Abstract
Members of the 4.1 superfamily of proteins, including ezrin, moesin, merlin, and willin regulate many normal physiologic processes such as cellular shape, motility, and proliferation. In addition, they contribute both to tumor development and tumor progression. We reported previously that strong cytoplasmic ezrin expression was independently associated with poorer patient survival. One hundred and thirty-one histologically confirmed primary head and neck squamous cell carcinomas were examined prospectively for cancer progression and survival at a large health care center in the Bronx, NY, USA. Immunohistochemical analysis of ezrin, moesin, merlin, and willin expression in tissue microarray samples of primary head and neck squamous cell carcinoma revealed a significant association of increased cytoplasmic ezrin with poor cancer survival. Global RNA analyses suggest that cancers with high cytoplasmic ezrin have a more invasive phenotype. This study supports our previous findings associating cytoplasmic ezrin with more aggressive behavior and poorer outcome and indicates the need for a multi-institutional study to validate the use of cytoplasmic ezrin as a biomarker for treatment planning in head and neck squamous cell carcinoma.
Keywords: Head and neck cancer, Ezrin, Moesin, Willin, Merlin, Immunohistochemistry, Survival
Introduction
Squamous cell carcinoma of the head and neck (HNSCC) is a disease of considerable morbidity and mortality with approximately 50,000 new cases and 11,000 cancer deaths in the United States annually [1]. Previously, we found altered RNA expression of moesin correlating with HNSCC tumor progression [2]. Moesin is a member of the 4.1 superfamily of proteins which are identified by the presence of a 4.1 ezrin, radixin moesin (FERM) domain which can bind both proteins and lipids, and as such, they can function in many normal physiologic processes including cell shape and motility, proliferation and development. In some cases, this family of proteins directly link transmembrane proteins to the cytoskeleton or link kinase and/or phosphatase enzymatic activity to the plasma membrane. Specifically, they have now been shown to be involved in the control of several different signal transduction pathways, including RhoA, Hedgehog, membrane receptor (e.g. Patched, CD43 or CD44) signalling (reviewed in [3]) and, more recently, Hippo pathway signalling [4–6]. In addition to the many binding partners and multiple pathways they control, FERM proteins themselves can be controlled by post-translational modification, such as phosphorylation, and are differentially expressed in normal murine and human tissues [7, 8].
Altered expression of a subset of 4.1 family proteins is believed to contribute to carcinogenesis and metastasis, as exemplified by the following: merlin has been shown to function as a tumor suppressor [9]; ezrin is believed to play a role in the development of metastasis [10, 11]; moesin has been implicated in oral squamous cell carcinomas [12–14]; and willin has been shown to antagonize some of the functions of the YAP oncogene [6]. We have shown previously that a high level of cytoplasmic ezrin correlates or that high levels of cytoplasmic ezrin correlate with poor survival in head and neck squamous cell carcinoma [15], and recent studies show that increased expression of ezrin is also associated with poor clinical outcome in a variety of human cancers [16–21].
In this study, we compare the protein expression of four members of the 4.1. superfamily: ezrin, moesin, merlin, and willin in primary HNSCC and relate the expression of these cytoplasmic proteins with clinical outcome in a prospective cohort of HNSCC patients. We find that high levels of ezrin and moesin expression are associated with poor cancer survival. Subsequent analyses of global RNA expression suggest that a complex interaction of multiple signaling pathways may be driving the observed progression to poorer clinical outcomes.
Materials and Methods
Study Design
This project followed an Institution Review Board approved protocol and is HIPPA-compliant. This cohort consisted of 131 consecutive, prospectively collected primary HNSCC from 128 patients treated at Montefiore Medical Center (MMC) from 2002 onward. Demographic data, detailed smoking and alcohol information, treatment details, and follow-up data were collected prospectively and entered into a clinical database. Snap frozen samples from the primary carcinoma, and adjacent mucosa were collected at initial diagnosis and treatment. Extracted RNA from these samples was used for global expression analysis. The corresponding matched formalin-fixed paraffin embedded (FFPE) tumor samples were retrieved from the pathology files from primary tumor and adjacent mucosa and used to produce tissue microarrays (TMA).
Tissue Microarray Analysis
Tissue microarray blocks were constructed from FFPE tissue using a semi-automatic tissue arrayer (Chemicon) and 1.0 mm cores, which were represented by three or more cores from the areas of interest. Microarray sections were deparaffinized, rehydrated, and washed in TBS (SIGMA tris buffer, T6664).
Slides were pretreated with 0.3% H2O2 for 10 min, (or DAKO s2001) and washed in TBS. Antigen retrieval was performed using pH 6.0 10 mM sodium citrate buffer (Vector, H3300) in a steamer for 20 min, then cooled for 30 min at RT. Slides were blocked in 5% normal goat serum/2% BSA for 1 h at RT before incubating with the following antibodies: ezrin (1/100, Neomarkers, 3C12), willin (1/100 gift from FGM), merlin (1/100, NF-2, Santa Cruz, sc331), and moesin (1/400, Neomarkers, 3837) overnight at 4°C diluted in blocking solution. Slides were washed 4 times, 3 min each with TBS before applying biotin labeled secondary antibody (goat anti-mouse DAKO, E0433(ezrin, moesin) and goat anti-rabbit DAKO, E0432 (willin, merlin)) at 1/500 for 1 h at RT. Slides were washed and incubated for 30 min with the avidin–biotin–HRP complex as directed by DAKO (ABC-HRP K0355). Slides were washed in TBS and DAB (VECTOR, SK 4100) applied for 3 min (merlin) and 1.5 min (ezrin, willin, moesin).
Two pathologists (MBG, DB), blinded to outcome and tissue source, read the microarrays, and achieved a consensus regarding staining pattern and intensity. The level of staining intensity was recorded for the nucleus, cytoplasm, and the cell membrane. Level of expression was scored according to the strongest intensity that comprised at least 10% of each core on a scale of 0 to 3. Mean staining scores were then calculated across multiple core replicates from the same primary tumor biopsy or resection specimen and by cell location (cytoplasm, nucleus and membrane).
