Abstract
CCL11/eotaxin-1 is a potent eosinophilic CC chemokine expressed by primary human fibroblasts. The combination of TGF-β1 and IL-13 synergistically increases CCL11 expression, but the mechanisms behind the synergy are unclear. To address this, human airway fibroblast cultures from normal and asthmatic subjects were exposed to IL-13 alone or TGF-β1 plus IL-13. Transcriptional (nuclear run-on) and post-transcriptional (mRNA stability) assays confirmed that transcriptional regulation is critical for synergistic expression of CCL11. TGF-β1 plus IL-13 synergistically increased STAT-6 phosphorylation, nuclear translocation and binding to the CCL11 promoter as compared to IL-13 alone. STAT-6 siRNA significantly knocked down both STAT-6 mRNA expression and phosphorylation, and inhibited CCL11 mRNA and protein expression.
Regulation of the IL-4 receptor α (IL-4Rα) complex by TGF-β1 augmented IL-13 signaling by dampening IL-13 receptor α2 (IL-13Rα2) expression, overcoming IL-13's autoregulation of its pathway and enhancing the expression of CCL11. Our data suggest that TGF-β1 induced activation of the MEK-ERK pathway reduces IL-13Rα2 expression induced by IL-13. Thus, TGF-β1, a pleiotropic cytokine upregulated in asthmatic airways, can augment eosinophilic inflammation by interfering with IL-13's negative feedback autoregulatory loop under MEK/ERK dependent conditions.
Keywords: human airway fibroblasts, CCL11/Eotaxin-1, IL-13, TGF-β1, actinomycin D, STAT-6, MEK-ERK, CHIP, siRNA, qRT-PCR, IL-13Rα
Introduction
IL-13 is a Th2-derived cytokine implicated in allergic airway responses and asthma pathogenesis. TGF-β1, a pleiotropic cytokine, is associated with a wide range of activities from immune function to cell differentiation, wound healing and repair (1–4). As an immune-inhibitory cytokine, TGF-β1 reduces IL-13 production in T cells, reversing allergen-induced airway hyperreactivity and inflammation (3). In contrast, the effects of TGF-β1 in fibroblasts are generally pro-fibrotic, both IL-13 and TGF-β are increased in asthma and associated with eosinophil inflammation and fibrosis (5–7).
IL-4/-13 increase CCL11/eotaxin-1, a potent eosinophilic CC chemokine, expression in primary human fibroblasts through IL-4 receptor α (IL-4Rα)/STAT-6 dependent mechanisms (8–10). CCL11 can contribute to inflammation in asthma through activation of CCR3 on eosinophils, leukocytes and mast cells (11). In animal models of pulmonary allergic inflammation, eosinophil accumulation is controlled by CCL11 generation (12, 13). Several cytokines, including TNF-α, IL-1β and TGF-β, synergize with IL-4/-13 to increase CCL11 expression in airway fibroblasts (8, 10, 14), smooth muscle cells (15, 16) and keratocytes (17). The synergies between IL-4 and TNF-α involve overlapping consensus binding sites for NF-κB and STAT-6 in the CCL11 promoter in human airway epithelial cells and fibroblasts (10, 18). In contrast, TGF-β does not activate NF-κB, such that the mechanisms behind the synergistic increase in CCL11 with IL-13 are not clear (15). However, engagement of both the IL-4Rα and the TGF-β receptor complex activate ERK/MAPK suggesting possible synergistic pathways (19, 20). In addition, IL-13 downregulates activation of its own receptor complex through increased expression of its decoy receptor, IL-13Rα2 (21–23). Therefore, in addition to synergistic activation of signaling molecules, suppression of IL-13Rα2 by TGF-β1 could play a role in the synergistic increase of CCL11.
We therefore hypothesized that the synergistic increase in CCL11 following addition of TGF-β1 to IL-13 was transcriptionally regulated, with enhanced STAT-6 phosphorylation due to greater and more sustained IL-4Rα activation. We further hypothesized this sustained IL-4Rα activation was secondary to increased IL-13Rα1 in the face of decreased IL-13Rα2 expression, and that these changes in receptor balance were due to effects of TGF-β1 on the MEK/ERK pathway.
Materials and Methods
Materials
DMEM, penicillin, streptomycin and gentamicin were purchased from Life Technologies (Rockville, MD) and fetal bovine serum (FBS) was purchased from Gemini (Woodland, CA); recombinant human IL-13 and TGF-β1 were purchased from R & D System Inc. (Minneapolis, MN). Trizol was from Invitrogen Life Technologies (Carlsbad, CA). Anti-STAT-6 antibody, anti-IL-13Rα1 and anti-IL-13Rα2 antibody and Protein G Plus-Agarose immunoprecipitation reagent were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). p-STAT-6 antibody and PD98059 were purchased from Cell Signaling Technologies, Inc. (Danvers, MA). DPERK and β-actin antibodies, protease inhibitor cocktail and Cell-Lytic™ Nuclear™ extraction kit were purchased from Sigma (St. Louis, MO). Actinomycin D (Act D) was purchased from VMW (West Chester, PA). U0126 was purchased from Calbiochem (La Jolla, CA). Oligo primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). Fluorescein goat anti-rabbit IgG and rabbit anti-goat IgG were purchased from VECTOR Laboratories Inc. (Burlingame, CA). Supersignal West Femto Maximum sensitivity substrate and Sonic dismembrator model 100 was purchased from Fisher Scientific (Pittsburgh, PA). The Fuji Film Image Reader LAS-3000 was used to detect specific bands for Western blot. Geneclean kit was purchased from Qbiogene Inc. (Morgan Ivine, CA). TransIT-siQuest™ reagent was purchased from Mirus Bio Corporation (Madison, WI). Assay-on-Demand (AOD) and STAT-6 siRNA were purchased from Applied Biosystems (Foster City, CA), and quantitative real-time PCR (qRT-PCR) was performed using ABI 7900 HT Real-Time PCR system.
Fibroblast culture
Endobronchial biopsies were obtained from normal control and asthmatic subjects. As previously reported CCL11 production in response to IL-13 or the combination of IL-13 plus TGF-β1 did not vary between fibroblasts from asthmatics and normal controls and therefore the cells were used interchangeably (24). Primary fibroblasts were grown in 10% FBS DMEM and used at the 3rd or 4th passage. 80% confluent fibroblasts were serum deprived for 24 h, then stimulated with IL-13 (30ng/ml) or TGF-β1 (5ng/ml) plus IL-13 in the presence of specific chemical inhibitors or target siRNA according to the experimental protocols.
Measurement of gene expression by qRT-PCR
CCL11, STAT-6, IL-13Rα1 and IL-13Rα2 mRNA expression was determined by reverse transcription followed by qRT-PCR according to the protocol (8). Assay IDs as follows: CCL11: Hs00237013_m1; STAT-6: Hs00598625_m1; IL-13Rα1: Hs00609817; IL-13Rα2: Hs00152924. VIC-labeled human GAPDH primers and probe were performed as internal control as previously described (24). All target gene mRNA expression was indexed to GAPDH.
