Abstract
Diamond-Blackfan anemia (DBA) is an inherited form of pure red cell aplasia that usually presents in infancy or early childhood and is associated with congenital malformations in ~30-50% of patients. DBA has been associated with mutations in nine ribosomal protein (RP) genes in about 53% of patients. We completed a large scale screen of 79 RP genes by sequencing 16 RP genes (RPL3, RPL7, RPL8, RPL10, RPL14, RPL17, RPL19, RPL23A, RPL26, RPL27, RPL35, RPL36A, RPL39, RPS4X, RPS4Y1, and RPS21) in 96 DBA probands. We identified a de novo two-nucleotide deletion in RPL26 in one proband associated with multiple severe physical abnormalities. This mutation gives rise to a remarkable ribosome biogenesis defect that affects maturation of both the small and the large subunits. We also found a deletion in RPL19 and missense mutations in RPL3 and RPL23A, which may be variants of unknown significance. Together with RPL5, RPL11, and RPS7, RPL26 is the fourth ribosomal protein regulating p53 activity that is linked to DBA.
Keywords: Diamond-Blackfan anemia, ribosomal protein genes, RPL26, ribosome biogenesis
INTRODUCTION
Diamond-Blackfan anemia (DBA; MIM# 105650) is an inherited form of pure red cell aplasia [Diamond LK, 1938] that usually presents in infancy or early childhood and affects approximately 5 to 7 children per million live births, equally in both genders [Ball et al., 1996; Campagnoli et al., 2004; Willig et al., 1999b]. The disease is most often characterized by a mild-to-severe macrocytic anemia, a normocellular bone marrow with selective erythroid hypoplasia, and occasional neutropenia and/or thrombocytosis [Alter and Young, 1998; Vlachos et al., 2008]. Laboratory findings such as increased mean corpuscular volume (MCV), elevated erythrocyte adenosine deaminase activity (eADA), and hemoglobin F (HbF) are observed in a majority of DBA patients [Glader et al., 1983; Vlachos et al., 2008; Willig et al., 1999a]. Growth retardation and congenital anomalies, especially of the head, neck, upper limbs and urinary system, are present in approximately 30-50% of patients, indicating that DBA is a broad disorder of development [Alter and Young, 1998; Ball et al., 1996; Vlachos et al., 2008; Vlachos et al., 2001; Willig et al., 1999b]. Although most cases are sporadic, about 40-45% are familial and show autosomal, and commonly dominant inheritance [Orfali et al., 2004]. The anemia is initially responsive to prednisone in approximately 60-70% of patients; however, to maintain normal erythropoiesis, a majority of these patients require continued low dose steroid treatment [Alter and Young, 1998; Vlachos and Muir, 2010]. Patients who do not respond to steroid therapy are usually transfusion-dependent, although responses to androgen and Interleukin-3 (IL-3) have been reported [Ball et al., 1995; Dunbar et al., 1991; Gillio et al., 1993]. Bone marrow transplantation can be curative for transfusion-dependent patients if a histocompatible bone marrow donor is available [Alter, 1998; Vlachos et al., 2008; Vlachos and Muir, 2010]. Reports in the literature reveal an increase in the incidence of malignancies, particularly acute myelogenous leukemia and solid tumors, in patients with DBA [Aquino and Buchanan, 1996; Janov et al., 1996; Lipton et al., 2001; van Dijken and Verwijs, 1995].
DBA has been associated with heterozygous mutations in nine ribosomal protein genes; in six small subunit ribosomal protein genes RPS7 (MIM# 612563), RPS10 (MIM# 603632), RPS17 (MIM# 180472), RPS19 (MIM# 603474), RPS24 (MIM# 602412), and RPS26 (MIM# 603701), and in three large subunit ribosomal protein genes RPL5 (MIM# 612561), RPL11 (MIM# 612562), and RPL35A (MIM# 180468) [Cmejla et al., 2007; Doherty et al., 2010; Draptchinskaia et al., 1999; Farrar et al., 2008; Gazda et al., 2006; Gazda et al., 2008]. Mutations in these genes have been reported in about 53% of DBA patients indicating that DBA is a disorder of ribosomal biogenesis and/or function. All these ribosomal protein genes are necessary for production of the small or the large subunits. Mutations in DBA usually result in haploinsufficiency of the protein, which causes a defect in ribosome biogenesis that can be detected in patients’ total RNA [Choesmel et al., 2007; Doherty et al., 2010; Farrar et al., 2008; Flygare et al., 2007; Gazda et al., 2008; Idol et al., 2007]; there is evidence for a dominant negative mechanism that may explain the pathogenesis in some patients with point mutations [Devlin et al.]. DBA is to date the only congenital disease associated with ribosomal protein gene mutations, while haploinsufficiency for ribosomal protein gene RPS14 has been implicated in the pathophysiology of the 5q-syndrome, a subtype of acquired myelodysplastic syndrome [Ebert et al., 2008].
The pathophysiological mechanisms of DBA remain elusive. Perdahl et al. demonstrated increased apoptosis in DBA erythroid progenitor cells [Perdahl et al., 1994]. Increased apoptosis has also been shown in hematopoietic cell lines and in bone marrow cells with deficiency of RPS19 and RPL35A [Farrar et al., 2008; Miyake et al., 2008]. Haploinsufficiency of ribosomal proteins in mice does not recapitulate the hematological phenotype observed in humans, but rather induces various phenotypes which are suppressed by impairment of p53, suggesting that imbalance of the p53 family proteins is involved in DBA [McGowan et al., 2008]. Similar observations were made in zebrafish treated with morpholinos specific to RPS19, which displayed abnormal embryogenesis and anemia [Danilova et al., 2008]. Interestingly, it has been demonstrated that erythroid progenitor cells may be more affected by p53 activation upon decreased expression of the RPS14 and RPS19 genes, as compared with other hematopoietic lineages [Dutt et al., 2011], which provides a rationale for the specific sensitivity of this lineage in DBA. Several ribosomal proteins can up-regulate p53 activity by sequestering Hdm2, the E3 ubiquitin ligase that targets p53 to the proteasome, or by favoring p53 translation [Chen et al., 2007; Dai and Lu, 2004; Dai et al., 2004; Lohrum et al., 2003; Ofir-Rosenfeld et al., 2008; Zhang et al., 2010]. Haploinsufficiency of a ribosomal protein perturbs pre-rRNA maturation and incorporation of neo-synthesized ribosomal proteins into nascent pre-ribosomes, thereby increasing the pool of free ribosomal proteins available to stimulate p53 activation. In addition, defects in ribosome production may also stimulate the synthesis of ribosomal proteins [Fumagalli et al., 2009].
