Abstract
The ability of two or more cells of the same type to fuse has been utilized in metazoans throughout evolution to form many complex organs, including skeletal muscle, bone and placenta. Contemporary studies demonstrate fusion of cells of the same type confers enhanced function. For example, when the trophoblast cells of the placenta fuse to form the syncytiotrophoblast, the syncytiotrophoblast is better able to transport nutrients and hormones across the maternal-fetal barrier than unfused trophoblasts1-4. More recent studies demonstrate fusion of cells of different types can direct cell fate. The "reversion" or modification of cell fate by fusion was once thought to be limited to cell culture systems. But the advent of stem cell transplantation led to the discovery by us and others that stem cells can fuse with somatic cells in vivo and that fusion facilitates stem cell differentiation5-7. Thus, cell fusion is a regulated process capable of promoting cell survival and differentiation and thus could be of central importance for development, repair of tissues and even the pathogenesis of disease.
Limiting the study of cell fusion, is lack of appropriate technology to 1) accurately identify fusion products and to 2) track fusion products over time. Here we present a novel approach to address both limitations via induction of bioluminescence upon fusion (Figure 1); bioluminescence can be detected with high sensitivity in vivo8-15. We utilize a construct encoding the firefly luciferase (Photinus pyralis) gene placed adjacent to a stop codon flanked by LoxP sequences.When cells expressing this gene fuse with cells expressing the Cre recombinase protein, the LoxP sites are cleaved and the stop signal is excised allowing transcription of luciferase. Because the signal is inducible, the incidence of false-positive signals is very low. Unlike existing methods which utilize the Cre/LoxP system16, 17, we have incorporated a "living" detection signal and thereby afford for the first time the opportunity to track the kinetics of cell fusion in vivo.
To demonstrate the approach, mice ubiquitously expressing Cre recombinase served as recipients of stem cells transfected with a construct to express luciferase downstream of a floxed stop codon. Stem cells were transplanted via intramyocardial injection and after transplantation intravital image analysis was conducted to track the presence of fusion products in the heart and surrounding tissues over time. This approach could be adapted to analyze cell fusion in any tissue type at any stage of development, disease or adult tissue repair.
Keywords: Bioengineering, Issue 59, Cell fusion, stem cell, fusogen, cre recombinase, biophotonic imaging, cellular transplantation
Protocol
1. Donor Cell Transfection
Harvest mesenchymal stem cells (MSCs, derived from H1 embryonic stem cells kindly donated by Dr. Peiman Hematti; alternatively, any cell type of any species hypothesized to fuse in vivo could be employed) when 70 - 80% confluent with 1X trypsin (Mediatech, Manassas VA) for 5 min. Inactivate trypsin with α-MEM complete medium (antibiotic free, Invitrogen, Carlsbad CA)18. Centrifuge at 300 x g for 5 min.
Carefully aspirate supernatant and re-suspend pellet in 1 mL of 1X PBS and count cells using a hemacytometer.
Transfer 1.5 x 106 cells to a 1.5 mL centrifuge tube. Centrifuge at 300 x g for 5 min.
Carefully aspirate supernatant. Resuspend pellet in 300 μL of R Buffer (Neon Transfection System, Invitrogen) and 6 μg (2 μg / 5.0 x 105 cells) of p231 pCMVe-betaAc-STOP-luc (Addgene, Cambridge, MA). Place 3 mL of E Buffer (Invitrogen) into the electroporation docking port per manufacturer's protocol (Neon Transfection System, Invitrogen).
Transfer cell-plasmid solution to a 100 μL Neon pipet tip and electroporate with a pulse duration of 20 ms and a magnitude of 1500 volts. Place electroporated cells into a 15 mL conical tube containing 9.7 mL α-MEM complete medium.
Repeat step 1.5 two additional times and pool transfected cells to yield a total volume of 10 mL. Add the 10 mL cell suspension (1.5 x 106 cells) to a T175 flask containing 10 mL α-MEM complete medium. Cell viability after electroporation is approximately 30%, to yield approximately 4.5 x 105 viable cells per T175.
Change α-MEM complete medium 24 hours following transfection.