In vitro PCR based nuclear run-on assay
PCR based nuclear run-on experiments were performed on intact nuclei isolated from IL-13 and TGF-β1 plus IL-13 stimulated fibroblasts (24 h). Intact nuclei (suspended in 200 μl) were equally split into 2 aliquots. To one aliquot, 0.5mM each of ATP, CTP, GTP and UTP were added (+NTPs); same volume of H2O (no NTPs control) were added to the second aliquot (−NTPs). Following 0.5 h incubation at 30°C in 20% glycerol, 30mM Tris-HCl, pH 8.0, 2.5mM MgCl2, 150mM KCl, 1mM DTT, and 40 units of RNasin (Promega, Madison, WI), Trizol was added and nuclear RNA isolated. RNA samples were digested with DNase I, reverse transcription and qRT-PCR performed according to the method described previously (24).
CCL11 mRNA stability
Fibroblasts stimulated with IL-13 or TGF-β1 plus IL-13 for 24 h, were treated with transcriptional inhibitor Act D (5μM) to terminate all gene transcription process. The fibroblasts were harvested at 0, 8 and 24 h, RNA extracted and qRT-PCR performed to quantify the remaining of CCL11 mRNA at different time period after blocking the gene transcription.
SDS-PAGE and Western blot analysis
Fibroblasts were exposed to IL-13 or TGF-β1 plus IL-13 at various time points. The preparation of whole cell lysates, SDS-PAGE and Western blot analysis was performed as previously described (24). The blots were incubated overnight at 4°C with p-STAT-6, DPERK, IL-13Rα1 or IL-13Rα2 antibody. HRP-conjugated secondary antibody (F(ab)2) was added and specific bands semi-quantified by densitometry. Equal loading was demonstrated by detecting total STAT-6, ERK or the housekeeping gene (β-actin or GAPDH) and the final assessment was evaluated as ratio of target protein to total protein or housekeeping gene. Similarly, both PD98059 and U0126 were applied to study the effects of MEK-ERK on phosphorylation of STAT-6, and expression of IL-13Rα1, IL-13Rα2 and CCL11.
Immunofluorescence staining of STAT-6
Fibroblasts plated on 4-well chamber were treated with IL-13 and TGF-β1 plus IL-13 for 0.25 and 15 h. The cells were fixed in 4% paraformaldehyde, blocked with appropriate blocking serum, then incubated overnight at 4°C with primary anti-STAT-6 antibody (rabbit, 1:50) followed by FITC-conjugated secondary antibody (1:50) for 1 h at room temperature. After washing with 0.5% TBS, the stained cells were mounted with DAKO fluorescent mounting medium and viewed under microscope.
Chromatin immunoprecipitation of STAT-6
Chromatin immunoprecipitation (CHIP) experiments were performed according to the published method (25, 26). Fibroblasts stimulated for 15 h with IL-13 or TGF-β1 plus IL-13 were fixed with 1% paraformaldehyde for 10 min at 37°C. After washing with cold PBS containing protease inhibitor cocktail (1:200), cells were scraped and chromatin DNA sheared using Sonic Dismembrator. The supernatants were centrifuged (13000g for 10 min at 4°C) and a portion saved for DNA input. The supernatant was incubated with STAT-6 antibody (1:100) to determine STAT-6 binding activity while anti-IgG (1:100) served as a negative control. After rotating overnight at 4°C, protein G plus-agarose was added to the protein-antibody complex, rotated at 4°C for 3 h and the immunoprecipitates separated by centrifugation (1000g for 5 min at 4°C). The pellet was washed with low salt buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl, pH 8.1 and 150mM NaCl), high salt buffer (0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-HCl, pH 8.1 and 500mM NaCl), LiCl wash buffer (0.25M LiCl, 1% NP-40, 1% Deoxycholate, 1mM EDTA and 10mM Tris-HCl, pH8.1) and finally 1XTris/EDTA buffer (10mM Tris-HCl and 1mM EDTA, pH 8.0). The protein-antibody complex was eluted for 20 min at room temperature with elution buffer (1% SDS, 0.1M NaHCO3 and 200mM NaCl), centrifuged at 13200rpm for 5 min at 4°C. To reverse the cross-linking of DNA, the elutes were incubated for 4 h at 65°C, followed by treatment with proteinase K at 45°C for 1 h (same treatment for DNA input samples). DNA was immediately recovered by Geneclean kit. The pellets were resuspended in 40 μl of nuclease-free H2O. PCR was carried out for 48 cycles using the specific primers (see below) for STAT-6 binding site with 3 μl of sample DNA solution, and PCR products were separated on 1.5% agarose gels in 1x Tris-acetate/EDTA.
Sense: TTGACTTCCTAGGATCTGGA (−146 to −127)
Anti-sense: TTTGGCGTGAGAAGGTGGT (+16 to +34)
Effects of STAT-6 siRNA on CCL11 expression
Cultured human airway fibroblasts were seeded in 6 or 24 well plates and incubated overnight in 10% DMEM to reach 50–60% confluency the following day. Medium was removed to leave 250 μl (24 well plate) or 1250 μl (6 well plate), and cells transfected with 5nM STAT-6 siRNA in the presence of TransIT-siQUEST™ reagent according to the protocol. After 48 h, cells were serum deprived for 24 h and stimulated with IL-13 or TGF-β1 plus IL-13 for 0.25 and 15 h (cell lysate collection), and 24 h (RNA harvest), and 48 h (supernatant collection). Gene knockdown of STAT-6 and its impact on CCL11 were evaluated by qRT-PCR, Western blot and ELISA.
Supernatant CCL11 protein measurement by ELISA
Sandwich ELISAs measured CCL11 in the supernatant (8). CCL11 antibody and standard were purchased from R&D Systems (Minneapolis, MN). Assay sensitivity ranged from 10 to 20 pg/ml.
Statistical analysis
Normally distributed data were expressed as mean ± SEM of at least 3 independent experiments. Natural log-transformation was done for non-normally distributed data. Graphic representation of log-transformed data was done by reconverting back to the original scale. Paired t-tests were used to assess the significant differences between the paired samples. A p value of less than 0.05 was considered statistically significant.
Results
Subject characteristics
In total, fibroblasts from 35 subjects with asthma and normal control subjects were obtained. Although the groups differed by age, asthma severity, FEV1% Predicted and inhaled/oral corticosteroid use, CCL11 mRNA/protein did not differ by groups or correlate with any of these parameters at baseline or with stimulation. For more details of subject characteristics, see Table S1. Fibroblasts from different subjects were therefore used interchangeably as previously described (24, 27). Although a total of 35 subjects took part in the study, sample sizes and subject used varied for each experimental system. Considering the variations of the multi-passage cultured fibroblasts, only 3rd or 4th generation fibroblasts were used in the experiments.