Here, we present the completion of the systematic sequencing of the ribosomal protein genes in a cohort of 96 patients. This work adds a tenth ribosomal protein gene mutated in a DBA, namely RPL26 (MIM# 603704), and identifies rare polymorphisms of uncertain pathogenic significance in three other genes. RPL26 mutation leads to a global pre-ribosomal RNA (pre-rRNA) maturation defect, which mainly affects formation of the large subunit, but also perturbs processing of the precursors to the 18S rRNA. Interestingly, RPL26 is a positive regulator of p53 activity, like RPL5, RPL11 and RPS7, which are also mutated in DBA.
MATERIALS AND METHODS
Patients
Ninety-six families participated in the study, the majority of whom were included in previous reports [Doherty et al., 2010; Farrar et al., 2008; Gazda et al., 2006; Gazda et al., 2008]. Twelve of them were multiplex DBA families,and 84 comprised of only one affected individual. Informed consent was obtained from all patients and their family members under a protocol at Children’s Hospital Boston (Boston, MA, USA). The diagnosis of DBA in all probands was based on the findings of normochromic anemia, elevated erythrocyte adenosine deaminase activity (eADA), reticulocytopenia and a low number or lack of erythroid precursors in the bone morrow, often associated with congenital malformations. None of the 96 probands was previously found to have a pathogenic mutation and one proband had a variant of unknown significance in RPS15 [Gazda et al., 2008].
DNA isolation and genomic DNA sequencing
Genomic DNA was isolated from blood samples using the Nucleic Acid Isolation System QuickGene – 610L (AutoGen, Inc., Holliston, MA, USA) according to the manufacturer’s instructions. To complete our large scale screen of RP genes in a DBA population, genomic DNA samples from 96 unrelated DBA probands enrolled in the study were amplified by polymerase chain reaction (PCR) and sequenced for mutations in 16 RP genes: RPL14, RPL7, RPL8, RPL35, RPL19, RPL23A, RPL26, RPL27, RPL17, RPS21, RPL3, RPL10, RPL36A, RPL39, RPS4X and RPS4Y1, on chromosomes 3, 8, 9, 17, 18, 20, 22, X and Y. In total, together with the 63 RP genes sequenced previously [Doherty et al., 2010; Farrar et al., 2008; Gazda et al., 2006; Gazda et al., 2008; Gazda et al., 2004], we screened all 79 RP genes in this cohort. Primers were designed using Primer3 software to amplify the coding exons and intron/exon boundaries of the above genes. PCR products, between 200 and 600 base pairs, were robotically prepared using a Hamilton MicroLab StarPLUS (Hamilton Company, Reno, NV), purified using Exo-SAP reagent (USB Corporation, Cleveland, OH) and sequenced using an Applied Biosystems 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). The chromatograms were analyzed using Sequencher software version 4.7 (Gene Codes Corporation, Ann Arbor, MI). Duplicate, independent PCR products were sequenced to confirm the observed nucleotide changes in the probands. DNA samples from at least 200 control individuals, i.e. 400 chromosomes were sequenced to determine whether the observed sequence variations were nonpathogenic variants. Subsequently, DNA samples from family members were sequenced to determine whether the mutation co-segregated with the DBA phenotype within the pedigree.
Tissue culture and RNA isolation
Epstein-Barr Virus (EBV)-transformed lymphoblastoid cell lines (LCLs) were cultured in RPMI 1640 medium supplemented with 15% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin and 0.292 mg/ml L-glutamine (all from Invitrogen, Carlsbad, California) at 37°C in 5% CO2. Total RNAs were isolated from LCLs using RNA isolation kit (Qiagen, Valencia, CA, USA).
Western blotting
Western blotting of protein lysate from lymphoblastoid cell lines from the six family members with the RPL19 mutation was performed as previously described [Gazda et al., 2004]. The RPL19 wt and mutated proteins were detected with the rabbit polyclonal anti-RPL19 antibody (ProteinTech Group, Inc, Chicago, IL) and anti-rabbit IgG (H+L) HRP conjugated antibodies (Bio-Rad, Hercules, CA). Proteins were visualized by enhanced chemiluminescence (Super Signal West Pico Chemiluminescent Substrate, Thermo Scientific, Rockford, IL) on the Gel Doc XR + System (Bio-Rad) using Quantity One 1-D Analysis Software (Bio-Rad). The p53 and GAPDH proteins were detected in lymphoblastoid cell lines from control individuals and RPL26 mutated patient using rabbit polyclonal anti-p53 and anti-GAPDH antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and anti-rabbit IgG (H+L) HRP conjugated antibodies (Bio-Rad, Hercules, CA). The proteins were visualized as above.
Cell culture and siRNAs
Human cervical carcinoma HeLa cells were grown in Dulbecco’s Modified Eagle Medium (DMEM), supplemented with 10% fetal calf serum (FCS) and 1 mM sodium pyruvate (Invitrogen, Paisley, UK). The 21 nt siRNA duplexes with a 3′ dTdT overhang corresponding to L3 mRNA (5′-GACCATGGAGGAGAAGAAA-3′), L19 mRNA (5′-GACCAATGAAATCGCCAAT-3′), L26 mRNA (5′-GTCCAGGTTTACAGGAAGA-3′) and L11 mRNA (5′-AAGGTGCGGGAGTATGAGTTA-3′), were purchased from Eurogentec (Seraing, Belgium) and diluted to 100 μM. After washing the cells once in DMEM (without FCS), 10 μL of the siRNA solution were added on ice to 107 HeLa cells in suspension in 200 μL of the same medium. Electro-transformation was performed at 250 V and at 975 μF with a Gene Pulser (BioRad, Hercules, CA) in a cuvette with a 4-mm inter-electrode distance. The cells were diluted in 20 mL DMEM supplemented with FCS and plated in a 140-cm2 Petri dish. Control samples were electro-transformed with a scrambled siRNA or without siRNA, which lead to similar results.
