Abstract
Tens of thousands of lymphoblastoid cell lines (LCLs) have been established by the research community, providing nearly unlimited source material from samples of interest. LCLs are used to address questions in population genomics, mechanisms of disease, and pharmacogenomics. Thus, it is of fundamental importance to define the extent of chromosomal variation in LCLs. We measured variation in genotype and copy number in multiple LCLs derived from peripheral blood mononuclear cells (PBMCs) of single individuals as well as two comparison groups: (1) three types of differentiated cell lines (DCLs) and (2) triplicate HapMap samples. We then validated and extended our findings using data from a large study consisting of samples from blood or LCLs. We observed high concordances between genotypes and copy number estimates within all sample groups. While the genotypes of LCLs tended to faithfully reflect the genotypes of PBMCs, 13.7% (4 of 29) of immortalized cell lines harbored mosaic regions greater than 20 megabases which were not present in PBMCs, DCLs, or HapMap replicate samples. We created a list of putative LCL-specific changes (affecting regions such as immunoglobulin loci) that is available as a community resource.
Keywords: lymphoblastoid cell lines, genotyping, microarrays, SNP, copy number variation
Introduction
Lymphoblastoid cell lines (LCLs) represent one of the most commonly used sources of biological material for genetic and cellular studies (Sie, et al., 2009). LCLs are routinely used to characterize genetic variation in samples from individuals with disease, for population genomics studies such as the HapMap project, and for other applications ranging from pharmacogenomics to gene expression (Altshuler, et al., 2010; Cheung, et al., 2003; Kalman, et al., 2009; Welsh, et al., 2009). With the advent of next-generation sequencing, whole exome and whole genome sequencing have been performed on genomic DNA from LCLs. For example, the 1000 Genomes project, one of the earliest projects to sequence large numbers of genomes, has included LCLs (Durbin, et al., 2010). Approximately two thirds of the anticipated 2,500 samples to be sequenced by that project are from LCLs.
LCLs are most commonly established by EBV infection of PBMCs using phytohemagglutinin as a mitogen. An outstanding question is the effect of EBV transformation on the stability of genomic DNA, including effects on genotype and copy number. EBV, a gamma herpesvirus, is maintained as an episome and is often associated with mononucleosis, nasopharyngeal carcinoma, Burkitt's lymphoma, gastric carcinoma, and post-transplant lymphoproliferative disease. EBV is implicated in promoting proliferation of tumor cells, as well as regulating DNA damage repair. Genomic instability is often characteristic of EBV-associated tumors (Kamranvar, et al., 2007). There is also evidence to support EBV-mediated induction of DNA damage, modulation of DNA repair, and inactivation of cell cycle checkpoints (Gruhne, et al., 2009; Wu, et al., 2010).
While EBV immortalization is a widespread laboratory practice, little is known about the frequencies and types of genomic instabilities and structural variations common to LCLs immortalized by EBV infection. Copy number variation was assessed in 270 LCLs from the HapMap project, and 30 cell lines were reported (of 268) having chromosomal abnormalities likely to be culture-induced (Redon, et al., 2006). After removing these, they further examined genotype data in CNV regions of father/mother/child trios consistent with somatic mutation (based on the occurrence of SNP alleles not present in either parent). This analysis suggested that 0.5% of CNVs could be attributed to somatic mutation. Conrad et al. assessed male and female germline mutation rates by sequencing genomes obtained from LCLs from two parent/offspring trios (Conrad, et al., 2011). They reported 35 and 49 de novo mutations in two offspring from trios, and about 20-fold more non-germline de novo mutations that arose either as somatic mutations or in transformed LCLs. Genome-wide association studies by the Wellcome Trust Case Control Consortium also found systematic differences in array signal intensity based on DNA source (Redon, et al., 2006). As noted by the 1000 Genomes Project, false positive rates from cell line mutations are likely to confound measurement of de novo mutation rates (Durbin, et al., 2010). Therefore, it is of interest to characterize the nature and extent of chromosomal variation in LCLs to better inform the interpretation of LCL genotyping and genome sequencing studies. Additionally, for functional studies utilizing LCLs, it is important to assess the fidelity of LCLs relative to the blood cells from which they are derived to gauge how closely the LCLs resemble their in vivo counterparts.
In previous studies, several groups have addressed related questions. Simon-Sanchez et al. (Simon-Sanchez, et al., 2007) assayed ≈400,000 single nucleotide polymorphisms (SNPs) in 276 EBV-immortalized LCLs derived from elderly subjects, finding ≈10% with regions of homozygosity >5 Mb and ≈67% with structural genomic alterations (two thirds of which did not intersect previously known variants). For five samples, regions of homozygosity were confirmed to also occur in corresponding blood-derived samples. Two individuals had deletions in LCL but not blood in the immunoglobulin lambda gene cluster of chromosome 22q11.2, a region also found to be altered in LCLs by Sebat et al. (Sebat, et al., 2004). In another study, Herbeck et al. (Herbeck, et al., 2009) compared genotypes in EBV-immortalized LCLs and PBMCs and found few significant differences.
In this study, we addressed the extent of genotypic and chromosomal copy number variability in LCLs relative to their primary PBMCs. Thus we assessed the effects of immortalization on chromosomal stability. We studied multiple LCLs derived from a given individual in order to assess differences in independently established LCLs from the same individual. To provide a baseline for the extent of genomic changes we studied, in parallel, both differentiated cell types (DCLs) derived from a given individual, and replicate HapMap samples. We then characterized chromosomal changes in LCLs and PBMC samples from a large genome-wide association study (GWAS), the Gene Environment Association Studies (GENEVA) project Study of Addiction: Genetics and Environment (SAGE) data set (Cornelis, et al., 2010).We found that multiple LCLs derived from a given individual were very similar in genotype and copy number. The magnitude of variation observed in LCLs relative to blood was comparable to that observed between differentiated cell types from the same individual, as well as replicate HapMap samples. However, there were notable occurrences of somatic changes including long stretches of homozygosity and regions of deletions and amplifications, some of which were mosaic.
Materials and Methods
Coriell multiple LCLs
All studies were performed with informed consent and approval of an Institutional Review Board (convened by the Coriell Institute for Medical Research [CIMR]) as well as approval of a Johns Hopkins IRB for analyses performed there. Six vials of blood were obtained during a single blood draw from each of six different individuals. From each individual, PBMCs were isolated from one vial of blood, and blood from each of the 5 remaining tubes was immortalized with EBV to establish independent LCLs, which were then frozen after one or two passages. A total of 29 LCLs were established within the NIGMS Human Genetic Cell Repository at the CIMR: five LCLs for individuals 1–5, and four LCLs from individual 6. DNA was isolated from each of the six PBMC samples to obtain samples (designated B1–B6 where B denotes blood) from cells that had not been immortalized or cultured. DNA was also isolated from each of the 29 LCLs to generate samples from populations of cells that were independently immortalized and cultured. These 29 samples were designated with labels such as L34 to indicate the fourth LCL established from individual 3, corresponding to PBMC DNA sample B3; sample characteristics and mappings of sample designations to Coriell identifiers are listed in Supp. Table S1. Each of the 35 DNA samples was genotyped on the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix 6.0) platform at the CIMR.