Global RNA Expression
In an initial attempt to characterize the tumor phenotype(s) associated with cytoplasmic ezrin expression in HNSCC, we performed whole genome expression analyses on 74 fresh, frozen primary HNSCC tumor samples (10 with high cytoplasmic ezrin and 64 low cytoplasmic ezrin expression as previously determined by IHC in the corresponding FFPE samples). Total RNA was prepared using TRIzol [22]. For each RNA sample, linear amplification and biotin-labeling of total RNA (500 ng) was carried out using the Illumina TotalPrep RNA Amplification Kit (Ambion). Whole-genome expression analysis was carried out by hybridization of amplified RNA to an Illumina HumanHT-12 v3 Expression BeadChip. With this beadchip, we interrogated greater than 48,000 probes per sample; targeting genes and known alternative splice variants from the RefSeq database release 17 and UniGene build 188. Controls for each RNA sample (greater than 1,000 bead types) confirmed sample RNA quality, labeling reaction success, hybridization stringency, and signal generation. The raw data were normalized using quantile–quantile normalization. Even though our samples had low background levels, to minimize the probability of spurious noise generating a false positive, we chose to process the data by requiring a selected feature to have expression levels greater than the 4th quartile of the samples designated background probes in at least 30% of the samples. This reduced our feature set to 9719 elements. Normalized RNA expression data were log2-transformed and a two-tailed t test was performed for each feature.
Statistical Analyses
Contingency tables were generated between high and low ezrin, moesin, willin, and merlin expression and gender, age, race, tumor site, smoking, AJCC TNM stage, nodal status at diagnosis and history of prior primaries. For efficiency and purposes of statistical validation, cases were classified as having strong to weak or no expression based on our previous findings on an earlier population of HNSCC patients [15]. Tumors with strong cytoplasmic ezrin expression, defined as having ≥2.5 mean staining, were classified as positive. Due to smaller numbers of +3 cytoplasmic staining tumors with moesin, willin, and merlin, positive expression for these were based on a +2 cut-off. Various cut-offs (e.g., >0, ≥1 and ≥1.5) were also tested for membranous and nuclear expression. Differences in expression with respect to clinical and demographic characteristics at diagnosis were assessed by two-tailed Chi-square or Fisher exact tests. Time to clinical outcome event was measured from treatment start to the first instance of a local or regional recurrence (LRR) or distant metastases and cancer death, or to the last recorded follow-up visit date for censored subjects. Survival curves were estimated using Kaplan–Meier analyses. Cumulative probability (i.e., 1-survival function) curves for disease progression were generated based on first incidence of LRR or distant metastasis.
We estimated the relative hazard (HR) of each outcome of interest for cytoplasmic ezrin, moesin, willin, and merlin expression by multivariable Cox proportional hazards regression. The potential for confounding was examined for all clinicopathologic factors: age, gender, race, ethnicity, smoking history, alcohol consumption, tumor anatomic site, TNM stage, treatment modality, detection of human papillomavirus (HPV), and method of specimen procurement (biopsy vs. surgical or laser resection). Presence of HPV DNA and p16 expression in tumors were tested for a subset of cases using previously described protocols [23]; tumors not tested were listed as unknown in the multivariable analyses. Empirical confounders were selected based on a ten percent change-in-estimate criterion [24] and subsequently controlled for in all models. We also conducted an exhaustive search for significant predictors. The resulting final multivariable regression models for disease-specific survival were stratified on tumor site, primary treatment, race, HPV, and specimen procurement method, and adjusted further for age, ethnicity, ever smoking status, alcohol drinking, prior primaries, and tumor stage. This allowed a different baseline hazard for each confounder combination without violating proportional hazards assumptions. Proportional hazards assumptions were tested for all multivariable regression models and were not found to be violated. The FERM specific HRs for LRR and distant metastases were stratified on tumor site, primary treatment, race, and HPV, and adjusted further for age, ethnicity, ever smoking status, alcohol drinking, prior primaries, and tumor stage. To test the null hypothesis that the regression coefficient was equal to zero, p-values based on the Wald Chi-square test were computed. Interactions between FERM proteins, and by sub-cellular location, were also examined by generating a cross-product term between each paired combination.
Associated Functional Analyses
For selected RNA, data were analyzed through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). The Functional Analysis of RNA identified the biological functions and/or diseases that were most significant to the data set in Table 5. Right-tailed Fisher’s exact test was used to calculate P values determining the probability that each biological function and/or disease assigned to that data set is due to chance alone. The functional analyses for selected processes related to cancer, proliferation, differentiation, invasion, and cell death are presented in Table 6.
Table 5.