Analysis of ribosomes by sucrose density gradient centrifugation
Forty-eight hours after transfection, HeLa cells were treated with 100 μg/ml cycloheximide (Sigma) for 10 min. The cells were successively washed at 4°C in the presence of cycloheximide with DMEM, PBS, and Buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl). They were then mechanically disrupted with a Dounce homogenizer in Buffer A containing 0.5 mM DTT. After centrifugation (4°C, 1000×g, 10 min), the upper cytoplasmic fractions were kept. Fractions containing 1 mg proteins were loaded on 10-50% (w/w) sucrose gradients, prepared with a Gradient Master former (BioComp Instruments, Fredericton, NB, Canada). The tubes were centrifuged at 4°C and at 36 000 rpm for 105 min in a SW41 rotor (Optima L100XP ultracentrifuge; Beckman Coulter, Villepinte, France). The gradient fractions were collected at OD254 nm with a Foxy Jr. gradient collector (Teledyne Isco, Lincoln, NE).
Extraction and analysis of pre-rRNAs by Northern blot
Total RNAs were purified from transfected HeLa cells with the Trizol reagent. After alcohol precipitation, RNA pellets were dissolved in formamide, quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and diluted to 1 mg/mL. RNAs (3 μg/well) were separated on a 1% agarose gel prepared with Tri/Tri buffer (30 mM tri-ethanolamine, 30 mM tricine, pH 7.9) containing 1.2% formaldehyde, and run in Tri/Tri buffer at 140 V. Alternatively, small pre-rRNA species were analyzed on 6% denaturing polyacrylamide gels [Gazda et al., 2008]. RNAs were then transferred to a Hybond N+ nylon membrane (GE Healthcare, Orsay, France) and fixed by UV-cross-linking. Membranes were hybridized as described previously [O’Donohue et al., 2010]. The probes used in the present study were 18S (5′-TTTACTTCCTCTAGATAGTCAAGTTCGACC-3′), 5′ITS1 (5′-CCTCGCCCTCCGGGCTCCGTTAATGATC-3′), ITS1-721 (5′-GGAGCGGAGTCCGCGGTG-3′), ITS2 (an equal amount of 5′-CTGCGAGGGAACCCCCAGCCGCGCA-3′ and 5′-GCGCGACGGCGGACGACACCGCGGCGTC-3′), 5′ITS2 (5′-GGGGCGATTGATCGGCAAGCGACGCTC-3′), 5.8S (5′-CAATGTGTCCTGCAATTCAC-3′), 28S (5′-CCCGTTCCCTTGGCTGTGGTTTCGCTAGATA-3′) and 7SK (an equal amount of 5′-CATGGAGCGGTGAGGGAGGA-3′ and 5′-GTGTCTGGAGTCTTGGAAGC-3′). After hybridization, the membranes were washed twice for 10 min at room temperature in 2× SSC, 0.1% SDS, and once in 1× SSC, 0.1% SDS. Labeled RNA signals were acquired with a FLA2000 PhosporImager (Fuji, Stamford, CT) and quantified with the ImageGauge software.
Accession Numbers
The GenBank accession number for human RPs are: NM_000987.3 for RPL26, NM_000981.3 for RPL19, NM_000967.3 for RPL3 and NM_000984.5 for RPL23A. In all genes presented here, nucleotide numbering reflects cDNA numbering with position +1 corresponding to the A of the ATG translation initiation codon. Mutation descriptions on the protein level consider the initiator methionine as codon 1 and have been checked using the Mutalyzer program (http://www.LOVD.nl/mutalyzer/). Variants were submitted to an RPL26 mutation database (http://www.lovd.nl/RPL26).
RESULTS
DNA sequencing analysis
We completed our large scale screen of 79 RP genes in our DBA patient cohort by sequencing 16 RP genes, RPL14, RPL7, RPL8, RPL35, RPL19, RPL23A, RPL26, RPL27, RPL17, RPS21, RPL3, RPL10, RPL36A, RPL39, RPS4X and RPS4Y1, on chromosomes 3, 8, 9, 17, 18, 20, 22, X and Y for mutations in 96 DBA probands. We identified a de novo heterozygous sequence change in RPL26 in one proband with severe multiple physical malformations and responsive to steroid treatment (both patient’s parents were tested and they are mutation free). The sequence change is the deletion of two nucleotides in exon 2 of the RPL26 gene (c. 120_121delGA) causing a frameshift at codon 40 and premature termination codon (p.Lys41ValfsX12). We also found a deletion of two nucleotides in exon 6 in the RPL19 gene (c. 562_563delTT) in one proband who is transfusion-dependant, was born with cleft palate and has a short stature. This sequence change causes a frameshift at codon 188 and a new stop codon 21 amino acids behind the wt stop codon (p.Leu188IlefsX30). We also sequenced DNA samples from this proband’s clinically unaffected parents and three siblings (with normal MCV and eADA), and identified the same sequence change in the mother and the brother. In addition, we identified two missense changes in two genes: RPL3 (c.32A>G) in a steroid-dependent proband, causing His11Arg substitution, and RPL23A (c.296T>C) in another steroid-dependent proband and her transfusion-dependent brother, resulting in Ile99Thr substitution. The same sequence changes were found in the first proband’s clinically unaffected father and second proband’s clinically unaffected mother, respectively.