Harvest transfected cells when 70 - 80% confluent (~2 - 3 days after electroporation). Perform cell count using a hemacytometer and resuspend the cells at a concentration of 1.0 x 106 cells/50 μL of α-MEM complete medium. Minimize time cells spend in suspension to reduce cell death prior to injection.
2. Intramyocardial Injection
Induce anesthesia by isoflurane (Phoenix Pharmaceuticals, Inc., St. Joseph, MO) on transgenic mice engineered to constitutively express Cre recombinase in every cell (B6.C-Tg(CMV-cre)1Cgn/J, Jackson Laboratory, Bar Harbor, ME).
Remove hair from chest region using hair clippers or chemical hair remover.
Intubate with an 18 gauge catheter (Becton Dickinson & Co, Franklin Lakes NJ) and place on mouse ventilator at 120 - 130 breaths per minute with a stroke volume of 150 μL.
Make lateral incision across the fourth intercostal space thereby producing a thoracotomy.
Visualizing the heart, make two 25 μL injections of transfected cell suspension using a 1 mL syringe (Temuro Medical Corporation, Somerset, NJ) and 28 gauge needle (Becton Dickinson & Co, Franklin Lakes NJ). To ease the intramyocardial injection and prevent excessive damage to the organ, bend the needle head ~90 degrees.
Following injection, use absorbable sutures (e.g., vicryl) to close the ribs and muscle layers. Suture skin closed using 4-0 nylon or silk.
Allow mouse to recover from anesthesia and extubate.
Control groups should include Cre mice receiving medium injections only, Cre mice receiving the same concentration of untransfected cells and wild type mice receiving transfected cells (alternatively, Cre mice receiving transfected cells not prone to fuse).
3. Biophotonic Imaging
Five to fifteen minutes before imaging, intraperitoneally (IP) inject 10 μL per gram of mouse body weight of 15 mg/mL D-Luciferin (Caliper Life Sciences, Hopkinton, MA).
Induce anesthesia on mice via isoflurane at 4% for induction and 1 - 2% for maintenance.
Place mice supine in imaging box with facemask administering 1 - 2% isoflurane for maintenance anesthesia (Xenogen Biophotonic Imaging System, Hopkinton, MA). Several mice can be imaged concurrently. Image sham control mouse with experimental mice for easy comparison of luminescent signal.
Using Living Image software (Xenogen), set appropriate exposure time (typically 60 sec, see Results). Set area of image to fit mice and keep area consistent throughout imaging to prevent changes in sensitivity. Set subject height at 4.5 cm.
Acquire luminescence intensity signal corresponding to mouse or mice within the view field and save unmodified image files. Process images to remove background signal corresponding to a control or unmanipulated mouse. Intensity values above background correspond to fused cells within the animal. Intensity analysis can be conducted using Living Image software (Xenogen) or open source image analysis software. Typically, a region of interest (ROI) is selected to compare intensity data between animals and experiments.
4. Representative Results
To determine the sensitivity of the Xenogen Biophotonic Imaging System, a cell line which constitutively expresses luciferase (231-LUC-D3H1, Xenogen) was delivered to the myocardium of C57/Bl6 mice (Jackson Laboratory). Cells were injected at concentrations of 1 x 106, 1 x 103, or 1 x 101 cells. Six hours after cell delivery, mice were injected intraperitoneally with luciferin and imaged using the Xenogen system. A specific signal could be detected with 1,000 cells (2 of 6 mice imaged, Figure 2), but detection was more reliable with 10,000 cells (6 of 6 mice imaged). Importantly, this study also served to establish a rough correlation between number of luciferase-expressing cells and signal intensity.
To demonstrate the utility of the described protocol in detecting and tracking cell fusion, MSCs were transfected with the LoxP-Stop-LoxP-Luciferase plasmid (Addgene) and delivered to the myocardium of Cre-expressing mice. Approximately one week after cell delivery, mice were first imaged using the Xenogen system without D-luciferin injection. As expected, without the enzymatic substrate, no intensity signal was detected (Figure 3). Next, D-luciferin was injected intraperitoneally and a signal corresponding to luciferase intensity and thus cell fusion was detected in two of four mice tested. A similar signal was detected one week later (Figure 3), suggesting MSC-coupled fusion products can be maintained in vivo. In this case, the study was terminated to allow for evaluation of heart and surrounding tissue, but one could envision longer-term analyses and more frequent imaging to track the maintenance, proliferation and perhaps migration of fusion products in mice.
