Abstract
Purpose
To investigate the role of WW Domain Containing Transcription Regulator (WWTR1/TAZ) protein in regulating the proliferation of normal human conjunctiva epithelial cells and epithelial cells from pterygium tissue.
Methods
Conjunctiva epithelial cells were cultured in keratinocytes growth medium and treated with transformation growth factor β (TGFβ) to analyze the expression and translocation of TAZ protein by immunostaining and BrdU analysis. Immortalized conjunctiva epithelial cells (NHC) were treated with TGFβ, targeting siRNA, TGFβ receptor antibody or TGFβ receptor inhibitor, to study the involvement of TAZ and TGFβ signaling pathway in conjunctiva cell proliferation by cell adhesion assay. Conjunctiva tissues from a normal human eye and an eye with pterygium disease were collected for histological analyses and western blot to evaluate the TAZ protein expression in vivo.
Results
TAZ expression was upregulated in mitotic conjunctiva epithelial cells, proliferating conjunctiva epithelial cells, TGFβ treated conjunctiva epithelial cells and human pterygium epithelium. TAZ siRNA induced less conjunctiva epithelial cell growth. Moreover, TGFβ receptor antibody and TGFβ receptor inhibitor rescued this anti-proliferative effect of TAZ siRNA.
Conclusions
TAZ is involved in human conjunctiva epithelial cells proliferation via regulating TGFβ signaling pathway.
Introduction
Pterygium is a common ocular disease characterized by a fibrovascular membrane advancing on the corneal surface. Pterygium is caused by the abnormal growth and differentiation of the conjunctival epithelial cells of the corneal limbus [1,2]. Previous studies have shown an overexpression of transforming growth factor β (TGFβ) signaling in pterygia tissue when compared with normal conjunctiva. Therefore, the proliferation of conjunctiva epithelium in pterygium may be attributed to the activities of TGFβ.
TGFβ regulates important biologic functions, including cell growth, differentiation, migration and apoptosis, which have been extensively investigated and reviewed [3-5]. In the mature mammalian epithelium, TGFβ signal induces cell cycle arrest, apoptosis, cell adhesion and cytokine secretion [6-8]. Biologic signals for TGFβ are transduced through transmembrane serine/threonine kinase receptors to a family of intracellular mediators known as mothers against decapentaplegic homolog (Smad) proteins [9]. Smad proteins are activated to either propagate or inhibit the TGFβ signal by interacting with other modulators. The TGFβ superfamily receptors have been found to have a distinct and specific distribution in the conjunctiva epithelium, indicating that conjunctiva epithelial cells respond to TGFβ cytokine and that TGFβ may have important autocrine and/or paracrine roles in the growth and metabolism of ocular tissues in vivo [10,11].
WW Domain Containing Transcription Regulator (WWTR1/TAZ) protein is a transcriptional co-activator, which contains a 14–3-3 binding fragment, a single WW domain and a PDZ-binding motif [12]. Mutations of TAZ protein have been reported to be related to a dysfunction of caspase activities in mammalian cells [13,14]. Recently, TAZ was reported to be essential in regulating hippo pathway and β-catenin/wnt pathway during organ size control, tissue regeneration and stem cell self-renewal [15-17]. Additionally, the WW domain of the TAZ protein was found to be able to bind with the PPXY motif of the transcription factor runt-related transcription factor 2 (RUNX2 [18-20]. This opens up the possibilities that other PPXY sequences containing transcription factors, such as myocyte enhancer factor 2B (MEF2B), Smad, and sex determining region Y-box (SOX) family molecules, are also possible binding candidates of TAZ [20]. Subsequently it was found that TAZ binds with Smad 2/3 to shuttle the TGFβ stimulated nucleus translocation of Smad proteins to regulate human embryonic stem cell proliferation [21]. Currently, there are no reports in the literature regarding the study of TAZ in ocular tissues.
In the present study, we sought to investigate the involvement of TAZ protein and TGFβ signaling in regulating conjunctival epithelial cell proliferation. Moreover, we aimed to investigate the expression and location of TAZ protein in human conjunctiva epithelium and epithelium from pterygium samples.
Methods
Human conjunctiva and pterygium cell isolation and culture
All human tissue related studies were performed in accordance with the tenets of the Declaration of Helsinki and the study protocol was approved by the Institutional Review Board of the Singapore Eye Research Institute and Singapore National Eye Centre. Informed written consent was obtained from each participant.