GENEVA SAGE
We obtained GENEVA SAGE data from the database of Genotypes and Phenotypes (dbGaP) at the National Center for Biotechnology Information (NCBI) with approval from a National Human Genome Research Institute data access committee. GENEVA SAGE data consisted of 4,032 samples genotyped on the Illumina Human 1M platform. DNA was derived from either whole blood or LCL. Within this data set there were 196 pairwise comparisons indicative of multiple samples from the same subject (51 from SAGE, 145 from HapMap controls). These comparisons, which included those between blood-derived samples, LCLs, and blood versus LCLs, were used as replicates.
Differentiated cell lines (DCLs)
Discarded human foreskins were obtained from Cooper Hospital in Camden, New Jersey. Fibroblast (F), keratinocyte (K), and melanocyte (M) cell lines were established in the NIGMS Human Genetic Cell Repository at the CIMR from the same foreskin specimen (n=9 individuals, n=27 samples). The resulting DCLs were designated with labels such as 3F, 3K, and 3M for the three DCLs derived from individual 3.
We established F, K, and M cell cultures from single neonatal foreskins as follows. Foreskin was placed in a 60 mm dish containing antibiotic wash (D-PBS + 20 μg/ml gentamicin [Invitrogen #15710-064] and 1 μg/ml fungizone® [Invitrogen #15290-018 or equivalent, 250μg/ml each of amphotericin B and sodium deoxycholate]) for 1 hr. Following removal of fat and connective tissue, skin was transferred to 6.25 ml 1× dispase at 4°C overnight. To establish keratinocyte and melanocyte cultures, the epidermal layer was peeled from the dermis using a forceps, transferred to a 60 mm dish containing phosphate-buffered saline (PBS) and then incubated in 5 ml 0.05% trypsin/0.53 mM EDTA for 10 min. Trypsin was neutralized using 10 ml soybean trypsin inhibitor. After filtration through a 70 micron mesh screen, the suspension was centrifuged (200 × g, 5 min, 15–20°C) in two tubes and the pellets were resuspended in 5 ml MGM-4 (a melanocyte medium including growth factors; Lonza catalog #CC-3249) with 10 μg/ml gentamicin (for melanocytes) or 5 ml CnT-07 (an epidermal progenitor cell medium; CELLnTEC Advanced Cell Systems #CnT-07, Zen Bio Inc.) with 10 μg/ml gentamicin (for keratinocytes). Suspensions were placed in collagen IV-coated T25 flasks and incubated at 37°C/5% CO2. Fibroblasts were established from dermis by finely mincing dermis using cross scalpels, transferring chunks to T25 flasks, adding 5–6 ml fibroblast growth medium with 10 μg/ml gentamicin, and incubating at least 24 hours. From days 3–7, primary cultures of keratinocytes were fed with CnT-07 and gentamicin for 1 to 3 days, and then fed every two days until expansion; a similar procedure was used for primary cultures of melanocytes, substituting MGM-4; and fibroblasts were fed with 15% fetal bovine serum in DMEM:HG-12 and gentamicin for 5–7 days after plating. Fibroblasts were eliminated from melanocyte cultures using geneticin (Invitrogen #10131-035).
Keratinocyte, melanocyte and fibroblast cultures were characterized by immunocytochemistry according to standard protocols as described. Fibroblasts were labeled using a monoclonal anti-fibroblast (clone TE-7, Millipore/Fisher #CBL271MI) at 1:200 dilution with AF633-conjugated goat anti-mouse IgG as a secondary antibody. gp100 (HMB45; 1:100 dilution) was used to label the surface of melanocytes, with AF488-conjugated goat anti-mouse IgG (1:200 dilution) as a secondary antibody. In some cases, monoclonal anti-MiTF (1:25 dilution) was used to label melanocyte nuclei, with AF488-conjugated goat anti-mouse IgG as a secondary. Anti-pan-cytokeratin-488 antibody (1:50 dilution) labels keratinocytes specifically. At least 100 positively staining cells were scored for each culture. Sample characteristics and mappings of sample designations to Coriell identifiers are listed in Supp. Table S2. Note that sample 2M consisted of only 30% melanocytes(with the remainder likely consisting of fibroblasts), and sample 3K included 62% keratinocytes (with the remaining 38% consisting of melanocytes). DNA was isolated from each specimen and genotyped on the Affymetrix 6.0 platform for SNP and copy number variation (CNV) analysis.
Technical replicates
Technical replicates consisted of 18 samples (triplicate samples from each of 6 HapMap individuals) obtained from the Gene Expression Omnibus (GEO) at NCBI (series GSE25893). These samples were genotyped on the same Affymetrix 6.0 platform at the Centre for Applied Genomics (TCAG) as part of a recent copy number variation assessment study (Pinto, et al., 2011).
Assessment of data quality
SNPs were excluded from analysis at thresholds of > 0%, >50%, >90%, >95%, and >99% call rate. Pairwise IBS distance matrices between genotypes of LCL, DCL, and replicate samples were calculated using PLINK (Purcell, et al., 2007).These methods corresponded to those of Herbeck et al. (Herbeck, et al., 2009) who also characterized variation in LCLs.
Computational analyses of chromosomal changes
The quality of SNP data was assessed using Affymetrix Genotyping Console software. This included median absolute pairwise distance (MAPD) values that were all below a threshold of 0.3, indicating negligible noise in the experiments for copy number analysis.
SNP genotype data were analyzed for identity-by-state (IBS) using SNPduo and SNPduo++ software (Roberson and Pevsner, 2009). The results of these analyses were analyzed using Partek Genomics Suite software version 6.5 (Partek, Inc. St. Louis, MO). We further used SNPtrio (Ting, et al., 2007) and pediSNP (Ting, et al., 2009) to evaluate genotypic changes. Pairwise distances between samples were calculated using PLINK (Purcell, et al., 2007).
Copy number changes were analyzed using Affymetrix Power Tools (Affymetrix, Inc. Santa Clara CA) and PennCNV-Affy (Wang, et al., 2007) using default settings, to obtain B allele frequencies (BAFs) and logR ratio. X chromosome pseudoautosomal regions (NCBI36 chrX:1–2,766,639 and chrX:154,583,754–154,913,754) were excluded from analysis (Flaquer, et al., 2008). CNVineta (Wittig, et al., 2010) was used for further analysis of copy number segmentation, including association tests and generation of heat maps to assess quality of CNV calling. Filtering was applied as specified in the CNVineta package to remove outlier samples containing an excessive number of CNV calls before CNVineta association testing. This did not significantly change the mean number of CNV calls in either case (LCL) or control (blood) groups (data not shown).