Case report (the patient with the RPL26 frameshift mutation)
A 31 year-old primigravida presented at 30 weeks gestation for evaluation of an abnormal prenatal scan. Pre-natal imaging showed an absent left kidney and foreshortened arms bilaterally with abnormal angulation of the wrists. Possible diagnoses of Fanconi anemia, VACTERL and thrombocytopoenia absent radius were considered. A female infant was delivered via C-section at a local hospital at 37 weeks gestation weighting five pounds five ounces, and was noted to have an isolated bilateral cleft palate, an absent external auditory meatus on the right, and a narrowed meatus on the left. In addition, she had a bicuspid aortic valve and agenesis of the left kidney. On the right side, there was a single bone in the forearm, likely an ulna, and, on the left, synostosis of a shortened radius and ulna. Both thumbs were absent, and she had three digits on each hand. The lower eyelid on the left was incomplete. The baby was anemic at birth with a hemoglobin of 8.5 g/dl and was transfused twice in the perinatal period. A DEB test for Fanconi anemia was negative. The child was discharged home. By seven weeks of age the child was noted to again be anemic and profoundly neutropenic with a hemoglobin of 4.1 g/dl, an absolute neutrophil count (ANC) of 0.2k/μl, and an absolute reticulocyte count of 57/μl. A bone marrow examination showed a cellularity of 40% with markedly decreased erythropoiesis with only rare erythroblasts and maturing forms with megaloblastoid changes. There were marked granulocytic hypoplasia with left shifted maturation and increased blasts (9%). In light of the significant neutropenia in addition to anemia, the patient received a trial of steroids with good response, and the steroids were weaned as tolerated. The child has not required a transfusion since she was three months of age, and is maintained on a minimal dose of steroids at the age of three and a half years, with occasional neutropenia. The child continues to grow parallel to the standard growth curve for both height and weight, but is just below the fifth percentile for both at age three and a half years. Height is 90 cm (fiftieth percentile is 98 cm) and weight 12 kg (fiftieth percentile is 15 kg). Her weight for height is between the fifth and tenth percentile.
RPL19 protein analysis in lymphoblastoid cell lines from the family with c. 562_563delTT
We performed Western blotting using protein lysates from lymphoblastoid cell lines from the proband with sequence change, c. 562_563delTT, in RPL19 and his five first-degree relatives (his clinically unaffected mother and brother also carry the same sequence change). This sequence change causes a frameshift at codon 188 and a new stop codon 21 amino acids behind the wt stop codon (p.Leu188IlefsX30). Thus, the aberrant protein (216-amino-acid long with estimated molecular weight ~ 25.6 kDa) is predicted to be 20 amino acids longer than the wt protein (196-amino-acid long with estimated molecular weight ~ 23.47 kDa). As predicted, we found, in addition to the wt RPL19 protein, the mutated protein approximately ~2 kDa larger than the wt protein in all three individuals with the sequence change but not in the rest of the family (Figure 1).
Figure 1. Detection of RPL19 mutant form by Western blot.
Immunoblot shows RPL19 protein in all family members (with and without deletion (c. 562_563delTT) in the RPL19 gene). RPL19 mutant protein was detected in the probands and his clinically unaffected mother and brother with RPL19 deletion. Mut, mutation; Pt, patient; M*, mother; B*, brother; F, father; Sa, sister a; Sb, sister b.
Polysome analysis and pre-rRNA maturation assays in families with mutations in RPL26, RPL19 and RPL3
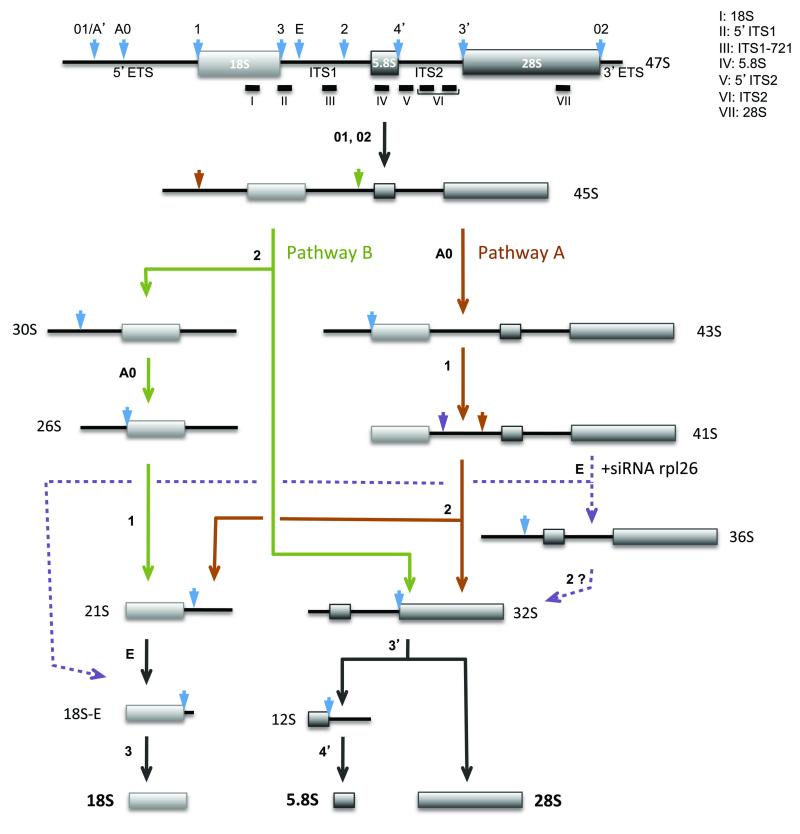
The 18S, 5.8S and 28S rRNAs are the end products of the maturation of the pre-ribosomal RNA (pre-rRNA) through endo- or exonucleolytic processing steps. The order of these processing events may vary, leading to the formation of different intermediates (Figure 2). All of the ribosomal proteins involved in DBA so far have proved essential in ribosome production, and pre-rRNA processing defects can be detected in patient cells.
Figure 2. Pre-rRNA processing in human cells.
The early 47S precursor is processed along different pathways depending on the order of the cleavages. Given the higher abundance of 30S pre-rRNA relative to the 41S species, cleavage of the ITS1 at site 2 appears to occur most frequently before processing of the 5′-ETS. Arrowheads indicate the pre-rRNA endonucleolytic cleavage sites, although some processing steps involve exonucleases rather than endonucleases. Dashed lines indicate the hypothetical pathway induced by RPL26 depletion.