Pterygium biopsies were collected from patients undergoing routine surgery for pterygium removal. A small piece of normal conjunctiva (1×3 mm) was removed from the super bulbar region, 10–15 mm from the limbus. For cell isolation and culture, tissue biopsies were briefly rinsed with phosphate buffer saline, followed by stirring in buffer containing 30 mM HEPES, 4 mM glucose, 3 mM KCL, 1 mM Na2HPO4 and 1.2% dispase. Epithelial cells were collected by centrifugation and cultured in serum-free keratinocyte growth medium (KGM) with supplements of bovine pituitary and epithelial growth factor (EGF). For TGFβ stimulation experiments, conjunctiva epithelial cells were cultured in KGM medium without supplements for 2 days before treatment.
5-Bromo-2’-deoxyuridine (BrdU) assay
Immortalized normal human conjunctiva cells (NHC cells) were cultured in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal calf serum and grown on round shape glass coverslips, which were inserted on the bottom of the 24 well plate dishes. BrdU (10 µm) was added into the culture medium for 24 h before fixation with 4% paraformaldehyde. After treatment with 2 N hydrochloric acid (HCL) for 15 min, cells were stained with monoclonal anti-BrdU (Sigma, St. Louis, MO) and polyclonal anti-TAZ (Santa Cruz Biotechnology, Santa Cruz, CA). 4',6-diamidino-2-phenylindole (DAPI) was applied in the mounting medium as a nucleus counter stain. Images of the cells after staining were taken by LSM510 confocal microscope (Carl Zeiss, Jena, Germany) and the fluorescent intensity of the defined area was measured by Image J (NIH) software. Briefly, the staining pictures were converted into monochrome images and measured with the intensity tools. The intensities of the cells at cell cycle resting stage or TGFβ non-treated cells were defined as “1.” The fluorescence intensities of testing cells were normalized to the readings of cells at control group and defined as “relative fluorescence intensity.”
SiRNA transfection and commassie brilliant blue staining
TAZ siRNA (Santa Cruz) is a combination of three siRNA sequences targeting the human TAZ gene (NM_013595): A; GUA CUU CCU CAA UCA CAU Att, UAU GUG AUU GAG GAA GUA Ctt: B; CUA GGA AGG CGA UGA AUC Att, UGA UUC AUC GCC UUC CUA Gtt: C; GGA UGU AGC CAU GAC CUU Att, UAA GGU CAU GGC UAC AUC Ctt. Both TAZ siRNA and mock siRNA (Santa Cruz Biotechnology) were transiently transfected into NHC cells with lipofectamine 2000 (Invitrogen, Carlsbad, CA) as a carrier, according to the protocol provided by the manufacturer. NHC cells were collected at 24 h after transfection for further processing. For cell density analysis, NHC cells were stained with 0.5% commassie brilliant blue before scanning. The scanning pictures were converted into monochrome images and measured with the intensity measurement tools of Image J (NIH) software. The averaged intensities of the scanning images for non-treated specimens were defined as “1.” The intensities of the scanning images for treated specimens were normalized to the readings of control group and defined as “relative cell density.”
Transcriptional response assay
For the luciferase reporter assay, NHC were grown on Costar® white 96-well dish and transfected with 10–4-cyclin E-lux cDNA (addgene plasmid 8458) with lipofectamineTM 2000 (Invitrogen). 10–4-cyclin E–lux (50 ng) was transfected into NHC cells with a combination of mock siRNA or siTAZ. Luciferase activity in cell lysate was measured using the Bright-GloTM luciferase assay system (Promega, Madison, WI) in a TecanTM Xfluor 4 Geniospro V4.5 luminometer (Tecan, Seestrasse, Männedorf). Data were collected from 3 repeats.
Immunofluorescence
NHC cell line or primary human conjunctiva epithelial cells after treatment were fixed with 4% paraformaldehyde and washed by phosphate buffer saline with 0.2% triton-x 100. Normal conjunctiva or pterygium tissue after dissection was fixed with 4% paraformaldehyde before cryopreserving with 20% sucrose. Eight µm thickness sections were collected with a cryostat (Leica, Wetzlar, Germany). The primary antibodies for immunostaining include polyclonal anti-TAZ and anti-Smad4 (Cell signaling Technology, Danvers, MA). Secondary antibodies include Alexflour 488 or Alexflour 555 conjugated goat anti-mouse or goat anti-rabbit secondary antibody (Invitrogen). Images of the specimens after staining were taken by LSM510 confocal microscope (Carl Zeiss, Jena, Germany).