For all samples, large mosaic abnormalities were detected by visual inspection of B Allele Frequency (BAF) and Mosaic Alteration Detection (MAD) software (Gonzalez, et al., 2011) with a false discovery rate (FDR) of.001 (a=0.8, T=8, minLength=25 markers). Percent mosaicism was estimated for each abnormal region by reflecting the B Allele Frequency (BAF) about 0.5 and applying the formula [(observed median BAF ÷ expected BAF) −1] to data points < 0.95, where expected BAF = 0.5.
Results
Quality control and sample genotype concordance rates
We assessed data quality in samples of Coriell PBMCs and their corresponding LCLs. We obtained multiple tubes of blood from six apparently healthy volunteers during the same blood draw. We froze the PBMCs isolated from one tube of blood and established four or five LCLs by independent EBV transformation of blood from each of the remaining tubes. We extracted DNA from each cell type and sample and performed genotyping on high density SNP microarrays to assess both genotype and copy number changes. We performed parallel analyses on the PBMC vs. LCL data set and two control data sets: DCLs, to evaluate variation between primary cell types within individuals, and technical replicates in triplicate of HapMap individuals. Genotype concordance was determined by calculating pairwise distance between the genotypes of samples with PLINK software. Extremely high concordance was seen between sample genotypes in all groups. PBMC vs. LCL comparisons had a mean and standard deviation of 0.005 ± 0.001 (Supp. Figure S1A), while the control group means ranged from approximately 0.0002 (DCLs) to 0.002 (HapMap replicates). Since these distance estimates could be influenced by genotyping quality (Herbeck, et al., 2009), we filtered SNPs by progressively including only those with high call rates ranging from 50% to 99%. This filtering had a negligible effect on the pairwise distance comparisons between PBMC and LCLs or the control groups (Supp. Figure S1). These results suggest that technical variations between samples from the same individual were extremely low, allowing us to characterize genotypic differences as a function of transformation.
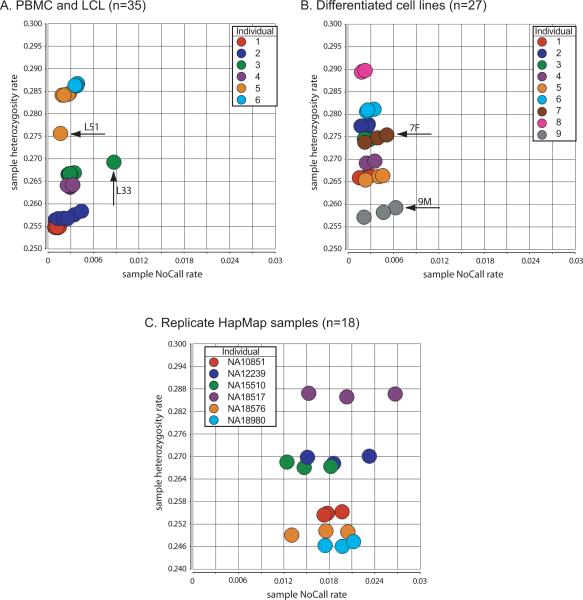
The measurement of genotyping NoCall rates, combined with heterozygosity rates for each sample, provides a useful method to identify outliers from each group that reflect chromosomal genotype variation. Average NoCall rates for genotyping experiments were extremely low: 0.28% ± 0.15 for PBMC and LCL (mean ± standard deviation, n=35), 0.28% ± 0.12 for DCLs (n=27), and 1.83% ± 0.35 (n=18) for HapMap replicates (Figure 1A–C, x-axes). A plot of NoCall rate for each PBMC and LCL sample versus autosome-wide heterozygosity rate showed that only one LCL sample had a relatively elevated NoCall rate (L33, i.e. LCL sample 3 from individual 3)(Figure 1A). Each sample had a characteristic heterozygosity rate, with samples 1–4 from Caucasian individuals having lower percent heterozygosity than samples 5 and 6 derived from African-American individuals (Figure 1A, y-axis). Sample L51 (i.e. LCL 1 from individual 5) had a markedly reduced heterozygosity rate relative to B5 (PBMC sample from individual 5) and the other LCLs derived from that individual (Figure 1A, arrow; described in detail below). This difference in heterozygosity was reflected in a relatively low genotypic concordance of 98.95% between L51 and B5 (Table 1). For DCLs and the replicate HapMap samples there were no comparable abnormalities in heterozygosity rate (Figure 1B,C).
Figure 1.
Plots of heterozygosity versus NoCalls. We measured sample heterozygosity rates (y-axis) compared to sample NoCall rates (x-axis) for (A) Coriell PBMC and LCL samples, (B) Coriell differentiated cell lines, and (C) HapMap replicate samples. Several outliers are indicated, having relative differences in heterozygosity and/or NoCall rates.
Table 1.
Concordance of autosomal genotypes between samples derived from same individual (n=6 individuals, n=4 or n=5 pairwise comparisons between each individual's PBMC genotypes and corresponding LCLs)
| IID 1 | IID 2 | IBS0 | IBS1 | IBS2 | Concordance | Observed abnormalities |
|---|---|---|---|---|---|---|
| B1 | L11 | 0 | 295 | 868741 | 99.97 | |
| B1 | L12 | 0 | 234 | 869219 | 99.97 | |
| B1 | L13 | 0 | 232 | 869276 | 99.97 | |
| B1 | L14 | 0 | 221 | 869233 | 99.97 | Segmental mosaic LOH chromosome 6q |
| B1 | L15 | 0 | 254 | 869032 | 99.97 | |
| B2 | L21 | 2 | 226 | 869202 | 99.97 | |
| B2 | L22 | 4 | 905 | 866577 | 99.90 | |
| B2 | L23 | 2 | 514 | 868272 | 99.94 | |
| B2 | L24 | 3 | 1861 | 864834 | 99.78 | |
| B2 | L25 | 3 | 471 | 867890 | 99.95 | |
| B3 | L31 | 0 | 156 | 869198 | 99.98 | |
| B3 | L32 | 0 | 398 | 868375 | 99.95 | |
| B3 | L33 | 0 | 2973 | 861518 | 99.66 | Mosaic loss of X chromosome; confirmed by G-banded karyotyping of 50 cells: mos45, X[36]/46,XX[14] |
| B3 | L34 | 0 | 172 | 869043 | 99.98 | |
| B3 | L35 | 0 | 140 | 869113 | 99.98 | |
| B4 | L41 | 1 | 350 | 868675 | 99.96 | Mosaic loss of X chromosome |
| B4 | L42 | 0 | 321 | 868658 | 99.96 | |
| B4 | L43 | 1 | 349 | 868322 | 99.96 | Segmental mosaic LOH on chromosome 11q |
| B4 | L44 | 0 | 213 | 869023 | 99.98 | |
| B4 | L45 | 0 | 364 | 868180 | 99.96 | |
| B5 | L51 | 4 | 9094 | 859130 | 98.95 | Segmental mosaic LOH on chromosome 4q |
| B5 | L52 | 0 | 299 | 867808 | 99.97 | |
| B5 | L53 | 0 | 476 | 866902 | 99.95 | |
| B5 | L54 | 1 | 528 | 866816 | 99.94 | |
| B5 | L55 | 0 | 330 | 867559 | 99.96 | |
| B6 | L61 | 1 | 317 | 868008 | 99.96 | |
| B6 | L62 | 0 | 322 | 867869 | 99.96 | |
| B6 | L63 | 0 | 218 | 868196 | 99.97 | |
| B6 | L64 | 0 | 322 | 868079 | 99.96 | |
| Mean | 99.91 |
For the five entries having observed abnormalities, explanations are provided.
