Abstract
The cuticle of C. elegans is a highly resistant structure that surrounds the exterior of the animal1-4. The cuticle not only protects the animal from the environment, but also determines body shape and plays a role in motility4-6. Several layers secreted by epidermal cells comprise the cuticle, including an outermost lipid layer7.
Circumferential ridges in the cuticle called annuli pattern the length of the animal and are present during all stages of development8. Alae are longitudinal ridges that are present during specific stages of development, including L1, dauer, and adult stages2,9. Mutations in genes that affect cuticular collagen organization can alter cuticular structure and animal body morphology5,6,10,11. While cuticular imaging using compound microscopy with DIC optics is possible, current methods that highlight cuticular structures include fluorescent transgene expression12, antibody staining13, and electron microscopy1. Labeled wheat germ agglutinin (WGA) has also been used to visualize cuticular glycoproteins, but is limited in resolving finer cuticular structures14. Staining of cuticular surface using fluorescent dye has been observed, but never characterized in detail15. We present a method to visualize cuticle in live C. elegans using the red fluorescent lipophilic dye DiI (1,1'-dioctadecyl-3,3,3',3'-tetramethylindocarbocyanine perchlorate), which is commonly used in C. elegans to visualize environmentally exposed neurons. This optimized protocol for DiI staining is a simple, robust method for high resolution fluorescent visualization of annuli, alae, vulva, male tail, and hermaphrodite tail spike in C. elegans.
Keywords: Developmental Biology, Issue 59, Cuticle, alae, annuli, C. elegans, DiI, lipid staining, live stain
Protocol
1. Preparation of DiI stain
Prepare a stock solution of 20 mg/mL DiI (Biotium, Inc., Hayward, CA) in DMF. DiI is light sensitive, so protect DiI from light by wrapping in foil.
Create a working dilution of DiI by adding 0.6 μL DiI stock to 399.4 μL M9 for each population. This should give a final working dilution of 30 μg/mL DiI in M9. This can be scaled up for staining multiple populations simultaneously. Shield DiI from light by wrapping the tube(s) in foil.
2. Preparation of nematodes
Use a 60 mm plate containing a population of uncontaminated nematodes. Wash animals from plate using a solution of 0.5% Triton X-100 in M9 buffer by gently swirling liquid in a circular motion across the surface of the plate to loosen all larval and adult animals. Transfer wash into a sterile 1.5 mL tube.
Immediately spin down the animals at 2000 rpm for 30 sec. Remove and discard as much supernatant as possible without disturbing the mass of animals at the bottom of the tube.
To reduce residual Triton X-100, rinse animals using M9 buffer, spin, and remove supernatant. Repeat this step once.
Add 400 μL of working DiI solution in M9 to the tube and vortex briefly to resuspend animals in the solution.
Shake tube at 20°C horizontally for 3 hours at 350 rpm in a light-protected environment. If desired, animals can be incubated up to 16 hours for staining.
To reduce the amount of unbound dye, spin down the animals at 2000 rpm for 20 sec. Remove and discard as much supernatant as possible without disturbing the mass of animals.
Resuspend animals in 400 μL M9 buffer and pour liquid onto a bacteria-free portion of a NGM agar plate seeded with OP50 E. coli. Allow animals to recover at least 30 minutes. During the recovery time the animals should crawl away from the DiI staining liquid and onto the food. This step reduces background fluorescence from free DiI.
3. Mounting and observing specimens
Melt 4% agar in water using an autoclave or a microwave.
Create reusable spacers, which can be used to ensure uniform thickness of the agar pad, by layering two pieces of lab tape on a glass slide. Make two spacer slides total.
Arrange a clean glass slide between two spacer slides. Pipette about 150 μL (four drops) of molten 4% agar onto the center of the clean glass slide. Quickly cover the molten agar using an additional slide to form an agar pad. Carefully remove the cover slide, keeping the pad centered on the top of the mounting slide.
Pipette about 5 μL of nematode anesthetic (100 μM - 1 mM levamisole, for example) onto the pad.
Mount 8 - 12 animals in the anesthetic and cover with a microscope coverslip.
Observe animals using a compound or confocal microscope fitted with at least a 40x objective and a DSRed/TRITC (or other compatible) filter. The fluorescence excitation maximum of DiI is 549 nm and its emission maximum is 565 nm for bound dye (Biotium, Inc., Hayward, CA).
4. Representative Results
DiI stains the cuticle of wild-type and mutant C. elegans. The cuticular surface contains annuli separated by circumferential furrows and, in some stages, longitudinal ridges called alae. Each developmental stage has cuticular structures with distinct compositions2. Ridges or furrows of both alae and annuli fluorescently stain, depending on surface composition, throughout larval and adult stages and remain visible up to a day after recovery using this method. Background fluorescent speckles are sometimes observed (Figures 2F, G), but not routinely (Figure 1, Figure 2A-E, H, I). All images were taken with spinning disk confocal or, when noted, widefield (wf) compound microscopy.