Immunoblotting
Conjunctiva epithelial cells were plated on a 9 cm × 9 cm culture dish till 80% to 90% confluence. After treatment, the cells were collected and lysed in RIPA buffer (50 mM Tris-HCL, 1 mM EDTA, 0.25% sodium deoxycholate, 150 mM sodium chloride, and 1% NP-40) containing 1 mM DTT, protease inhibitor cocktail, 1 mM Na3VO4, and 1 mM PMSF. A total of 30 µg of each protein was loaded for SDS–PAGE gel analysis. Primary antibodies for western blot were anti-TAZ (Santa Cruz Biotechnology) and anti-β-actin (Sigma, St. Louis, MO).
Secondary antibodies for western blot included horseradish peroxidase conjugated sheep anti-mouse/rabbit IgG (GE Healthcare, Buckinghamshire, England).
Results
Expression of TAZ protein in proliferating human conjunctiva epithelial cells
We performed TAZ immunostaining on primary normal human conjunctiva epithelial cells to investigate the role of TAZ protein in cell proliferation. Nucleus DAPI counterstaining was applied to indicate the cell dividing stage. We observed that the fluorescence intensity of TAZ protein was strongly upregulated in dividing cells compared with that of neighbor cells at the resting stage (Figure 1A). A total of 120 cells from three independent repeats were collected for analysis. The averaged fluorescence intensity of TAZ protein in mitotic cells was around 5.2±0.4 fold higher than that of the resting cells (Figure 1B). Upregulation of TAZ protein expression was also observed in proliferating cells during other cell cycle phases, which were reflected by the DAPI staining pattern (Figure 2). We treated conjunctiva epithelial cells with 2N HCL for 15 min (Figure 2A) or 40 min (Figure 2B) to allow for permeabilization of TAZ antibody. We found that in dividing conjunctiva keratinocyte, TAZ protein concisely co-localizes with DAPI, an indicator for DNA. We did transcriptional response assay to determine the involvement of TAZ in cell cycling/proliferation, which was accessed by transcriptional activities of 10–4-cyclin E promoter. 10–4-cyclin E promoter is required for the mammalian cell transition from G1 to S phase. TAZ siRNA induced significant less transcriptional activities of 10–4-cyclin E promoter in NHC cells and this effect is dosage dependent (Figure 2C).
Figure 1.

TAZ protein expression is upregulated in mitotic cells. A: Conjunctiva epithelial cells were stained with anti-TAZ antibody. TAZ: red; DAPI: blue. Higher magnification image is shown in lower row. Immunofluorescence signal of TAZ protein is significantly higher in a dividing cell (white arrow) compared with that of a neighbor cell (white star). Scale bar is 20 µm. B: Relative quantitative red fluorescence intensity of total 30 dividing cells and 90 neighbor cells were measured. Data were represented as mean±se ** p<0.05, one-way ANOVA (ANOVA).
Figure 2.

TAZ protein expression is upregulated in mitotic cells at various cell cycle stage. A and B: Conjunctiva epithelial cells were stained by TAZ (red) after treating with hydrochloride for 15 min (A) or 40 min (B). Counterstaining with DAPI (blue) was performed as an indicator of cells at different cell cycle stages. White arrows indicate the cells which were undergoing cell division. Scale bar is 20 µm. C: TAZ siRNA induced less 10–4-cyclin E promoter transcription activities in NHC cell lines in a dosage dependent manner. Data have been represented as mean±se from three replicates.
BrdU analysis was performed to further examine whether TAZ protein is upregulated in proliferating conjunctiva epithelial cells. We observed that in BrdU positive cells, TAZ protein was found clustering in the nucleus of the cells, while in cells that were BrdU negative, TAZ protein was predominantly expressed in the cytoplasm (Figure 3). Moreover, the fluorescence intensity of TAZ signal in BrdU positive cells was also observed to be much higher than that of BrdU negative cells. This result is consistent with our previous observation that TAZ protein was highly upregulated during epithelial cell division and proliferation; therefore, the activation and translocation of TAZ protein are closely associated with epithelial cell proliferation.
Figure 3.