Identity-by-state (IBS) provides a useful measure of genetic relatedness. We analyzed genotype calls in pairwise comparisons of all samples and measured IBS2 (two shared alleles, i.e. AA/AA or BB/BB in samples 1/2), IBS1 (one shared allele, e.g. AA/AB), or IBS0 (zero shared alleles, i.e. AA/BB or BB/AA). As expected, pairwise comparisons were characterized by extensive IBS2 sharing and only limited IBS0 or IBS1 (Table 1). For comparisons between PBMC and LCLs, the pairwise concordance rate ranged from 98.95 to 99.97% (mean 99.91%; Table 1). This was comparable to concordances observed in differentiated cell types from the same individual (n=9 individuals, three cell lines each), which ranged from 99.70 to 99.98% (mean 99.90%; Supp. Table S3). Concordance rates between replicate HapMap samples were slightly lower, due to an overall increase in the number of NoCalls (Supp. Table S4).
Variation in genotype calls and assessment of mosaicism
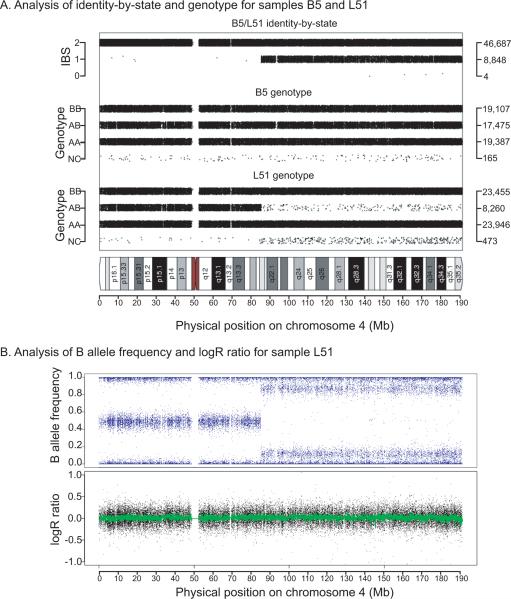
Several of the pairwise comparisons had particularly high IBS1 measurements, including B3/L33, B4/L41 and B5/L51. We used SNPduo software (Roberson and Pevsner, 2009) to identify IBS sharing across all chromosomes for these samples. This revealed expected IBS2 sharing for most chromosomes. For PBMC sample B5 compared to one of its five derived LCLs (sample L51), we observed a region of 100 Mb on chromosome 4q, extending to the telomere, characterized by IBS1 sharing (Figure 2A), consistent with its reduced heterozygosity rate plotted in Figure 1A. This region was confirmed by MAD analysis and visual inspection of the B Allele Frequency (BAF) and was determined to be a region of mosaic UPD in 75% of cells (see Methods). Further mosaicism analysis revealed mosaic UPD in the entire chromosome 6q arm (12% abnormal cells) of sample L14 (Supp. Figure S2A), a 20 Mb region (38% abnormal cells) of mosaic UPD in chromosome 11q of sample L43 (Supp. Figure S2B), and the mosaic loss of the X chromosome of LCL samples L33 and L41 (Supp. Figure S2C,D). For cell line L33 we confirmed the mosaic deletion by G-banded karyotyping of 50 cells, with karyotype mos45, X[36]/46,XX[14] (data not shown). We did not detect mosaic abnormalities in either the DCL or HapMap replicate data sets based on MAD and visual analyses.
Figure 2.
Analysis of genotype and copy number changes in LCL individual sample L51 relative to its parental PBMC DNA sample, B5. (A) Plot of chromosome 4 using SNPduo software (Roberson and Pevsner, 2009). Top panel shows identity-by-state including a region of IBS2 (typical of replicate or identical samples) extending from 0–90 Mb followed by a region of IBS1 extending for 100 Mb (from 90 Mb to 190 Mb). Middle panel: genotypes for B5 showing typical patterns of BB, AB, AA, and NoCalls (NC). Lower panel: genotypes for L51 showing prominent decrease in heterozygous (AB) calls from 90 to 190 Mb, with an increase in the NoCall rate. (B) B allele frequency for sample L51 revealed a typical pattern (0–90 Mb) corresponding to BB, AB, and AA genotypes at y-axis values of 1.0, 0.5, and 0 respectively. From 90–190 Mb a shift in the pattern occurred resulting in four bands and indicated a mosaic abnormality. Lower panel: logR ratio, reflecting copy number, indicated that L51 had no gain or loss of chromosomal material.
Variation in copy number
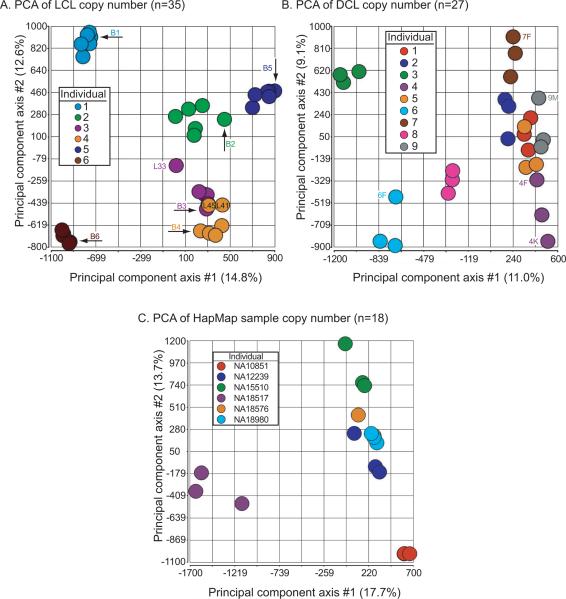
We analyzed copy number variation (CNV) in multiple lymphoblastoid cell lines, differentiated cells, and HapMap replicate samples. We used principal components analysis (PCA) to visualize the relatedness between copy number values across samples for 2,765,691 markers (both SNPs and nonpolymorphic markers from the Affymetrix 6.0 microarray, spanning all autosomes). For PBMC and LCL samples, we observed that each group (a PBMC sample and the derived LCLs) formed a cluster (Figure 3A). These clusters showed good cohesion, suggesting that the genome-wide copy number data were similar, with substantial similarity within a group and separation between groups. The first principal component axis (PC1) accounted for 14.8% of the variance, a relatively low value, suggesting that the overall data quality were good (without notable outliers). L33, an LCL having a mosaic loss of the X chromosome (Supp. Figure S2C), was separated from other members of its group. Note that the mosaicism affecting sample L51 did not involve copy number changes (Figure 2B), and sample L51 remained close to its group in PCA space.