RNA expression of tumor related genes by high versus low cytoplasmic ezrin protein expression in HNSCC
| Gene symbol | Median RNA expression | High/low ezrin expression ratio* | P valuea | Function | |
|---|---|---|---|---|---|
| High ezrin group | Low ezrin group | ||||
| TGFBR2 | 1603 | 880 | 1.82 | 0.00546 | Proliferation |
| MAP2K1 | 1006 | 658 | 1.53 | 0.00106 | Proliferation |
| PTPRK | 911 | 611 | 1.49 | 0.03787 | Proliferation |
| PTPRE | 637 | 444 | 1.43 | 0.00768 | Proliferation |
| PTEN | 682 | 490 | 1.39 | 0.00111 | Proliferation |
| RAB11FIP2 | 377 | 274 | 1.38 | 0.00200 | Proliferation |
| RAF1 | 615 | 454 | 1.35 | 0.00441 | Proliferation |
| KIF2C | 284 | 390 | 0.73 | 0.04469 | Proliferation |
| CEBPA | 467 | 665 | 0.70 | 0.03164 | Proliferation |
| CDT1 | 242 | 368 | 0.66 | 0.02036 | Proliferation |
| RAB3IP | 712 | 1084 | 0.66 | 0.00055 | Proliferation |
| E2F2 | 453 | 732 | 0.62 | 0.02644 | Proliferation |
| MKNK2 | 254 | 420 | 0.61 | 0.00002 | Proliferation |
| PITX1 | 3738 | 6854 | 0.55 | 0.04464 | Proliferation |
| SOX2 | 850 | 1564 | 0.54 | 0.04859 | Proliferation |
| ISG15 | 698 | 1365 | 0.51 | 0.01200 | Proliferation |
| SERPINE2 | 8937 | 2817 | 3.17 | 0.00006 | Invasion |
| EFNB2 | 2222 | 936 | 2.37 | 0.00470 | Invasion |
| ETS1 | 1902 | 922 | 2.06 | 0.00278 | Invasion |
| NET1 | 3397 | 2017 | 1.68 | 0.00546 | Invasion |
| ITGB1 | 2693 | 1654 | 1.63 | 0.00210 | Invasion |
| PLAT | 330 | 484 | 0.68 | 0.00413 | Invasion |
| CELSR3 | 182 | 270 | 0.67 | 0.00859 | Invasion |
| TMPRSS4 | 197 | 309 | 0.64 | 0.00009 | Invasion |
| CX3CL1 | 303 | 479 | 0.63 | 0.03848 | Invasion |
| CRB3 | 239 | 321 | 0.74 | 0.01760 | Differentiation |
| DTX2 | 1510 | 2049 | 0.74 | 0.02208 | Differentiation |
| DHCR24 | 498 | 684 | 0.73 | 0.02243 | Differentiation |
| CNN2 | 1143 | 1590 | 0.72 | 0.04610 | Differentiation |
| FXYD3 | 175 | 248 | 0.71 | 0.00220 | Differentiation |
| CLDN7 | 801 | 1187 | 0.68 | 0.04948 | Differentiation |
| NOTCH1 | 647 | 1105 | 0.59 | 0.02216 | Differentiation |
| ELF3 | 510 | 877 | 0.58 | 0.04982 | Differentiation |
| KRT15 | 780 | 2453 | 0.32 | 0.02801 | Differentiation |
| BIRC2 | 1673 | 1141 | 1.47 | 0.00282 | Cell death |
| TGM2 | 481 | 241 | 1.99 | 0.00087 | Cell death |
| TNFRSF21 | 4472 | 2691 | 1.66 | 0.00487 | Cell death |
| TNFRSF1A | 1918 | 1354 | 1.42 | 0.01322 | Cell death |
| CCDC50 | 1258 | 908 | 1.39 | 0.00353 | Cell death |
| MAP3K7 | 1098 | 797 | 1.38 | 0.00498 | Cell death |
| EDARADD | 223 | 322 | 0.69 | 0.00009 | Cell death |
| P8 | 231 | 336 | 0.69 | 0.02489 | Cell death |
| NOL3 | 174 | 264 | 0.66 | 0.00867 | Cell death |
| PDZK1IP1 | 250 | 431 | 0.58 | 0.02960 | Cell death |
* RNA expression ratio between high/low cytoplasmic ezrin HNSCC
aP value by two-tailed t-test using log2 transformed expression data
Table 6.
IPA identified functional categories of differentially expressed genes based on high versus low cytoplasmic ezrin
| Function | Molecules | # molecules | P value |
|---|---|---|---|
| Cell death | BIRC2, CEBPA, CX3CL1, DHCR24, E2F2, EFNB2, ETS1, ITGB1, MAP2K1, MAP3K7, NOL3, NOTCH1, PLAT, PTEN, RAF1, SERPINE2, TGFBR2, TGM2, TNFRSF21, TNFRSF1A | 20 | 1.54E−10 |
| Proliferation | CEBPA, CNN2, DHCR24, E2F2, EFNB2, ELF3, ETS1, ISG15, ITGB1, KIF2C, MAP2K1, MAP3K7, NOTCH1, PDZK1IP1, PLAT, PTEN, PTPRE, PTPRK, RAF1, SERPINE2, SOX2, TGFBR2, TGM2, TNFRSF21, TNFRSF1A | 25 | 4.71E−10 |
| Invasion | CNN2, CX3CL1, EFNB2, ETS1, ITGB1, MAP2K1, MAP3K7, NOTCH1, PLAT, PTEN, PTPRK, RAF1, SERPINE2, TGFBR2, TGM2, TNFRSF21, TNFRSF1A | 17 | 5.47E−08 |
| Differentiation | BIRC2, CEBPA, CX3CL1, DHCR24, EFNB2, ITGB1, MAP3K7, NOTCH1, PITX1, PTEN, SOX2, TGFBR2, TGM2, TNFRSF1A | 14 | 1.01E−07 |
| Cancer | BIRC2, CEBPA, CX3CL1, DHCR24, E2F2, EFNB2, ETS1, FXYD3, ISG15, ITGB1, MAP2K1, NOTCH1, PLAT, PTEN, PTPRE, RAF1, SERPINE2, SOX2, TGFBR2, TMPRSS4, TNFRSF1A | 21 | 1.80E−04 |
Results
Patient Cohort
One hundred and thirty-one histologically confirmed HNSCC primaries were examined from a total of 128 patients undergoing primary treatment at MMC. The mean age of the patients was 62 years (±12.4 standard deviation) with a range of 25–91 years. The majority of cases were male (70%) and smokers (82%; Table 1). Mean follow-up time was 52.4 months (for a cohort total of 6,866 months combined). During follow-up, 65 cases (54%) died after an average of 25.6 months following diagnosis, of which 22 (33.8%) were directly attributed to their cancer. Thirty-two cases (24.4%) developed a recurrence and/or distant metastasis following treatment after an average period of 16.5 months.
Table 1.
Clinicopathologic characteristics of HNSCC (n = 131)
| Clinicopathologic characteristic | Category | N (%)* |
|---|---|---|
| Age at diagnosis | <60 | 49 (37%) |
| ≥60 | 82 (63%) | |
| Gender* | Female | 39 (30%) |
| Male | 89 (70%) | |
| Smoking* | Never | 23 (18%) |
| Ever | 105 (82%) | |
| Sitea | Lip/oral cavity | 43 (33%) |
| Pharynx | 53 (40%) | |
| Larynx | 35 (27%) | |
| Tissue examined | Biopsies | 46 (35%) |
| Surgical resection | 60 (46%) | |
| Laser resection | 25 (19%) | |
| Stageb | I | 15 (11%) |
| II | 15 (11%) | |
| III | 27 (21%) | |
| IV | 73 (56%) | |
| T statusb | 1 | 25 (19%) |
| 2 | 35 (27%) | |
| 3 | 30 (23%) | |
| 4 | 40 (31%) | |
| N statusb | 0 | 55 (42%) |
| 1 | 20 (15%) | |
| 2 | 52 (40%) | |
| 3 | 3 (2%) | |
| Primary Treatment modalityc | Surgical resection only | 39 (30%) |
| Surgery plus therapy | 56 (43%) | |
| Chemo/radiation | 33 (25%) | |
| Index primary examineda | First | 113 (86%) |
| Second | 18 (14%) | |
| P16 expressiond | Negative | 20 (19%) |
| Positive | 86 (81%) | |
| HPV DNAd | Negative | 75 (75%) |
| Positive | 25 (25%) |
* Total based on 128 patients (123 with ezrin, moesin or merlin staining, and 122 with willin staining)
aThree patients had multiple primary tumors involving more than one location
bTNM staging was not performed on one primary
cThree patients refused treatment
dTotal based on 106 with p16 immunohistochemistry and 100 with HPV in situ hybridization results
Eighteen (14%) patients had a previous history of cancer. History of prior primaries was adjusted for in all multivariable models. We also conducted sensitivity analyses excluding these cases; no differences in association were observed after restriction.