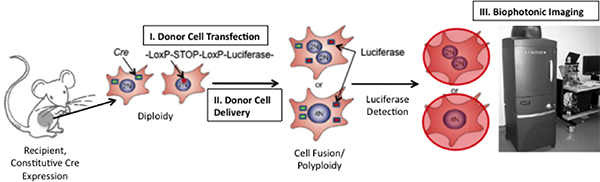 Figure 1. Schematic of Technique to Detect Cell Fusion In Vivo. If fusion between Cre-expressing mouse cells and transplanted cells expressing a floxed luciferase plasmid occurs, luciferase will be expressed. Luciferase can be detected by injecting the enzymatic substrate, D-luciferin, into the mouse and then imaging the mouse using a Xenogen Biophotonic Imaging System (Adapted from 19)
Figure 1. Schematic of Technique to Detect Cell Fusion In Vivo. If fusion between Cre-expressing mouse cells and transplanted cells expressing a floxed luciferase plasmid occurs, luciferase will be expressed. Luciferase can be detected by injecting the enzymatic substrate, D-luciferin, into the mouse and then imaging the mouse using a Xenogen Biophotonic Imaging System (Adapted from 19)
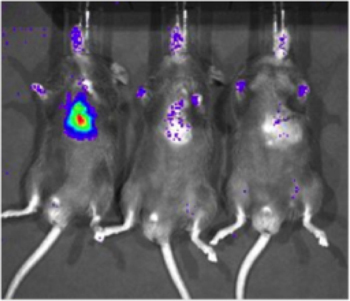 Figure 2. Sensitivity of detection of luciferase-expressing cells in cardiac tissue with biophotonic imaging . A cell line which constitutively expresses luciferase (231-LUC-D3H1, Xenogen) was delivered to the intramyocardial space of C57/Bl6 mice at various total cell numbers. Representative images of mice receiving 1 x 106, 1 x 103 and 1 x 101 cells (left to right) are shown, imaging was conducted approximately 6 hours after injection.
Figure 2. Sensitivity of detection of luciferase-expressing cells in cardiac tissue with biophotonic imaging . A cell line which constitutively expresses luciferase (231-LUC-D3H1, Xenogen) was delivered to the intramyocardial space of C57/Bl6 mice at various total cell numbers. Representative images of mice receiving 1 x 106, 1 x 103 and 1 x 101 cells (left to right) are shown, imaging was conducted approximately 6 hours after injection.
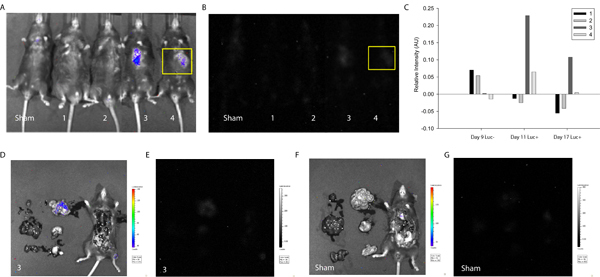 Figure 3. Quantification of In Vivo Luminescence Indicative of Cell Fusion. MSCs were transfected with the LoxP-Stop-LoxP-Luciferase plasmid and delivered to the myocardium of Cre-expressing mice. Approximately one week and two weeks after cell delivery, Cre mice were imaged using the Xenogen Biophotonic Imaging System to measure the intensity of luminescence indicative of cell fusion. (A) Overlay of photograph and intensity of luminescence of sham and mice 1-4 (left to right) 17 days after cell delivery. (B) Intensity of luminescence of sham and mice 1-4 (left to right) 17 days after cell delivery. A region of interest was selected (yellow) corresponding to the injection site and intensity levels were determined using ImageJ (free source) software20. (C) Intensity of luminescence was normalized to the same region of interest on sham mouse for all experimental conditions. At one week, mice 3 and 4 showed positive luminescence signal suggesting spontaneous fusion of a mouse cell and transplanted MSC. The signal persisted in mouse 3 at two weeks. To determine organ-specific localization of the signal corresponding to mouse 3, the thoracic cavity was exposed and primary organs excised and imaged. (D) Overlay of photograph and intensity of luminescence of mouse 3. Note localization of intensity signal in the small intestine. (E) Intensity of luminescence of mouse 3. (F) Overlay of photograph and intensity of luminescence of sham mouse. (G) Intensity of luminescence of sham mouse.