Figure 3.
Principal components analysis of copy number data from (A) PBMC and LCL samples (n=35 samples derived from six individuals) (B) differentiated cell lines (n=27 samples derived from nine individuals), and (C) HapMap replicate samples (n=18 samples derived from six individuals). Values on the principal component axes correspond to percent of the variance explained.
We analyzed DCL copy number data by PCA and again observed clear evidence for nine clustered groups (corresponding to the nine individuals) with modestly more separation of the three cell types (fibroblast, keratinocyte, melanocyte)(Figure 3B). The percent of variance captured along PC1 (11.0%) was comparable to that observed in PBMC and LCL data. For the HapMap replicates, samples from each of the six individuals also formed cohesive clusters (Figure 3C). Taken together, the PCA results suggested that there was more variability between than within sets of related samples.
We assessed specific chromosomal loci of copy number variants (CNVs) in each sample of each copy number data set by defining segments and regions. We defined segments based on PennCNV segmentation output (n ≥ 25 SNPs), and we defined the broader category of CNV regions as consisting of intersecting segments (the regions had a range of 1 to 61 segments). We tabulated the number of samples with CNV segments that occurred in each common region, across the entire genome. In the majority of instances the CNV regions consisted of five or six samples, corresponding to a particular CNV occurring in all samples derived from one individual. There were only rare examples of CNVs involving fewer than 5 or 6 samples. There were seven regions in PBMC-derived or LCL cells that were at least 50 kb in length and occurred in over half of all samples (Table 2). These regions included three loci harboring immunoglobulin genes (on chromosomes 2, 7, and 14). The result of the same analysis of the DCL data set is available in Supp. Table S5.
Table 2.
Common CNV regions in PBMC/LCL (variant in > 50% of samples & size > 50 kb)
| Index | Chr | Region start | Region end | Size (bp) | Number of genes | Ig overlap | # CNV | # CNV DGV | #CNV GS/B n=2514 | #CNV GS/LCL n=1335 | p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 88,914,239 | 89,282,353 | 368,114 | 5 | 13 IgK | 30 | 114 | 0 | 0 | NA |
| 2 | 3 | 163,995,351 | 164,108,689 | 113,338 | 1 | 18 | 39 | 0 | 1 | 0.34 | |
| 3 | 7 | 142,001,624 | 142,203,712 | 202,088 | 58 | 52 TCR | 20 | 56 | 16 | 21 | 0.0082 |
| 4 | 8 | 7,011,977 | 7,869,464 | 857,487 | 31 | 22 | 128 | 6 | 10 | 0.031 | |
| 5 | 8 | 39,354,760 | 39,506,122 | 151,362 | 13 | 18 | 38 | 22 | 14 | 0.60 | |
| 6 | 14 | 105,289,630 | 106,269,389 | 979,759 | 28 | 9 IgH | 28 | 349 | 13 | 105 | 1.34e- |
| 7 | 17 | 41,709,662 | 42,120,174 | 410,512 | 15 | 24 | 78 | 171 | 70 | 0.059 |
Abbreviations: Chr, chromosome; Number of genes refers to UCSC Genes track (hg18) from the UCSC Genome Browser; Ig overlap, number of immunoglobulin elements in the region based on BioMart at Ensembl (build GRCh37.p3); IgK, immunoglobulin kappa; TCR, T-cell receptor; IgH, immunoglobulin heavy chain; # CNV, number of copy number variants observed in this study; # CNV DGV, number of copy number variants present in the Database of Genomic Variants; #CNV GS/B, number of autosomal copy number variants in GENEVA SAGE samples derived from blood; #CNV GS/LCL, number of autosomal copy number variants in GENEVA SAGE samples derived from LCL; p-value, result of two-sided Fisher's Exact test on GENEVA SAGE samples (case=LCL derived samples, control=blood derived samples).
To visualize variability in the numbers and types of CNVs in our three data sets, between samples and across individuals, we plotted deletions, amplifications, and regions of homozygosity by chromosomal position (Figure 4). We observed several categories of CNV: (1) Variant regions (i.e. containing deletions or amplifications) that were conserved between cell types (PBMC and LCL). Examples were evident on chromosomes 1, 5, 8, 12, 15 and 17 for individual 2 (Figure 4, second data column). (2) Variant regions in which the copy number state differed between PBMC and LCL samples. For example, chromosome 2 for all six individuals had amplifications in PBMCs and deletions across all LCL samples. (3) Variant regions that occurred most commonly, listed in Table 2, are indicated (Figure 4, column labeled “Table 2 index”). (4) In some instances a deletion or amplification occurred in only a subset of samples for a given individual. For example, inspection of chromosome 9 shows that CNVs occurred in just one LCL (for individuals 1, 2 and 5) and three of the six PBMC samples (samples B4, B5, and B6). We also plotted regions of homozygosity (>98% homozygous genotype calls spanning ≥50 SNPs). A prominent region of homozygosity was evident on chromosome 4q of sample L51 (Figure 4, fifth data column), as described above (Figure 2). Regions of homozygosity tended to be conserved across all samples from the same individual (e.g. see individuals 1 and 2 on chromosome 10).
Figure 4.
Ideogram representation of CNV segmentation (segments >25 SNPs and >50 kb in length) from six individuals (see six data columns to the left), differentiated cell lines (nine central data columns), and HapMap replicate samples (six data columns to the right). Data for the six PBMC derived samples are shaded pink. Genomic coordinates are decreasing on the y-axis, with centromeres indicated by dashed horizontal lines. Each horizontal bar corresponds to a segment having copy number loss (colored red) or copy number gain (shaded blue), relative to the reference. Segments shaded gray represent regions that were >98% homozygous. The thickness of each bar corresponds to the size of the segment. Note that regions defined in Table 2 are indicated in the column labeled Table 2 index. Top rows indicate means and standard deviations of the Jaccard similarity coefficients for groups of samples within individuals.
Analysis of CNVs in the differentiated cell lines and the HapMap replicates also revealed a variety of amplifications and deletions (Figure 4). The mean and standard deviation per sample of CNVs in the multiple LCLs (13.8 +/− 3.0) was less than that of PBMCs (25.8 +/− 3.9), DCLs (21.3 +/− 2.7), and HapMap replicates (15.5 +/− 4.2). We also plotted regions of homozygosity (Figure 4), to show possible UPD events. The most notable instance was on chromosome 4 of sample L51.
We quantified the extent of concordance between LCL samples as plotted in Figure 4. The concordance between CNV calls from technical replicates is a measure of the reproducibility of CNV calling. Pinto et al. recently demonstrated that reproducibility is significantly affected by DNA quality, genotyping platform, and the algorithm applied to CNV detection. The Jaccard similarity coefficient describes the concordance between two sets of CNV intervals (A,B), given by . For comparisons between identical sets of interval data, this relationship reduces to 1. For our LCL, DCL, and HapMap data sets, medians ± S.D. are 0.56±0.04, 0.70±0.07, and 0.70±0.06 respectively. Values for individual comparisons are shown in Figure 4.
