrousov I, Narvskaia O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim MZ, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Rusch Gerdes S, Sajduda A, Samper S, Shemyakin I, Singh UB, Somoskovi A, Skuce R, van Soolingen D, Streicher EM, Suffys PN, Tortoli E, Tracevska T, Vincent V, Victor TC, Warren R, Yap SF, Zaman K, Portaels F, Rastogi N, Sola C. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:6–23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadmus S, Palmer S, Okker M, Dale J, Gover K, Smith N, Jahans K, Hewinson RG, Gordon SV. Molecular analysis of human and bovine tubercle bacilli from a local setting in Nigeria. J. Clin. Microbiol. 2006;44:29–34. doi: 10.1128/JCM.44.1.29-34.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castets M, Sarrat H. Bacteriologic aspects of mycobacteria isolated at Dakar in 1967. Bull. Soc. Med. Afr. Noire Lang Fr. 1968;13:463–469. (In French) [PubMed] [Google Scholar]
- Collins CH, Yates MD, Grange JM. Subdivision of Mycobacterium tuberculosis into five variants for epidemiological purposes: methods and nomenclature. J. Hyg. (Lond) 1982;89:235–242. doi: 10.1017/s0022172400070765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong BC, Antonio M, Gagneux S. Mycobacterium africanum - review of an important cause of human tuberculosis in West Africa. PLoS Negl. Trop. Dis. 2010;4(9):e744. doi: 10.1371/journal.pntd.0000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diguimbaye C, Hilty M, Ngandolo R, Mahamat HH, Pfyffer GE, Baggi F, Tanner M, Schelling E, Zinsstag J. Molecular characterization and drug resistance testing of Mycobacterium tuberculosis isolates from Chad. J. Clin. Microbiol. 2006;44:1575–1577. doi: 10.1128/JCM.44.4.1575-1577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterbrook PJ, Gibson A, Murad S, Lamprecht D, Ives N, Ferguson A, Lowe O, Mason P, Ndudzo A, Taziwa A, Makombe R, Mbengeranwa L, Sola C, Rastogi N, Drobniewski F. High rates of clustering of strains causing tuberculosis in Harare, Zimbabwe: a molecular epidemiological study. J. Clin. Microbiol. 2004;42:4536–4544. doi: 10.1128/JCM.42.10.4536-4544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frothingham R, Strickland PL, Bretzel G, Ramaswamy S, Musser JM, Williams DL. Phenotypic and genotypic characterization of Mycobacterium africanum isolates from West Africa. J. Clin. Microbiol. 1999;37:1921–1926. doi: 10.1128/jcm.37.6.1921-1926.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frottier J, Eliaszewicz M, Arlet V, Gaudillat C. Infections caused by Mycobacterium africanum. Bull. Acad. Natl. Med. 1990;174:29–33. [PubMed] [Google Scholar]
- García de Viedma D, Mokrousov I, Rastogi N. Innovations in the molecular epidemiology of tuberculosis. Enferm. Infecc. Microbiol. Clin. 2011;29(Suppl 1):8–13. doi: 10.1016/S0213-005X(11)70012-X. [DOI] [PubMed] [Google Scholar]
- Godreuil S, Torrea G, Terru D, Chevenet F, Diagbouga S, Supply P, Van de Perre P, Carriere C, Banuls AL. First molecular epidemiology study of Mycobacterium tuberculosis in Burkina Faso. J. Clin. Microbiol. 2007;45:921–927. doi: 10.1128/JCM.01918-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenheit R, Ghebremichael S, Svensson J, Rabna P, Colombatti R, Riccardi F, Couvin D, Hill V, Rastogi N, Koivula T, Källenius G. The Guinea–Bissau family of Mycobacterium tuberculosis complex revisited. PLoS One. 2011;6(4) doi: 10.1371/journal.pone.0018601. e18601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas WH, Bretzel G, Amthor B, Schilke K, Krommes G, Rusch-Gerdes S, Sticht-Groh V, Bremer HJ. Comparison of DNA fingerprint patterns of isolates of Mycobacterium africanum from East and West Africa. J. Clin. Microbiol. 1997;35:663–666. doi: 10.1128/jcm.35.3.663-666.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans PW, Messadi F, Guebrexabher H, van Soolingen D, de Haas PE, Heersma H, de Neeling H, Ayoub A, Portaels F, Frommel D. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia, Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J. Infect. Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc. Natl. Acad. Sci. USA. 2004;101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffner SE, Svenson SB, Norberg R, Dias F, Ghebremichael S, Kallenius G. Biochemical heterogeneity of Mycobacterium tuberculosis complex isolates in Guinea–Bissau. J. Clin. Microbiol. 1993;31:2215–2217. doi: 10.1128/jcm.31.8.2215-2217.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källenius G, Koivula T, Ghebremichael S, Hoffner SE, Norberg R, Svensson E, Dias F, Marklund BL, Svenson SB. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea–Bissau. J. Clin. Microbiol. 1999;37:3872–3878. doi: 10.1128/jcm.37.12.3872-3878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, van Agderveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden JDA. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulenga C, Shamputa IC, Mwakazanga D, Kapata N, Portaels F, Rigouts L. Diversity of Mycobacterium tuberculosis genotypes circulating in Ndola, Zambia. BMC Infect. Dis. 2010;10:177. doi: 10.1186/1471-2334-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niobe-Eyangoh SN, Kuaban C, Sorlin P, Cunin P, Thonnon J, Sola C, Rastogi N, Vincent V, Gutierrez MC. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 2003;41:2547–2553. doi: 10.1128/JCM.41.6.2547-2553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parwati I, van Crevel R, van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect. Dis. 2010;10:103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- Rastogi N, Sola C. Molecular evolution of the Mycobacterium tuberculosis complex. In: Palomino JC, Leao S, Ritacco V, editors. Tuberculosis 2007: from basic science to patient care. 2007. pp. 53–91. Amedeo Online Textbooks, http://www.tuberculosistextbook.com/index.htm. [Google Scholar]
- Siddiqi SH, Rüsch-Gerdes S. Prepared for the Foundation for Innovative New Diagnostics. Geneva, Switzerland: 2006. Jul, Procedure Manual For BACTEC MGIT 960™ TB System. Mycobacteria Growth Indicator Tube (MGIT) Culture and Drug Susceptibility Demonstration Projects. 2006, FIND Diagnostics, http://www.finddiagnostics.org/export/sites/default/resource-centre/find_documentation/pdfs/mgit_manual_nov_2007.pdf. [Google Scholar]
- Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, Schecter GF, Daley CL, Schoolnik GK. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. N. Engl. J. Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 2001;39:3563–3571. doi: 10.1128/JCM.39.10.3563-3571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamura M, Mizuno S, Toyama H. Taxonomic studies on the Mycobacterium tuberculosis series. Microbiol. Immunol. 1985;29:285–299. doi: 10.1111/j.1348-0421.1985.tb00827.x. [DOI] [PubMed] [Google Scholar]
- van Embden JDA, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM, Small PM. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 2001;249:1–26. doi: 10.1046/j.1365-2796.2001.00772.x. [DOI] [PubMed] [Google Scholar]
- van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, Qing HZ, Enkhsaikan D, Nymadawa P, van Embden JD. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhomwegen J, Kwara A, Martin M, Gillani FS, Fontanet A, Mutungi P, Crellin J, Obaro S, Gosciminski M, Carter EJ, Rastogi N. Impact of immigration on the molecular epidemiology of tuberculosis in Rhode Island. J. Clin. Microbiol. 2011;49:834–844. doi: 10.1128/JCM.01952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne LG. Microbiology of tubercle bacilli. Am. Rev. Respir. Dis. 1982;125:31–41. doi: 10.1164/arrd.1982.125.3P2.31. [DOI] [PubMed] [Google Scholar]
- Wayne LG, Kubica GP. The mycobacteria. In: Sneath PHA, Holt JG, editors. Bergey’s Manual of Systemic Bacteriology. Baltimore: The Williams & Wilkins; 1986. pp. 1435–1457. [Google Scholar]
- WHO. Global Tuberculosis Control: WHO Report, 2010. Geneva, Switzerland: WHO Publications; 2010. http://www.who.int/tb/publications/global_report/en/ [Google Scholar]
- Wieten G, Haverkamp J, Groothuis DG, Berwald LG, David HL. Classification and identification of Mycobacterium africanum by pyrolysis mass spectrometry. J. Gen. Microbiol. 1983;129:3679–3688. doi: 10.1099/00221287-129-12-3679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.