Expression of FERM Proteins
Table 2 summarizes the mean expression patterns of the FERM proteins for this cohort. With respect to anatomic site, strong ezrin expression was observed more often in tumors of the pharynx although the difference was not significant (P = 0.689), whereas strong cytoplasmic moesin (P = 0.326), willin (P = 0. 024), and merlin (P = 0. 218) expression were found most often in oral cavity tumors. There were no significant differences in FERM expression between advanced and lower TNM stage tumors, except for merlin, which was lower in metastatic tumors (P = 0.037; Table 3). Somewhat higher cytoplasmic ezrin expression was observed in tumors from younger patients (P = 0.092; Fig. 1 histology photomic), and lower cytoplasmic willin expression was detected among HPV positive tumors (P = 0.011).
Table 2.
FERM expression patterns of HNSCC
| FERM protein | Expression | Cytoplasmic | Membranous | Nuclear | |||
|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | ||
| Ezrin (n = 130)* | 0–0.49 | 6 | 5 | 56 | 43 | 114 | 88 |
| 0.5–1.49 | 47 | 36 | 36 | 28 | 2 | 2 | |
| 1.5–2.49 | 59 | 45 | 23 | 18 | 2 | 2 | |
| 2.5–3 | 18 | 14 | 15 | 12 | 1 | 1 | |
| Moesin (n = 130) | 0–0.49 | 33 | 25 | 101 | 78 | 129 | 99 |
| 0.5–1.49 | 57 | 44 | 14 | 11 | 1 | 1 | |
| 1.5–2.49 | 34 | 26 | 9 | 7 | 0 | 0 | |
| 2.5–3 | 6 | 5 | 6 | 5 | 0 | 0 | |
| Willin (n = 124) | 0–0.49 | 18 | 15 | 96 | 77 | 25 | 20 |
| 0.5–1.49 | 59 | 48 | 12 | 10 | 26 | 21 | |
| 1.5–2.49 | 38 | 31 | 10 | 8 | 43 | 35 | |
| 2.5–3 | 9 | 7 | 6 | 5 | 30 | 24 | |
| Merlin (n = 123) | 0–0.49 | 6 | 5 | 105 | 85 | 60 | 49 |
| 0.5–1.49 | 46 | 37 | 10 | 8 | 21 | 17 | |
| 1.5–2.49 | 48 | 39 | 6 | 5 | 24 | 20 | |
| 2.5–3 | 23 | 19 | 2 | 2 | 18 | 15 | |
* Ezrin nuclear staining was not discernable for 11 tumors
Table 3.
Distribution of FERM cytoplasmic expression in HNSCC
| Characteristic at diagnosis | Ezrin (n = 130) | Moesin (n = 130) | Willin (n = 124) | Merlin (n = 123) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <2.5 | 2.5/3 | P valuec | <2 | 2/3 | P valuec | <2 | 2/3 | P valuec | ≤2 | >2 | P valuec | |
| Age | ||||||||||||
| <60 | 39 (80%) | 10 (20%) | 0.092 | 42 (86%) | 7 (14%) | 0.787 | 35 (74%) | 12 (26%) | 0.957 | 39 (83%) | 8 (17%) | 0.299 |
| ≥60 | 73 (90%) | 8 (10%) | 68 (84%) | 13 (16%) | 57 (74%) | 20 (26%) | 57 (75%) | 19 (25%) | ||||
| Gender* | ||||||||||||
| Female | 34 (89%) | 4 (11%) | 0.582 | 35 (92%) | 3 (8%) | 0.177 | 29 (76%) | 9 (24%) | 0.768 | 31 (86%) | 5 (14%) | 0.295 |
| Male | 71 (84%) | 14 (16%) | 69 (81%) | 16 (19%) | 62 (74%) | 22 (26%) | 63 (78%) | 18 (22%) | ||||
| Smoking* | ||||||||||||
| Never | 21 (95%) | 1 (5%) | 0.192 | 20 (91%) | 2 (9%) | 0.521 | 19 (86%) | 3 (14%) | 0.188 | 18 (82%) | 4 (18%) | 1.000 |
| Ever | 84 (83%) | 17 (17%) | 84 (83%) | 17 (17%) | 72 (72%) | 28 (28%) | 76 (80%) | 19 (20%) | ||||
| Tumor site | ||||||||||||
| Lip/oral cavity | 38 (88%) | 5 (12%) | 0.689 | 34 (79%) | 9 (21%) | 0.326 | 25 (60%) | 17 (40%) | 0.029 | 29 (69%) | 13 (31%) | 0.233 |
| Pharynx | 43 (83%) | 9 (17%) | 44 (85%) | 8 (15%) | 39 (80%) | 10 (20%) | 40 (83%) | 8 (17%) | ||||
| Larynx | 31 (89%) | 4 (11%) | 32 (91%) | 3 (9%) | 28 (85%) | 5 (15%) | 27 (82%) | 6 (18%) | ||||
| TNM Stagea | ||||||||||||
| I–III | 51 (91%) | 5 (9%) | 0.149 | 49 (88%) | 7 (13%) | 0.409 | 36 (71%) | 15 (29%) | 0.366 | 37 (73%) | 14 (27%) | 0.230 |
| IV | 60 (82%) | 13 (18%) | 60 (82%) | 13 (18%) | 56 (78%) | 16 (22%) | 58 (82%) | 13 (18%) | ||||
| Nodal Statusa | ||||||||||||
| Negative | 48 (89%) | 6 (11%) | 0.429 | 46 (85%) | 8 (15%) | 0.854 | 40 (77%) | 12 (23%) | 0.642 | 35 (69%) | 16 (31%) | 0.037 |
| Positive | 63 (84%) | 12 (16%) | 63 (84%) | 12 (16%) | 52 (73%) | 19 (27%) | 60 (85%) | 11 (15%) | ||||
| Prior primaries | ||||||||||||
| No (1st primary) | 94 (84%) | 18 (16%) | 0.075 | 95 (85%) | 17 (15%) | 1.000 | 81 (75%) | 27 (25%) | 0.594 | 85 (79%) | 22 (21%) | 0.335 |