Transcriptional and post-transcriptional regulation of CCL11 in the presence of TGF-β1 and IL-13
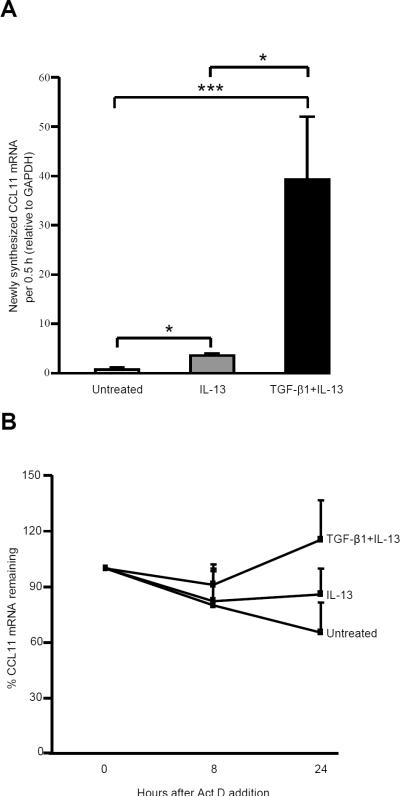
As previously reported (8), TGF-β1 plus IL-13 synergistically increased CCL11 mRNA (24 h) and protein (48 h) compared to IL-13 alone while TGF-β1 alone had no effect. To confirm that the synergistic effects of TGF-β1 and IL-13 on CCL11 occurred at the transcriptional level, an in vitro nuclear run-on assay was performed. Both IL-13 and TGF-β1 plus IL-13 significantly increased the nuclear transcriptional activity of CCL11 following addition of nucleotides. However, TGF-β1 plus IL-13 increased CCL11 mRNA 8-fold compared to IL-13 alone (Fig. 1A).
FIGURE 1.
TGF-β1 plus IL-13 synergistically increased transcriptional activity of CCL11 and modestly stabilized CCL11 mRNA in primary human airway fibroblasts. A, In vitro PCR based nuclear run-on assay was performed after 24 h stimulation with IL-13 or TGF-β1 plus IL-13. Nuclei were isolated and newly synthesized CCL11 mRNA measured after incubation with NTPs. RNA was isolated and qRT-PCR performed. Net newly synthesized CCL11 mRNA levels are shown as mean ± SEM (n=4). * p<0.05, *** p<0.0001. B, CCL11 mRNA stability was performed by measuring the remaining CCL11 mRNA (after 24 h transcription) following addition of Act D (5 μM) at 0, 8 and 24 h (n=4). Results are expressed as the percentage of each condition at the time Act D was added.
As post-transcriptional mechanisms for CCL11 have also been identified (28), CCL11 mRNA stability was investigated by adding Act D 24 h after stimulation with IL-13 or TGF-β1 plus IL-13. CCL11 mRNA was measured at 0, 8 and 24 h following Act D. Slightly more CCL11 mRNA remained (~10%) at 8 h in the presence of TGF-β1 plus IL-13 compared to IL-13 alone. This difference increased to 20% at 24 h (Fig. 1B). Thus, compared to the 8-fold increase in transcription, a much smaller effect of mRNA stability on overall CCL11 mRNA levels was observed (Fig. 1A, 1B).
The addition of TGF-β 1 t o I L-13 induces a synergistic increase in STAT-6 phosphorylation which only occurs 6 h or more post stimulation
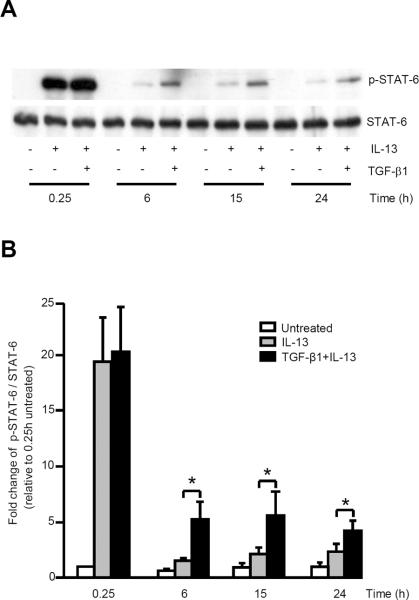
Both IL-13 alone and in combination with TGF-β1 were previously reported to similarly induce STAT-6 phosphorylation at early time points (8). In contrast, using a supersignal ECL reagent, at all later time points (6, 15 and 24 h) the combination of TGF-β1 and IL-13 synergistically increased STAT-6 phosphorylation over that of IL-13 alone, although the signals were overall weaker than those at the earliest time point (Fig. 2A, 2B). TGF-β1 alone had no effect on STAT-6 phosphorylation (8).
FIGURE 2.
TGF-β1 plus IL-13 synergistically increased STAT-6 phosphorylation in primary human airway fibroblasts. 80% confluent fibroblasts were serum-deprived, then stimulated with IL-13 or TGF-β1 plus IL-13 at 0.25, 6, 15 and 24 h, cell lysates harvested and Western blot performed. A, Representative Western blot of p-STAT-6 and STAT-6 (n=4). B, Quantitative comparison of p-STAT-6 as compared with total STAT-6 in response to IL-13 alone and TGF-β1 plus IL-13. Data are expressed as mean ± SEM from 4 different subjects. * p<0.05.
STAT-6 nuclear translocation is increased in the presence of TGF-β1 plus IL-13
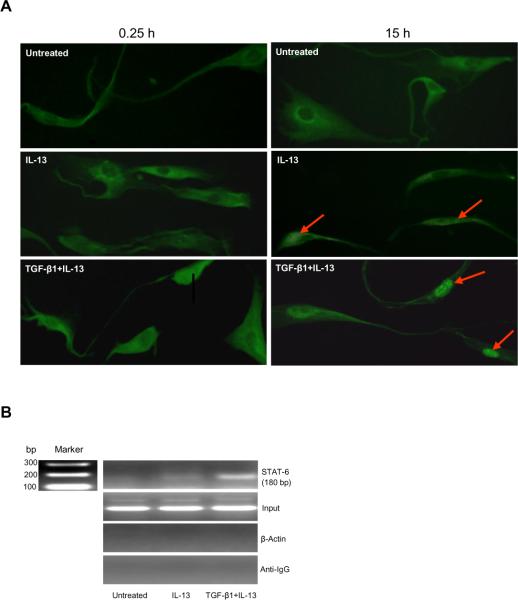
To determine whether increased STAT-6 phosphorylation was accompanied by enhanced nuclear translocation, immunoflurescence studies of fibroblasts were performed. At 0.25 h, minimal nuclear STAT-6 translocation was observed in the presence of IL-13 or TGF-β1 plus IL-13 (Figure 3A). At 15 h, unstimulated fibroblasts exhibited diffuse cytoplasmic staining without STAT-6 nuclear translocation. Modest translocation was observed in the presence of IL-13 alone, while TGF-β1 plus IL-13 induced marked nuclear translocation of STAT-6 translocation (Fig. 3A). Phosphorylated STAT-6 homodimerizes before translocating to the nucleus suggesting there is a small delay between these two events as has been previously reported (29).
FIGURE 3.
