Mitochondria are the bioenergetic centers of eukaryotic cells that produce ATP through oxidative phosphorylation. A byproduct of oxidative phosphorylation is the generation of reactive oxygen species (ROS). Cancer cells exhibit high basal levels of oxidative stress due to activation of oncogenes, loss of tumor suppressors, and the effects of the tumor microenvironment [1]. A large body of evidence suggests important roles for oxygen free radicals in the mutagenesis that drives carcinogenesis [2], the expansion of tumor clones, and the acquisition of malignant properties [3].
Several studies have reported that a high frequency of clonal and, therefore, selected mitochondrial mutations are present in a variety of human tumors [4]–[15]. Mitochondrial DNA (mtDNA) mutations in cancer cells include intragenic deletions, missense and chain-termination point mutations, and alterations of homopolymeric sequences that result in frameshift mutations [16], [17]. The biological impact of a given mutation may vary, depending on the proportion of mutant mtDNAs carried by the cell. The assumption from these studies is that this high frequency of clonal mutations arises from the ROS produced in mitochondria by the escape of oxygen free radicals during oxidative phosphorylation, and that these mutations play a role in driving cancer (Figure 1). Therefore, genomic instability of mitochondria was thought to be a hallmark of cancer.
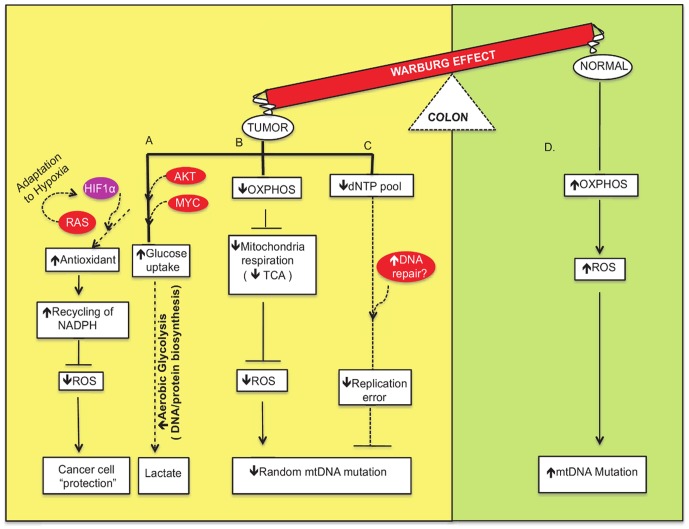
Figure 1. Network modeling of the interconnections among the crucial factors involved in metabolic flow and signaling pathways of the Warburg effect to “protect” the cancer cells.
(A) Cancer cells make use of their nutrient-rich environment by taking in glucose and converting it into molecular precursors by aerobic glycolysis (shown in yellow). This is mediated by activation of proto-oncogenes such as AKT and MYC and other genes in important growth factor signaling pathways. Moreover, RAS-mediated activation of HIF1 induces adaptation to hypoxic environments and promotes “niches” that are conducive to cancer cells. In addition, the use of antioxidants and recycling of NADPH as defense mechanisms to sequester ROS favor the survival of cancer cells. (B) Oxidative phosphorylation (OXPHOS) impairment leads to crippled mitochondrial respiration. The dysfunctional TCA cycle generates fewer reactive oxygen species that may or may not induce DNA damage, and this subsequently leads to fewer mitochondrial DNA mutations. (C) Efficient repair of mtDNA leads to fewer mutations and less mitochondrial dysfunction. (D) In contrast, normal, low proliferative cells utilize OXPHOS and generate ROS that induce mtDNA damage and increase mutation frequency (shown in green).
Cancer arises from a mutator phenotype and it has been established that the random mutation rate of the nuclear DNA of tumors is quite high [18]. However, little is known about the random mutation rates of mitochondria derived from tumors. In this issue of PLoS Genetics, Ericson and colleagues [19] report their striking finding that the random mutation frequency of colorectal tumor mtDNA is significantly lower than that of nuclear DNA or of mtDNA in the surrounding normal tissue. These results were obtained using the random mutation capture assay that was developed to measure random, and not clonal, mutations [19]. This group also shows that the mitochondria from the colon tumors use aerobic glycolysis rather than oxidative phosphorylation to generate energy for the cells. Importantly, this metabolic shift from oxidative phosphorylation to aerobic glycolysis is likely to produce fewer ROS that damage mtDNA and lead to mutagenesis, mitochondrial dysfunction, and cell death.