Copy number analysis of LCL versus blood samples in a large GWAS
In addition to the three data sets described above, we introduced a fourth data set, consisting of a large GWAS. The purpose of including these data was to compare chromosomal copy number between a large number of blood samples (n=2,514) and LCLs (n=1,335).
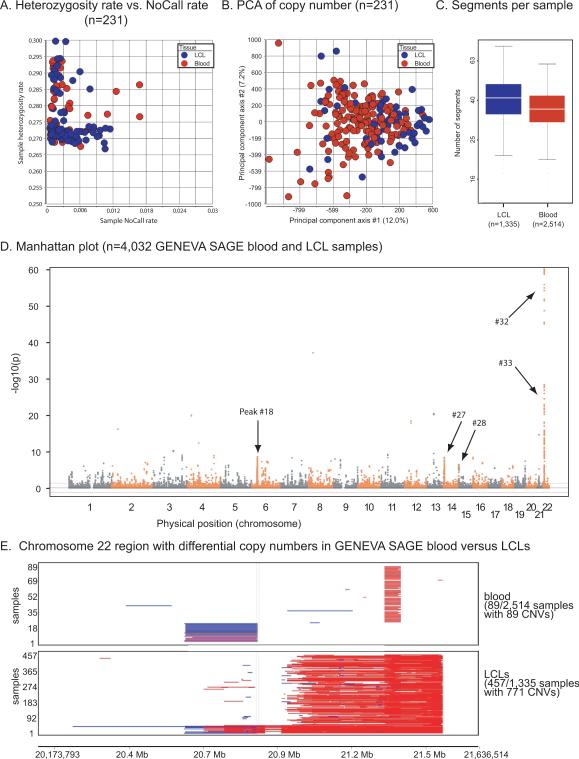
To assess data quality, we analyzed a subset of 231 replicate samples within the GENEVA SAGE project. 231 samples formed 195 pairwise replicate sets, including groups of replicate samples derived from blood-blood comparisons (n=30), LCL-LCL (n=156), or blood-LCL (n=9). For each of these groups, the pairwise distances were extremely small (mean values of 7.9e–05, 2.0e–04, 9.9e–05 respectively)(data not shown). These distances were even smaller than those reported for the other data sets (Supp. Figure S1), possibly due to the use of the Illumina Human1M genotyping platform. Heterozygosity and NoCall rates for the 231 individuals were comparable to those of our previous data sets, with no appreciable differences between samples derived from blood or LCLs (Figure 5A). PCA of logR ratio estimates of copy number did not reveal overall differences between blood-derived and LCL samples (Figure 5B).
Figure 5.
Analysis of CNVs in blood samples and LCLs from the GENEVA SAGE data set. (A) Plot of heterozygosity rate (y-axis) compared to sample NoCall rate (x-axis) for GENEVA SAGE replicate samples. (B) Principal components analysis of copy number data from GENEVA SAGE replicate samples. (C) Boxplot of the number of segments per sample after filtering using CNVineta software. (D) Manhattan plot showing association of CNV regions with blood samples or LCLs (y-axis; −log10 of probability value) as a function of chromosomal position (x-axis). Note that peak numbers for significant regions are found in Supp. Table S3. Several peaks are indicated: peak 41 (chr6:32,066,939–32,114,701 encompassing 12 genes such as TNXB), peak 72 (chr14:40,739,852–40,739,853 having no annotated genes), peak 73 (chr14:105,268,029–105,397,778 having 17 genes including immunoglobulin loci for IGHA2, IGHE, IGHG1, and IGHD), peak 84 (chr22:21,028,552–21,443,164 having 17 genes including immunoglobulin loci), and peak 85 (chr22:21,543,587–21,570,027 having four genes including immunoglobulin loci). (E) Visualization of a region showing the greatest difference between LCLs and blood samples (see panel D, peak at chromosome 22). The number of samples (y-axis) is plotted by chromosomal position (x-axis, chr22:20,745,308–21,609,299). Each horizontal bar in the plot corresponds to a CNV segment in a single sample from the GENEVA SAGE data set.
We used the R package CNVineta to analyze variation in copy number across samples in the GENEVA SAGE data set. There were more CNV segments per sample on average for cell line derived samples than whole blood derived samples, but the differences were not statistically significant (Figure 5C). Segments with greater than five markers and an average marker distance of 4 kb or less were included in subsequent analysis. Results from logistic regression analysis of case and control (cell line derived and whole blood derived) samples revealed 26 regions across the genome having (−log10p > 5) (Figure 5D). Each of these regions represented a locus having significantly different number of CNVs in cell lines relative to whole blood samples. Theseregions (Table 3) included a locus of 415 kb on chromosome 22 that had a dramatic increase in CNVs in the LCL samples (Figure 5E). This locus includes immunoglobulin lambda genes.
Table 3.
Regions associated with CNVs based on CNVineta analysis of GENEVA SAGE
| Peak# | Chromosome | Start-End | Length | Minimum p-value | Number of Genes |
|---|---|---|---|---|---|
| 1 | 1 | 187897346–187952120 | 54,774 | 2.09E-10 | 0 |
| 2 | 1 | 194989670–195077621 | 87,951 | 1.18E-12 | 4 |
| 3 | 2 | 89029231–89060957 | 31,726 | 3.93E-08 | 2 |
| 4 | 2 | 89132524–89320349 | 187,825 | 2.08E-08 | 2 |
| 5 | 2 | 89603161–89726605 | 123,444 | 1.18E-11 | 3 |
| 6 | 2 | 89810766–89885025 | 74,259 | 1.71E-12 | 1 |
| 7 | 3 | 163992511–164101579 | 109,068 | 6.79E-14 | 1 |
| 8 | 3 | 165387064–165498710 | 111,646 | 8.54E-09 | 0 |
| 9 | 3 | 166758469–166797775 | 39,306 | 2.74E-09 | 0 |
| 10 | 3 | 90359201–90437709 | 78,508 | 4.36E-12 | 0 |
| 11 | 4 | 34469747–34499424 | 29,677 | 3.16E-12 | 0 |
| 12 | 4 | 69085989–69117497 | 31,508 | 6.25E-10 | 1 |
| 13 | 5 | 104501925–104562047 | 60,122 | 2.22E-09 | 0 |
| 14 | 5 | 105415344–105443057 | 27,713 | 4.08E-11 | 0 |
| 15 | 5 | 28842013–28912873 | 70,860 | 1.92E-13 | 0 |
| 16 | 5 | 29457378–29488308 | 30,930 | 4.74E-09 | 0 |
| 17 | 6 | 103261441–103294189 | 32,748 | 1.91E-08 | 0 |
| 18 | 6 | 32066939–32114701 | 47,762 | 2.22E-11 | 12 |
| 19 | 6 | 32555416–32664508 | 109,092 | 2.82E-12 | 8 |
| 20 | 6 | 79029649–79090197 | 60,548 | 1.59E-09 | 0 |
| 21 | 9 | 43702737–43730292 | 27,555 | 1.89E-08 | 1 |
| 22 | 11 | 38249818–38354482 | 104,664 | 3.70E-17 | 0 |
| 23 | 11 | 48663034–48716940 | 53,906 | 4.09E-10 | 0 |
| 24 | 13 | 56537521–56585977 | 48,456 | 2.51E-07 | 0 |
| 25 | 13 | 56659471–56790893 | 131,422 | 3.48E-22 | 0 |
| 26 | 13 | 70999362–71033579 | 34,217 | 1.18E-13 | 3 |
| 27 | 14 | 105268029–105397778 | 129,749 | 1.39E-34 | 17* |
| 28 | 14 | 105529364–106174705 | 645,341 | 3.83E-54 | 6 |
| 29 | 16 | 18206435–18262224 | 55,789 | 1.93E-10 | 0 |
| 30 | 16 | 33889259–33928458 | 39,199 | 1.55E-09 | 0 |
| 31 | 17 | 16664267–16698162 | 33,895 | 2.92E-07 | 1 |
| 32 | 22 | 21028552–21443164 | 414,612 | 1.59E-35 | 17* |
| 33 | 22 | 21543587–21570027 | 26,440 | 8.34E-10 | 4* |
Regions greater than 25 kb and comprised of markers significantly associated with CNVs (−log10 p > 5) are listed. Regions having a length of one are represented by only one single significant marker. Minimum p-values are the most significant p-value in each region. Number of genes refers to the UCSC Genes track from the UCSC Genome Browser (hg18).