TGF-β1 plus IL-13 synergistically increased STAT-6 nuclear translocation and promoter binding in primary human airway fibroblasts. A, Immunofluorescence of STAT-6 in the presence of media alone, IL-13 and TGF-β1 plus IL-13 at 15 h. The arrows show STAT-6 in the nucleus (n=3). B, CHIP analysis showed significant binding of STAT-6 to the CCL11 promoter in the presence of IL-13 alone and TGF-β1 plus IL-13. After stimulation for 15 h, fibroblasts were fixed and chromatin DNA isolated, incubated with STAT-6 antibody to form a protein-antibody complex, the cross-link reversed, protein removed and PCR performed to measure STAT-6 binding (n=3).
TGF-β1 plus IL-13 enhances STAT-6-CCL11 promoter binding in vivo
To confirm enhanced binding of STAT-6 to the CCL11 promoter, CHIP was performed. Sequential CHIPs were prepared with chromatin from fibroblasts stimulated with IL-13 or TGF-β1 plus IL-13 for 15 h when the synergy of STAT-6 phosphorylation and translocation was most strongly observed. These were probed with a highly specific anti-STAT-6 antibody. Control CHIPs were performed by adding anti-IgG antibody or amplifying with β-actin primers. The chromatin used in this procedure was below 1 kb. Oligonucleotides containing a STAT-6 fragment in the CCL11 promoter were amplified using traditional PCR. A stronger STAT-6 binding band was observed in TGF-β1 plus IL-13 stimulated fibroblasts, compared to IL-13 alone (Fig. 3B).
STAT-6 siRNA inhibits STAT-6 mRNA expression and phosphorylation of STAT-6 protein while decreasing CCL11 expression
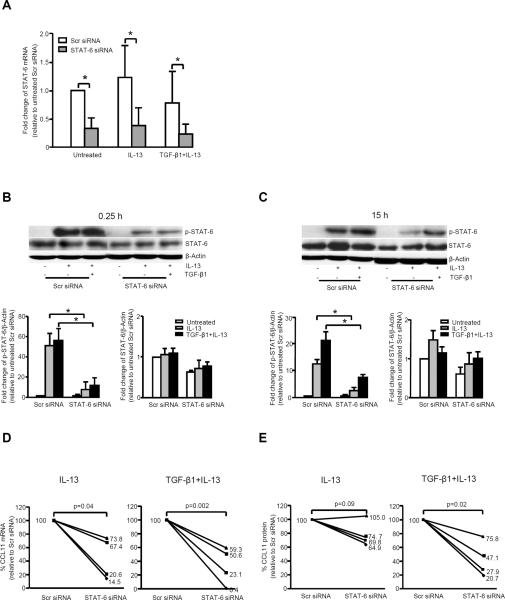
To confirm the critical nature of STAT-6 to the synergistic increase in CCL11, the effect of STAT-6 siRNA on CCL11 expression was evaluated. STAT-6 siRNA significantly knocked down STAT-6 mRNA, protein and phosphorylation, although the impact on phospho-STAT-6 was greater than the effect on total STAT-6 at both 0.25 and 15 h stimulation with either IL-13 or TGF-β1 plus IL-13 (Fig. 4A, 4B, 4C).
FIGURE 4.
Knock down of STAT-6 mRNA expression and phosphorylation with subsequent decrease in CCL11 mRNA/protein in the presence of IL-13 and TGF-β1 plus IL-13. Fibroblasts were transfected with STAT-6 siRNA for 48 h, serum deprived, stimulated for 0.25 and 15 h to collect cell lysates to measure p-STAT-6/STAT-6, and harvested at 24 h by adding Trizol to measure STAT-6 and CCL11 mRNA, and collect supernatant at 48 h to measure CCL11 protein. A, STAT-6 siRNA knocked down STAT-6 mRNA expression under all conditions (n=4). * p<0.05. B and C, STAT-6 siRNA significantly decreased phospho-STAT-6 levels, with lesser effects on total STAT-6, in the presence of IL-13 and TGF-β1 plus IL-13 at 0.25 h (B) and 15 h (C). Densitometry data are expressed as mean ± SEM (n=4). * p<0.05. D, STAT-6 siRNA decreased CCL11 mRNA expression in the presence of IL-13 (p=0.04), with a greater decrease in the presence of TGF-β1 plus IL-13 (p=0.002) (n=4). The percentage of CCL11 mRNA or protein relative to scramble siRNA is shown. E, CCL11 protein expression was knocked down by STAT-6 siRNA in the presence of TGF-β1 plus IL-13 (p=0.02), with less effect on IL-13 alone (p=0.09) (n=4).
48 h of transient transfection with STAT-6 siRNA as compared to scramble siRNA, followed by 24 h of stimulation with IL-13 or TGF-β1 plus IL-13 consistently inhibited CCL11 mRNA (Fig. 4D). CCL11 mRNA expression (as percentage of the scramble siRNA) was 44 ± 31% for IL-13 (p=0.04) and 33 ± 27% for TGF-β1 plus IL-13 (p=0.002). CCL11 protein expression was 79 ± 9% of scramble siRNA for IL-13 alone (p=0.09) and 43 ± 12% for TGF-β1 plus IL-13 (p=0.02) (Fig. 4E). The stronger inhibition of STAT-6 siRNA on TGF-β1 plus IL-13 induced CCL11 expression confirms that STAT-6 plays a critical role in the synergistic expression of CCL11.
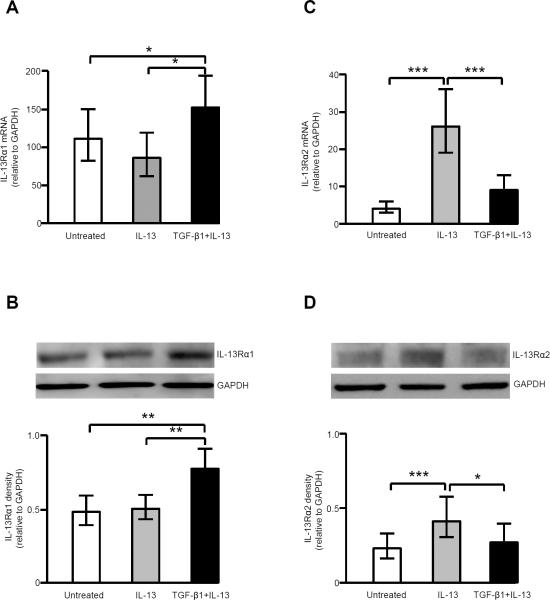
TGF-β1 plus IL-13 increases IL-13Rα1, while decreasing IL-13-induced expression of IL-13Rα2
IL-4Rα was constitutively expressed in human airway fibroblasts in the presence of IL-13, TGF-β1 or the combination (data not shown). The addition of TGF-β1 to IL-13 significantly increased IL-13Rα1 mRNA and protein expression, while IL-13 alone had no effect (Fig. 5A, 5B). In contrast, IL-13 itself significantly increased IL-13Rα2 mRNA and protein expression, while the addition of TGF-β1 downregulated IL-13-induced IL-13Rα2 mRNA and protein expression (Fig. 5C, 5D).
FIGURE 5.