Seven decades ago, Warburg discovered that mitochondria in cancer cells metabolize glucose by aerobic glycolysis and suggested that this was a result of impaired mitochondrial function that contributes to tumorigenesis [20], [21]. Recently, Thompson and colleagues suggested that mitochondrial function itself is not impaired in cancer cells that metabolize glucose by aerobic glycolysis, and that this type of anabolic metabolism is critical for the production of essential cellular building blocks including dNTPs, amino acids, and lipids [22] (Figure 1). Growth factor signaling by activated AKT, Myc, and other proto-oncogenes results in altered mitochondrial metabolism [22]. For example, a majority of human tumors harbor mutations in the AKT gene, and activated AKT enhances glucose uptake, allowing cells to maintain a higher than adequate level of ATP [23]. What is unknown is whether the colon tumors that were characterized by the Bielas group [19] harbor activated proto-oncogenes. Nevertheless, the picture that now emerges is that aerobic glycolysis enhances growth of cancer cells by anabolic metabolism without producing high levels of ROS that could inactivate the power supply of the cell.
Other explanations for the low frequency of random mitochondrial mutations in colon tumors include highly efficient DNA repair (for excellent review see [24]) and the coupling of antioxidant proteins to the NADPH/NADP balance (Figure 1). Recent studies have demonstrated that mtDNA is repaired by a variety of mechanisms, including short- and long-patch base excision repair, mismatch repair, and homologous recombination [24]–[27]. The sanitation of dNTPs is also likely to result in fewer mutations. Antioxidant proteins may also play a role in the low random mutation rate observed in mitochondria. Proteins such as reduced glutathione (GSH) or thioredoxin that are closely coupled to the NADPH/NADP balance can inactivate ROS [28], [29]. The presence of NADPH and other free radical scavengers may be more pronounced in the colorectal tumor environment to neutralize the impact of oxidative stress (Figure 1).
What are the implications of the findings of Ericson et al. [19] for future cancer therapy strategy or biomarker discovery? Cancer cells exhibit increased uptake of glucose, which increases the bioenergetic competence required of malignant cells. This metabolic feature has led to the hypothesis that inhibition of glycolysis may abolish ATP and important precursor generation in cancer cells and thus may preferentially kill the malignant cells [30], [31]. Nuclear genetic and epigenetic changes have been the cornerstone of such studies. In addition to these strategies, it is likely that mutated genes that function to alter mitochondrial metabolic competence in tumors, including HIF-1 and MYC, will emerge as new drug targets and molecular markers of prognosis and responses to therapy. In addition, targeting mtDNA repair proteins could serve as a potential alternative approach to kill cancer cells. The work of the Ericson et al. [19] adds significantly to our understanding of the roles of mitochondria in supporting the growth of tumors. In combination with recent findings regarding mitochondrial metabolism in cancer cells, this groundbreaking finding suggests that novel drugs that target the powerhouse of cancer cells are likely to make a significant impact on cancer treatment.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by NIH P01CA129186 (JBS) and NCI 1 K01 CA15485401A (DK). The funders had no role in the preparation of the article.
References
- 1.Cairns RA, Harris I, McCracken S, Mak TW. Cancer cell metabolism. Cold Spring Harb Symp Quant Biol. 2011 doi: 10.1101/sqb.2011.76.012856. E-pub ahead of print 12 December 2011. [DOI] [PubMed] [Google Scholar]
- 2.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 3.Moolgavkar SH, Luebeck EG. Multistage carcinogenesis and the incidence of human cancer. Genes Chromosomes Cancer. 2003;38:302–306. doi: 10.1002/gcc.10264. [DOI] [PubMed] [Google Scholar]
- 4.Abu-Amero KK, Alzahrani AS, Zou M, Shi Y. High frequency of somatic mitochondrial DNA mutations in human thyroid carcinomas and complex I respiratory defect in thyroid cancer cell lines. Oncogene. 2005;24:1455–1460. doi: 10.1038/sj.onc.1208292. [DOI] [PubMed] [Google Scholar]
