Abstract
Intercontinental disjunctions between tropical regions, which harbor two-thirds of the flowering plants, have drawn great interest from biologists and biogeographers. Most previous studies on these distribution patterns focused on woody plants, and paid little attention to herbs. The Orchidaceae is one of the largest families of angiosperms, with a herbaceous habit and a high species diversity in the Tropics. Here we investigate the evolutionary and biogeographical history of the slipper orchids, which represents a monophyletic subfamily (Cypripedioideae) of the orchid family and comprises five genera that are disjunctly distributed in tropical to temperate regions. A relatively well-resolved and highly supported phylogeny of slipper orchids was reconstructed based on sequence analyses of six maternally inherited chloroplast and two low-copy nuclear genes (LFY and ACO). We found that the genus Cypripedium with a wide distribution in the northern temperate and subtropical zones diverged first, followed by Selenipedium endemic to South America, and finally conduplicate-leaved genera in the Tropics. Mexipedium and Phragmipedium from the neotropics are most closely related, and form a clade sister to Paphiopedilum from tropical Asia. According to molecular clock estimates, the genus Selenipedium originated in Palaeocene, while the most recent common ancestor of conduplicate-leaved slipper orchids could be dated back to the Eocene. Ancestral area reconstruction indicates that vicariance is responsible for the disjunct distribution of conduplicate slipper orchids in palaeotropical and neotropical regions. Our study sheds some light on mechanisms underlying generic and species diversification in the orchid family and tropical disjunctions of herbaceous plant groups. In addition, we suggest that the biogeographical study should sample both regional endemics and their widespread relatives.
Introduction
Tropical regions harbor almost two-thirds of the flowering plants [1], [2], where intercontinental disjunctions occur commonly within and among plant genera due to Gondwana breakup, immigration from the Laurasian tropics and transoceanic dispersal [3], [4]. Compared with the Southern Hemisphere biogeography, whether vicariance or long distance dispersal has played a more important role during and after the fragmentation of Gondwana (160-30 Mya) [5], [6], biogeography of the Northern Hemisphere is more complex because of not only the impact of climatic and geological changes [7]–[9], but also the frequent migration by the North Atlantic land bridge and the Bering land bridge in the Tertiary [10]–[14]. A series of studies have suggested the boreotropical region as a corridor for the migration of thermophilic groups, such as Magnoliaceae [15], [16], Alangiaceae [17], Burmanniaceae [18], Altingiaceae [19], and Malpighiaceae [20]. However, most of them focused on woody plants, and paid little attention to herbs, which have shorter life histories, higher rates of molecular evolution [21], and much fewer fossils due to differential leaf and pollen production [22]. It would be of great interest to investigate the biogeographical history of herbaceous plant groups showing tropical disjunct distributions.
On the other hand, owing to the occurrence of a series of climatic oscillations and geographic events in the past 65 Mya [12], [13], [23]–[25], plants not only experienced expansion and contraction of their ranges [26]–[30], but also diversified to adapt to new niches [31]–[35]. It may explain why Wing [36] detected a mixture of tropical and temperate elements in the Eocene floras of the Rocky Mountains. Lavin & Luckow [37] and Wen [38] proposed that the study of disjunctions in temperate groups should include their subtropical and tropical relatives, and vice versa.
Orchidaceae is one of the largest families of flowering plants, accounting for approximately 10% of seed plants [39]. All orchids are herbaceous, of which about 73% are epiphytic or lithophytic [39]. According to fossil records, a fossil orchid with its pollinator in particular, the common ancestor of modern orchid lineages could be dated back to the late Cretaceous [40]–[42], although the radiation of most clades of the Orchidaceae occurred in the Tertiary. The subfamily Cypripedioideae (slipper orchids) is one of the monophyletic groups of Orchidaceae [43]–[48], including all the species with a pouchlike lip, two fertile stamens, a shield-like staminode and a synsepal composed of the fused lateral sepals [49]. There are almost 200 species of slipper orchids (http://apps.kew.org/wcsp/), belonging to five accepted genera, i.e., Cypripedium, Mexipedium, Paphiopedilum, Phragmipedium and Selenipedium [50]. The attractive flowers of slipper orchids make them have high ornamental and commercial values, and hold a special place in the hearts of botanists and hobbyists [51]. Also, this group is the most studied among all orchids due to its distinctive features [52]–[59]. Dressler [60] even considered that this group could have an unusual way of specialization given its unique flower morphology.
Pfitzer [61] and Atwood [62] investigated the relationships of slipper orchids based on morphological data, then Albert [63] based on both morphology and the chloroplast rbcL gene, and Cox et al. [64] using nuclear ribosomal DNA internal transcribed spacers (nrDNA ITS). Besides, several phylogenetic studies of Orchidaceae sampled slipper orchids [43], [46]–[48], [65]. All the previous studies strongly support the monophyly of slipper orchids, but have not reached a consensus about the intergeneric relationships, and in particular the published chloroplast DNA (cpDNA) phylogenies have low resolution or incomplete sampling in this orchid clade [43], [46], [47], [58].
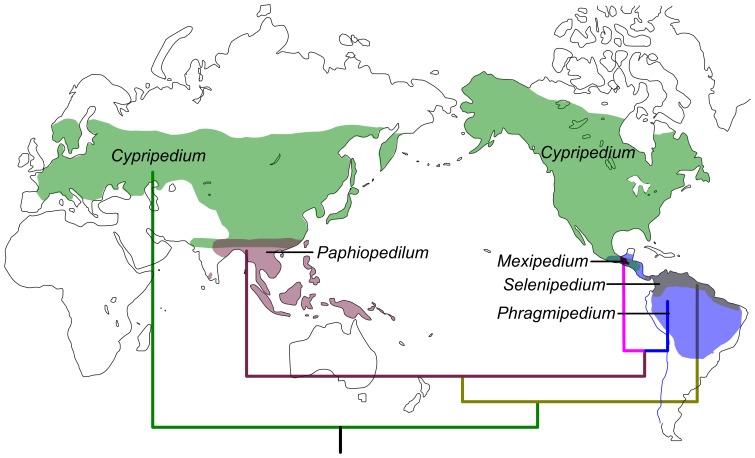
The slipper orchids are widely distributed in temperate to tropical regions of Eurasia and America. The genus Cypripedium occurs in temperate and subtropical areas of the North Hemisphere, with some species extending to tropical North America. The two conduplicate-leaved genera Mexipedium and Phragmipedium and the plicate-leaved genus Selenipedium are restricted to the neotropics, whereas Paphiopedilum is confined to the palaeotropics (Fig. 1). Atwood [62] and Albert [63] supported the boreotropical hypothesis [66], and considered that fragmentation of continents and the following climatic cooling in the Ice Ages caused the present disjunct distribution of slipper orchids. While the ITS analysis supports southern North America/Mesoamerica as the origin center of slipper orchids [64], the sister relationship between Mexipedium and Paphiopedilum revealed in the low copy nuclear Xdh gene phylogeny [48], although with weak support and based on a limited sampling, seems to suggest a long distance dispersal from palaeotropical to neotropical regions. Therefore, the biogeographical history of slipper orchids is far from being resolved.
Figure 1. The distribution of slipper orchids modified from Pridgeon et al. [165].
Shaded areas show the current species distribution, with different colors to represent the five genera. The tree topology indicates the phylogenetic relationships of slipper orchids reconstructed in this study.
It has been widely recognized that the use of multiple genes is helpful for the accuracy of phylogenetic and biogeographical reconstruction (e.g. [67], [68]). In addition to the widely used cpDNA markers such as rbcL, matK, ndhF and ycf2 [69]–[72], more and more studies indicate that ycf1, one of the two longest coding genes of cpDNA, has great potential in plant phylogenetic reconstruction [73]–[75]. Meanwhile, single or low copy nuclear genes are increasingly used in plant phylogenetic studies due to their rapid evolutionary rates and biparental inheritance [76]–[80]. For instance, LFY, which is involved in regulating flower meristem identity and flowering time [81]–[84], has been successfully used as a single copy gene to investigate intra- and inter-generic relationships [68], [85]–[88], and allopolyploid speciation [89]. Also, the ACO gene, which encodes the ACC oxidase enzyme to catalyze the last step of ethylene biosynthesis in plants [90], is important for flower development, fruit ripening, and responses to biotic and abiotic stresses [91]. This gene may also exist as a single locus in slipper orchids according to the result of 3′-RACE.
In the present study, we aim to reconstruct the phylogeny of slipper orchids with multiple coding chloroplast and low copy nuclear genes. In addition, we intend to estimate divergence times of the five genera of slipper orchids, and to explore their biogeographical history, particularly the disjunction between neotropical and palaeotropical regions. This study may also shed some light on the mechanisms underlying the diversification of Orchidaceae.
Materials and Methods
Ethics statement
No specific permits were required for the described field studies.
Plant sampling
We sampled 31 species, which represent all five genera of the subfamily Cypripedioideae and cover seven sections of Paphiopedilum and four sections of Phragmipedium. In the genus Cypripedium, 16 species of nine sections were collected from eastern Asia and North America. Owing to the rarity and difficulty in collection, this study only sampled one individual of Selenipedium, a genus with five accepted species that are morphologically similar and endemic to the tropical regions of Central and South America [92]. In addition, four species representing three genera of the two subfamilies Apostasioideae and Vanilloideae were chosen as outgroups, since previous studies showed that Apostasioideae and Vanilloideae are sister to slipper orchids plus the other monandrous orchids [44], [48]. The origins of the materials are shown in Supplementary Table S1.