TGF-β1 reduced IL-13-induced increase in IL-13Rα2 mRNA and protein expression, but augmented IL-13Rα1 mRNA and protein expression. A & B, IL-13Rα1 mRNA (n=11) (A) and protein expression (n=12) (B) in the presence of IL-13 and TGF-β1 plus IL-13. * p<0.05; ** p<0.01. C & D, IL-13Rα2 mRNA (n=24) (C) and protein expression (n=12) (D) in the presence of IL-13 and TGF-β1 plus IL-13. * p<0.05; *** p<0.001.
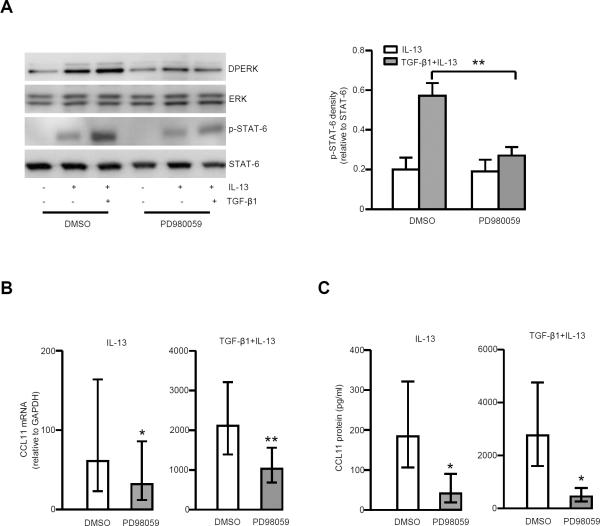
MEK/ERK inhibition decreases STAT-6 phosphorylation and CCL11 mRNA/protein expression
The MEK inhibitor PD98059 (25 μM) decreased STAT-6 phosphorylation in the presence of TGF-β1 plus IL-13, but not IL-13 alone, although ERK phosphorylation was observed under both conditions (Fig. 6A). CCL11 mRNA and protein were concomitantly inhibited by PD98059 with the stronger inhibition in the presence of TGF-β1 plus IL-13. The average inhibition of CCL11 mRNA was 35 ± 36% for IL-13 alone, and 44 ± 17% for TGF-β1 plus IL-13; the average inhibition of CCL11 protein was 62 ± 40% for IL-13 alone, and 70 ± 29% for TGF-β1 plus IL-13 (Fig. 6B, 6C). A second MEK-ERK inhibitor (U0126, 10 μM) duplicated those of PD98059 (Fig. S1).
FIGURE 6.
MEK-ERK inhibitor, PD98059 (25 μM) decreased CCL11 mRNA and protein expression through down-regulation of STAT-6 phosphorylation. Fibroblasts were serum deprived, then pretreated for 1 h with PD98059 followed by stimulation with IL-13 and TGF-β1 plus IL-13 for 24 h to measure CCL11 mRNA and 48 h to measure CCL11 protein. A, PD98059 decreased phosphorylation of ERK and STAT-6 at 15 h, especially in the presence of TGF-β1 plus IL-13 (n=5). Right-hand side shows the relative density of p-STAT-6/STAT-6 in the presence of IL-13 and TGF-β1 plus IL-13, either with or without PD98059. ** p<0.01. B, PD98059 significantly decreased CCL11 mRNA expression in the presence of IL-13 and TGF-β1 plus IL-13 (n=9). * p<0.05; ** p<0.01. C, PD98059 significantly decreased CCL11 protein in the presence of IL-13 and TGF-β1 plus IL-13 (n=5). * p<0.05.
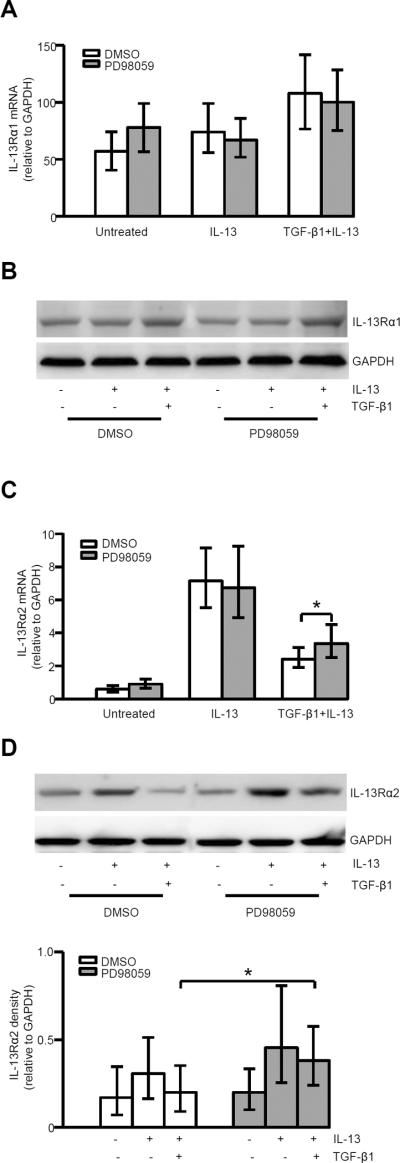
MEK/ERK inhibition increases IL-13Rα2 mRNA and protein expression
MEK/ERK inhibition had no effect on IL-13Rα1 mRNA and protein expression under all conditions (Fig. 7A, 7B). However, MEK/ERK inhibition by PD98059 increased IL-13Rα2 mRNA and protein expression in the presence of TGF-β1 plus IL-13, but not for IL-13 alone (Fig. 7C, 7D).
FIGURE 7.
MEK/ERK inhibition reversed the TGF-β1 inhibition of IL-13-induced IL-13Rα2 expression. A, Effects of PD98059 on IL-13Rα1 mRNA expression at 24 h in the presence of IL-13 and TGF-β1 plus IL-13 (n=8). B, Representative Western blot of IL-13Rα1 (n=4); C, PD98059 increased IL-13Rα2 mRNA expression at 24 h in the presence of TGF-β1 plus IL-13 (n=8). * p<0.05. D, Representative Western blot of IL-13Rα2 and the relative density of IL-13Rα2/GAPDH (n=5). * p<0.05
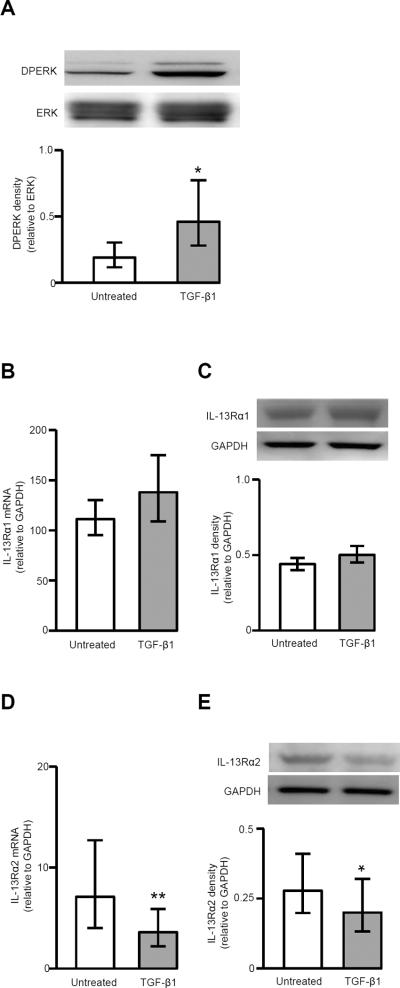
Effects of TGF-β1 on ERK phosphorylation and expression of IL-13Rα1 and 2
To further investigate the effects of TGF-β1, ERK activation and expression of IL-13Rα1 and 2 were measured in the presence of TGF-β1 alone. TGF-β1 significantly increased ERK phosphorylation at 24 h (Fig. 8A) and decreased IL-13Rα2 mRNA and protein expression (Fig. 8D, 8E). TGF-β1 had no obvious effect on IL-13Rα1 mRNA and protein expression (Fig. 8B, 8C).