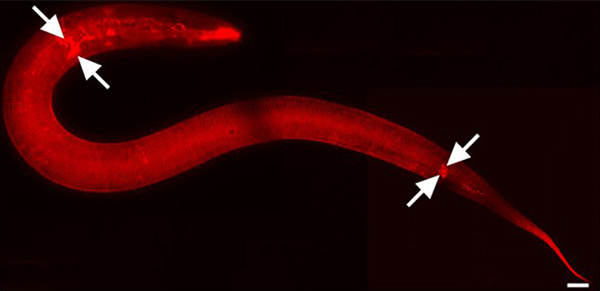 Figure 1. DiI stains cuticle and environmentally exposed neurons. L2 stage animal. 630x magnification. Mosaic image parts were captured using iVision-Mac software (BioVision Technologies, Exton, PA). Images were joined using Adobe Photoshop CS3 (Adobe Systems, Inc., San Jose, CA). Scale bar = 10 μm. DiI also fluorescently stains amphid and phasmid sensory neurons in the head and tail respectively (arrows mark some).
Figure 1. DiI stains cuticle and environmentally exposed neurons. L2 stage animal. 630x magnification. Mosaic image parts were captured using iVision-Mac software (BioVision Technologies, Exton, PA). Images were joined using Adobe Photoshop CS3 (Adobe Systems, Inc., San Jose, CA). Scale bar = 10 μm. DiI also fluorescently stains amphid and phasmid sensory neurons in the head and tail respectively (arrows mark some).
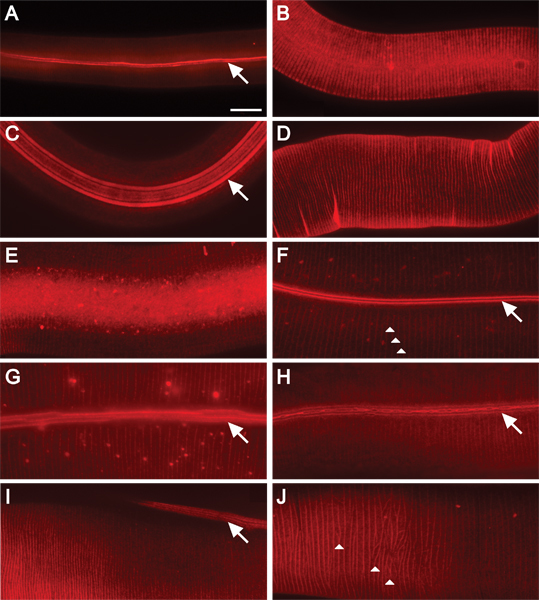 Figure 2. DiI fluorescently stains C. elegans cuticle at all stages of post-embryonic development. 630x magnification. Scale bar = 10 μm. Staining of wild-type animals in A) L1; B) L2; C) dauer; D) L3; E) L4; and F) adult stages. The alae and annular ridges are fluorescently stained in L1 and dauer animals (A, C). DiI stains annular ridges in L2 animals (B). Annular furrows stain in L3 and L4 animals (D, E). The furrows of the alae and annuli are stained in adult animals (F-H). Alae are composed of two, five, or three ridges (in L1, dauer, or adult animals, respectively) that run the length of the animal (arrows)16. Annuli create circumferential ridges around the animal (arrowheads, F and J). The cuticle of adult mutant animals display moderate cuticular organization defects (G-H). G) Ridges of alae are discontinuous (wf). H) Supernumerary alae ridges are fused and branched or bifurcated (wf). Collagen gene mutants exhibit alae and annular organization defects (I-J). I) In transgenic animals overexpressing pRF4 (rol-6(su1006)), ridges of alae lie at an angle to the length of the animal. J) Annuli in transgenic animals overexpressing pRF4 (rol-6(su1006)) display an irregular pattern.
Figure 2. DiI fluorescently stains C. elegans cuticle at all stages of post-embryonic development. 630x magnification. Scale bar = 10 μm. Staining of wild-type animals in A) L1; B) L2; C) dauer; D) L3; E) L4; and F) adult stages. The alae and annular ridges are fluorescently stained in L1 and dauer animals (A, C). DiI stains annular ridges in L2 animals (B). Annular furrows stain in L3 and L4 animals (D, E). The furrows of the alae and annuli are stained in adult animals (F-H). Alae are composed of two, five, or three ridges (in L1, dauer, or adult animals, respectively) that run the length of the animal (arrows)16. Annuli create circumferential ridges around the animal (arrowheads, F and J). The cuticle of adult mutant animals display moderate cuticular organization defects (G-H). G) Ridges of alae are discontinuous (wf). H) Supernumerary alae ridges are fused and branched or bifurcated (wf). Collagen gene mutants exhibit alae and annular organization defects (I-J). I) In transgenic animals overexpressing pRF4 (rol-6(su1006)), ridges of alae lie at an angle to the length of the animal. J) Annuli in transgenic animals overexpressing pRF4 (rol-6(su1006)) display an irregular pattern.