DNA extraction, PCR amplification, cloning and sequencing
Total DNA was extracted from silica gel-dried leaves using a modified cetyltrimethylammonium bromide (CTAB) protocol [93] or Plant Genomic DNA Kit (Tiangen Biotech Co.). We screened eight chloroplast coding genes (accD, rbcL, matK, rpoC1, rpoC2, ycf1, ycf2, and ndhF) and two low-copy nuclear genes (LFY and ACO). The LFY gene was amplified with the forward primer LFYE1jF (5′-TGAGGGAGGAGGAGGTSGACGAYATGAT-3′) located at the first exon and the reverse primer LFYE3kR (5′-AGATBGAGAGGCGSGGATGSGCGTT GAA-3′) at the third exon, and the ACO gene with ACOE1aF (5′-GCNTGYGAGAA CTGGGGHTTCTTYGAG-3′) and ACOE2aR (5′-ATGGTCTTCATGGCCTCAA ACCT-3′). All the four primers were designed based on the sequences available in the public databases. However, the two LFY primers did not work in Phragmipedium besseae, Paphiopedilum delenatii, P. vietnamense, and some species of Cypripedium, and thus another reverse primer LFYE2S5R at the second exon was further designed. The details of other primers are shown in Supplementary Table S2. Although the ndhF gene is conservatively located at the small single copy (SSC) region of the published chloroplast genomes, e.g. Oryza sativa [94], Amborella trichopoda [95], Nymphaea alba [96] and Acorus calamus [97], it was reported to have been lost in the sequenced chloroplast genomes of the four orchids Phalaenopsis aphrodite [98], Oncidium Gower Ramsey [99], Rhizanthella gardneri [100] and Neottia nidus-avis [101]. Therefore, we tried to amplify the ndhF gene with primers trnNguu and trnLuag that are located in its two flanking regions, and ndhFcF and ndhFaR in its coding regions, respectively.
Amplification reactions were conducted in a Tgradient Thermocycler (Biometra) or a Mastercycler (Eppendorf, Hamburg, Germany) in a volume of 25 µL containing 10–50 ng DNA template, 200 µmol/L of each dNTP, 6.25 pmol of each primer pair, and 0.75 U of Taq DNA polymerase (TakaRa Biotech Co., Dalian, China). PCR cycles are as follows: for the chloroplast genes, 4 min at 70°C, 4 cycles of 2 min at 94°C, 30 s at 51°C, and 1–3 min at 72°C, followed by 36 cycles of 30 s at 94°C, 30 s at 53°C, and 1–3 min at 72°C, with a final elongation for 10 min at 72°C; for the nuclear genes, 4 min at 70°C, 4 cycles of 2 min at 94°C, 30 s at 57°C, and 5 min at 68°C, followed by 36 cycles of 30 s at 94°C, 30 s at 60°C, and 5 min at 68°C, with a final extension for 15 min at 68°C. PCR products were separated by 1.5% agarose gel electrophoresis and purified with a Gel Band Purification Kit (TIANgel Midi Purification Kit). The purified PCR products of the chloroplast genes were directly sequenced with the PCR primers and the internal primers designed in this study (Supplementary Table S2). For the nuclear genes, the purified PCR products were cloned with pGEM-T® Easy Vector System II (Promega). Twelve clones were picked for each sample, and 4–6 of them with correct insertion (determined by digestion with EcoR I) were sequenced with primers T7 and SP6 and the internal primers designed in this study (Supplementary Table S2). After precipitation with 95% EtOH, 3 M NaAc and 125 mM EDTA, the sequencing products were separated on an ABI PRISM 3730XL DNA analyzer (Applied Biosystems). The sequences reported in this study are deposited in GenBank under accession numbers JN181400–JN181549 and JQ182152–JQ182298 (Supplementary Table S1).
Data analysis
The ContigExpress program of the Vector NTI Suite 6.0 (Informax Inc.) was used to assemble sequences from different primers. Sequence alignments were made with BioEdit 7.0 [102] and refined manually. Nucleotide diversity (Pi) was estimated using DnaSP version 5.0 [103]. Indels were coded using GapCoder [104], with a ‘1’ for present, ‘0’ for missing, and ‘-’ for inapplicable. The unalignable regions of the rpoC1 intron were excluded from our analyses. The incongruence length difference (ILD) test [105] was used to assess the congruence between different datasets. Phylogenetic analyses based on maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) were performed with PAUP version 4.0b10 [106], PhyML 2.4.4 [107] and MrBayes 3.1.2 [108], respectively. In the MP and ML analyses, the missing data were coded by “?”, while it was excluded from the BI analysis. The MP analysis used a heuristic search with 1000 random addition sequence replicates, tree-bisection-reconnection (TBR) and MULTREES on, and branch support was evaluated by bootstrap analysis [109] of 1000 replicates using the same heuristic search settings. The evolutionary models for the ML and BI analyses were determined by Modeltest 3.07 [110] and MrModeltest v2.2 [111], respectively (Table 1). The ML analysis used the GTR model and a BIONJ tree as a starting point, and branch support was estimated by bootstrap analysis [109] of 1000 replicates. For the Bayesian inference, one cold and three incrementally heated Markov chain Monte Carlo (MCMC) chains were run for 1,000,000 cycles and repeated twice to avoid spurious results. One tree per 100 generations was saved. The first 300 samples for each run were discarded as burn-in to ensure that the chains had become stationary. Phylogenetic inferences were made based on the trees sampled after generation 30,000.
Table 1. Results of Model test and MrModel test.
| Model test | MrModel test | |||
| AIC | hLRTs | AIC | hLRTs | |
| combined cpDNA | TVM+I+G | TVM+I+G | GTR+I+G | GTR+I+G |
| ACO | K81uf+G | K80+G | GTR+G | SYM+G |
| LFY | GTR+G | TrN+G | GTR+G | GTR+G |
| combined nuclear DNA | GTR+G | GTR+G | GTR+G | GTR+G |
| cpDNA+nuclear DNA | TVM+I+G | TVM+I+G | GTR+I+G | GTR+I+G |
Molecular dating is very helpful to interpret plant distribution patterns [112], [113]. The likelihood ratio test (LRT) was used to test the rate constancy among lineages [114]. Log likelihood ratios of the chosen model with and without an enforced molecular clock were compared. The degree of freedom is equivalent to the number of terminal taxa minus two [115]. Significance was assessed by comparing two times the log likelihood difference to a chi-square distribution. Due to the lack of fossil evidence for the subfamily Cypripedioideae, we first performed a family-level analysis to get a more reliable estimate of the divergence times in Orchidaceae by integrating all the three available fossils of the family. The analysis was based on the combined matK and rbcL gene sequences and a sampling following the latest angiosperm phylogeny APG III [116]. In addition to the sequences of the slipper orchids and their close relatives determined in the present study, the matK and rbcL sequences of 165 taxa were downloaded from GenBank (see Supplementary Table S3), which represent 119 genera of Orchidaceae, 24 genera of non-orchid Asparagales, 5 genera of Commelinids, and 3 genera of Liliales as outgroups. The final data matrix comprised 200 taxa, which are many more than that sampled in previous studies. Divergence times were estimated using the nonparametric rate smoothing (NPRS) [117] and penalized-likelihood (PL) [118] implemented in the program r8s v1.71 [119], and Bayesian inference in BEAST v1.5.4 [120]. Four calibration points were used for age estimation, including Dominican amber (15–20 Mya) as a minimum age constraint for the Goodyerinae [40], macrofossils of Dendrobium (20–23 Mya) and Earina (20–23 Mya) [41] as the lower bound of the two genera following Gustafsson et al. [42], the age of the oldest known Asparagales (93–105 Mya) as the minimum age of the root of the tree, and the age of the oldest known fossil monocot as the maximum age at the root of the tree (110–120 Mya) [121] following the phylogenetic placement of Ramírez et al. [40]. In the r8s analysis, the oldest and youngest ages of the fossils were used separately. In the BEAST analysis, the age was estimated with the tree priors set as follows: i) age for the Goodyerinae (a monophyletic subtribe) as uniform distribution with a lower bound of 15 Mya and an upper bound of 120 Mya; ii) age for both Dendrobium and Earina as uniform distribution (lower bound: 20 Mya; upper bound: 120 Mya); iii) age for the root of the tree with a normal prior distribution as 106.5±8.21 Mya (95% CI: 93–120 Mya) [42]. The above 200-taxa analysis showed that the age estimates by NPRS and PL are very close, but are older than that by BEAST (see Results section). Considering that the use of multiple gene sequences could yield a more accurate time estimation when a constant diversification rate among lineages is violated [122], we conducted a further analysis for the slipper orchids (35 taxa, including outgroups) using combined six chloroplast genes (combined cpDNA, including matK, rbcL, rpoC1, rpoC2, ycf1, and ycf2) with NPRS and PL methods. We did not use the BEAST estimate in this analysis due to its wide confidence interval. The crown ages of Cypripedioideae were set to 64±4 Mya (oldest age) and 58±4 Mya (youngest age), according to the result of PL analysis on the 200-taxa dataset. For the PL method, a cross-validation procedure was used to determine the most likely smoothing parameter. To calculate the standard errors, one hundred bootstrapped trees with fixed topology were generated with PAUP version 4.0b10. In the Bayesian analysis, divergence times were estimated with a log normal relaxed molecular clock using the Yule model of speciation. We ran 20,000,000 generations of Markov chain Monte Carlo (MCMC), and sampled every 2000 generations, with a burn-in of 1000 trees. The MCMC output analysis was conducted with TreeAnnotator v1.5.4, and the chronological phylogeny was displayed by FigTree v1.3.1.
The ancestral distribution of slipper orchids was reconstructed with S-DIVA 1.9 beta [123], [124], and Lagrange [125], [126]. S-DIVA complements DIVA, and considers the phylogenetic uncertainty in DIVA optimization. We used the randomly sampled 9000 post-burnin trees derived from the BEAST analysis for ancestral area reconstruction. In contrast, as a likelihood-based method under the dispersal-extinction-cladogenesis model, Lagrange enables the estimation of ancestral states, and calculates the probabilities of the most-likely areas at each node. Based on the present distribution of slipper orchids, we directly divided it into two geographical areas, Old World and New World. The biogeographical data were coded based directly on the distribution of the studied species, and the distribution of outgroups was excluded due to its wideness.