FIGURE 8.
Effects of TGF-β1 on phosphorylation of ERK and expression of IL-13Rα1 and IL-13Rα2. Fibroblasts were serum-deprived, stimulated with TGF-β1 for 24 h, ERK phosphorylation, IL-13Rα1/2 mRNA and protein were measured. A, Representative Western blot of DPERK and ERK and the ratio of DPERK/ERK by densitometry (n=7). * p<0.05. Effects of TGF-β1 on IL-13Rα1 mRNA (n=10) (B) and protein (n=9) (C) expression. Effects of TGF-β1 on IL-13Rα2 mRNA (n=16) (D) and protein (n=9) (E) expression. * p<0.05; ** p<0.01.
Discussion
Airway inflammation has long been a hallmark of asthma (30). Many studies support a role for eosinophils in asthma, including recent clinical studies reporting benefit of a monoclonal antibody to the Th2/eosinophilic cytokine IL-5 in eosinophil associated asthma (31, 32). Interestingly, the response to anti-IL-5 is not complete and eosinophils remain in the tissue (33). CCL11, a potent eosinophilic chemoattractant produced by resident airway cells including fibroblasts, is likely to also contribute to this eosinophilic inflammation (5, 34, 35).
IL-4 and IL-13 increase CCL11 expression due to activation of STAT-6 pathways (9, 36, 37). However, various factors, including IL-1β, TNF-α and TGF-β1 in combination with IL-4 or -13, all markedly enhance CCL11 expression over the Th2 cytokine alone (8, 10, 14, 38–40) and potentially amplify eosinophil related inflammation and remodeling (11). Yet, the mechanisms for this synergistic production of CCL11, especially as it related to TGF-β1, remain poorly understood.
To address the potential mechanisms, an in vitro nuclear run-on assay first confirmed the predominant role of TGF-β1 in combination with IL-13, to enhance the transcription, as compared to mRNA stabilization, in the synergistic increase of CCL11. STAT-6 is believed to be the major transcriptional factor involved in the regulation of IL-13 induced CCL11 expression (9, 10, 37, 41), STAT-6 phosphorylation was only observed at early time points in the presence of IL-13 and TGF-β1 plus IL-13, but not with TGF-β1 stimulation alone (8). In contrast, the current data show that, at later time points, STAT-6 phosphorylation and nuclear translocation in the presence of TGF-β1 plus IL-13 was consistently greater than that of IL-13 alone.
CHIP further confirmed that, compared to IL-13 alone, TGF-β1 plus IL-13 generated stronger STAT-6 binding to the CCL11 promoter using the primers based on the more prominent proximal STAT-6 binding region (10, 42). Transient transfection of STAT-6 siRNA more strongly knocked down STAT-6 phosphorylation than total STAT-6, as well as CCL11 mRNA and protein. From the disconnect between phospho- and total STAT-6 in the presence of STAT-6 siRNA, we speculate that STAT-6 phosphorylation may occur more prominently on newly formed STAT-6. These studies demonstrated that augmented STAT-6 phosphorylation, binding and transcriptional activation are all involved in the synergistic increase in CCL11 expression.
Activation of STAT-6 by IL-13 is mediated by the type II receptor complex, including IL-4Rα and IL-13Rα1. IL-13Rα2 is believed to be a decoy receptor induced on fibroblasts, smooth muscle cells, keratinocytes, and other cells by IL-13, IL-4, TNF-α and IFN-γ (43–45). Our data confirmed that IL-13 induced IL-13Rα2 mRNA and protein. However, TGF-β1 prevented IL-13-induced IL-13Rα2 expression, reducing the expected auto-downregulation of IL-13 on its pathways. To further augment type II receptor activation, TGF-β1 in combination with IL-13 also upregulated the IL-13Rα1 component of the receptor, while IL-13 alone had no effect. Although the magnitude of the effects is small, they are consistent and appear to have functional consequences. Thus, these results identify a highly novel mechanism by which TGF-β1, a factor found in abundance in asthmatic tissues, can enhance and sustain the impact of IL-13 on eosinophilic inflammation.
ERK activation is critical for CCL11 production from airway smooth muscle cells (36, 46, 47). In the current study, TGF-β1 and IL-13 enhanced ERK activation, while MEKERK inhibition decreased STAT-6 phosphorylation and CCL11 expression. Interestingly, MEK-ERK inhibition had a greater impact on TGF-β1 plus IL-13 induced CCL11 expression than on that from IL-13 alone, supporting its role in the synergistic increase in CCL11. Mechanistically, it is likely this greater effect on CCL11 expression is through TGF-β1/ERK induced decreases in IL-13Rα2 gene expression. Taken together, these findings support a potential role for MEK-ERK pathway in the pathogenesis and treatment of eosinophilic asthma.
TGF-β is abundant in human asthmatic airways, being produced by inflammatory cells, epithelial cells and the fibroblasts (6, 48). However, TGF-β1 is also commonly implicated in Treg-mediated suppression of allergic asthma, with their anti-inflammatory properties (49). Thus, whether TGF-β plays a positive or negative role in asthmatic inflammation has been highly controversial. Our results would strongly support a prominent pro-Th2 inflammatory role for TGF-β in the submucosal tissue compartment, with a unique ability of TGF-β to overcome the autoregulation of the type II receptor complex by IL-13 itself. Autoregulation of a cytokine pathway by the cytokine itself is well known and observed for other cytokines, including IL-1, IL-6 and IL-10 (50–52). However, very little work has been done on factors which may overcome this downregulation of cytokine signaling. Understanding these counterbalancing pathways further could have significant implications in understanding the persistence of inflammation in chronic diseases like asthma.
In conclusion, we have identified mechanisms by which TGF-β augments IL-13 induced CCL11 expression, through TGF-β1/ERK induced effects on the type II IL-4Rα receptor complex and IL-13Rα2 which lead to enhanced and sustained STAT-6 activation and CCL11 transcription. Further exploration of the mechanisms by which cytokines like TGF-β (and likely others) are able to overcome cytokine autoregulation could lead to a broader understanding of the mechanisms leading to the persistence of inflammation in asthma and other diseases.