| Yes | 18 (100%) | 0 (0%) | 15 (83%) | 3 (17%) | 11 (69%) | 5 (31%) | 11 (69%) | 5 (31%) | ||||
| P16 expressionb | ||||||||||||
| Negative | 18 (90%) | 2 (10%) | 0.731 | 18 (90%) | 2 (10%) | 0.514 | 12 (60%) | 8 (40%) | 0.118 | 16 (80%) | 4 (20%) | 0.579 |
| Positive | 72 (85%) | 13 (15%) | 69 (81%) | 16 (19%) | 64 (77%) | 19 (23%) | 59 (72%) | 23 (28%) | ||||
| HPV DNAb | ||||||||||||
| Negative | 65 (88%) | 9 (12%) | 0.733 | 61 (82%) | 13 (18%) | 0.754 | 51 (69%) | 23 (31%) | 0.011 | 54 (74%) | 19 (26%) | 0.678 |
| Positive | 21 (84%) | 4 (16%) | 22 (88%) | 3 (12%) | 22 (96%) | 1 (4%) | 16 (70%) | 7 (30%) | ||||
* Total based on 128 patients (123 with ezrin, moesin or merlin staining, and 122 with willin staining)
aTNM staging was not available for one primary
bTotal based on 106 primaries with p16 and 100 primaries with HPV results
cTwo-tailed P value for Chi-square/Fisher exact test using first row as reference
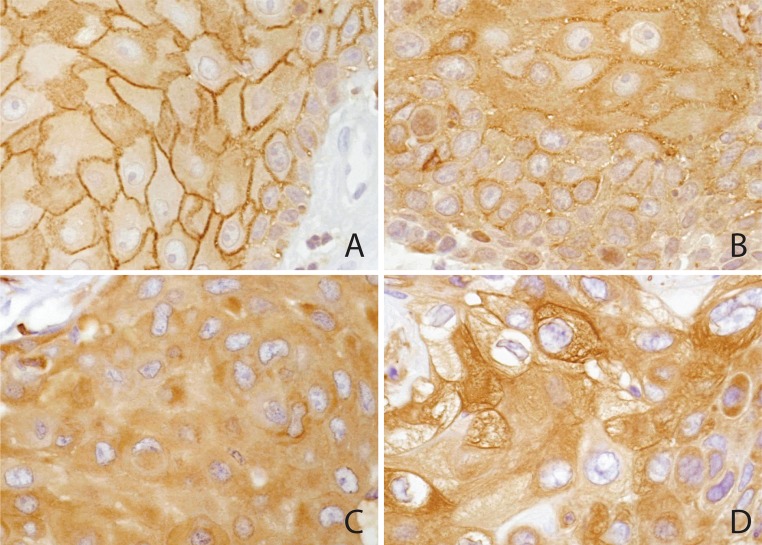
Fig. 1.
Cytoplasmic ezrin. a Ezrin expression in normal squamous mucosa. There is strong staining of the cell membranes of the stratum spinosum, but only faint uniform staining of the cytoplasm. Basal cells at the stromal interface are negative. b Squamous cell carcinoma demonstrating weak (+1) cytoplasmic ezrin localization. Prominent membrane localization is accentuated in the interior of the cell group, recapitulating the stratification of normal epithelium. c Squamous cell carcinoma demonstrating moderate (+2) cytoplasmic ezrin expression, with reduced membrane localization. d Squamous cell carcinoma demonstrating strong (+3) cytoplasmic ezrin expression. Many cells have intensely staining cytoplasmic filaments, while membrane localization is minimal. Two pathologists (MBG, DB), blinded to outcome and tissue source, read the microarrays and achieved a consensus regarding staining pattern and intensity. The level of cytoplasmic staining was scored according to the strongest intensity that comprised at least 10% of each core on a scale of 0–3. Mean staining scores were then calculated across multiple core replicates from the same primary tumor biopsy or resection specimen
Assessment of 5 year cancer survival by Kaplan–Meier univariate analyses showed significant positive (risk) associations with strong cytoplasmic (≥2.5) ezrin (log rank P = 0.0048; Fig. 2). A positive association was also suggested for LRR and incidence of distant metastases with increased cytoplasmic (≥2) moesin expression, although this was not significant (log rank P = 0.0713; Fig. 3).
Fig. 2.
Order of graphs clockwise from top left: a Ezrin expression; b Moesin expression, c Willin expression, and d Merlin expression. Survival time displayed in months truncated at 60 months for illustration. Censored cases shown by tick marks at last recorded follow-up
Fig. 3.
Order of graphs clockwise from top left: a Ezrin expression; b Moesin expression, c Willin expression, and d Merlin expression. Time to first LRR (or distant metastasis) displayed in months truncated at 60 months for illustration. Censored cases shown by tick marks at last recorded follow-up
Multivariate Cox regression analysis showed that strong cytoplasmic ezrin expression was significantly associated with a four-fold increased likelihood of cancer death (Table 4). In addition, we observed significantly higher hazards for combined cytoplasmic ezrin and moesin expression (adjusted HR for disease-specific survival = 6.9 [95% CI 2.2–20.8] and HR for LRR or distant metastasis = 2.7 [95% CI 1.1–6.3]). In contrast, cytoplasmic merlin and willin expression were not associated with cancer survival or disease progression. With respect to non-cytoplasmic expression, no significant associations were observed, although inverse associations with increased (≥2) willin, and merlin nuclear expression were suggested for overall and cancer mortality, and disease progression (not shown).