denotes regions inclusive of immunoglobulin loci.
In an analysis of CNVs in ~19,000 individuals, the Wellcome Trust Case Control Consortium (Redon, et al., 2006) measured genome-wide intensity data at several thousand polymorphic loci. They noted that samples were separable based on their origin (blood versus LCLs) based on PCA of intensity data. We plotted PCA based on intensity data for 106 SNPs spanning the chromosome 22 locus of Figure 5E (see Supp. Figure S3). This showed an overlapping profile for the majority of LCL- and blood-derived samples, with a large number of additional signals corresponding exclusively to LCL-derived samples.
We created a database of variants that were significantly associated with LCLs (from the GENEVA SAGE data set) in the form of a browser extensible data (.bed) file that is compatible with resources such as the UCSC Genome Browser (Hinrichs, et al., 2006))(Supp. File S1). For comparison we created.bed files representing the data in Figure 4 for LCLs, DCLs, and HapMap replicates (Supp. Files S2–S4).
Discussion
A major finding of this study was that multiple immortalized LCLs derived from a given individual were extremely similar in terms of genotype and copy number, compared to controls. Taking into account the technical performance of the Affymetrix 6.0 platform, 26 of the 29 Coriell LCLs (90%) were not significantly different from PBMCs derived from the same individual, as ascertained by SNP concordance. These 26 LCLs showed 99.94–99.98% concordance with PBMCs from the same individual (Table 1). Affymetrix states that genotyping results obtained using its SNP 6.0 platform are 99.9% reproducible, a finding confirmed by Nishida et al. (Nishida, et al., 2008), who similarly found an average concordance rate of 99.8% in SNP 6.0 data analyzed with the Affymetrix Birdseed algorithm. The remaining 3 LCLs had concordance rates of 99.78% (L24), 99.66% (L33), and 98.95% (L51) to PBMCs from the same subject, attributed to lower data quality in samples L33 and L24, as well as mosaic UPD on chromosome 4q in sample L51. Mosaic loss of the x chromosome occurs commonly in cultured lymphocytes (Guttenbach, et al., 1995).
Five of the 29 Coriell LCL samples harbored large mosaic abnormalities, while such abnormalities were not present in the DCLs or replicates. There are several possible sources for the introduction of mosaic abnormalities in LCLs. EBV infection may introduce genomic instability in newly established cell lines, or the conditions of cell culture may favor an increase in genomic instability or proliferation of a sub-population of variants pre-existing in the primary tissue. For example, Rodríguez-Santiago et al. have demonstrated the existence of mosaic abnormalities in 1.7% of buccal and blood samples (Rodriguez-Santiago, et al., 2010).Mosaic aneuploidy has been detected in 1% of 2,019 cases referred for clinical diagnostic testing, with abnormalities caused by meioitic or mitotic nondisjunction of both autosomes and sex chromosomes (Conlin, et al., 2010). Regardless of origin, the presence of mosaic abnormalities may result in a skewing of genome-wide allele frequencies by causing a reduction in heterozygous genotype calls while increasing NoCalls and homozygous calls. This could affect analyses utilizing identity-by-state (IBS) and identity-by-descent (IBD) estimations. The presence of mosaicism also indicates a propensity for the introduction or propagation of abnormalities in LCLs during cell culture.
The design of the Coriell data sets allowed us to perform a direct comparison of the concordance between a matched primary/immortalized data set and a matched DCL data set, providing a unique window to distinguish between common cell culture-induced and transformation-induced alterations. The prominent regions of copy number change in LCLs, found in over half of the samples and spanning at least 50 kb, included intersections with the three main loci harboring immunoglobulin genes on chromosomes 2, 7, and 14 (Table 2). These included genes encoding VDJ segments and immunoglobulin heavy and light chains. Thus we interpret such variants to represent possible LCL-specific alterations rather than natural variation. Other commonly occurring regions matched CNV regions reported in dozens or even hundreds of samples from the Database of Genomic Variants (DGV)(Zhang, et al., 2006). This likely reflects common variation in our samples.
A wide range of CNV concordances with values as high as 70% has been reported for replicate samples from the same individual on the Affymetrix 6.0 genotyping platform (Pinto, et al., 2011). In the present study, the CNV concordance rates among DCLs were also 70% (Figure 4). For LCLs, the CNV concordance rate was slightly lower (56%). This lower value may be attributed to greater variation inherent in LCLs, although these concordance rates were derived from a relatively small number of samples (n=35 LCLs). For this reason we complemented studies of the Coriell LCLs with analyses of a large GWAS from GENEVA SAGE, in which we compared whole blood to LCL samples from thousands of individuals, as well as a series of several hundred replicates. The application of CNVineta to this large data set allowed us to assign p-values to CNV regions enriched in LCLs and address LCL-specific changes with more confidence. As with the Coriell data set, we observed large differences in copy number at immunoglobulin loci (including chromosomes 2, 6, 14 and 22); see Figures 5D and 5E for examples of events encompassing the HLA, immunoglobulin kappa, and immunoglobulin lambda regions. Of the regions associated with LCLs in GENEVA SAGE, three (including two immunoglobulin regions) are also represented in Coriell LCLs at a significance threshold of 0.05 (Table 2). These regions, including the chromosome 22q11.2 immunoglobulin lambda region (Sebat, et al., 2004), have been previously reported as variable in LCLs (Simon-Sanchez, et al., 2007). This provided some validation of our ability to find LCL-specific changes in GENEVA SAGE.