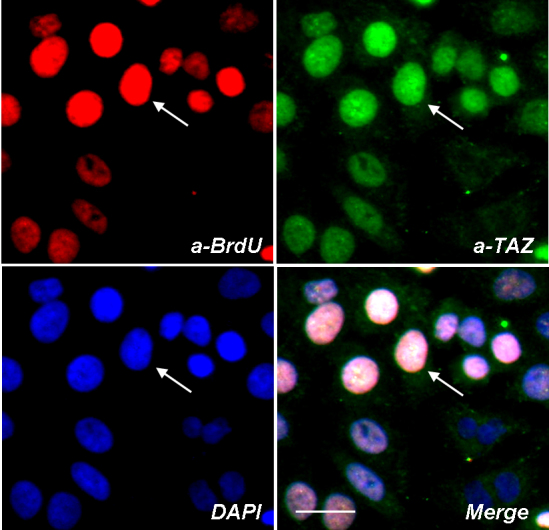
TAZ protein clusters at the nucleus of proliferating epithelial cells. Conjunctiva epithelial cells were labeled for BrdU (red) and TAZ (green), DAPI was applied as nucleus counter staining dye (blue). White arrows indicate a BrdU positive epithelial cell also stained by high intensity of TAZ protein. Scale bar is 20 µm.
TAZ regulates TGFβ signaling
It was recently reported that TAZ is responsive to TGFβ cytokine, forming a complex with Smad protein at the cytoplasm, which facilitated the nucleus translocation of Smad proteins in human embryonic stem cells [21]. In our study, we aimed to investigate whether the TAZ signal in human conjunctiva epithelial cells is also responsive to TGFβ. Our data showed that the fluorescence intensity of endogenous TAZ signal in epithelial cells that were treated with TGFβ is higher than that of cells without TGFβ treatment (Figure 4A). Smad4 staining was simultaneously performed as an indicator of the cells that were responsive to TGFβ. The relative quantitative fluorescence intensity in the nucleus of the epithelial cells was measured (Figure 4B). A total of 100 cells from three repeated experiments were analyzed. We found that after TGFβ treatment, fluorescent intensity of TAZ protein in the cell nucleus is about 2.5±0.3 times higher than that of untreated cells (Figure 4B). Western blot analysis further confirmed that the TAZ protein expression was upregulated in TGFβ treated cells (Figure 4C). Altogether, these results indicate that TAZ protein is responsive to TGFβ stimuli in human conjunctiva epithelial cells.
Figure 4.

TAZ is responsive to TGFβ signal in conjunctiva epithelial cells. A: Fluorescent signals of TAZ protein (green) and smad4 (red) were upregulated after TGFβ treatment. Scale bar is 20 µm. B: Relative quantitation of the green fluorescent intensity of TAZ protein. Data were represented as mean±se, * p<0.01, ANOVA. C: Cell lysate of the epithelial cells with or without TGFβ treatment were blotted by anti-TAZ. Expression of TAZ protein was upregulated in TGFβ treated cells.
TAZ regulates conjunctiva epithelial cell proliferation via TGFβ signaling
We initially did western blot and commassie brilliant blue staining to confirm the effect of TAZ siRNA. Western blot images (Figure 5A) indicated that there was down-regulation of TAZ gene expression by TAZ siRNA. Commassie brilliant blue staining images (Figure 5B) indicated that TAZ silencing induced lower conjunctiva cell attachment and cell growth. We then sought to investigate whether TAZ regulates the growth rate of NHC cells by interacting with a TGFβ signal. NHC is an immortalized cell line, which was derived from the normal human conjunctiva epithelium and shared a lot of characteristics with primary conjunctiva epithelial cells [22]. TGFβ receptor I antibody was added to block the signal transduction of TGFβ. We found that silencing of the TAZ gene significantly suppressed the proliferation rate of NHC cells (Figure 6A, “a”). However, this suppression was rescued by the treatment of anti-TGFβ Receptor I antibody (10 µg/ml, Figure 6A, “b”). Our data indicated that TAZ regulated the epithelial cell proliferation by interacting with the TGFβ signal. NHC cells were also treated with the specific TGFβ receptor inhibitor, SB431542. Silencing of the TAZ gene suppressed the cell growth rate of NHC cells and this effect was significantly rescued by SB431542 (10 nm, Figure 6B).
Figure 5.

TAZ siRNA induced a depletion of TAZ protein in NHC keratinocytes. A: Anti-TAZ blotting of the NHC cells which were treated by 15 PM TAZ siRNA or mock siRNA. B: Pictures show representative fields of cells treated by mock or TAZ targeting siRNA. NHC cells were stained by commassie brilliant blue. Scale bar is 10 um.