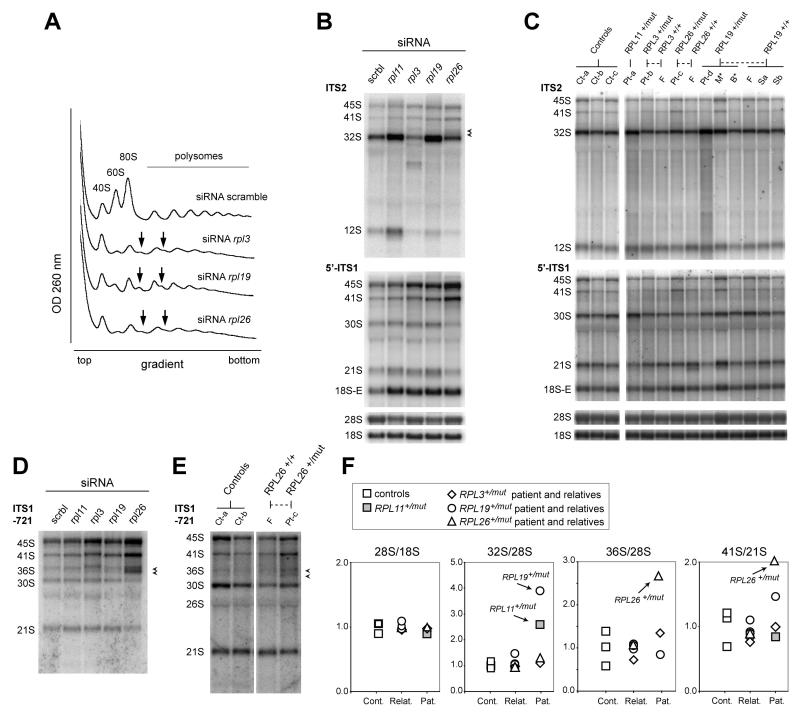
In order to determine the role of RPL3, RPL19 and RPL26 in pre-rRNA maturation and 60S subunit production, we designed siRNAs to knock down their synthesis in HeLa cells. Using quantitative PCR, we found that the siRNAs effectively reduced the amount of the mRNAs encoding the three proteins by 90%, 94% and 96% respectively. Interfering with the synthesis of any of the three proteins resulted in a strong defect in 60S subunit production: in sucrose gradient analysis of cytoplasmic ribosomes, the amount of free 60S subunits dropped, together with the 80S monosomes and the polysomes (Figure 3A). In addition, shoulders in the polysome peaks indicated the presence of half-mers, which correspond to 40S subunits positioned on start codons that fail to recruit 60S subunits. Pre-rRNA analysis by Northern-blot revealed three different phenotypes (Figure 3B). Upon RPL3 knockdown, we observed a general decrease of the amount of pre-60S specific RNAs and the presence of an unusual precursor migrating between the 32S and the 12S pre-rRNAs. No evidence for this phenotype was found in the RPL3+/mut patient (Figure 3C), suggesting that this mutation did not strongly affect RPL3 function in ribosome biogenesis. RPL19 depleted HeLa cells accumulated precursors to the 28S and 5.8S rRNAs, especially the 32S pre-rRNA (Figure 3B). Accordingly, we observed higher levels of 32S pre-rRNA in lymphoblasts from the RPL19+/mut patient (Figure 3C and 3F), whereas the mother and the brother harboring the same mutation in RPL19 did not display this phenotype. This result indicates that DBA is linked to a ribosome biogenesis defect in this patient, but that the mutation of RPL19 is not sufficient to explain this phenotype. Interestingly, the C-terminus of RPL19 extends out of the 60S subunit and is not involved in the interaction of this protein with ribosomal RNA [Ben-Shem et al., 2010] (Supp. Figures S1A and S1B). Finally, depletion of RPL26 with siRNAs in HeLa cells resulted in diminished accumulation of 32S pre-rRNA (Figure 3B, top panel). Interestingly, the pattern of the precursors to the 18S rRNA was also significantly altered, with lower proportions of 30S and 21S pre-rRNAs and a high level of 41S pre-rRNA (Figure 3B, lower panel). In addition, a probe hybridizing to the middle of the ITS1 (ITS1-721 probe) showed an increase in 36S pre-rRNA, as well as another precursor running just below (Figure 3D, arrows). This doublet was also visible with the ITS2 probe (Figure 3B). We interpret this peculiar phenotype as a defect in ITS1 cleavage at site 2, which normally separates the initial 90S pre-ribosomal particle into pre-40S and pre-60S particles. Strikingly, this doublet was found in the RPL26+/mut patient (Figure 3E). The 41S/21S pre-rRNA ratio was also much higher in this patient (Figure 3E and 3F), consistent with the pattern observed in RPL26 depleted cells (Figure 3D). This unique phenotype, not observed in the other patients or in controls, indicates that RPL26 function in ribosome biogenesis is affected.
Figure 3. Pre-rRNA processing in DBA patients.
(A) Sucrose gradient analysis of cytoplasmic ribosomes extracted from HeLa cells treated with siRNAs against RPL3, RPL19 and RPL26. The decrease of the free 60S ribosomal subunit and the formation of half-mers (arrows) indicate a strong defect in 60S subunit production. (B) Northern blot analysis of pre-rRNAs in HeLa cells upon depletion of RPL3, RPL19 and RPL26. siRNAs targeting RPL11, another 60S subunit protein involved in DBA, were used as a comparison [Doherty et al., 2010]. Precursors to the 18S rRNA are detected with the 5′-ITS1 probe, while the ITS2 probe reveals precursors to the 60S subunit RNAs. Arrowheads indicate the 36S pre-rRNA and a shorter precursor detected in RPL26 depleted cells (see panel D). (C) Northern blot analysis of pre-rRNAs in DBA patients and their relatives. RNAs were extracted from lymphoblastoid cells. Controls correspond to non-DBA individuals. RNAs from a DBA harboring a mutation in RPL11 were included as a comparison. These results were reproduced on three independent northern blots. (D) A probe hybridizing to the core of the ITS1 reveals accumulation of 36S pre-rRNA together wih a shorter precursor (arrowheads). (E) A similar doublet is detected in cells from the RPL26 mutated patient. Similar results were obtained on two independent northern blots. (F) Quantification of the Northern blot data by phosphorimager. For each ratio, the values were divided by the means of the values obtained for the controls. Measurements were performed three times on independent Northern blots with similar results, except for the 36S/18S ratio which was only measured on one blot. This ratio takes into account both the 36S pre-rRNA and the band migrating right below (see panel E).