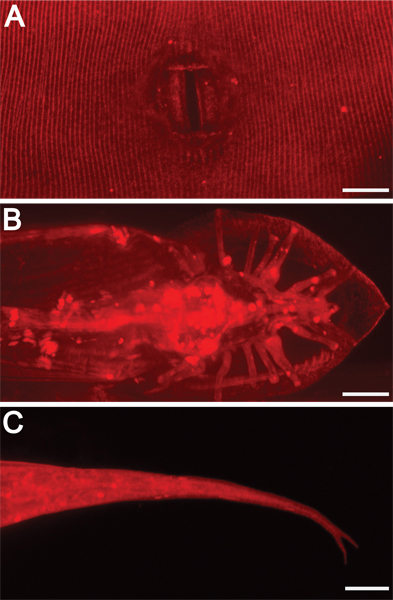 Figure 3. External morphological structures are illuminated by DiI staining. 630x magnification. Scale bars = 10 μm. DiI also highlights other exterior features, including A) adult hermaphrodite vulva, B) adult male tail rays and fan, and C) hermaphrodite tail spike (forked in this mutant background).
Figure 3. External morphological structures are illuminated by DiI staining. 630x magnification. Scale bars = 10 μm. DiI also highlights other exterior features, including A) adult hermaphrodite vulva, B) adult male tail rays and fan, and C) hermaphrodite tail spike (forked in this mutant background).
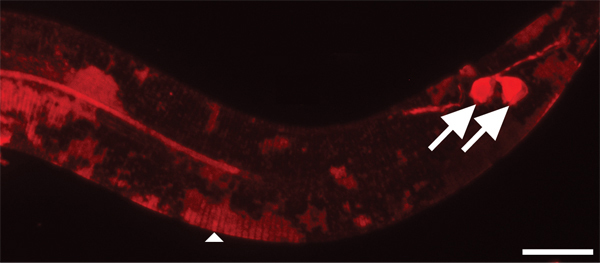 Figure 4. Cuticle takes longer to stain with DiI than environmentally exposed neurons. 630x magnification. Scale bar = 10 μm. After two hours of staining, amphid (not shown) and phasmid sensory neurons (arrows) are sufficiently stained. In contrast, the cuticle of younger animals is only partially stained in patches (arrowhead).
Figure 4. Cuticle takes longer to stain with DiI than environmentally exposed neurons. 630x magnification. Scale bar = 10 μm. After two hours of staining, amphid (not shown) and phasmid sensory neurons (arrows) are sufficiently stained. In contrast, the cuticle of younger animals is only partially stained in patches (arrowhead).
| Wash | Stain solution (+ DiI) | Incubation time | Cuticle stained |
| M9 + 0.5% Triton X-100 | M9 + 0.5% Triton X-100 | 2 hrs | no |
| M9 + 0.5% Triton X-100 | M9 + 0.5% Triton X-100 | 3 hrs | no |
| M9 + 0.5% Triton X-100 | M9 | 2 hrs | partial |
| M9 + 0.5% Triton X-100 | M9 | 3 hrs | yes |
| M9 + 0.5% Triton X-100 | H20 | 2 hrs | partial |
| M9 + 0.5% Triton X-100 | H20 | 3 hrs | yes |
Table 1. Cuticular staining under different conditions. Various incubation solutions and times were tested to optimize cuticular staining in animals. H20, sterile distilled water. Partial staining indicates patchy staining of larval cuticle (Figure 4), though adult cuticle stains consistently.