Figure 3. Quantification of In Vivo Luminescence Indicative of Cell Fusion. MSCs were transfected with the LoxP-Stop-LoxP-Luciferase plasmid and delivered to the myocardium of Cre-expressing mice. Approximately one week and two weeks after cell delivery, Cre mice were imaged using the Xenogen Biophotonic Imaging System to measure the intensity of luminescence indicative of cell fusion. (A) Overlay of photograph and intensity of luminescence of sham and mice 1-4 (left to right) 17 days after cell delivery. (B) Intensity of luminescence of sham and mice 1-4 (left to right) 17 days after cell delivery. A region of interest was selected (yellow) corresponding to the injection site and intensity levels were determined using ImageJ (free source) software20. (C) Intensity of luminescence was normalized to the same region of interest on sham mouse for all experimental conditions. At one week, mice 3 and 4 showed positive luminescence signal suggesting spontaneous fusion of a mouse cell and transplanted MSC. The signal persisted in mouse 3 at two weeks. To determine organ-specific localization of the signal corresponding to mouse 3, the thoracic cavity was exposed and primary organs excised and imaged. (D) Overlay of photograph and intensity of luminescence of mouse 3. Note localization of intensity signal in the small intestine. (E) Intensity of luminescence of mouse 3. (F) Overlay of photograph and intensity of luminescence of sham mouse. (G) Intensity of luminescence of sham mouse.
Discussion
The method described here allows, for the first time, discrete identification and temporal analysis of cell fusion in organisms, including small animals. The approach combines Cre-LoxP recombination with subsequent biophotonic image analysis. The approach is amenable to tracking not only cell-cell fusion, but also virus-cell fusion and so could prove useful for tracking viral infections. Image analysis is rapid and it is possible to image multiple small animals simultaneously. Detection of fusion is limited by the frequency of fusion of particular cell partners in their corresponding microenvironments and by the efficiency of transfection of the LoxP-Stop-LoxP-Luciferase plasmid. Thus, optimum results would be obtained with fusion partners containing an integrated form of the LoxP-Stop-LoxP-Luciferase sequence. In addition, the organisms must be stationary to image and so anesthesia is required for small animals.
Disclosures
No conflicts of interest declared.
Acknowledgments
The authors would like to thank Dr. Peiman Hemmati (Department of Medicine, University of Wisconsin-Madison) for generously providing the H1 MSCs, and Dr. Tim Hacker, Dr. Gouqing Song and Ms. Jill Koch of the University of Wisconsin Cardiovascular Physiology Core Facility for performing mouse surgeries. This work was supported by the National Science Foundation through a Graduate Research Fellowship to Brian Freeman and NIH R21 HL089679.