Results
Sequence characterization
Six chloroplast genes (matK, rbcL, rpoC1, rpoC2, ycf1, and ycf2) were successfully amplified and directly sequenced for all samples except the cloning of ycf1 from Vanilla planifolia. The amplification of accD failed in one of the outgroups, and thus this gene was excluded from further analysis. The PCR products of primers trnN/trnL had great length variation in slipper orchids, ranging from ∼1400 bp to ∼6000 bp, which, together with the amplification results of primers ndhFcF/ndhFaR, suggests that the ndhF gene has been completely lost in Mexipedium and the studied species of Phragmipedium (see Supplementary Table S4). Hence, this gene was also excluded from the phylogenetic analysis. The amplified matK region includes the complete matK coding sequence and ∼180 bp of the trnK intron. It is interesting that only a pseudogene of matK, with a frameshift mutation and an early stop codon, was obtained from Vanilla sp. Although we tried to clone the PCR product and to amplify the gene with redesigned primers specific to Vanilla, the functional copy of matK was still not found. Actually, several previous studies have reported that the functional matK gene does not occur in some orchids [127]–[129]. The matK pseudogene of Vanilla sp. was finally used in the phylogenetic analysis, since it only differs from the sequence of its congeneric species in several nucleotide substitutions and three nontriplet indels (5 bp insertion, 13 bp insertion, and 4 bp deletion). The amplification products of the rpoC1 gene cover about 1300 bp coding and about 800 bp intron sequences. The direct sequencing chromatogram of ycf1 from Vanilla planifolia showed double-peaks, and therefore we cloned the purified PCR product. Consequently, we obtained two distinct sequences of ycf1 from the species, both of which can be successfully translated. We chose the ycf1 copy that shows a higher similarity with the other outgroup species. A summary of the sequences that we used is shown in Table 2. Among the chloroplast genes, ycf1 is the most variable and parsimony-informative.
Table 2. Sequence information of the genes used in the present study.
| Genes | Length (bp) | Alignment Length (bp) | Pi | Parsimony-informative sites | ||||
| Within Cypripedioideae | Entire dataset | Within Cypripedioideae | Entire dataset | Within Cypripedioideae | Entire dataset | Within Cypripedioideae | Entire dataset | |
| mat K | 1492–1518 | 1470–1523 | 1545 | 1581 | 0.04238 | 0.05872 | 175 | 302 |
| rbc L | 1266 | 1266 | 1266 | 1266 | 0.01132 | 0.01679 | 42 | 90 |
| rpo C1 | 2003–2096 | 1962–2096 | 2056* | 2090* | 0.01959* | 0.02892* | 91* | 210* |
| rpo C2 | 2616–2661 | 2616–2691 | 2712 | 2811 | 0.02393 | 0.03884 | 159 | 415 |
| ycf 1 | 1570–1690 | 1405–1690 | 1831 | 1870 | 0.05108 | 0.07255 | 200 | 357 |
| ycf 2 | 1449–1512 | 1449–1644 | 1575 | 1836 | 0.00566 | 0.01809 | 17 | 157 |
| combined cpDNA | 10431–10627 | 10400–10627 | 10985* | 11454* | 0.02570* | 0.03875* | 684* | 1531* |
| ACO Exon | 780–795 | 780–795 | 798 | 801 | 0.08798 | 0.11352 | 204 | 303 |
| LFY Exon | 912–933 | 912–945 | 942 | 978 | 0.08745 | 0.11450 | 172 | 239 |
| combined nuclear DNA | 1704–1725 | 1704–1734 | 1740 | 1776 | 0.08965 | 0.11381 | 306 | 439 |
| Total | 12141–12323 | 12134–12375 | 12683* | 13173* | 0.03349* | 0.04854* | 915* | 1736* |
The unalignable regions of the rpoC1 intron were excluded from our analyses.
The LFY gene of the slipper orchids amplified with primers LFYE1jF and LFYE3kR ranges from 1853 bp to 3717 bp in length, including partial sequences of exon 1 (258–270 bp) and exon 3 (234 bp), and complete sequences of exon2 (417–432 bp) and the two introns. In the three species Phragmipedium besseae, Paphiopedilum delenatii and P. vietnamense, the LFY gene amplified with primers LFYE1jF and LFYE2S5R includes partial sequence of exon1 and almost the whole length of exon2 and intron 1. Unfortunately, none of the two primer pairs worked in the five species of Cypripedium (C. californicum, C. candidum, C. farreri, C. debile and C. palangshanense). The intron sequences cannot be reliably aligned among the five genera of slipper orchids, and thus were excluded from our analyses. Except the failure of PCR amplification in Phragmipedium besseae and Neuwiedia singapureana, we got the ACO gene from all the other samples of slipper orchids, which ranges from 909 bp to 2178 bp in length. After excluding the introns, due to the difficulty in aligning, the coding region of ACO ranges from 780 bp to 795 bp in length (Table 2). The ACO gene has four exons and three introns except the loss of the second intron in the two genera Mexipedium and Phragmipedium, Apostasia sp. and two species of Cypripedium (C. fasciculatum and C. palangshanense), and the loss of the third intron in the two species of Vanilla.
Phylogenetic analysis, molecular dating and ancestral area reconstruction
Since the plastid genome behaves as a single locus, we directly combined the six chloroplast genes into a single dataset (combined cpDNA) for phylogenetic analysis. The MP analysis generated 60 equally most parsimonious trees (MPTs), with tree length = 4620 steps, consistency index (CI) = 0.78, and rentention index (RI) = 0.84. The ML and Bayesian trees of the combined cpDNA are nearly identical to the MP trees in topology except the slight difference in interspecific relationships of Cypripedium and the weak bootstrap support for the position of Selenipedium in the MP trees. The ML tree is shown in Supplementary Fig. S1. The nuclear gene analyses generated 1807 MPTs for ACO (tree length = 859 steps, CI = 0.67, RI = 0.85), and 80 MPTs for LFY (tree length = 966 steps, CI = 0.70, RI = 0.86). Also, the MP trees of the nuclear genes are identical to the ML and Bayesian trees in topology except a minor difference in the Cypripedium clade (see ML trees in Supplementary Figs. S2, S3).
Since the ILD test did not detect significant incongruence between the two nuclear genes (p = 0.69) and between combined cpDNA and nuclear DNA (p = 0.50), we further conducted phylogenetic analyses using the two combined datasets. As a result, 27 MPTs were generated for the combined nuclear genes (tree length = 1621 steps, CI = 0.71, RI = 0.80), and 6 MPTs were generated for the combined cp- and nuclear DNA (tree length = 5284 steps, CI = 0.79, RI = 0.84), respectively. The ML and Bayesian trees generated based on the two combined datasets show the same intergeneric relationships of slipper orchids as in the MP trees (see ML trees in Supplementary Fig. S4; Fig. 2).
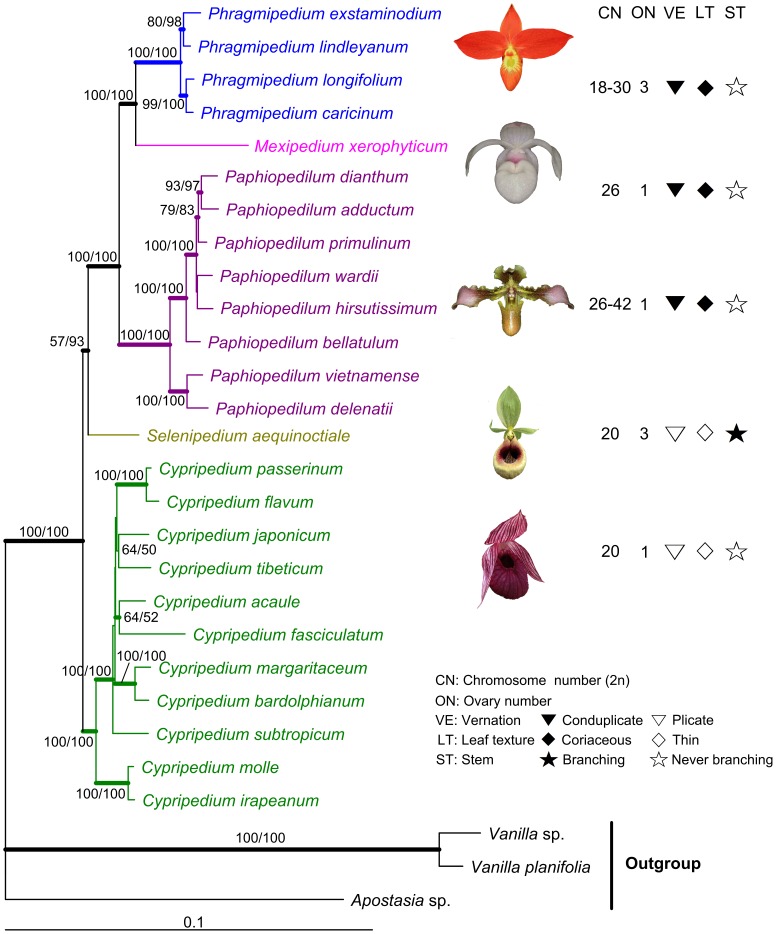
Figure 2. The ML tree of slipper orchids constructed based on the combined cpDNA+nuclear genes.
Numbers above branches indicate the bootstrap values ≥50% for the MP and ML analyses, respectively. Bayesian posterior probabilities (≥0.90) are shown in bold lines. Symbols on the right indicate the distribution of some important characters of slipper orchids.
All gene trees generated in the present study, either based on separate genes or on combined datasets (Fig. 2; Supplementary Figs. S1, S2, S3, S4), are consistent about the intergeneric relationships of slipper orchids. That is, the widespread Cypripedium diverged first, followed by Selenipedium from South America, and finally the three conduplicate genera. The monotypic genus Mexipedium is most closely related to the South American Phragmipedium, and the two New World genera form a clade sister to the Old World Paphiopedilum (Fig. 2; Supplementary Figs. S1, S2, S3, S4).