Supplementary Material
Acknowledgments
Supported by NIH grants HL-69174, AI-40600-15 and CTSI UL1-RR024153
Footnotes
This article has an online data supplement
References
- 1.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 2.Doherty T, Broide D. Cytokines and growth factors in airway remodeling in asthma. Curr Opin Immunol. 2007;19:676–680. doi: 10.1016/j.coi.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Hansen G, McIntire JJ, Yeung VP, Berry G, Thorbecke GJ, Chen L, DeKruyff RH, Umetsu DT. CD4(+) T helper cells engineered to produce latent TGF-beta1 reverse allergen-induced airway hyperreactivity and inflammation. J Clin Invest. 2000;105:61–70. doi: 10.1172/JCI7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wahl SM, Hunt DA, Wong HL, Dougherty S, McCartney-Francis N, Wahl LM, Ellingsworth L, Schmidt JA, Hall G, Roberts AB, et al. Transforming growth factor-beta is a potent immunosuppressive agent that inhibits IL-1-dependent lymphocyte proliferation. J Immunol. 1988;140:3026–3032. [PubMed] [Google Scholar]
- 5.Fulkerson PC, Fischetti CA, Hassman LM, Nikolaidis NM, Rothenberg ME. Persistent effects induced by IL-13 in the lung. Am J Respir Cell Mol Biol. 2006;35:337–346. doi: 10.1165/rcmb.2005-0474OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balzar S, Chu HW, Silkoff P, Cundall M, Trudeau JB, Strand M, Wenzel S. Increased TGF-beta2 in severe asthma with eosinophilia. J Allergy Clin Immunol. 2005;115:110–117. doi: 10.1016/j.jaci.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 7.Wills-Karp M, Chiaramonte M. Interleukin-13 in asthma. Curr Opin Pulm Med. 2003;9:21–27. doi: 10.1097/00063198-200301000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Wenzel SE, Trudeau JB, Barnes S, Zhou X, Cundall M, Westcott JY, McCord K, Chu HW. TGF-beta and IL-13 synergistically increase eotaxin-1 production in human airway fibroblasts. J Immunol. 2002;169:4613–4619. doi: 10.4049/jimmunol.169.8.4613. [DOI] [PubMed] [Google Scholar]
- 9.Matsukura S, Stellato C, Georas SN, Casolaro V, Plitt JR, Miura K, Kurosawa S, Schindler U, Schleimer RP. Interleukin-13 upregulates eotaxin expression in airway epithelial cells by a STAT6-dependent mechanism. Am J Respir Cell Mol Biol. 2001;24:755–761. doi: 10.1165/ajrcmb.24.6.4351. [DOI] [PubMed] [Google Scholar]
- 10.Hoeck J, Woisetschlager M. STAT6 mediates eotaxin-1 expression in IL-4 or TNF-alpha-induced fibroblasts. J Immunol. 2001;166:4507–4515. doi: 10.4049/jimmunol.166.7.4507. [DOI] [PubMed] [Google Scholar]
- 11.Puxeddu I, Bader R, Piliponsky AM, Reich R, Levi-Schaffer F, Berkman N. The CC chemokine eotaxin/CCL11 has a selective profibrogenic effect on human lung fibroblasts. J Allergy Clin Immunol. 2006;117:103–110. doi: 10.1016/j.jaci.2005.08.057. [DOI] [PubMed] [Google Scholar]
- 12.Humbles AA, Conroy DM, Marleau S, Rankin SM, Palframan RT, Proudfoot AE, Wells TN, Li D, Jeffery PK, Griffiths-Johnson DA, Williams TJ, Jose PJ. Kinetics of eotaxin generation and its relationship to eosinophil accumulation in allergic airways disease: analysis in a guinea pig model in vivo. J Exp Med. 1997;186:601–612. doi: 10.1084/jem.186.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Wang D, Griffiths-Johnson DA, Wells TN, Williams TJ, Jose PJ, Jeffery PK. Eotaxin protein and gene expression in guinea-pig lungs: constitutive expression and upregulation after allergen challenge. Eur Respir J. 1997;10:1946–1954. doi: 10.1183/09031936.97.10091946. [DOI] [PubMed] [Google Scholar]
- 14.Terada N, Hamano N, Nomura T, Numata T, Hirai K, Nakajima T, Yamada H, Yoshie O, Ikeda-Ito T, Konno A. Interleukin-13 and tumour necrosis factor-alpha synergistically induce eotaxin production in human nasal fibroblasts. Clin Exp Allergy. 2000;30:348–355. doi: 10.1046/j.1365-2222.2000.00750.x. [DOI] [PubMed] [Google Scholar]
- 15.Matsukura S, Odaka M, Kurokawa M, Kuga H, Homma T, Takeuchi H, Notomi K, Kokubu F, Kawaguchi M, Schleimer RP, Johnson MW, Adachi M. Transforming growth factor-beta stimulates the expression of eotaxin/CC chemokine ligand 11 and its promoter activity through binding site for nuclear factor-kappaB in airway smooth muscle cells. Clin Exp Allergy. 2010;40:763–771. doi: 10.1111/j.1365-2222.2010.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuyderduyn S, Hiemstra PS, Rabe KF. TGF-beta differentially regulates TH2 cytokine-induced eotaxin and eotaxin-3 release by human airway smooth muscle cells. J Allergy Clin Immunol. 2004;114:791–798. doi: 10.1016/j.jaci.2004.06.037. [DOI] [PubMed] [Google Scholar]
- 17.Kumagai N, Fukuda K, Ishimura Y, Nishida T. Synergistic induction of eotaxin expression in human keratocytes by TNF-alpha and IL-4 or IL-13. Invest Ophthalmol Vis Sci. 2000;41:1448–1453. [PubMed] [Google Scholar]
- 18.Matsukura S, Stellato C, Plitt JR, Bickel C, Miura K, Georas SN, Casolaro V, Schleimer RP. Activation of eotaxin gene transcription by NF-kappa B and STAT6 in human airway epithelial cells. J Immunol. 1999;163:6876–6883. [PubMed] [Google Scholar]
- 19.Liu Y, Meyer C, Muller A, Herweck F, Li Q, Mullenbach R, Mertens PR, Dooley S, Weng HL. IL-13 Induces Connective Tissue Growth Factor in Rat Hepatic Stellate Cells via TGF-{beta}-Independent Smad Signaling. J Immunol. 2011;187:2814–2823. doi: 10.4049/jimmunol.1003260. [DOI] [PubMed] [Google Scholar]
- 20.Pannu J, Nakerakanti S, Smith E, ten Dijke P, Trojanowska M. Transforming growth factor-beta receptor type I-dependent fibrogenic gene program is mediated via activation of Smad1 and ERK1/2 pathways. J Biol Chem. 2007;282:10405–10413. doi: 10.1074/jbc.M611742200. [DOI] [PubMed] [Google Scholar]
- 21.Zheng T, Zhu Z, Liu W, Lee CG, Chen Q, Homer RJ, Elias JA. Cytokine regulation of IL-13Ralpha2 and IL-13Ralpha1 in vivo and in vitro. J Allergy Clin Immunol. 2003;111:720–728. doi: 10.1067/mai.2003.1383. [DOI] [PubMed] [Google Scholar]