References
- Bernirschke KKP. Pathology of the Human Placenta. New York: Springer; 2000. [Google Scholar]
- Hoshina M, Boothby M, Boime I. Cytological localization of chorionic gonadotropin alpha and placental lactogen mRNAs during development of the human placenta. J. Cell. Biol. 1982;93:190–198. doi: 10.1083/jcb.93.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen M, Redman CW, Wilkins T, Sargent IL. Trophoblast deportation in human pregnancy--its relevance for pre-eclampsia. Placenta. 1999;20:531–539. doi: 10.1053/plac.1999.0422. [DOI] [PubMed] [Google Scholar]
- Redman CW, Sargent IL. Placental debris, oxidative stress and pre-eclampsia. Placenta. 2000;21:597–602. doi: 10.1053/plac.2000.0560. [DOI] [PubMed] [Google Scholar]
- Ogle BM. Spontaneous fusion of cells between species yields transdifferentiation and retroviral transfer in vivo. FASEB. J. 2004;18:548–550. doi: 10.1096/fj.03-0962fje. [DOI] [PubMed] [Google Scholar]
- Nygren JM. Bone marrow-derived hematopoietic cells generate cardiomyocytes at a low frequency through cell fusion, but not transdifferentiation. Nat. Med. 2004;10:494–501. doi: 10.1038/nm1040. [DOI] [PubMed] [Google Scholar]
- Nygren JM. Myeloid and lymphoid contribution to non-haematopoietic lineages through irradiation-induced heterotypic cell fusion. Nat. Cell. Biol. 2008;10:584–592. doi: 10.1038/ncb1721. [DOI] [PubMed] [Google Scholar]
- Kutschka I. Adenoviral human BCL-2 transgene expression attenuates early donor cell death after cardiomyoblast transplantation into ischemic rat hearts. Circulation. 2006;114:I174–I180. doi: 10.1161/CIRCULATIONAHA.105.001370. [DOI] [PubMed] [Google Scholar]
- Min JJ. In vivo bioluminescence imaging of cord blood derived mesenchymal stem cell transplantation into rat myocardium. Ann. Nucl. Med. 2006;20:165–170. doi: 10.1007/BF03027425. [DOI] [PubMed] [Google Scholar]
- Malstrom SE, Tornavaca O, Meseguer A, Purchio AF, West DB. The characterization and hormonal regulation of kidney androgen-regulated protein (Kap)-luciferase transgenic mice. Toxicol. Sci. 2004;79:266–277. doi: 10.1093/toxsci/kfh125. [DOI] [PubMed] [Google Scholar]
- Weir LR. Biophotonic imaging in HO-1.luc transgenic mice: real-time demonstration of gender-specific chloroform induced renal toxicity. Mutat. Res. 2005;574:67–75. doi: 10.1016/j.mrfmmm.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Rajashekara G, Glover DA, Banai M, O'Callaghan D, Splitter GA. Attenuated bioluminescent Brucella melitensis mutants GR019 (virB4), GR024 (galE), and GR026 (BMEI1090-BMEI1091) confer protection in mice. Infect. Immun. 1090;74:2925–2936. doi: 10.1128/IAI.74.5.2925-2936.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL. Noninvasive biophotonic imaging for monitoring of catheter-associated urinary tract infections and therapy in mice. Infect. Immun. 2005;73:3878–3887. doi: 10.1128/IAI.73.7.3878-3887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan PL, Youngblood RC, Harvill J, Willard ST. Photonic monitoring in real time of vascular endothelial growth factor receptor 2 gene expression under relaxin-induced conditions in a novel murine wound model. Ann. N.Y. Acad. Sci. 2005;1041:398–414. doi: 10.1196/annals.1282.061. [DOI] [PubMed] [Google Scholar]
- Zhu L. Non-invasive imaging of GFAP expression after neuronal damage in mice. Neurosci. Lett. 2004;367:210–212. doi: 10.1016/j.neulet.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Noiseux N. Mesenchymal stem cells overexpressing Akt dramatically repair infarcted myocardium and improve cardiac function despite infrequent cellular fusion or differentiation. Mol. Ther. 2006;14:840–850. doi: 10.1016/j.ymthe.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Ajiki T. Composite tissue transplantation in rats: fusion of donor muscle to the recipient site. Transplant Proc. 2005;37:208–209. doi: 10.1016/j.transproceed.2004.12.134. [DOI] [PubMed] [Google Scholar]
- Trivedi P, Hematti P. Derivation and immunological characterization of mesenchymal stromal cells from human embryonic stem cells. Exp. Hematol. 2008;36:350–359. doi: 10.1016/j.exphem.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle BM, Cascalho M, Platt JL. Biological implications of cell fusion. Nat. Rev. Mol. Cell. Biol. 2005;6:567–575. doi: 10.1038/nrm1678. [DOI] [PubMed] [Google Scholar]
- Collins TJ. ImageJ for microscopy. BioTechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]