- 5.Alonso A, Martin P, Albarran C, Aquilera B, Garcia O, et al. Detection of somatic mutations in the mitochondrial DNA control region of colorectal and gastric tumors by heteroduplex and single-strand conformation analysis. Electrophoresis. 1997;18:682–685. doi: 10.1002/elps.1150180504. [DOI] [PubMed] [Google Scholar]
- 6.Liu VW, Shi HH, Cheung AN, Chiu PM, Leung TW, et al. High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Res. 2001;61:5998–6001. [PubMed] [Google Scholar]
- 7.Montanini L, Regna-Gladin C, Eoli M, Albarosa R, Carrara F, et al. Instability of mitochondrial DNA and MRI and clinical correlations in malignant gliomas. J Neurooncol. 2005;74:87–89. doi: 10.1007/s11060-004-4036-5. [DOI] [PubMed] [Google Scholar]
- 8.Nagy A, Wilhelm M, Kovacs G. Mutations of mtDNA in renal cell tumours arising in end-stage renal disease. J Pathol. 2003;199:237–242. doi: 10.1002/path.1273. [DOI] [PubMed] [Google Scholar]
- 9.Nomoto S, Yamashita K, Koshikawa K, Nakao A, Sidransky D. Mitochondrial D-loop mutations as clonal markers in multicentric hepatocellular carcinoma and plasma. Clin Cancer Res. 2002;8:481–487. [PubMed] [Google Scholar]
- 10.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Cespedes M, Ahrendt SA, Piantadosi S, Rosell R, Monzo M, et al. Chromosomal alterations in lung adenocarcinoma from smokers and nonsmokers. Cancer Res. 2001;61:1309–1313. [PubMed] [Google Scholar]
- 12.Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res. 2002;62:972–976. [PubMed] [Google Scholar]
- 13.Wu CW, Yin PH, Hung WY, Li AF, Li SH, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer. 2005;44:19–28. doi: 10.1002/gcc.20213. [DOI] [PubMed] [Google Scholar]
- 14.Modica-Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion. 2004;4:755–762. doi: 10.1016/j.mito.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 15.Singh KK, Kulawiec M, Still I, Desouki MM, Geradts J, et al. Inter-genomic cross talk between mitochondria and the nucleus plays an important role in tumorigenesis. Gene. 2005;354:140–146. doi: 10.1016/j.gene.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 16.Copeland WC, Wachsman JT, Johnson FM, Penta JS. Mitochondrial DNA alterations in cancer. Cancer Invest. 2002;20:557–569. doi: 10.1081/cnv-120002155. [DOI] [PubMed] [Google Scholar]
- 17.Carew JS, Huang P. Mitochondrial defects in cancer. Mol Cancer. 2002;1:9. doi: 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bielas JH, Loeb LA. Quantification of random genomic mutations. Nat Methods. 2005;2:285–290. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 19.Ericson NG, Kulawiec M, Vermulst M, Sheahan K, O'Sullivan J, et al. Decreased mitochondrial DNA mutagenesis in human colorectal cancer. PLoS Genet. 2012;8:e1002689. doi: 10.1371/journal.pgen.1002689. doi: 10.1371/journal.pgen.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 21.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 22.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 24.Liu P, Demple B. DNA repair in mammalian mitochondria: Much more than we thought? Environ Mol Mutagen. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- 25.Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Fukui H, Moraes CT. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum Mol Genet. 2009;18:1028–1036. doi: 10.1093/hmg/ddn437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraytsberg Y, Schwartz M, Brown TA, Ebralidse K, Kunz WS, et al. Recombination of human mitochondrial DNA. Science. 2004;304:981. doi: 10.1126/science.1096342. [DOI] [PubMed] [Google Scholar]
- 28.Hamanaka RB, Chandel NS. Cell biology. Warburg effect and redox balance. Science. 2011;334:1219–1220. doi: 10.1126/science.1215637. [DOI] [PubMed] [Google Scholar]
- 29.Sattler UG, Mueller-Klieser W. The anti-oxidant capacity of tumour glycolysis. Int J Radiat Biol. 2009;85:963–971. doi: 10.3109/09553000903258889. [DOI] [PubMed] [Google Scholar]
- 30.Izyumov DS, Avetisyan AV, Pletjushkina OY, Sakharov DV, Wirtz KW, et al. “Wages of fear”: transient threefold decrease in intracellular ATP level imposes apoptosis. Biochim Biophys Acta. 2004;1658:141–147. doi: 10.1016/j.bbabio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Pinedo C, Ruiz-Ruiz C, Ruiz de Almodovar C, Palacios C, Lopez-Rivas A. Inhibition of glucose metabolism sensitizes tumor cells to death receptor-triggered apoptosis through enhancement of death-inducing signaling complex formation and apical procaspase-8 processing. J Biol Chem. 2003;278:12759–12768. doi: 10.1074/jbc.M212392200. [DOI] [PubMed] [Google Scholar]