Figure 6.

TGFβ receptor antibody and TGFβ receptor inhibitor rescued the effect of TAZ siRNA on cell growth. A: NHC cells were treated with TGFβ receptor type I antibody together with mock siRNA or TAZ siRNA. Relative cell density was analyzed from three replicates. a: a significant reduction of cell density after TAZ siRNA treatment. “b”: the cell growth inhibition was rescued by TGFβ receptor I antibody treatment. B: NHC cells were treated with TGFβ receptor inhibitor (SB431542) together with mock siRNA or TAZ siRNA. Relative cell density was analyzed from three replicates. “a’”: a significant reduction of cell density after TAZ siRNA treatment. “b’”: cell growth inhibition was rescued by SB431542. Data were represented as mean±se (p<0.05).
Expression of TAZ in pterygium tissue
We then examined the expression of TAZ protein in human normal conjunctiva epithelium and pterygium conjunctiva epithelium. TAZ protein was blotted from the total lysate of the epithelial layers of conjunctiva tissues. We found that TAZ expression in pterygium tissue was much higher than that of normal tissue (Figure 7A). Intensity of the blotting bands showed that the expression level of TAZ protein in pterygium epithelium is around 10 times higher than that of the normal conjunctiva tissue (Figure 7B). Immunostaining of the pterygium conjunctiva tissue revealed that TAZ protein is highly clustering in the epithelial layer of conjunctiva tissue (Figure 7C).
Figure 7.

Location and expression of TAZ proteins in normal or pterygium conjunctiva epithelium. A: Total tissue lysate from normal or human conjunctiva tissue with pterygium disease were blotted with anti-TAZ (A). Relative intensity of the blots was measured from three replicates (B). C: Conjunctiva tissue was stained by anti-TAZ (red) and DAPI (blue). Merged images show that the TAZ protein is expressed at the epithelial layer of conjunctiva tissue. Scale bar is 10 µm.
Discussion
The TGFβ signaling pathway controls a diverse set of cellular activities and is regulated by various receptors and transcription factors. Here, we addressed the role of TAZ in the regulation of conjunctiva epithelial cell proliferation by interacting with the TGFβ signaling pathway. TAZ protein expression was found to be upregulated in the mitotic conjunctiva epithelial cells and pterygium epithelium, which is a novel observation and has never been reported before. Moreover, TGFβ receptor antibody and TGFβ receptor inhibitor partially rescued cell growth inhibition, which was induced by TAZ siRNA. Hence, TAZ is involved in regulating conjunctiva cell proliferation via the TGFβ pathway.
Previous studies revealed that Smad family protein functions by directly binding with DNA promoters and consensus fragments resulting in the activation of target genes [23-25]. A growing number of DNA binding factors, such as Fas-activated serine/threonine kinase (FAST), activator protein 1 (AP1)-containing elements and transcription factor E (TFE), as well as TAZ, have been identified to facilitate the Smad-induced gene activation [26]. In our study, we found the co-localization of TAZ protein with DAPI signal in mitotic cells, which indicated that TAZ is related to cell cycle regulation and TAZ protein is a transcriptional factor that may act by binding with Smad and DNA promoters or consensus after stimulation.
Several WW domain containing ubiquitin ligases have been identified to negatively regulate the TGFβ signal by targeting and degradation of Smad family proteins [26,27]. The HECT-type WW domain containing ubiquitin ligase, Smurf2, was reported to down-regulate the TGFβ signal by binding with the proline-rich domain of Smad7 and form a complex with TGFβ receptors [28-30]. Smurf2 was also reported to degrade the receptor binding Smad protein, Smad 1 [31]. Another receptor binding Smad protein, Smad2, was also found to be able to form assembly with Smurf2 and be degraded by Smurf2 [32,33], while the co-Smad, Smad4, can be targeted indirectly by Smurf2 and be degraded simultaneously with R-Smad and I-Smad [34]. The WW domain containing protein, TAZ, shares a similar structure with smurf2 [19]. TAZ functions by binding with Smad family proteins and TAZ antagonised the TGFβ induced cell growth inhibition. Altogether, these findings indicate that TAZ and smurf family proteins probably serve separate but perhaps overlapping functions. Hence, TAZ may be an ubiquitin co-activator that negatively regulates the TGFβ/Smad signaling pathway to regulate mammalian cell proliferation.