Table 4.
Association between FERM cytoplasmic expression and HNSCC prognosis
| Outcome | Ezrin | Moesin | Willin | Merlin | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | |
| Disease-specific survival* | 4.1 | (1.4–12.6) | 0.013 | 2.0 | (0.5–7.4) | 0.310 | 1.1 | (0.3–1.5) | 0.897 | 1.0 | (0.2–4.3) | 0.955 |
| LRR or Distant Metastasisa | 2.0 | (0.7–5.2) | 0.176 | 2.2 | (0.8–5.9) | 0.138 | 1.0 | (0.4–2.7) | 0.937 | 0.8 | (0.2–2.4) | 0.638 |
* Relative hazards (HR), 95% confidence intervals (CI) and p-values for FERM staining estimated by Cox regression stratified on tumor site, primary treatment, race, HPV and specimen procurement method, and adjusted further for age, ethnicity, ever smoking status, alcohol drinking, prior primaries and tumor stage
aHR, 95% CI and p-values stratified on tumor site, primary treatment, race and HPV, and adjusted further for age, ethnicity, ever smoking status, alcohol drinking, prior primaries and tumor stage. LRR Local or regional recurrence
RNA Expression in HNSCC with High Cytoplasmic Ezrin
Given the exploratory nature of these analyses, we did not correct for multiple comparisons. Nonetheless, we identified 842 features that had a P value less than 0.05 and 152 features that had a P value less than 0.005. In Table 5, we present median values of non-transformed data for biologically relevant genes, the ratio of the median between cancers expressing high versus low cytoplasmic ezrin and the p-value derived from the log2-transformed data.
Because we routinely perform global RNA analysis of primary HNSCC samples, we could analyze expression to correlate biologically significant differences in expression between cancers expressing high versus low cytoplasmic ezrin (Tables 5, 6). We analyzed genes based on differential expression in high versus low cytoplasmic ezrin cancers. A subset of genes with known biologic relevance to cancer that had statistically significant levels of expression in high versus low cytoplasmic ezrin cancers were analyzed using IPA software focusing on proliferation, epithelial differentiation, invasion, and cell death. Many genes were found in several functional categories (Tables 5, 6). The functional categorization of high cytoplasmic ezrin cancers favors gene expression patterns featuring a more invasive phenotype (P < 5.47E−08), inhibition of cell death (P < 1.54E−10), dampening of proliferation (P < 4.71E−10), decreased differentiation (P, 1.01E−07), and expression of cancer-associated genes (P < 1.80E−04), which is all consistent with poorer cancer survival.
Discussion
Members of the FERM family of proteins contribute to diverse physiologic functions through multiple signaling pathways [3]. We have shown previously that moesin, willin, and ezrin are expressed in the normal human oral mucosa as the cells differentiate [15]. In addition we showed that the abnormal localization of ezrin in the cytoplasm of HNSCC cells correlates with poor overall survival [15]. While the mechanism for cytoplasmic localization is unknown and whether cytoplasmic ezrin contributes to aggressive phenotype or is merely a biomarker for it, there is mounting evidence that expression of ezrin in several types of tumors correlates with poor outcome [16–21].
In an attempt to understand the relationship of cytoplasmic ezrin with aggressive cancer phenotype, we looked for correlations with RNA expression. While there are several potential interpretations of the data, we favor the pattern of gene expression for dampening of proliferation for the following several reasons: (1) genes associated with cell division are relatively low (KIF2C, CDT1, E2F2); (2) certain genes induce a nonproliferative state (TGFBR2) [25] or block internalization of EGFR (RAB11FIP2) [26], which favors motility over proliferation [27, 28]; and (3) increased phosphatase expression that can modulate pathway activation (PTPRK) [29]. With increased expression of MAP2K1 and RAF1 and decreased expression of PITX1, ISG15, CEBPA, MKNK2 and SOX2 [30–34], we believe that the phosphatases are particularly interesting because they serve to inactivate kinase cascades [35, 36] and can modulate pathway interaction, such as EGF and TGF-beta [29]. In addition, Notch1, which has been recently identified as a tumor suppressor for SCC, is decreased in high cytoplasmic ezrin tumors and may dampen kinase activity [37–39].
The pattern of gene expression in high cytoplasmic ezrin cancers supports an invasive phenotype. There is increased expression of genes driving migration and invasion (SERPINE2, EFNB2, ETS1, NET1, ITGB1) [40–42], and a decrease in differentiation associated genes that are required for epidermal development (e.g. tight junctions) (CLDN7, CRB3, DHCR24, CNN2) [43–45]. Finally, the pattern of gene expression in high cytoplasmic ezrin supports survival rather than cell death because of increased expression of anti-apoptotic genes (BIRC2, CCDC50, TGM2) [46, 47], decreased expression of pro-apoptotic genes (EDARADD) that mediate apoptosis and increased expression of genes that modulate pro-apoptotic pathways [48].
Recent work by Gunn-Moore and colleagues [6] shows that the Hippo signaling pathway can be activated or blocked by FERM proteins; willin and merlin lead to activation of the pathway while ezrin and moesin block activation. Activation results in a kinase cascade resulting in the phosphorylation of YAP which inactivates this transcription factor resulting in translocation to the cytoplasm and apoptosis. Non-phosphorylated YAP results in the expression of genes that promote cellular proliferation and block apoptosis in some systems and promote apoptosis in others [49]. We measured YAP and pYAP expression immunohistochemically in six total cases of high and low cytoplasmic ezrin cancers; both types of HNSCC expressed YAP and pYAP but no clear pattern distinguishing the two types of cancer could be found (data not shown). Because the pattern of FERM protein expression correlates clinical outcome with potential Hippo pathway activity—high cytoplasmic ezrin/moesin associated with poor cancer survival—and because active YAP is present in the nucleus, we cannot rule out some component of the hHppo pathway involvement in HNSCC.