DISCUSSION
Sequence changes in four RP genes, RPL26, RPL19, RPL3, and RPL23A, were identified in four DBA probands. In the case of RPL19, RPL3, and RPL23A, the mutations were found in clinically unaffected family members as well. The RPL26 gene is located on chromosome 17 and contains four exons with the start codon in exon 2. RPL26 encodes a 145-amino-acid long RPL26 protein, a component of the 60S ribosomal subunit. The de novo frameshift mutation (c. 120_121delGA) identified here is associated with severe multiple physical abnormalities and is predicted to create a 51-amino-acid long truncated protein which is unlikely to associate with the nascent ribosomal subunits. RPL26 is essential for the synthesis of the large ribosomal subunits: knockdown of RPL26 expression in HeLa cells resulted in low levels of free 60S subunit, formation of half-mers in the polysomes and a decrease in the amount of the precursors to large subunit rRNAs. Interestingly, depletion of RPL26 also has a significant impact on the 18S rRNA maturation pathway: pre-rRNA processing analysis by Northern blot strongly suggests a defect in cleavage of the ITS1 at site 2 that is not observed upon depletion of other large subunit ribosomal proteins mutated in DBA [Choesmel et al., 2008; Farrar et al., 2008]. This unique defect in the maturation of ribosomal RNA was unambiguously detected in the patient mutated in RPL26, while it was not observed in cells from the patient’s unaffected father. These findings indicate that the identified mutation in RPL26 is most likely a pathogenic mutation and causes DBA in the patient. This frameshift mutation in RPL26 presumably results in haploinsufficiency of the protein. First, the premature termination codon due to frame-shift is expected to induce nonsense mediated decay and prevent translation. Second, any synthesized protein would be missing around 90 amino-acids that ensure multiple contacts with the rRNA [Ben-Shem et al., 2010] (Supp. Figure S1C) and should be essential for RPL26 incorporation into the nascent ribosomal particles.
Remarkably, RPL26 was described as a regulator of p53 activity [Chen and Kastan, 2010; Ofir-Rosenfeld et al., 2008; Takagi et al., 2005; Zhang et al., 2010]. After RPL5, RPL11 and RPS7 [Gazda et al., 2008], RPL26 is yet another ribosomal protein involved in p53 regulation that is mutated in DBA. RPL26 binds the 5′-UTR of p53 mRNA and stimulates its translation [Chen and Kastan, 2010; Ofir-Rosenfeld et al., 2008; Takagi et al., 2005]. More recently, it was also proposed to be a negative regulator of Hdm2 [Zhang et al., 2010], the E3 ubiquitin ligase that promotes p53 degradation, as shown before for RPL5, RPL11 and RPS7. It is of note that this mutation of RPL26 is associated with severe skeletal defects, while patients with mutations in RPL5 and RPL11 also display developmental anomalies, especially cleft palates and triphalangeal thumbs. It is currently thought that the activation of p53 in response to a ribosomal stress caused by ribosomal protein haploinsufficiency is central to DBA physiopathological mechanisms. Indeed, we verified that p53 levels are higher in RPL26 mutated cells than in control cells (Supp.Figure S2). How loss of function of positive regulators of p53 fits with this hypothesis still calls for an answer. Redundant signaling pathways may be involved, as illustrated by the different modes of p53 regulation involving RPL11, which regulates p53 stability, and RPL26, which modulates both p53 synthesis and stability.
In contrast, sequence changes in RPL19, RPL3, and RPL23A are present in the probands and in their clinically unaffected family members. The RPL19 gene is located on chromosome 17 and contains six exons with the start codon in exon 1. It encodes a 196-amino-acid long RPL19 protein, a component of the 60S ribosomal subunit. The frameshift mutation (c. 562_563delTT) was identified in one proband and in his clinically unaffected mother and brother. This sequence change causes a frameshift at codon 188 and a new stop codon 21 amino acids behind the wt stop codon (p.Leu188IlefsX30). The mutation is predicted to result in the creation of a mutated protein 20 amino-acids longer than the wt protein. Accordingly, we detected a ~2kD larger aberrant protein in the proband, as well as in his clinically unaffected mother and brother who carry the mutation, but not in his father and two sisters who are mutation free. The wild-type and mutated forms were detected in similar amounts, which indicates that they are equally stable. This strongly suggests that the mutated form is incorporated into ribosomal subunits like wild-type RPL19 and might be functional. Interestingly, pre-rRNA processing of the large subunit is affected in the proband, but not in his mother and his brother carrying the RPL19 mutation. The abnormal pre-rRNA processing in the proband resembles the pattern of the rRNA processing in RPL19 depleted HeLa cells, but is also similar to the defects in DBA cells with RPL5 and RPL11 mutations [Gazda et al., 2008]. This patient’s DNA was screened by direct sequencing for mutations in all 79 RP genes, and no mutation was found in any of these genes including RPL5 and RPL11, although a large deletion in one of these genes in the proband cannot be excluded as it would not be detected by direct sequencing. Silent mutations in relatives of DBA patients were reported before in the case of RPS19 [Willig et al., 1999a]. However, these mutated, but unaffected, individuals displayed high levels of eADA, while the mother and the brother carrying the mutation in RPL19 display normal levels of ADA and a normal MCV. Therefore, we conclude that the frameshift mutation (c. 562_563delTT) in RPL19 is most likely a rare variant of unknown significance (not seen in 200 control individuals) that does not strongly affect RPL19 function. It is possible however that RPL19 mutation contributes to the phenotype in combination with a second mutation still to be found in the proband, or that a modifier gene suppresses RPL19 mutation pathogenic effect in the two other family members with the RPL19 mutation. We identified a missense mutation in the RPL3 gene (c.32A>G) in one proband and his clinically unaffected father causing His11Arg substitution. The RPL3 gene is located on chromosome 22 and comprises two isoforms, which contain ten exons. In both isoforms the start codon is located in exon 1. Isoform a encodes a 403-amino-acid, and isoform b, a 354-amino-acid long RPL3 protein, a component of the large ribosomal subunit. While RPL3 depletion in HeLa cells strongly affected the processing and the stability of the large subunit precursors, we did not detect similar defects in cells from the patient and his father, which indicates that the mutation does not affect RPL3 function in the ribosome assembly process. This suggests that the identified mutation in RPL3 is most likely a rare variant of unknown significance. We also found a missense mutation in the RPL23A gene (c.296T>C) resulting in Ile99Thr substitution in one proband and in her clinically unaffected mother. The RPL23A gene is located on chromosome 17 and contains 5 exons with the start codon in exon 1. RPL23A encodes a 156 amino-acid long protein, a component of the large ribosomal subunit. This missense change in RPL23A was not identified in ~200 control samples, and the family has one more child with DBA who has the same missense change, therefore this sequence change may be a pathogenic mutation present in the patient, her brother and her clinically unaffected mother. However, analysis of the RPL23A missense mutation (c.296T>C) was predicted to be benign when analyzed using PolyPhen software (http://genetics.bwh.harvard.edu/pph/).