Based on our study of PBMCs and corresponding LCLs, we conclude that LCLs are generally able to faithfully reflect the genotype and copy number of PBMCs from which they are derived given the resolution of genotyping platforms and concordance between CNV calls in matched samples. However, the occurrence of large regions of mosaic UPD or aneuploidy, in 4 out of 29 LCL samples (13.7%), suggests that it is appropriate to routinely characterize LCLs via SNP array genotyping or other methods before performing further studies such as whole genome sequencing, assaying gene expression, pharmacogenomic investigations, or other applications. The list of putative LCL-specific changes (Supp. File S1) resulting from our analysis of GENEVA SAGE may prove useful for these types of studies. It will also be of interest to characterize chromosomal alterations that occur as a function of increasing passage number.
Supplementary Material
Acknowledgments
We thank Yue Yu for help with data analysis, and Drs. Sarah Wheelan and Robert Scharpf for helpful discussions. We thank members of the SAGE project including Drs. Laura Bierut, Cathy Laurie, Sherri Fisher, and Bruce Weir, for generously sharing data, providing helpful comments, and interpreting results. Funding support for SAGE was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of GENEVA under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by NCBI. Support for collection of data sets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The data sets used for the analyses described in this manuscript were obtained from dbGaP (accession number phs000092.v1.p).
Grant sponsor: JP was supported by NIH grant HD24061. Z.T., N.G., C.M.B., and D.S.B. were supported by NIGMS HGCR contract HHS-N-263-2009-00026-C.
References
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–5. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, Zackai EH, Deardorff MA, Krantz ID, Hakonarson H, Spinner NB. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19:1263–75. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, Keebler JE, DePristo MA, Lindsay SJ, Zhang Y, Casals F, Idaghdour Y, Hartl CL, Torroja C, Garimella KV, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–4. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis MC, Agrawal A, Cole JW, Hansel NN, Barnes KC, Beaty TH, Bennett SN, Bierut LJ, Boerwinkle E, Doheny KF, et al. The Gene, Environment Association Studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol. 2010;34:364–72. doi: 10.1002/gepi.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaquer A, Rappold GA, Wienker TF, Fischer C. The human pseudoautosomal regions: a review for genetic epidemiologists. Eur J Hum Genet. 2008;16:771–9. doi: 10.1038/ejhg.2008.63. [DOI] [PubMed] [Google Scholar]
- Gonzalez JR, Rodriguez-Santiago B, Caceres A, Pique-Regi R, Rothman N, Chanock SJ, Armengol L, Perez-Jurado LA. A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC bioinformatics. 2011;12:166. doi: 10.1186/1471-2105-12-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhne B, Sompallae R, Masucci MG. Three Epstein-Barr virus latency proteins independently promote genomic instability by inducing DNA damage, inhibiting DNA repair and inactivating cell cycle checkpoints. Oncogene. 2009;28:3997–4008. doi: 10.1038/onc.2009.258. [DOI] [PubMed] [Google Scholar]
- Guttenbach M, Koschorz B, Bernthaler U, Grimm T, Schmid M. Sex chromosome loss and aging: in situ hybridization studies on human interphase nuclei. Am J Hum Genet. 1995;57:1143–50. [PMC free article] [PubMed] [Google Scholar]
- Herbeck JT, Gottlieb GS, Wong K, Detels R, Phair JP, Rinaldo CR, Jacobson LP, Margolick JB, Mullins JI. Fidelity of SNP array genotyping using Epstein Barr virus-transformed B-lymphocyte cell lines: implications for genome-wide association studies. PLoS One. 2009;4:e6915. doi: 10.1371/journal.pone.0006915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–8. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman L, Wilson JA, Buller A, Dixon J, Edelmann L, Geller L, Highsmith WE, Holtegaard L, Kornreich R, Rohlfs EM, et al. Development of genomic DNA reference materials for genetic testing of disorders common in people of ashkenazi jewish descent. J Mol Diagn. 2009;11:530–6. doi: 10.2353/jmoldx.2009.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamranvar SA, Gruhne B, Szeles A, Masucci MG. Epstein-Barr virus promotes genomic instability in Burkitt's lymphoma. Oncogene. 2007;26:5115–23. doi: 10.1038/sj.onc.1210324. [DOI] [PubMed] [Google Scholar]
- Nishida N, Koike A, Tajima A, Ogasawara Y, Ishibashi Y, Uehara Y, Inoue I, Tokunaga K. Evaluating the performance of Affymetrix SNP Array 6.0 platform with 400 Japanese individuals. BMC Genomics. 2008;9:431. doi: 10.1186/1471-2164-9-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Darvishi K, Shi X, Rajan D, Rigler D, Fitzgerald T, Lionel AC, Thiruvahindrapuram B, Macdonald JR, Mills R, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol. 2011;29:512–20. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Pevsner J. Visualization of shared genomic regions and meiotic recombination in high-density SNP data. PLoS One. 2009;4:e6711. doi: 10.1371/journal.pone.0006711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Santiago B, Malats N, Rothman N, Armengol L, Garcia-Closas M, Kogevinas M, Villa O, Hutchinson A, Earl J, Marenne G, et al. Mosaic uniparental disomies and aneuploidies as large structural variants of the human genome. Am J Hum Genet. 2010;87:129–38. doi: 10.1016/j.ajhg.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sie L, Loong S, Tan EK. Utility of lymphoblastoid cell lines. J Neurosci Res. 2009;87:1953–9. doi: 10.1002/jnr.22000. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, de Vrieze FW, Peckham E, Gwinn-Hardy K, et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- Ting JC, Roberson ED, Currier DG, Pevsner J. Locations and patterns of meiotic recombination in two-generation pedigrees. BMC Med Genet. 2009;10:93. doi: 10.1186/1471-2350-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JC, Roberson ED, Miller ND, Lysholm-Bernacchi A, Stephan DA, Capone GT, Ruczinski I, Thomas GH, Pevsner J. Visualization of uniparental inheritance, Mendelian inconsistencies, deletions, and parent of origin effects in single nucleotide polymorphism trio data with SNPtrio. Hum Mutat. 2007;28:1225–35. doi: 10.1002/humu.20583. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M, Mangravite L, Medina MW, Tantisira K, Zhang W, Huang RS, McLeod H, Dolan ME. Pharmacogenomic discovery using cell-based models. Pharmacol Rev. 2009;61:413–29. doi: 10.1124/pr.109.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig M, Helbig I, Schreiber S, Franke A. CNVineta: a data mining tool for large case-control copy number variation datasets. Bioinformatics. 2010;26:2208–9. doi: 10.1093/bioinformatics/btq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Liu MT, Chang YT, Fang CY, Chou SP, Liao HW, Kuo KL, Hsu SL, Chen YR, Wang PW, et al. Epstein-Barr virus DNase (BGLF5) induces genomic instability in human epithelial cells. Nucleic Acids Res. 2010;38:1932–49. doi: 10.1093/nar/gkp1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res. 2006;115:205–14. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.