The LRT test rejected a clock-like evolution of combined matK+rbcL (δ = 1815.1177, df = 198, P<0.001) and combined six chloroplast genes (δ = 5839.3257, df = 33, P<0.001). Therefore, we used NPRS and PL in r8s and Bayesian methods to estimate the divergence times. The family-level analysis (200 taxa) showed that the crown ages of Orchidaceae and its five subfamilies are older than the estimates by previous studies [40], [42], although the BEAST estimates showed a wide range (Table 3, Fig. 3). It is interesting that the crown ages of the subfamily Cypripedioideae estimated by NPRS and PL in the present study are very close, not as in Ramírez et al. [40] that obtained very different estimates by the two methods. This implys that a good sampling is important for molecular dating. The divergence times within Cypripedioideae estimated from the combined six chloroplast genes are generally congruent with those from the family-level analysis (Table 3). According to the age estimate, the genus Selenipedium originated in Palaeocene, while the most recent common ancestors of conduplicate slipper orchids (Mexipedium, Phragmipedium and Paphiopedilum) and of Cypripedium could be dated back to the Eocene (Table 3, Figs. 3, 4). Since the divergence times estimated with NPRS and PL are very close (Table 3), and thus only the PL estimates were used in the discussion. The ancestral area reconstruction suggests a New World origin or a wide ancestral distribution of slipper orchids, and indicates that vicariance is responsible for the disjunct distribution of conduplicate slipper orchids in palaeotropical and neotropical regions (Fig. 4).
Table 3. Estimated divergence times (Mya) derived from BEAST and r8s.
| Node | matK+rbcL | combined six chloroplast genes | |||||||
| BEAST | r8s | r8s | |||||||
| Median | Oldest ages | Youngest ages | Oldest ages | Youngest ages | |||||
| (95% HPD) | NPRS | PL | NPRS | PL | NPRS | PL | NPRS | PL | |
| Family Orchidaceae | 87 (73–102) | 88±3 | 89±2 | 81±2 | 82±2 | — | — | — | — |
| Subfam. Apostasioideae | 43 (25–64) | 52±4 | 50±4 | 48±3 | 45±4 | — | — | — | — |
| Subfam. Vanilloideae | 66 (52–81) | 74±3 | 74±3 | 67±3 | 68±3 | — | — | — | — |
| Subfam. Cypripedioideae | 43 (32–56) | 64±4 | 64±4 | 59±4 | 58±4 | — | — | — | — |
| Cypr | 33 (23–45) | 57±4 | 56±4 | 52±4 | 51±5 | 53.5±13.7 | 53.6±13.8 | 48.8±12.5 | 48.9±12.5 |
| PaSe | — | — | — | — | — | 60.4±15.4 | 60.4±15.4 | 55.1±14.0 | 55.1±14.0 |
| PaMe | 33 (23–43) | 50±4 | 49±4 | 46±4 | 45±4 | 46.8±12.0 | 46.5±11.9 | 42.7±10.9 | 42.3±10.9 |
| PhMe | 27 (18–37) | 44±4 | 43±4 | 41±4 | 39±4 | 43.1±11.1 | 42.7±11.0 | 39.3±10.1 | 38.9±10.0 |
| Paph | 18 (11–26) | 28±4 | 27±4 | 26±4 | 25±4 | 24.2±6.4 | 22.2±5.9 | 22.1±5.9 | 20.0±5.4 |
| Phra | 15 (8–21) | 26±4 | 25±4 | 24±4 | 23±4 | 25.9±7.0 | 24.4±6.7 | 23.6±6.4 | 22.1±6.1 |
| Subfam. Orchidoideae | 63 (51–75) | 65±3 | 67±3 | 60±3 | 62±3 | — | — | — | — |
| Subfam. Epidendroideae | 55 (42–68) | 73±3 | 74±3 | 67±3 | 68±3 | — | — | — | — |
Figure 3. Fossil-calibrated molecular chronogram of the family Orchidaceae based on combined matK+rbcL sequences.
Red circles indicate age-constrained nodes, and arrows indicate the crown ages of the five subfamilies of Orchidaceae.
Figure 4. Chronogram of slipper orchids inferred from the combined six chloroplast genes, and ancestral area reconstruction.
The crown age of slipper orchids was set as a calibration point for time estimation. Two areas were defined: (A) Old World and (B) New World. The ancestral areas with the highest probabilitiy are shown above (S-DIVA) and below (Lagrange) the branches with pie charts.
Discussion
Phylogeny and evolution of the slipper orchids
In previously reported phylogenies of slipper orchids, the main discrepancies are phylogenetic positions of Selenipedium and Mexipedium. Atwood [62] proposed Selenipedium be merged into Cypripedium. The morphological study [61] and the combination analysis of morphological and rbcL data [63] as well as the nrDNA ITS tree [64] indicate that Selenipedium is basal to the other slipper orchids, whereas the phylogenies based on the low copy nuclear gene Xdh [48], atpB [130] and the combined matK+rbcL [47] suggest a basal position of Cypripedium. On the other hand, nrDNA ITS [64] and cpDNA [43], [46] trees supports a sister relationship between the two North American genera Mexipedium and Phragmipedium, whereas the Xdh tree indicates that Mexipedium is most closely related to the Old World Paphiopedilum [48].
Like the unstable phylogenetic position, Selenipedium also has a very interesting morphology. This genus has fragrant and crustose seeds like Vanilla, but has the same chromosome number (2n = 20) [131], valvate sepal aestivation, and leaf vernation and texture as Cypripedium (Fig. 2), and even shares some anatomical features with Cypripedium irapeanum and C. californicum. In addition, the three-locular ovary and the multi-flower inflorescence with one flower opening at a time in Selenipedium seem to be primitive features [51], [62], [92]. Moreover, Selenipedium is similar to the conduplicate-leaved genera in having persistent perianth [62].
Mexipedium is a monotypic genus endemic to Oaxaca of Mexico. Albert and Chase [50] established this genus, to which the species initially published as Phragmipedium xeropedium was transferred [132]. Similar to the situation in Selenipedium, the genus Mexipedium not only shares characters with Phragmipedium (e.g. valvate sepal aestivation), but also with Paphiopedilum (e.g. unilocular ovary). Due to the limited markers used, the phylogenetic position of Mexipedium was not consistent among several previous molecular phylogenetic studies [43], [46], [48], [64].
The phylogenetic relationships among the genera of slipper orchids are relatively well resolved in the present study, given the topological consistency among the gene trees generated either from cpDNA or from the low copy nuclear genes (Fig. 2; Supplementary Figs. S1, S2, S3, S4). We found that Cypripedium diverged first, followed by Selenipedium, and finally the three conduplicate genera, although the sister relationship between Selenipedium and the conduplicate genera is not very strongly supported (Fig. 2). That is, the plicate-leaved genera could be more primitive, while the conduplicate-leaved genera are more advanced. We also found that the two New World genera Mexipedium and Phragmipedium are most closely related and form a clade sister to the Old World Paphiopedilum (Fig. 2; Supplementary Figs. S1, S2, S3, S4). Moreover, the close relationship between the two neotropical conduplicate genera is corroborated by the shared loss of the ndhF gene. Based on the combined chloroplast and nuclear gene phylogeny (Fig. 2), in slipper orchids, the coriaceous conduplicate leaf has a single origin, but ovary number is not phylogenetically informative.
Biogeography of the slipper orchids: Implications for the evolution of Orchidaceae
The biogeographical history of slipper orchids is of great interest, but still remains controversial. Atwood [62] and Albert [63] put forward that slipper orchids were once widely distributed in North America/Asia, and that its current disjunct distribution was shaped by the separation of continents and the climatic cooling in the Ice Ages. Cox et al. [64] suggested southern North America/Mesoamerica as the origin center of slipper orchids based on the nrDNA ITS analysis. However, the reconstruction of biogeographical history should be based on a solid phylogeny, divergence time estimation and ancestral area reconstruction. In the relatively well-resolved phylogeny of slipper orchids reconstructed in the present study, Cypripedium, a genus with a wide distribution in temperate and subtropical North Hemisphere, is basal to the other genera. Also, the PL estimate suggests a Palaeocene origin of Selenipedium, while the most recent common ancestors of conduplicate slipper orchids and of Cypripedium could be dated back to the Eocene (Table 3, Figs. 3, 4). Although no available fossils of slipper orchids can be used for time calibration, the time estimates from combined matK+rbcL using other orchid fossils as calibration points are generally congruent with those from the combined six chloroplast genes using a secondary calibration point. It is well known that the climatic cooling or oscillation since Eocene/Oligocene [133], [134] has led to great changes in plant distribution patterns. Therefore, although southern North America/Mesoamerica has three out of the five genera (Cypripedium, Phragmipedium and Mexipedium) of slipper orchids, this region is very likely a museum rather than a cradle for the diversity. In fact, Phragmipedium is mainly distributed in South America. The ancestral area reconstruction also suggests that the common ancestor of slipper orchids occurred in the New World or had a wide distribution in both Old and New Worlds (Fig. 4).
The Isthmus of Panama had served as a corridor for flora and fauna exchange between North America and South America before 3–3.5 Mya, which may explain the distribution of Selenipedium and Phragmipedium in South America. For instance, pollen records and vertebrate fossils from the Caribbean region indicate that the GAARlandia land bridge had connected North and South America during Eocene-Oligocene (35-33 Mya) [135]. In addition, Iturralde-Vinent & MacPhee [135] and Pennington & Dick [136] both suggested the existence of a land bridge between the two continents in Miocene. Furthermore, the study of the palm tribe Chamaedoreeae also supports the Middle Eocene and Miocene migrations of plants between North and South America [137].
It is very interesting that the Old World Paphiopedilum is sister to a clade comprising the two New World genera Mexipedium and Phragmipedium (Fig. 2; Supplementary Figs. S1, S4), suggesting a vicariant differentiation of the conduplicate genera between the Old World and New World tropics. The three conduplicate genera occur in both the Northern and Southern Hemispheres, also including South America and a part of Southeast Asia from the Gondwanaland [138], [139]. According to many previous studies on other plant groups, the neotropical and palaeotropical disjunction could be explained by: (1) Gondwana breakup [140], [141], (2) trans-Pacific long distance dispersal [142], [143], and (3) fragmentation of the boreotropical flora [37], [66]. However, the first two hypotheses are not suitable for the conduplicate slipper orchids, although they can not be completely ruled out.