Discussion
The DiI staining method presented here allows for a relatively quick and convenient way to visualize the cuticle in C. elegans. By repurposing and optimizing a method commonly used to image environmentally exposed sensory neurons15,17, DiI can be used to fluorescently stain both alae and annular structures (Figures 1 and 2), as well as the vulva, male tail, and hermaphrodite tail spike (Figure 3). We have found that the incubation solution and time influence the ability of DiI to consistently stain the cuticle (Table 1, Figure 4). The method for DiI staining environmentally exposed neurons uses a two-hour incubation time in M9 with Triton X-10015. An initial wash of M9 with the surfactant Triton X-100 helps remove contamination from the cuticular surface and prevents the animals from clumping. However, incubating the animals three hours in a staining solution in the same buffer prevents the dye from staining the cuticle (Table 1). This may be caused by lipids on the nematode surface being stripped off by the longer treatment in the detergent solution. Incubation of animals in water with DiI stains the cuticle, indicating that the M9 salt solution is not required for DiI staining of cuticle. Although a water-based protocol is recommended for neuronal DiI staining15, we find that animals treated this way often appear unhealthy. Washing animals briefly in 0.5% Triton X-100 in M9, followed by a three-hour incubation in staining solution made with M9, provides consistent staining of cuticular structures and maintains the well-being of the animals.
Animals can be rescued after observation and maintained, permitting direct downstream analyses. This method is a convenient tool that can be used in many studies, including, but not limited to, cuticular secretion and organization, epidermal cell development, heterochronic gene pathways, and nematode evolution.
Disclosures
We have nothing to disclose.
Acknowledgments
We wish to thank S. Taneja-Bageshwar, K. Beifuss, S. Kedroske, and H-C. Hsiao for helpful discussions. This work was funded by start-up funds from the TAMHSC Department of Molecular and Cellular Medicine. The compound scope and spinning disk were purchased with funds provided by the department and the TAMHSC Office of the Dean. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources. pRF4 (rol-6(su1006)) was a gift of A. Fire.
References
- Cox GN, Kusch M, Edgar RS. Cuticle of Caenorhabditis elegans: its isolation and partial characterization. The Journal of Cell Biology. 1981;90:7–17. doi: 10.1083/jcb.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GN, Staprans S, Edgar RS. The cuticle of Caenorhabditis elegans. II. Stage-specific changes in ultrastructure and protein composition during postembryonic development. Dev. Biol. 1981;86:456–470. doi: 10.1016/0012-1606(81)90204-9. [DOI] [PubMed] [Google Scholar]
- Hall D, Altun Z. C. elegans Atlas. Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- Page AP, Johnstone IL. The C. elegans Research Community. WormBook; 2007. The cuticle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM, Johnson JJ, Edgar RS, Basch C, Roberts S. The sqt-1 gene of C. elegans encodes a collagen critical for organismal morphogenesis. Cell. 1988;55:555–565. doi: 10.1016/0092-8674(88)90214-0. [DOI] [PubMed] [Google Scholar]
- Mende Nvon, Bird DM, Albert PS, Riddle DL. dpy-13: a nematode collagen gene that affects body shape. Cell. 1988;55:567–576. doi: 10.1016/0092-8674(88)90215-2. [DOI] [PubMed] [Google Scholar]
- Blaxter ML. Cuticle surface proteins of wild type and mutant Caenorhabditis elegans. The Journal of Biological Chemistry. 1993;268:6600–6609. [PubMed] [Google Scholar]
- Costa M, Draper BW, Priess JR. The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev. Biol. 1997;184:373–384. doi: 10.1006/dbio.1997.8530. [DOI] [PubMed] [Google Scholar]
- Sapio MR, Hilliard MA, Cermola M, Favre R, Bazzicalupo P. The Zona Pellucida domain containing proteins, CUT-1, CUT-3 and CUT-5, play essential roles in the development of the larval alae in Caenorhabditis elegans. Dev. Biol. 2005;282:231–245. doi: 10.1016/j.ydbio.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Johnstone IL. Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends Genet. 2000;16:21–27. doi: 10.1016/s0168-9525(99)01857-0. [DOI] [PubMed] [Google Scholar]
- Kramer JM, French RP, Park EC, Johnson JJ. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol. Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein MC. Caenorhabditis elegans exoskeleton collagen COL-19: an adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev. Dyn. 2003;226:523–539. doi: 10.1002/dvdy.10259. [DOI] [PubMed] [Google Scholar]
- McMahon L, Muriel JM, Roberts B, Quinn M, Johnstone IL. Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Molecular Biology of the Cell. 2003;14:1366–1378. doi: 10.1091/mbc.E02-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link CD, Ehrenfels CW, Wood WB. Mutant expression of male copulatory bursa surface markers in Caenorhabditis elegans. Development. 1988;103:485–495. doi: 10.1242/dev.103.3.485. [DOI] [PubMed] [Google Scholar]
- Tong YG, Burglin TR. Conditions for dye-filling of sensory neurons in Caenorhabditis elegans. J. Neurosci. Methods. 2010;188:58–61. doi: 10.1016/j.jneumeth.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Singh RN, Sulston JE. Some observations on moulting in Caenorhabditis elegans. Nematologica. 1978;24:63–71. [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK. Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics. 1998;148:187–200. doi: 10.1093/genetics/148.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]