In summary, we completed large-scale sequencing of 79 ribosomal protein genes and identified a frameshift de novo mutation in RPL26 in one proband. This report brings the total number of known DBA associated RP genes to ten: RPS19 (~25% of the patients), RPS24 (~2%), RPS17 (~1%), RPL35A (~3.5%), RPL5 (~6.6%), RPL11 (~4.8%), RPS7 (~1%), RPS10 (~6.4%), RPS26 (~2.6%), and RPL26 (~1%) [Doherty et al., 2010]. In total, these mutations account for approximately 53.9% of DBA patients. Characteristic defects in pre-rRNA maturation observed in cells from patients indicate that mutations in these genes have a direct effect on the protein function in ribosome biogenesis. We also found seven rare variants in RPS15, RPS27A, RPL3, RPL9, RPL19, RPL23A and RPL36 [Doherty et al., 2010] which could not be convincingly linked to pre-rRNA maturation defects. For example, as described above, pre-rRNA processing was normal in the family members with mutations in RPL3 and RPL19. Thus, without an alternative molecular phenotype to evaluate the loss-of-function of a ribosomal protein, these sequence changes can only be classified as rare variants of unknown significance. Further work is needed to understand whether these rare variants contribute in any away to the pathological mechanism of DBA, possibly in combination with other gene modifications.
Supplementary Material
Acknowledgments
We thank Marie Arturi for stimulating DBA research collaboration; and all of the physicians and DBA patients for participating in the study.
Grant Sponsor This work was supported by grants from the Diamond-Blackfan anemia Foundation (HTG), The Manton Center for Orphan Disease Research (HTG), NIH R01HL107558 (HTG), NIH R01AR044345 (AHB), NIH R01HL079571 (AV, EA, JML), the Centers for Disease Control and Prevention (AV), the Pediatric Cancer Foundation (JML) and the Agence Nationale de la Recherche (ANR-RIBOCRASH program) (PEG). DNA sequencing was performed by the Children’s Hospital Boston Program in Genomics and the Molecular Genetics Core Facility supported by the Developmental Disabilities Research Center (NIH P30 HD18655) and the Harvard Neuromuscular Disease Project (NIH P50 NS040828).
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
REFERENCES
- Alter BP. Bone marrow transplant in Diamond-Blackfan anemia. Bone Marrow Transplant. 1998;21(9):965–6. doi: 10.1038/sj.bmt.1701243. [DOI] [PubMed] [Google Scholar]
- Alter BP, Young NS. The bone marrow failure syndromes. In: Nathan DG, Orkin HS, editors. Hematology of Infancy and Childhood. Saunders; Philadelphia, PA: 1998. pp. 237–335. [Google Scholar]
- Aquino VM, Buchanan GR. Osteogenic sarcoma in a child with transfusion-dependent Diamond-Blackfan anemia. J Pediatr Hematol Oncol. 1996;18(2):230–2. doi: 10.1097/00043426-199605000-00030. [DOI] [PubMed] [Google Scholar]
- Ball SE, McGuckin CP, Jenkins G, Gordon-Smith EC. Diamond-Blackfan anaemia in the U.K.: analysis of 80 cases from a 20-year birth cohort. Br J Haematol. 1996;94(4):645–53. doi: 10.1046/j.1365-2141.1996.d01-1839.x. [DOI] [PubMed] [Google Scholar]
- Ball SE, Tchernia G, Wranne L, Bastion Y, Bekassy NA, Bordigoni P, Debre M, Elinder G, Kamps WA, Lanning M, et al. Is there a role for interleukin-3 in Diamond-Blackfan anaemia? Results of a European multicentre study. Br J Haematol. 1995;91(2):313–8. doi: 10.1111/j.1365-2141.1995.tb05295.x. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330(6008):1203–9. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- Campagnoli MF, Garelli E, Quarello P, Carando A, Sv SV, Nobili B, Dl DL, Pecile V, Zecca M, Dufour C, Ramenghi U, Dianzan I. Molecular basis of Diamond-Blackfan anemia: new findings from the Italian registry and a review of the literature. Haematologica. 2004;89(4):480–9. [PubMed] [Google Scholar]
- Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26(35):5029–37. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- Chen J, Kastan MB. 5′-3′-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev. 2010;24(19):2146–56. doi: 10.1101/gad.1968910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109(3):1275–83. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choesmel V, Fribourg S, Aguissa-Toure AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17(9):1253–63. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28(12):1178–82. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279(43):44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24(17):7654–68. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228–37. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- Devlin EE, Dacosta L, Mohandas N, Elliott G, Bodine DM. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood. 2010;116(15):2826–35. doi: 10.1182/blood-2010-03-275776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond LK BK. Hypoplastic anemia. Am J Dis Child. 1938;56:464–467. [Google Scholar]
- Doherty L, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Clinton C, Schneider HE, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Glader B, Arceci RJ, Farrar JE, Atsidaftos E, Lipton JM, Gleizes PE, Gazda HT. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2010;86(2):222–8. doi: 10.1016/j.ajhg.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169–75. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Dunbar CE, Smith DA, Kimball J, Garrison L, Nienhuis AW, Young NS. Treatment of Diamond-Blackfan anaemia with haematopoietic growth factors, granulocyte-macrophage colony stimulating factor and interleukin 3: sustained remissions following IL-3. Br J Haematol. 1991;79(2):316–21. doi: 10.1111/j.1365-2141.1991.tb04540.x. [DOI] [PubMed] [Google Scholar]
- Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, Kutok JL, Ebert BL. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117(9):2567–76. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, Golub TR. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008;451(7176):335–9. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC, Jr., Meltzer P, Esposito D, Beggs AH, Schneider HE, Grabowska A, Ball SE, Niewiadomska E, Sieff CA, Vlachos A, Atsidaftos E, Ellis SR, Lipton JM, Gazda HT, Arceci RJ. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112(5):1582–92. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109(3):980–6. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11(4):501–8. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, Niewiadomska E, Da Costa L, Tchernia G, Niemeyer C, Meerpohl JJ, Stahl J, Schratt G, Glader B, Backer K, Wong C, Nathan DG, Beggs AH, Sieff CA. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79(6):1110–8. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83(6):769–80. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Zhong R, Long L, Niewiadomska E, Lipton JM, Ploszynska A, Zaucha JM, Vlachos A, Atsidaftos E, Viskochil DH, Niemeyer CM, Meerpohl JJ, Rokicka-Milewska R, Pospisilova D, Wiktor-Jedrzejczak W, Nathan DG, Beggs AH, Sieff CA. RNA and protein evidence for haplo-insufficiency in Diamond-Blackfan anaemia patients with RPS19 mutations. Br J Haematol. 2004;127(1):105–13. doi: 10.1111/j.1365-2141.2004.05152.x. [DOI] [PubMed] [Google Scholar]
- Gillio AP, Faulkner LB, Alter BP, Reilly L, Klafter R, Heller G, Young DC, Lipton JM, Moore MA, O’Reilly RJ. Treatment of Diamond-Blackfan anemia with recombinant human interleukin-3. Blood. 1993;82(3):744–51. [PubMed] [Google Scholar]
- Glader BE, Backer K, Diamond LK. Elevated erythrocyte adenosine deaminase activity in congenital hypoplastic anemia. N Engl J Med. 1983;309(24):1486–90. doi: 10.1056/NEJM198312153092404. [DOI] [PubMed] [Google Scholar]
- Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, Bessler M, Mason PJ. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis. 2007;39(1):35–43. doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Janov AJ, Leong T, Nathan DG, Guinan EC. Diamond-Blackfan anemia. Natural history and sequelae of treatment. Medicine (Baltimore) 1996;75(2):77–8. doi: 10.1097/00005792-199603000-00004. [DOI] [PubMed] [Google Scholar]
- Lipton JM, Federman N, Khabbaze Y, Schwartz CL, Hilliard LM, Clark JI, Vlachos A. Osteogenic sarcoma associated with Diamond-Blackfan anemia: a report from the Diamond-Blackfan Anemia Registry. J Pediatr Hematol Oncol. 2001;23(1):39–44. doi: 10.1097/00043426-200101000-00009. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3(6):577–87. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40(8):963–70. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake K, Utsugisawa T, Flygare J, Kiefer T, Hamaguchi I, Richter J, Karlsson S. Ribosomal protein S19 deficiency leads to reduced proliferation and increased apoptosis but does not affect terminal erythroid differentiation in a cell line model of Diamond-Blackfan anemia. Stem Cells. 2008;26(2):323–9. doi: 10.1634/stemcells.2007-0569. [DOI] [PubMed] [Google Scholar]
- O’Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol. 2010;190(5):853–66. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32(2):180–9. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orfali KA, Ohene-Abuakwa Y, Ball SE. Diamond Blackfan anaemia in the UK: clinical and genetic heterogeneity. Br J Haematol. 2004;125(2):243–52. doi: 10.1111/j.1365-2141.2004.04890.x. [DOI] [PubMed] [Google Scholar]
- Perdahl EB, Naprstek BL, Wallace WC, Lipton JM. Erythroid failure in Diamond-Blackfan anemia is characterized by apoptosis. Blood. 1994;83(3):645–50. [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123(1):49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- van Dijken PJ, Verwijs W. Diamond-Blackfan anemia and malignancy. A case report and a review of the literature. Cancer. 1995;76(3):517–20. doi: 10.1002/1097-0142(19950801)76:3<517::aid-cncr2820760324>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Vlachos A, Ball S, Dahl N, Alter BP, Sheth S, Ramenghi U, Meerpohl J, Karlsson S, Liu JM, Leblanc T, Paley C, Kang EM, Leder EJ, Atsidaftos E, Shimamura A, Bessler M, Glader B, Lipton JM. Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. Br J Haematol. 2008;142(6):859–76. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos A, Klein GW, Lipton JM. The Diamond Blackfan Anemia Registry: tool for investigating the epidemiology and biology of Diamond-Blackfan anemia. J Pediatr Hematol Oncol. 2001;23(6):377–82. doi: 10.1097/00043426-200108000-00015. [DOI] [PubMed] [Google Scholar]
- Vlachos A, Muir E. How I treat Diamond-Blackfan anemia. Blood. 2010;116(19):3715–23. doi: 10.1182/blood-2010-02-251090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willig TN, Draptchinskaia N, Dianzani I, Ball S, Niemeyer C, Ramenghi U, Orfali K, Gustavsson P, Garelli E, Brusco A, Tiemann C, Perignon JL, Bouchier C, Cicchiello L, Dahl N, Mohandas N, Tchernia G. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: wide variations in phenotypic expression. Blood. 1999a;94(12):4294–306. [PubMed] [Google Scholar]
- Willig TN, Niemeyer CM, Leblanc T, Tiemann C, Robert A, Budde J, Lambiliotte A, Kohne E, Souillet G, Eber S, Stephan JL, Girot R, Bordigoni P, Cornu G, Blanche S, Guillard JM, Mohandas N, Tchernia G, DBA group of Societe d’Hematologie et d’Immunologie Pediatrique (SHIP), Gesellshaft fur Padiatrische Onkologie und Hamatologie (GPOH), and the European Society for Pediatric Hematology and Immunology (ESPHI) Identification of new prognosis factors from the clinical and epidemiologic analysis of a registry of 229 Diamond-Blackfan anemia patients. Pediatr Res. 1999b;46(5):553–61. doi: 10.1203/00006450-199911000-00011. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Yuan Y, Zhang W, Guan W, Wu Z, Jin C, Chen H, Zhang L, Yang X, He F. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 2010;38(19):6544–54. doi: 10.1093/nar/gkq536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.