First, the crown age of slipper orchids was dated back to Palaeocene (Table 3; Fig 3), which is much younger than the time of Gondwana breakup, and slipper orchids do not occur in Australia and Africa. Therefore, the present distribution pattern of slipper orchids cannot be attributed to the Gondwana breakup. Second, trans-Pacific long distance dispersal is not supported by the reciprocal monophyly of the conduplicate slipper orchids from both sides of the Pacific Ocean, particularly the monophyly of the New World conduplicate slipper orchids comprising the two genera Mexipedium and Phragmipedium, and not by the divergence time estimation. That is, the conduplicate genera have a crown age of 42.3±10.9 Mya (youngest age) to 46.5±11.9 Mya (oldest age, in the Eocene), but the most recent common ancestors of Paphiopedilum and Phragmipedium are dated back to 22.2±5.9 Mya (oldest age) and 24.4±6.7 (oldest age) Mya, respectively (Table 3, Fig. 4). The estimated divergence times suggest an early origin for each of the conduplicate genera but a much later diversification or the extinction of ancient species within the genera. It is very likely that vicariant differentiation is responsible for the disjunct distribution of the conduplicate genera between the Old World and New World tropics. That is, the ancestor of the conduplicate slipper orchids could have a continuous distribution in the boreotropics, and migrated southwards to both sides of the Pacific Ocean due to the climate cooling in the late Cenozoic [23], [134], and then evolved into separate genera. Although the seeds of orchids are tiny [144], which may facilitate long distance dispersal, Moles et al. [145] found that seed size is more associated with growth form than with dispersal syndrome. In fact, boreotropical vicariance was also reported in Persea [146] and Parthenocissus [147]. Additionally, the existence of a boreotropical flora is supported by many other plant biogeographic studies, such as in Burmanniaceae [18], Chamaedoreeae [137], Rubiaceae [148], and Annonaceae [149]. The high latitude of the Bering land bridge made it a barrier for the migration of thermophilic plants but still a corridor for the exchange of temperate plants like Cypripedium. According to the divergence times and distributions of different lineages of Cypripedium, multiple events of vicariance and dispersal between East Asia and North America could have occurred in the genus from middle to late Tertiary (Fig. 4).
The phylogenetic and biogeographic history of slipper orchids revealed in the present study may shed some lights on the evolution of Orchidaceae, one of the largest families of angiosperms with ∼850 genera and ∼25,000 species recorded [39]. A series of studies have investigated the mechanisms underlying the high diversity of orchids, such as epiphytism and pollinator specialization [150], deceptive pollination [151], mycorrhizal fungi [152], crassulacean acid metabolism [153], and reduction of evolutionary constraints on the class B floral homeotic genes [154]. However, the previous studies mainly focused on the key characters of orchids, and paid little attention to the impacts of climatic oscillations and geological events, which are important driving forces of speciation [155]–[157].
In Cypripedium, the basal clade of slipper orchids (Fig. 2; Supplementary Figs. S1, S4), the most ancestral species are distributed in subtropical Mexico (Fig. 2; Supplementary Fig. S1), although most species of the genus are confined to the temperate Northern Hemisphere. Interestingly, the basal species of Paphiopedilum, a mainly tropical genus, also occur in the subtropics (southwest China and Vietnam) (Fig. 2; Supplementary Figs. S1, S4). That is, although the largest two genera of slipper orchids (Cypripedium and Paphiopedilum) have very different distributions, both of them seem to have an origin in the subtropics. This may suggest that their high species diversity and present wide distribution, either in temperate or in tropical regions, were developed to adapt to new niches created by climatic oscillations in the late Cenozoic. Actually, according to anatomical structures, plicate (Cypripedium) and conduplicate (Paphiopedilum) leaves can really adapt to different environments [158].
Moreover, previous biogeographical studies of orchids mainly focused on some endemic genera, e.g. Bromheadia and Holcoglossum in Southeast Asia [159], [160], Antilles in the neotropics [161], and Caladenia in Australia [162], except a couple of them that dealt with widely distributed genera, e.g. Vanilla [163] and Polystachya [164]. In the present study, we sampled all five genera of slipper orchids, including both endemic and widespread ones, and found the vicariant differentiation of the conduplicate genera between the Old World and New World tropics. Obviously, to interpret the nearly cosmopolitan distribution of Orchidaceae (except poles and deserts) [39], the future biogeographical study of orchids should include both regional endemics and their widespread relatives, which will be also helpful to achieve a widely-accepted classification of orchids, particularly at the genus level.
Supporting Information
The ML tree of the slipper orchids constructed based on the combined six chloroplast genes. Numbers above branches indicate bootstrap values ≥50% for the MP and ML analyses, respectively. Bayesian posterior probabilities (≥0.90) are shown in bold lines.
(TIF)
The ML tree of the slipper orchids constructed based on the nuclear ACO gene. Numbers above branches indicate bootstrap values ≥50% for the MP and ML analyses, respectively. Bayesian posterior probabilities (≥0.90) are shown in bold lines. Numbers following the species names are the clone numbers.
(TIF)
The ML tree of the slipper orchids constructed based on the nuclear LFY gene. Numbers above branches indicate bootstrap values ≥50% for the MP and ML analyses, respectively. Bayesian posterior probabilities (≥0.90) are shown in bold lines. Numbers following the species names are the clone numbers.
(TIF)
The ML tree of the slipper orchids constructed based on the combined nuclear genes. Numbers above branches indicate bootstrap values ≥50% for the MP and ML analyses, respectively. Bayesian posterior probabilities (≥0.90) are shown in bold lines.
(TIF)
Sources of materials.
(DOC)
PCR (P) and sequencing (S) primers used in this study.
(DOC)
GenBank accession numbers of taxa used in this study.
(DOC)
Amplification results of the ndh F gene with different primer pairs in the present study.
(DOC)
Acknowledgments
The authors thank Dr. Peter Bernhardt of Saint Louis University, Dr. Gerardo A. Salazar Chávez of Universidad Nacional Autonoma de Mexico, and Dr. Marcin Górniak of Gdansk University for providing key samples. We also thank Ms. Blanche Wagner (Missouri Botanical Garden), Drs. Fu-Sheng Yang (Institutite of Botany, CAS), Yu Zhang (Beijng Botanical Garden) and Edith Kapinos (Royal Botanical Garden, Kew) for their kind help in sample collection; Drs. Jin-Hua Ran and Zu-Yu Yang for their help in data analysis; Drs. Yang Liu, Victor Albert and Mary Stiffler for their help in reference collection; Drs. Jun Shi, Ying Liu, Phillip Cribb, Holger Perner and Kohji Karasawa for photographs used in this study. We also thank Ms. Wan-Qing Jin and Rong-Hua Liang for their assistance in DNA sequencing. We also thank the Academic Editor and the two anonymous reviewers for their insightful comments and suggestions on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Natural Science Foundation of China (Grant No. 30730010), and the Chinese Academy of Sciences (the 100-Talent Project). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Prance GT. Floristic inventory of the tropics: Where do we stand? Ann Mo Bot Gard. 1977;64:659–684. [Google Scholar]
- 2.Raven PH. Tropical floristics tomorrow. Taxon. 1988;37:549–560. [Google Scholar]
- 3.Givnish TJ, Renner SS. Tropical intercontinental disjunctions: Gondwana breakup, immigration from the boreotropics, and transoceanic dispersal. Int J Plant Sci. 2004;165:S1–S6. [Google Scholar]
- 4.Thorne R. Tropical plant disjunctions: a personal reflection. Int J Plant Sci. 2004;165:S137–S138. [Google Scholar]
- 5.Upchurch P. Gondwanan break-up: legacies of a lost world? Trends Ecol Evol. 2008;23:229–236. doi: 10.1016/j.tree.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Crisp MD, Trewick SA, Cook LG. Hypothesis testing in biogeography. Trends Ecol Evol. 2011;26:66–72. doi: 10.1016/j.tree.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Hewitt GM. Post-glacial re-colonization of European biota. Biol J Linn Soc. 1999;68:87–112. [Google Scholar]
- 8.Milne RI, Abbott RJ. The origin and evolution of Tertiary relict floras. Adv Bot Res. 2002;38:281–314. [Google Scholar]
- 9.Milne RI. Northern Hemisphere plant disjunctions: A window on Tertiary land bridges and climate change? Ann Bot. 2006;98:465–472. doi: 10.1093/aob/mcl148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenna MC. Holarctic landmass rearrangement, cosmic events, and Cenozoic terrestrial organisms. Ann Mo Bot Gard. 1983;70:459–489. [Google Scholar]
- 11.McKenna MC. Cenozoic paleogeography of North Atlantic land bridges. In: Bott S, Saxov MHP, Talwani M, Thiede J, editors. Structure and development of the Greenland-Scotland Ridge: new methods and concepts. New York: Plenum Press; 1983. pp. 351–399. [Google Scholar]
- 12.Tiffney BH. The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. J Arnold Arbor. 1985;66:243–273. [Google Scholar]
- 13.Tiffney BH. Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. J Arnold Arbor. 1985;66:73–94. [Google Scholar]