Many tumor cells are resistant to TGFβ induced apoptotic effect. The mechanisms are reported to be associated with the mutation of the receptors or components of the Smad pathways [35,36]. TGFβ has a biphasic role in tumor genesis. In the initial phases it acts as a tumor suppressor, whereas during the late phases it has been shown to function as a tumor promoter [4]. TGFβ stimulates the de-differentiation of epithelial cells into malignant invasive and metastatic fibroblastic cells. This procedure was regulated by various co-factors. Recently, Yes-associated protein 1 (YAP1), a protein that shares its homological structure with TAZ, was reported to play a critical role in regulating the expression of cancer gene tumor protein 73 (p73) and in regulating tumor cell migration [37]. These reports indicate that other transcriptional co-factors, such as TAZ, may also interact with the TGFβ signal and regulate cancer cell migration and invasion. In our studies, we found that TAZ expression was upregulated in the epithelial tissue with pterygium disease compared with that of the normal conjunctiva tissue (Figure 6). The pterygium is studied as a benign tumor whose genotype is highly associated with the mutation of tumor suppressor gene p53 [38,39]. Our findings at least indicated that TAZ is possibly involved in the pathology of pterygium by regulating the TGFβ signal. Further mechanism and in vivo studies are necessary to clarify this.
In conclusion, we found that TAZ regulates human conjunctiva epithelial cell proliferation via inhibiting TGFβ signaling. We also found that TAZ is upregulated in pterygium tissue. This finding provides an important clue to the mechanisms of TGFβ signaling in the regulation of conjunctiva cell proliferation and pterygium progression. Furthermore, TAZ may also play an important role in the pathogenesis of other ocular diseases, such as TGFβ related fibrogenesis diseases, corneal wound healing and protein aggregation.
Acknowledgments
We could like to acknowledge the technical support from Dr Wu Hong, Singapore Eye Research Institute, Singapore.
References
- 1.Golu T, Mogoanta L, Streba CT, Pirici DN, Malaescu D, Mateescu GO, Mutiu G. Pterygium: histological and immunohistochemical aspects. Rom J Morphol Embryol. 2011;52:153–8. [PubMed] [Google Scholar]
- 2.Kase S, Takahashi S, Sato I, Nakanishi K, Yoshida K, Ohno S. Expression of p27(KIP1) and cyclin D1, and cell proliferation in human pterygium. Br J Ophthalmol. 2007;91:958–61. doi: 10.1136/bjo.2006.110387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 4.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–21. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 5.Ten Dijke P, Goumans MJ, Itoh F, Itoh S. Regulation of cell proliferation by Smad proteins. J Cell Physiol. 2002;191:1–16. doi: 10.1002/jcp.10066. [DOI] [PubMed] [Google Scholar]
- 6.Cui W, Fowlis DJ, Cousins FM, Duffie E, Bryson S, Balmain A, Akhurst RJ. Concerted action of TGF-beta 1 and its type II receptor in control of epidermal homeostasis in transgenic mice. Genes Dev. 1995;9:945–55. doi: 10.1101/gad.9.8.945. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Alexander V, Vijayachandra K, Bhogte E, Diamond I, Glick A. Conditional epidermal expression of TGFbeta 1 blocks neonatal lethality but causes a reversible hyperplasia and alopecia. Proc Natl Acad Sci USA. 2001;98:9139–44. doi: 10.1073/pnas.161016098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang XJ, Liefer KM, Tsai S, O'Malley BW, Roop DR. Development of gene-switch transgenic mice that inducibly express transforming growth factor beta1 in the epidermis. Proc Natl Acad Sci USA. 1999;96:8483–8. doi: 10.1073/pnas.96.15.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Attisano L. Mutations in the tumor suppressors Smad2 and Smad4 inactivate transforming growth factor beta signaling by targeting Smads to the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 2000;97:4820–5. doi: 10.1073/pnas.97.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obata H, Kaji Y, Yamada H, Kato M, Tsuru T, Yamashita H. Expression of transforming growth factor-beta superfamily receptors in rat eyes. Acta Ophthalmol Scand. 1999;77:151–6. doi: 10.1034/j.1600-0420.1999.770207.x. [DOI] [PubMed] [Google Scholar]
- 11.Pasquale LR, Dorman-Pease ME, Lutty GA, Quigley HA, Jampel HD. Immunolocalization of TGF-beta 1, TGF-beta 2, and TGF-beta 3 in the anterior segment of the human eye. Invest Ophthalmol Vis Sci. 1993;34:23–30. [PubMed] [Google Scholar]