We should note that despite using expert staining and evaluations, tests were performed on tissue microarrays, which do not reproduce exactly the clinical diagnostic setting. Although, multiple cores were selected by a pathologist for each tumor to reflect the diagnostic histopathology specimen, in a small percentage of cases results may differ for clinical tissue sections. Presently, TNM staging based on physical exam, imaging, and histopathology is used for initial treatment planning. Prognostic biomarkers that can distinguish aggressive phenotypes at initial diagnosis are needed. However, many potential biomarkers fail because appropriate confirming studies are not performed [50]. Our initial study of cytoplasmic ezrin in an independent population of HNSCC patients published in 2006 indicated the need for an intermediate level assessment presented here. The present study warrants a larger scale multi-institutional trial to confirm the use of FERM protein expression as prognostic biomarkers for HNSCC, and to test for site and treatment specific associations, which was not possible due to the inherent heterogeneity in management of HNSCC. In addition, our initial correlative findings with RNA expression provide new insight into the high cytoplasmic ezrin phenotype that will aid in future clinical studies as well as studies in model systems aimed at defining pathophysiologic mechanisms.
Acknowledgments
Contract grant sponsor: National Institutes of Health; Contract grant numbers: CA103547 (to MBP), CA115243 (to NFS), CA104402 (to TJB); Contract grant sponsor: UK Biotechnology and Biological Sciences Research Council (to FGM). The present study was supported by the Department of Pathology, Albert Einstein College of Medicine/Montefiore Medical Center. We thank the participants of this study; Catherine Sarta for her time and effort spent enrolling and following participants and with data entry, Gregory Rosenblatt for his assistance with data management and Dr. Joseph Locker for preparation of Fig. 1.
Conflict of interest
The authors disclose no conflict of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Belbin TJ, Singh B, Smith RV, et al. Molecular profiling of tumor progression in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2005;131:10–18. doi: 10.1001/archotol.131.1.10. [DOI] [PubMed] [Google Scholar]
- 3.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Natl Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem Sci. 2010;35:627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Bao Y, Hata Y, Ikeda M, Withanage K. Mammalian Hippo pathway: from development to cancer and beyond [in process citation] J Biochem. 2011;149:361–379. doi: 10.1093/jb/mvr021. [DOI] [PubMed] [Google Scholar]
- 6.Angus L, Moleirinho S, Herron LR, et al. Willin/FRMD6 expression activates the hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene. 2011. doi:10.1038/onc.2011.224. [DOI] [PubMed]
- 7.Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993;105:1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz-Albiez R, Merling A, Spring H, Moller P, Koretz K. Differential expression of the microspike-associated protein moesin in human tissues. Eur J Cell Biol. 1995;67:189–198. [PubMed] [Google Scholar]
- 9.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Natl Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 10.Hunter KW. Ezrin, a key component in tumor metastasis. Trends Mol Med. 2004;10:201–204. doi: 10.1016/j.molmed.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Heiska L, Melikova M, Zhao F, Saotome I, McClatchey AI, Carpen O. Ezrin is key regulator of Src-induced malignant phenotype in three-dimensional environment [epub ahead of print] [record supplied by publisher]. 2011 Oncogene. [DOI] [PubMed]
- 12.Ichikawa T, Masumoto J, Kaneko M, Saida T, Sagara J, Taniguchi S. Expression of moesin and its associated molecule CD44 in epithelial skin tumors. J Cutan Pathol. 1998;25:237–243. doi: 10.1111/j.1600-0560.1998.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi H, Sagara J, Masuumoto J, Kurita H, Kurashina K, Tokunaga J. Shifts in cellular localization of moesin in normal oral epithelium, oral epithelial dysplasia, verrucous carcinoma and oral squamous cell carcinoma. J Oral Pathol Med. 2003;32:344–349. doi: 10.1034/j.1600-0714.2003.00111.x. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi H, Sagara J, Kurita H, et al. Clinical significance of cellular distribution of moesin in patients with oral squamous cell carcinoma. Clin Cancer Res. 2004;10:572–580. doi: 10.1158/1078-0432.CCR-1323-03. [DOI] [PubMed] [Google Scholar]
- 15.Madan R, Brandwein-Gensler M, Schlecht NF, et al. Differential tissue and subcellular expression of ERM proteins in normal and malignant tissues: cytoplasmic ezrin expression has prognostic significance for head and neck squamous cell carcinoma. Head Neck. 2006;28:1018–1027. doi: 10.1002/hed.20435. [DOI] [PubMed] [Google Scholar]
- 16.Bruce B, Khanna G, Ren L, et al. Expression of the cytoskeleton linker protein ezrin in human cancers. Clin Exp Metastasis. 2007;24:69–78. doi: 10.1007/s10585-006-9050-x. [DOI] [PubMed] [Google Scholar]
- 17.Elzagheid A, Korkeila E, Bendardaf R, et al. Intense cytoplasmic ezrin immunoreactivity predicts poor survival in colorectal cancer. Hum Pathol. 2008;39:1737–1743. doi: 10.1016/j.humpath.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Kobel M, Langhammer T, Huttelmaier S, et al. Ezrin expression is related to poor prognosis in FIGO stage I endometrioid carcinomas. Mod Pathol. 2006;19:581–587. doi: 10.1038/modpathol.3800567. [DOI] [PubMed] [Google Scholar]
- 19.Mhawech-Fauceglia P, Dulguerov P, Beck A, Bonet M, Allal AS. Value of ezrin, maspin and nm 23-H1 protein expressions in predicting outcome of patients with head and neck squamous-cell carcinoma treated with radical radiotherapy. J Clin Pathol. 2007;60:185–189. doi: 10.1136/jcp.2006.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh CN, Pang ST, Chen TW, Wu RC, Weng WH, Chen MF. Expression of ezrin is associated with invasion and dedifferentiation of hepatitis B related hepatocellular carcinoma. BMC Cancer. 2009;9:233. doi: 10.1186/1471-2407-9-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Hu MY, Wu WZ, et al. The membrane-cytoskeleton organizer ezrin is necessary for hepatocellular carcinoma cell growth and invasiveness. J Cancer Res Clin Oncol. 2006;132:685–697. doi: 10.1007/s00432-006-0117-5. [DOI] [PubMed] [Google Scholar]