- 14.Hallam A. Oxford: Oxford University Press; 1995. An Outline of Phanerozoic Biogeography. [Google Scholar]
- 15.Azuma H, Garcia-Franco JG, Rico-Gray V, Thien LB. Molecular phylogeny of the Magnoliaceae: the biogeography of tropical and temperate disjunctions. Am J Bot. 2001;88:2275–2285. [PubMed] [Google Scholar]
- 16.Nie ZL, Wen J, Azuma H, Qiu YL, Sun H, et al. Phylogenetic and biogeographic complexity of Magnoliaceae in the Northern Hemisphere inferred from three nuclear data sets. Mol Phylogenet Evol. 2008;48:1027–1040. doi: 10.1016/j.ympev.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Feng CM, Manchester SR, Xiang QY. Phylogeny and biogeography of Alangiaceae (Cornales) inferred from DNA sequences, morphology, and fossils. Mol Phylogenet Evol. 2009;51:201–214. doi: 10.1016/j.ympev.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Merckx V, Chatrou LW, Lemaire B, Sainge MN, Huysmans S, et al. Diversification of myco-heterotrophic angiosperms: Evidence from Burmanniaceae. BMC Evol Biol. 2008;8:178. doi: 10.1186/1471-2148-8-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ickert-Bond SM, Wen J. Phylogeny and biogeography of Altingiaceae: Evidence from combined analysis of five non-coding chloroplast regions. Mol Phylogenet Evol. 2006;39:512–528. doi: 10.1016/j.ympev.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Davis CC, Bell CD, Mathews S, Donoghue MJ. Laurasian migration explains Gondwanan disjunctions: Evidence from Malpighiaceae. Proc Natl Acad Sci USA. 2002;99:6833–6837. doi: 10.1073/pnas.102175899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donoghue MJ. A phylogenetic perspective on the distribution of plant diversity. Proc Natl Acad Sci USA. 2008;105:11549–11555. doi: 10.1073/pnas.0801962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lidgard S, Crane PR. Angiosperm diversification and Cretaceous floristic trends: a comparison of palynofloras and leaf macrofloras. Paleobiology. 1990;16:77–93. [Google Scholar]
- 23.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 24.Zachos JC, Wara MW, Bohaty S, Delaney ML, Petrizzo MR, et al. A transient rise in tropical sea surface temperature during the Paleocene-Eocene thermal maximum. Science. 2003;302:1551–1554. doi: 10.1126/science.1090110. [DOI] [PubMed] [Google Scholar]
- 25.Spicer RA, Harris NBW, Widdowson M, Herman AB, Guo S, et al. Constant elevation of southern Tibet over the past 15 million years. Nature. 2003;421:622–624. doi: 10.1038/nature01356. [DOI] [PubMed] [Google Scholar]
- 26.Abbott RJ, Smith LC, Milne RI, Crawford RMM, Wolff K, et al. Molecular analysis of plant migration and refugia in the Arctic. Science. 2000;289:1343–1346. doi: 10.1126/science.289.5483.1343. [DOI] [PubMed] [Google Scholar]
- 27.Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, et al. Glacial refugia: Hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- 28.Yang FS, Li YF, Ding X, Wang XQ. Extensive population expansion of Pedicularis longiflora (Orobanchaceae) on the Qinghai-Tibetan Plateau and its correlation with the Quaternary climate change. Mol Ecol. 2008;17:5135–5145. doi: 10.1111/j.1365-294X.2008.03976.x. [DOI] [PubMed] [Google Scholar]
- 29.Cun YZ, Wang XQ. Plant recolonization in the Himalaya from the southeastern Qinghai-Tibetan Plateau: Geographical isolation contributed to high population differentiation. Mol Phylogenet Evol. 2010;56:972–982. doi: 10.1016/j.ympev.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Antonelli A, Verola CF, Parisod C, Gustafsson ALS. Climate cooling promoted the expansion and radiation of a threatened group of South American orchids (Epidendroideae: Laeliinae). Biol J Linn Soc. 2010;100:597–607. [Google Scholar]
- 31.Hughes C, Eastwood R. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc Natl Acad Sci USA. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valiente-Banuet A, Rumebe AV, Verdú M, Callaway RM. Modern Quaternary plant lineages promote diversity through facilitation of ancient Tertiary lineages. Proc Natl Acad Sci USA. 2006;103:16812–16817. doi: 10.1073/pnas.0604933103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakob SS, Ihlow A, Blattner FR. Combined ecological niche modelling and molecular phylogeography revealed the evolutionary history of Hordeum marinum (Poaceae) — niche differentiation, loss of genetic diversity, and speciation in Mediterranean Quaternary refugia. Mol Ecol. 2007;16:1713–1727. doi: 10.1111/j.1365-294X.2007.03228.x. [DOI] [PubMed] [Google Scholar]
- 34.Sapir Y, Moody ML, Brouillette LC, Donovan LA, Rieseberg LH. Patterns of genetic diversity and candidate genes for ecological divergence in a homoploid hybrid sunflower, Helianthus anomalus. Mol Ecol. 2007;16:5017–5029. doi: 10.1111/j.1365-294X.2007.03557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bittkau C, Comes HP. Molecular inference of a Late Pleistocene diversification shift in Nigella s. lat. (Ranunculaceae) resulting from increased speciation in the Aegean archipelago. J Biogeogr. 2009;36:1346–1360. [Google Scholar]
- 36.Wing SL. Eocene and Oligocene floras and vegetation of the Rocky Mountains. Ann Mo Bot Gard. 1987;74:748–784. [Google Scholar]
- 37.Lavin M, Luckow M. Origins and relationships of tropical North America in the context of the boreotropics hypothesis. Am J Bot. 1993;80:1–14. [Google Scholar]
- 38.Wen J. Evolution of eastern Asian and eastern north American disjunct distributions in flowering plants. Annu Rev Ecol Syst. 1999;30:421–455. [Google Scholar]
- 39.Roberts DL, Dixon KW. Orchids. Curr Biol. 2008;18:R325–R329. doi: 10.1016/j.cub.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 40.Ramírez SR, Gravendeel B, Singer RB, Marshall CR, Pierce NE. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature. 2007;448:1042–1045. doi: 10.1038/nature06039. [DOI] [PubMed] [Google Scholar]
- 41.Conran JG, Bannister JM, Lee DE. Earliest orchid macrofossils: Early Miocene Dendrobium and Earina (Orchidaceae: Epidendroideae) from New Zealand. Am J Bot. 2009;96:466–474. doi: 10.3732/ajb.0800269. [DOI] [PubMed] [Google Scholar]
- 42.Gustafsson AL, Verola C, Antonelli A. Reassessing the temporal evolution of orchids with new fossils and a Bayesian relaxed clock, with implications for the diversification of the rare South American genus Hoffmannseggella (Orchidaceae: Epidendroideae). BMC Evol Biol. 2010;10:177. doi: 10.1186/1471-2148-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cameron KM, Chase MW, Whitten WM, Kores PJ, Jarrell DC, et al. A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Am J Bot. 1999;86:208–224. [PubMed] [Google Scholar]
- 44.Cameron KM, Chase MW. Nuclear 18S rDNA sequences of Orchidaceae confirm the subfamilial status and circumscription of Vanilloideae. In: Wilson KL, Morrison DA, editors. Monocots: Systematics and Evolution. Collingwood: CSIRO publishing. 2000;457–464 [Google Scholar]
- 45.Freudenstein JV, Chase MW. Analysis of mitochondrial nad1b-c intron sequences in Orchidaceae: utility and coding of length-change characters. Syst Bot. 2001;26:643–657. [Google Scholar]
- 46.Cameron KM. Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Mol Phylogenet Evol. 2004;31:1157–1180. doi: 10.1016/j.ympev.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 47.Freudenstein JV, van den Berg C, Goldman DH, Kores PJ, Molvray M, et al. An expanded plastid DNA phylogeny of Orchidaceae and analysis of jackknife branch support strategy. Am J Bot. 2004;91:149–157. doi: 10.3732/ajb.91.1.149. [DOI] [PubMed] [Google Scholar]
- 48.Górniak M, Paun O, Chase MW. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Mol Phylogenet Evol. 2010;56:784–795. doi: 10.1016/j.ympev.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Lindley J. Ridgeways, London: W. NICOL, 60, PALL MALL; 1840. Genera and Species of Orchidaceous Plants. [Google Scholar]
- 50.Albert VA, Chase MW. Mexipedium: a new genus of slipper orchid (Cypripedioideae: Orchidaceae). Lindleyana. 1992;7:172–176. [Google Scholar]
- 51.Dressler RL. Massachusetts: Cambridge University Press, Cambridge; 1993. Phylogeny and Classification of the Orchid Family. [Google Scholar]
- 52.Bream GJ. Hildesheim: Brücke-Verlag Kurt Schmersow; 1988. Paphiopedilum. [Google Scholar]
- 53.Cribb P. Portland, Oregon: Timber Press; 1997. The Genus Cypripedium. [Google Scholar]
- 54.Cribb P. Kota Kinabalu and Kew: Natural History Publications; 1998. The Genus Paphiopedilum. [Google Scholar]
- 55.Averyanov L, Cribb P, Loc PK, Hiep NT. Royal Botanic Gardens, Kew: Compass Press Limited; 2003. Slipper Orchids of Vietnam. [Google Scholar]
- 56.Koopowitz H, Comstock J, Woodin C. Portland, Oregon: Timber Press, Inc; 2008. Tropical Slipper Orchids: Paphiopedilum and Phragmipedium Species and Hybrids. [Google Scholar]
- 57.Liu ZJ, Chen SC, Chen LJ, Lei SP. Beijing: Science Press; 2009. The Genus Paphiopedilum in China. [Google Scholar]
- 58.Li JH, Liu ZJ, Salazar GA, Bernhardt P, Perner H, et al. Molecular phylogeny of Cypripedium (Orchidaceae: Cypripedioideae) inferred from multiple nuclear and chloroplast regions. Mol Phylogenet Evol. 2011;61:308–320. doi: 10.1016/j.ympev.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 59.Lan TY, Albert VA. Dynamic distribution patterns of ribosomal DNA and chromosomal evolution in Paphiopedilum, a lady's slipper orchid. BMC Plant Biol. 2011;11:126. doi: 10.1186/1471-2229-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dressler RL. Massachusetts: Harvard University Press, Cambridge; 1981. Orchids: Natural History and Classification. [Google Scholar]