- 12.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, Yaffe MB. TAZ: a novel transcriptional co-activator regulated by interactions with 14–3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–91. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuijpers TW, Maianski NA, Tool AT, Becker K, Plecko B, Valianpour F, Wanders RJ, Pereira R, Van Hove J, Verhoeven AJ, Roos D, Baas F, Barth PG. Neutrophils in Barth syndrome (BTHS) avidly bind annexin-V in the absence of apoptosis. Blood. 2004;103:3915–23. doi: 10.1182/blood-2003-11-3940. [DOI] [PubMed] [Google Scholar]
- 14.Valianpour F, Mitsakos V, Schlemmer D, Towbin JA, Taylor JM, Ekert PG, Thorburn DR, Munnich A, Wanders RJ, Barth PG, Vaz FM. Monolysocardiolipins accumulate in Barth syndrome but do not lead to enhanced apoptosis. J Lipid Res. 2005;46:1182–95. doi: 10.1194/jlr.M500056-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Hergovich A, Hemmings BA. TAZ-mediated crosstalk between Wnt and Hippo signaling. Dev Cell. 2010;18:508–9. doi: 10.1016/j.devcel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, Wrana JL, Attisano L. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–91. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–83. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui CB, Cooper LF, Yang X, Karsenty G, Aukhil I. Transcriptional coactivation of bone-specific transcription factor Cbfa1 by TAZ. Mol Cell Biol. 2003;23:1004–13. doi: 10.1128/MCB.23.3.1004-1013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, Hopkins N, Yaffe MB. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–8. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 20.Hong JH, Yaffe MB. TAZ: a beta-catenin-like molecule that regulates mesenchymal stem cell differentiation. Cell Cycle. 2006;5:176–9. doi: 10.4161/cc.5.2.2362. [DOI] [PubMed] [Google Scholar]
- 21.Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–48. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 22.Diebold Y, Calonge M, Enriquez de Salamanca A, Callejo S, Corrales RM, Saez V, Siemasko KF, Stern ME. Characterization of a spontaneously immortalized cell line (IOBA-NHC) from normal human conjunctiva. Invest Ophthalmol Vis Sci. 2003;44:4263–74. doi: 10.1167/iovs.03-0560. [DOI] [PubMed] [Google Scholar]
- 23.Johnson K, Kirkpatrick H, Comer A, Hoffmann FM, Laughon A. Interaction of Smad complexes with tripartite DNA-binding sites. J Biol Chem. 1999;274:20709–16. doi: 10.1074/jbc.274.29.20709. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–8. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 25.Shi Y, Wang YF, Jayaraman L, Yang H, Massague J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94:585–94. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 26.Massagué J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–44. [PubMed] [Google Scholar]
- 27.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–93. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 28.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–80. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 29.Tajima Y, Goto K, Yoshida M, Shinomiya K, Sekimoto T, Yoneda Y, Miyazono K, Imamura T. Chromosomal region maintenance 1 (CRM1)-dependent nuclear export of Smad ubiquitin regulatory factor 1 (Smurf1) is essential for negative regulation of transforming growth factor-beta signaling by Smad7. J Biol Chem. 2003;278:10716–21. doi: 10.1074/jbc.M212663200. [DOI] [PubMed] [Google Scholar]
- 30.Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, Forman-Kay JD. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–62. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2001;98:974–9. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3:587–95. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- 33.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–22. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 34.Morén A, Imamura T, Miyazono K, Heldin CH, Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280:22115–23. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- 35.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–29. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 36.Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22–9. doi: 10.1016/s0959-437x(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 37.Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O'Neill E. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962–75. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai TJ, Tsai YY, Cheng YW, Chiang CC, Lee H, Chou MC, Chang JH. An association between BPDE-like DNA adduct levels and P53 gene mutation in pterygium. Mol Vis. 2006;12:1687–91. [PubMed] [Google Scholar]
- 39.Tsai YY, Cheng YW, Lee H, Tsai FJ, Tseng SH, Chang KC. P53 gene mutation spectrum and the relationship between gene mutation and protein levels in pterygium. Mol Vis. 2005;11:50–5. [PubMed] [Google Scholar]