- 22.Belbin TJ, Singh B, Smith RV, et al. Molecular profiling of tumor progression in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2005;131:10–18. doi: 10.1001/archotol.131.1.10. [DOI] [PubMed] [Google Scholar]
- 23.Schlecht NF, Brandwein-Gensler M, Nuovo GJ, et al. A comparison of clinically utilized human papillomavirus detection methods in head and neck cancer [epub ahead of print] [record supplied by publisher]. Mod Pathol. 2011. [DOI] [PMC free article] [PubMed]
- 24.Greenland S, Robins JM. Identifiability, exchangeability, and epidemiological confounding. Int J Epidemiol. 1986;15:413–419. doi: 10.1093/ije/15.3.413. [DOI] [PubMed] [Google Scholar]
- 25.Lucke CD, Philpott A, Metcalfe JC, et al. Inhibiting mutations in the transforming growth factor beta type 2 receptor in recurrent human breast cancer. Cancer Res. 2001;61:482–485. [PubMed] [Google Scholar]
- 26.Cullis DN, Philip B, Baleja JD, Feig LA. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J Biol Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- 27.Iyer AK, Tran KT, Griffith L, Wells A. Cell surface restriction of EGFR by a tenascin cytotactin-encoded EGF-like repeat is preferential for motility-related signaling. J Cell Physiol. 2008;214:504–512. doi: 10.1002/jcp.21232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haugh JM. Localization of receptor-mediated signal transduction pathways: the inside story. Mol Interv. 2002;2:292–307. doi: 10.1124/mi.2.5.292. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Baker D, Quan T, Baldassare JJ, Voorhees JJ, Fisher GJ. Receptor type protein tyrosine phosphatase-kappa mediates cross-talk between transforming growth factor-beta and epidermal growth factor receptor signaling pathways in human keratinocytes. Mol Biol Cell. 2010;21:29–35. doi: 10.1091/mbc.E09-08-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Knosel T, Ye F, Pacyna-Gengelbach M, Deutschmann N, Petersen I. Decreased PITX1 homeobox gene expression in human lung cancer. Lung Cancer. 2007;55:287–294. doi: 10.1016/j.lungcan.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Kolfschoten IG, Leeuwen B, Berns K, et al. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 32.Feng Q, Sekula D, Guo Y, et al. UBE1L causes lung cancer growth suppression by targeting cyclin D1. Mol Cancer Ther. 2008;7:3780–3788. doi: 10.1158/1535-7163.MCT-08-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett KL, Romigh T, Arab K, et al. Activator protein 2 alpha (AP2alpha) suppresses 42 kDa C/CAAT enhancer binding protein alpha (p42(C/EBPalpha)) in head and neck squamous cell carcinoma. Int J Cancer. 2009;124:1285–1292. doi: 10.1002/ijc.24087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knauf U, Tschopp C, Gram H. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol Cell Biol. 2001;21:5500–5511. doi: 10.1128/MCB.21.16.5500-5511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toledano-Katchalski H, Kraut J, Sines T, et al. Protein tyrosine phosphatase epsilon inhibits signaling by mitogen-activated protein kinases. Mol Cancer Res. 2003;1:541–550. [PubMed] [Google Scholar]
- 36.Kraut-Cohen J, Muller WJ, Elson A. Protein-tyrosine phosphatase epsilon regulates Shc signaling in a kinase-specific manner: increasing coherence in tyrosine phosphatase signaling. J Biol Chem. 2008;283:4612–4621. doi: 10.1074/jbc.M708822200. [DOI] [PubMed] [Google Scholar]
- 37.Lefort K, Mandinova A, Ostano P, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang NJ, Sanborn Z, Arnett KL, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma [in process citation] Proc Natl Acad Sci USA. 2011;108:17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tachibana M, Tonomoto Y, Hyakudomi R, et al. Expression and prognostic significance of EFNB2 and EphB4 genes in patients with oesophageal squamous cell carcinoma. Dig Liver Dis. 2007;39:725–732. doi: 10.1016/j.dld.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 41.Fujimoto J, Aoki I, Toyoki H, et al. Clinical implications of expression of ETS-1 related to angiogenesis in metastatic lesions of ovarian cancers. Oncology. 2004;66:420–428. doi: 10.1159/000079491. [DOI] [PubMed] [Google Scholar]
- 42.Lee J, Moon HJ, Lee JM, Joo CK. Smad3 regulates Rho signaling via NET1 in the transforming growth factor-beta-induced epithelial-mesenchymal transition of human retinal pigment epithelial cells. J Biol Chem. 2010;285:26618–26627. doi: 10.1074/jbc.M109.073155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemmers C, Michel D, Lane-Guermonprez L, et al. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell. 2004;15:1324–1333. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirza R, Hayasaka S, Takagishi Y, et al. DHCR24 gene knockout mice demonstrate lethal dermopathy with differentiation and maturation defects in the epidermis. J Invest Dermatol. 2006;126:638–647. doi: 10.1038/sj.jid.5700111. [DOI] [PubMed] [Google Scholar]
- 45.Fukui Y, Masuda H, Takagi M, Takahashi K, Kiyokane K. The presence of h2-calponin in human keratinocyte. J Dermatol Sci. 1997;14:29–36. doi: 10.1016/S0923-1811(96)00545-2. [DOI] [PubMed] [Google Scholar]
- 46.Kameda H, Watanabe M, Bohgaki M, Tsukiyama T, Hatakeyama S. Inhibition of NF-kappaB signaling via tyrosine phosphorylation of Ymer. Biochem Biophys Res Commun. 2009;378:744–749. doi: 10.1016/j.bbrc.2008.11.102. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi H, Wang HG. Tissue transglutaminase serves as an inhibitor of apoptosis by cross-linking caspase 3 in thapsigargin-treated cells. Mol Cell Biol. 2006;26:569–579. doi: 10.1128/MCB.26.2.569-579.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Cao N, Nantajit D, Fan M, Liu Y, Li JJ. Mitogen-activated protein kinase phosphatase-1 represses c-Jun NH2-terminal kinase-mediated apoptosis via NF-kappaB regulation. J Biol Chem. 2008;283:21011–21023. doi: 10.1074/jbc.M802229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehsanian R, Brown M, Lu H, et al. YAP dysregulation by phosphorylation or <Np63-mediated gene repression promotes proliferation, survival and migration in head and neck cancer subsets. Oncogene. 2010;29:6160–6171. doi: 10.1038/onc.2010.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]