- 61.Pfitzer EHH. Orchidaceae-Pleonandrae. In: Engler A, editor. Das Pflanzenreich. Leipzig: Engelmann; 1903. pp. 1–132. [Google Scholar]
- 62.Atwood JT. The relationships of the slipper orchids (subfamily Cypripedioideae, Orchidaceae). Selbyana. 1984;7:129–247. [Google Scholar]
- 63.Albert VA. Cladistic relationships of the slipper orchids (Cypripedioideae: Orchidaceae) from congruent morphological and molecular data. Lindleyana. 1994;9:115–132. [Google Scholar]
- 64.Cox AV, Pridgeon AM, Albert VA, Chase MW. Phylogenetics of the slipper orchids (Cypripedioideae, Orchidaceae): nuclear rDNA ITS sequences. Plant Syst Evol. 1997;208:197–223. [Google Scholar]
- 65.Freudenstein JV, Senyo DM, Chase MW. Mitochondrial DNA and relationships in the Orchidaceae. In: Wilson KL, Morrison DA, editors. Monocots: Systematics and Evolution. CSIRO publishing Collingwood; 2000. pp. 421–429. [Google Scholar]
- 66.Wolfe JA. Some aspects of plant geography of the Northern Hemisphere during the late Cretaceous and Tertiary. Ann Mo Bot Gard. 1975;62:264–279. [Google Scholar]
- 67.Rokas A, Carroll SB. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Mol Biol Evol. 2005;22:1337–1344. doi: 10.1093/molbev/msi121. [DOI] [PubMed] [Google Scholar]
- 68.Peng D, Wang XQ. Reticulate evolution in Thuja inferred from multiple gene sequences: Implications for the study of biogeographical disjunction between eastern Asia and North America. Mol Phylogenet Evol. 2008;47:1190–1202. doi: 10.1016/j.ympev.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 69.Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, et al. A DNA barcode for land plants. Proc Natl Acad Sci USA. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang HC, Moore MJ, Soltis PS, Bell CD, Brockington SF, et al. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci USA. 2009;106:3853–3858. doi: 10.1073/pnas.0813376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang JL, Sun GL, Zhang DM. Molecular evolution and phylogeny of the angiosperm ycf2 gene. J Syst Evol. 2010;48:240–248. [Google Scholar]
- 72.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neubig KM, Whitten WM, Carlsward BS, Blanco MA, Endara L, et al. Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Syst Evol. 2009;277:75–84. [Google Scholar]
- 74.Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84. doi: 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neubig KM, Abbott JR. Primer development for the plastid region ycf1 in Annonaceae and other magnoliids. Am J Bot. 2010;97:e52–e55. doi: 10.3732/ajb.1000128. [DOI] [PubMed] [Google Scholar]
- 76.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sang T. Utility of low-copy nuclear gene sequences in plant phylogenetics. Crit Rev Biochem Mol Biol. 2002;37:121–147. doi: 10.1080/10409230290771474. [DOI] [PubMed] [Google Scholar]
- 78.Small RL, Cronn RC, Wendel JF. Use of nuclear genes for phylogeny reconstruction in plants. Aust J Bot. 2004;17:145–170. [Google Scholar]
- 79.Whittall JB, Medina-Marino A, Zimmer EA, Hodges SA. Generating single-copy nuclear gene data for a recent adaptive radiation. Mol Phylogenet Evol. 2006;39:124–134. doi: 10.1016/j.ympev.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 80.Duarte JM, Wall PK, Edger PP, Landherr LL, Ma H, et al. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol. 2010;10:61. doi: 10.1186/1471-2148-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- 82.Nilsson O, Lee I, Blazquez MA, Weigel D. Flowering-time genes modulate the response to LEAFY activity. Genetics. 1998;150:403–410. doi: 10.1093/genetics/150.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, et al. The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science. 2005;308:260–263. doi: 10.1126/science.1108229. [DOI] [PubMed] [Google Scholar]
- 84.Moyroud E, Kusters E, Monniaux M, Koes R, Parcy F. LEAFY blossoms. Trends Plant Sci. 2010;15:346–352. doi: 10.1016/j.tplants.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 85.Oh SH, Potter D. Molecular phylogenetic systematics and biogeography of tribe Neillieae (Rosaceae) using DNA sequences of cpDNA, rDNA, and LEAFY. Am J Bot. 2005;92:179–192. doi: 10.3732/ajb.92.1.179. [DOI] [PubMed] [Google Scholar]
- 86.Oh SH, Potter D. Phylogenetic utility of the second intron of LEAFY in Neillia and Stephanandra (Rosaceae) and implications for the origin of Stephanandra. Mol Phylogenet Evol. 2003;29:203–215. doi: 10.1016/s1055-7903(03)00093-9. [DOI] [PubMed] [Google Scholar]
- 87.Grob GBJ, Gravendeel B, Eurlings MCM. Potential phylogenetic utility of the nuclear FLORICAULA/LEAFY second intron: comparison with three chloroplast DNA regions in Amorphophallus (Araceae). Mol Phylogenet Evol. 2004;30:13–23. doi: 10.1016/s1055-7903(03)00183-0. [DOI] [PubMed] [Google Scholar]
- 88.Wei XX, Yang ZY, Li Y, Wang XQ. Molecular phylogeny and biogeography of Pseudotsuga (Pinaceae): Insights into the floristic relationship between Taiwan and its adjacent areas. Mol Phylogenet Evol. 2010;55:776–785. doi: 10.1016/j.ympev.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 89.Kim ST, Sultan SE, Donoghue MJ. Allopolyploid speciation in Persicaria (Polygonaceae): Insights from a low-copy nuclear region. Proc Natl Acad Sci USA. 2008;105:12370–12375. doi: 10.1073/pnas.0805141105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.John P. Ethylene biosynthesis: the role of 1-aminocyclopropane- 1-carboxylate (ACC) oxidase, and its possible evolutionary origin. Physiol Plant. 1997;100:583–592. [Google Scholar]
- 91.Lin ZF, Zhong SL, Grierson D. Recent advances in ethylene research. J Exp Bot. 2009;60:3311–3336. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- 92.Cribb P. The genus Selenipedium. Curtis's Bot Mag. 2009;26:5–20. [Google Scholar]
- 93.Rogers SO, Bendich AJ. Extraction of DNA from plant tissues. Plant Mol Biol (Manual) 1988;A6:1–10. doi: 10.1007/BF00020088. [DOI] [PubMed] [Google Scholar]
- 94.Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 95.Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol Biol Evol. 2003;20:1499–1505. doi: 10.1093/molbev/msg159. [DOI] [PubMed] [Google Scholar]
- 96.Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. The chloroplast genome of Nymphaea alba: Whole-genome analyses and the problem of identifying the most basal angiosperm. Mol Biol Evol. 2004;21:1445–1454. doi: 10.1093/molbev/msh147. [DOI] [PubMed] [Google Scholar]
- 97.Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH. Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol. 2005;22:1813–1822. doi: 10.1093/molbev/msi173. [DOI] [PubMed] [Google Scholar]
- 98.Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- 99.Wu FH, Chan MT, Liao DC, Hsu CT, Lee YW, et al. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010;10:68. doi: 10.1186/1471-2229-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 2011;28:2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Logacheva MD, Schelkunov MI, Penin AA. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol Evol. 2011;3:1296–1303. doi: 10.1093/gbe/evr102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 103.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 104.Young ND, Healy J. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics. 2003;4:6. doi: 10.1186/1471-2105-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Farris JS, källerjö M, Kluge AG, Bult C. Testing significance of incongruence. Cladistics. 1995;10:315–319. [Google Scholar]
- 106.Swofford DL. Sunderland, MA: Sinauer Associates; 2002. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0 Beta. [Google Scholar]
- 107.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 108.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 109.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 110.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 111.Nylander JAA. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2004. MrModeltest v2. [Google Scholar]
- 112.Renner SS. Relaxed molecular clocks for dating historical plant dispersal events. Trends Plant Sci. 2005;10:550–558. doi: 10.1016/j.tplants.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 113.Yoder AD, Nowak MD. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu Rev Ecol Evol Systemat. 2006;37:405–431. [Google Scholar]
- 114.Felsenstein J. Phylogenies from molecular sequences: Inference and reliability. Ann Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 115.Sorhannus U, Bell CV. Testing for equality of molecular evolutionary rates: a comparison between a relative-rate test and a likelihood ratio test. Mol Biol Evol. 1999;16:849–855. [Google Scholar]
- 116.The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161:105–121. [Google Scholar]
- 117.Sanderson MJ. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- 118.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 119.Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 120.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Friis EM, Pedersen KR, Crane PR. Araceae from the Early Cretaceous of Portugal: Evidence on the emergence of monocotyledons. Proc Natl Acad Sci USA. 2004;101:16565–16570. doi: 10.1073/pnas.0407174101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang ZH, Yoder AD. Comparison of likelihood and Bayesian methods for estimating divergence times using multiple gene loci and calibration points, with application to a radiation of cute-looking mouse lemur species. Syst Biol. 2003;52:705–716. doi: 10.1080/10635150390235557. [DOI] [PubMed] [Google Scholar]
- 123.Yu Y, Harris AJ, He XJ. 2010. S-DIVA (Statistical Dispersal-Vicariance Analysis) version 1.9 beta. Available at http://mnh.scu.edu.cn/soft/blog/sdiva.
- 124.Yu Y, Harris AJ, He XJ. S-DIVA (Statistical Dispersal-Vicariance Analysis): a tool for inferring biogeographic histories. Mol Phylogenet Evol. 2010;56:848–850. doi: 10.1016/j.ympev.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 125.Ree RH, Moore BR, Webb CO, Donoghue MJ. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution. 2005;59:2299–2311. [PubMed] [Google Scholar]
- 126.Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 127.Kores PJ, Weston PH, Molvray M, Chase MW. Phylogenetic relationships within the Diurideae (Orchidaceae): inferences from plastid matK DNA sequences. In: Wilson KL, Morrison DA, editors. Monocots: Systematics and Evolution. Collingwood: CSIRO publishing; 2000. pp. 449–456. [Google Scholar]
- 128.Freudenstein JV, Senyo DM. Relationships and evolution of matK in a group of leafless orchids (Corallorhiza and Corallorhizinae; Orchidaceae: Epidendroideae). Am J Bot. 2008;95:498–505. doi: 10.3732/ajb.95.4.498. [DOI] [PubMed] [Google Scholar]
- 129.Kocyan A, de Vogel EF, Conti E, Gravendeel B. Molecular phylogeny of Aerides (Orchidaceae) based on one nuclear and two plastid markers: A step forward in understanding the evolution of the Aeridinae. Mol Phylogenet Evol. 2008;48:422–443. doi: 10.1016/j.ympev.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 130.Cameron KM. A comparison and combination of plastid atpB and rbcL gene sequences for inferring phylogenetic relationships within Orchidaceae. Aliso. 2006;22:447–464. [Google Scholar]
- 131.Karasawa K, Aoyama M, Ishida G. A karyomorphological study on Selenipedium aequinoctiale Garay, Orchidaceae. Bull Hiroshima Bot Gard 22/ 2005;23:1–4. [Google Scholar]
- 132.Soto MA, Salazar GA, Hágsater E. Phragmipedium xerophyticum, una nueva especie del sureste de México. Orquídea (México) 1990;12:1–10. [Google Scholar]
- 133.Zachos JC, Shackleton NJ, Revenaugh JS, Pälike H, Flower BP. Climate response to orbital forcing across the Oligocene-Miocene boundary. Science. 2001;292:274–278. doi: 10.1126/science.1058288. [DOI] [PubMed] [Google Scholar]
- 134.Zanazzi A, Kohn MJ, MacFadden BJ, Terry DO. Large temperature drop across the Eocene-Oligocene transition in central North America. Nature. 2007;445:639–642. doi: 10.1038/nature05551. [DOI] [PubMed] [Google Scholar]
- 135.Iturralde-Vinent MA, MacPhee RDE. Paleogeography of the Caribbean region: implications for Cenozoic biogeography. Bull Am Mus Nat Hist. 1999;238:1–95. [Google Scholar]
- 136.Pennington RT, Dick CW. The role of immigrants in the assembly of the South American rainforest tree flora. Phil Trans R Soc Lond B. 2004;359:1611–1622. doi: 10.1098/rstb.2004.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cuenca A, Asmussen-Lange CB, Borchsenius F. A dated phylogeny of the palm tribe Chamaedoreeae supports Eocene dispersal between Africa, North and South America. Mol Phylogenet Evol. 2008;46:760–775. doi: 10.1016/j.ympev.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 138.Heaney LR. A synopsis of climatic and vegetational change in Southeast Asia. Climatic Change. 1991;19:53–61. [Google Scholar]
- 139.Jokat W, Boebel T, König M, Meyer U. Timing and geometry of early Gondwana breakup. J Geophys Res. 2003;108:2428. [Google Scholar]
- 140.Kershaw P, Wagstaff B. The southern conifer family Araucariaceae: history, status, and value for paleoenvironmental reconstruction. Annu Rev Ecol Syst. 2001;32:397–414. [Google Scholar]
- 141.Ducousso M, Béna G, Bourgeois C, Buyck B, Eyssartier G, et al. The last common ancestor of Sarcolaenaceae and Asian dipterocarp trees was ectomycorrhizal before the India-Madagascar separation, about 88 million years ago. Mol Ecol. 2004;13:231–236. doi: 10.1046/j.1365-294x.2003.02032.x. [DOI] [PubMed] [Google Scholar]
- 142.Nathan R. Long-distance dispersal of plants. Science. 2006;313:786–788. doi: 10.1126/science.1124975. [DOI] [PubMed] [Google Scholar]
- 143.Michalak I, Zhang LB, Renner SS. Trans-Atlantic, trans-Pacific and trans-Indian Ocean dispersal in the small Gondwanan Laurales family Hernandiaceae. J Biogeogr. 2010;37:1214–1226. [Google Scholar]
- 144.Arditti J, Ghani AKA. Numerical and physical properties of orchid seeds and their biological implications. New Phytol. 2000;145:367–421. doi: 10.1046/j.1469-8137.2000.00587.x. [DOI] [PubMed] [Google Scholar]
- 145.Moles AT, Ackerly DD, Webb CO, Tweddle JC, Dickie JB, et al. A brief history of seed size. Science. 2005;307:576–580. doi: 10.1126/science.1104863. [DOI] [PubMed] [Google Scholar]
- 146.Li L, Li J, Rohwer JG, van der Werff H, Wang ZH, et al. Molecular phylogenetic analysis of the Persea group (Lauraceae) and its biogeographic implications on the evolution of tropical and subtropical Amphi-Pacific disjunctions. Am J Bot. 2011;98:1520–1536. doi: 10.3732/ajb.1100006. [DOI] [PubMed] [Google Scholar]
- 147.Nie ZL, Sun H, Chen ZD, Meng Y, Manchester SR, et al. Molecular phylogeny and biogeographic diversification of Parthenocissus (Vitaceae) disjunct between Asia and North America. Am J Bot. 2010;97:1342–1353. doi: 10.3732/ajb.1000085. [DOI] [PubMed] [Google Scholar]
- 148.Smedmark JEE, Eriksson T, Bremer B. Divergence time uncertainty and historical biogeography reconstruction – an example from Urophylleae (Rubiaceae). J Biogeogr. 2010;37:2260–2274. [Google Scholar]
- 149.Couvreur TLP, Pirie MD, Chatrou LW, Saunders RMK, Su YCF, et al. Early evolutionary history of the flowering plant family Annonaceae: steady diversification and boreotropical geodispersal. J Biogeogr. 2011;38:664–680. [Google Scholar]
- 150.Gravendeel B, Smithson A, Slik FJW, Schuiteman A. Epiphytism and pollinator specialization: drivers for orchid diversity? Phil Trans R Soc Lond B. 2004;359:1523–1535. doi: 10.1098/rstb.2004.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cozzolino S, Widmer A. Orchid diversity: an evolutionary consequence of deception? Trends Ecol Evol. 2005;20:487–494. doi: 10.1016/j.tree.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 152.Otero JT, Flanagan NS. Orchid diversity – beyond deception. Trends Ecol Evol. 2006;21:64–65. doi: 10.1016/j.tree.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 153.Silvera K, Santiago LS, Cushman JC, Winter K. Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiol. 2009;149:1838–1847. doi: 10.1104/pp.108.132555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mondragón-Palomino M, Theißen G. Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann Bot. 2009;104:583–594. doi: 10.1093/aob/mcn258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Rundle HD, Nosil P. Ecological speciation. Ecol Lett. 2005;8:336–352. [Google Scholar]
- 156.Ryan PG, Bloomer P, Moloney CL, Grant TJ, Delport W. Ecological speciation in South Atlantic island finches. Science. 2007;315:1420–1423. doi: 10.1126/science.1138829. [DOI] [PubMed] [Google Scholar]
- 157.Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- 158.Guan ZJ, Zhang SB, Guan KY, Li SY, Hu H. Leaf anatomical structures of Paphiopedilum and Cypripedium and their adaptive significance. J Plant Res. 2011;124:289–298. doi: 10.1007/s10265-010-0372-z. [DOI] [PubMed] [Google Scholar]
- 159.Repetur CP, van Welzen PC, de Vogel EF. Phylogeny and historical biogeography of the genus Bromheadia (Orchidaceae). Syst Bot. 1997;22:465–477. [Google Scholar]
- 160.Fan J, Qin HN, Li DZ, Jin XH. Molecular phylogeny and biogeography of Holcoglossum (Orchidaceae: Aeridinae) based on nuclear ITS, and chloroplast trnL-F and matK. Taxon. 2009;58:849–861. [Google Scholar]
- 161.Trejo-Torres JC, Ackerman JD. Biogeography of the Antilles based on a parsimony analysis of orchid distributions. J Biogeogr. 2001;28:775–794. [Google Scholar]
- 162.Phillips RD, Backhouse G, Brown AP, Hopper SD. Biogeography of Caladenia (Orchidaceae), with special reference to the South-west Australian Floristic Region. Aust J Bot. 2009;57:259–275. [Google Scholar]
- 163.Bouetard A, Lefeuvre P, Gigant R, Bory S, Pignal M, et al. Evidence of transoceanic dispersion of the genus Vanilla based on plastid DNA phylogenetic analysis. Mol Phylogenet Evol. 2010;55:621–630. doi: 10.1016/j.ympev.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 164.Russell A, Samuel R, Rupp B, Barfuss MHJ, Safran M, et al. Phylogenetics and cytology of a pantropical orchid genus Polystachya (Polystachyinae, Vandeae, Orchidaceae): evidence from plastid DNA sequence data. Taxon. 2010;59:389–404. [Google Scholar]
- 165.Pridgeon AM, Cribb JP, Chase WM, Rasmussen F. Volume 1: General Introduction, Apostasioideae, Cypripedioideae. Oxford: Oxford University Press; 1999. Genera Orchidacearum. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The ML tree of the slipper orchids constructed based on the combined six chloroplast genes. Numbers above branches indicate bootstrap values ≥50% for the MP and ML analyses, respectively. Bayesian posterior probabilities (≥0.90) are shown in bold lines.
(TIF)
The ML tree of the slipper orchids constructed based on the nuclear ACO gene. Numbers above branches indicate bootstrap values ≥50% for the MP and ML analyses, respectively. Bayesian posterior probabilities (≥0.90) are shown in bold lines. Numbers following the species names are the clone numbers.
(TIF)
The ML tree of the slipper orchids constructed based on the nuclear LFY gene. Numbers above branches indicate bootstrap values ≥50% for the MP and ML analyses, respectively. Bayesian posterior probabilities (≥0.90) are shown in bold lines. Numbers following the species names are the clone numbers.
(TIF)
The ML tree of the slipper orchids constructed based on the combined nuclear genes. Numbers above branches indicate bootstrap values ≥50% for the MP and ML analyses, respectively. Bayesian posterior probabilities (≥0.90) are shown in bold lines.
(TIF)
Sources of materials.
(DOC)
PCR (P) and sequencing (S) primers used in this study.
(DOC)
GenBank accession numbers of taxa used in this study.
(DOC)
Amplification results of the ndh F gene with different primer pairs in the present study.
(DOC)