- 22.Chiaramonte MG, Mentink-Kane M, Jacobson BA, Cheever AW, Whitters MJ, Goad ME, Wong A, Collins M, Donaldson DD, Grusby MJ, Wynn TA. Regulation and function of the interleukin 13 receptor alpha 2 during a T helper cell type 2-dominant immune response. J Exp Med. 2003;197:687–701. doi: 10.1084/jem.20020903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hecker M, Zaslona Z, Kwapiszewska G, Niess G, Zakrzewicz A, Hergenreider E, Wilhelm J, Marsh LM, Sedding D, Klepetko W, Lohmeyer J, Dimmeler S, Seeger W, Weissmann N, Schermuly RT, Kneidinger N, Eickelberg O, Morty RE. Dysregulation of the IL-13 receptor system: a novel pathomechanism in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2010;182:805–818. doi: 10.1164/rccm.200909-1367OC. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X, Trudeau JB, Schoonover KJ, Lundin JI, Barnes SM, Cundall MJ, Wenzel SE. Interleukin-13 augments transforming growth factor-beta1-induced tissue inhibitor of metalloproteinase-1 expression in primary human airway fibroblasts. Am J Physiol Cell Physiol. 2005;288:C435–442. doi: 10.1152/ajpcell.00035.2004. [DOI] [PubMed] [Google Scholar]
- 25.Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 26.Nelson J, Denisenko O, Bomsztyk K. The fast chromatin immunoprecipitation method. Methods Mol Biol. 2009;567:45–57. doi: 10.1007/978-1-60327-414-2_3. [DOI] [PubMed] [Google Scholar]
- 27.Kotaru C, Schoonover KJ, Trudeau JB, Huynh ML, Zhou X, Hu H, Wenzel SE. Regional fibroblast heterogeneity in the lung: implications for remodeling. Am J Respir Crit Care Med. 2006;173:1208–1215. doi: 10.1164/rccm.200508-1218OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atasoy U, Curry SL, Lopez de Silanes I, Shyu AB, Casolaro V, Gorospe M, Stellato C. Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J Immunol. 2003;171:4369–4378. doi: 10.4049/jimmunol.171.8.4369. [DOI] [PubMed] [Google Scholar]
- 29.Andrews RP, Ericksen MB, Cunningham CM, Daines MO, Hershey GK. Analysis of the life cycle of stat6. Continuous cycling of STAT6 is required for IL-4 signaling. J Biol Chem. 2002;277:36563–36569. doi: 10.1074/jbc.M200986200. [DOI] [PubMed] [Google Scholar]
- 30.Elias JA, Zhu Z, Chupp G, Homer RJ. Airway remodeling in asthma. J Clin Invest. 1999;104:1001–1006. doi: 10.1172/JCI8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haldar P, Brightling CE, Hargadon B, Gupta S, Monteiro W, Sousa A, Marshall RP, Bradding P, Green RH, Wardlaw AJ, Pavord ID. Mepolizumab and exacerbations of refractory eosinophilic asthma. N Engl J Med. 2009;360:973–984. doi: 10.1056/NEJMoa0808991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castro M, Mathur S, Hargreave F, Boulet LP, Xie F, Young J, Wilkins HJ, Henkel T, Nair P. Reslizumab for Poorly Controlled, Eosinophilic Asthma: A Randomized, Placebo-Controlled Study. Am J Respir Crit Care Med. 2011;184:1125–1132. doi: 10.1164/rccm.201103-0396OC. [DOI] [PubMed] [Google Scholar]
- 33.Flood-Page PT, Menzies-Gow AN, Kay AB, Robinson DS. Eosinophil's role remains uncertain as anti-interleukin-5 only partially depletes numbers in asthmatic airway. Am J Respir Crit Care Med. 2003;167:199–204. doi: 10.1164/rccm.200208-789OC. [DOI] [PubMed] [Google Scholar]
- 34.Mattoli S, Stacey MA, Sun G, Bellini A, Marini M. Eotaxin expression and eosinophilic inflammation in asthma. Biochem Biophys Res Commun. 1997;236:299–301. doi: 10.1006/bbrc.1997.6958. [DOI] [PubMed] [Google Scholar]
- 35.Ahrens R, Waddell A, Seidu L, Blanchard C, Carey R, Forbes E, Lampinen M, Wilson T, Cohen E, Stringer K, Ballard E, Munitz A, Xu H, Lee N, Lee JJ, Rothenberg ME, Denson L, Hogan SP. Intestinal macrophage/epithelial cell-derived CCL11/eotaxin-1 mediates eosinophil recruitment and function in pediatric ulcerative colitis. J Immunol. 2008;181:7390–7399. doi: 10.4049/jimmunol.181.10.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirst SJ, Hallsworth MP, Peng Q, Lee TH. Selective induction of eotaxin release by interleukin-13 or interleukin-4 in human airway smooth muscle cells is synergistic with interleukin-1beta and is mediated by the interleukin-4 receptor alpha-chain. Am J Respir Crit Care Med. 2002;165:1161–1171. doi: 10.1164/ajrccm.165.8.2107158. [DOI] [PubMed] [Google Scholar]
- 37.Yang M, Hogan SP, Henry PJ, Matthaei KI, McKenzie AN, Young IG, Rothenberg ME, Foster PS. Interleukin-13 mediates airways hyperreactivity through the IL-4 receptor-alpha chain and STAT-6 independently of IL-5 and eotaxin. Am J Respir Cell Mol Biol. 2001;25:522–530. doi: 10.1165/ajrcmb.25.4.4620. [DOI] [PubMed] [Google Scholar]
- 38.Chung KF, Patel HJ, Fadlon EJ, Rousell J, Haddad EB, Jose PJ, Mitchell J, Belvisi M. Induction of eotaxin expression and release from human airway smooth muscle cells by IL-1beta and TNFalpha: effects of IL-10 and corticosteroids. Br J Pharmacol. 1999;127:1145–1150. doi: 10.1038/sj.bjp.0702660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, Zhang Y, Elias JA. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Xia Y, Nguyen A, Lai YH, Feng L, Mosmann TR, Lo D. Effects of Th2 cytokines on chemokine expression in the lung: IL-13 potently induces eotaxin expression by airway epithelial cells. J Immunol. 1999;162:2477–2487. [PubMed] [Google Scholar]
- 41.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 42.Hein H, Schluter C, Kulke R, Christophers E, Schroder JM, Bartels J. Genomic organization, sequence, and transcriptional regulation of the human eotaxin gene. Biochem Biophys Res Commun. 1997;237:537–542. doi: 10.1006/bbrc.1997.7169. [DOI] [PubMed] [Google Scholar]
- 43.Yoshikawa M, Nakajima T, Tsukidate T, Matsumoto K, Iida M, Otori N, Haruna S, Moriyama H, Saito H. TNF-alpha and IL-4 regulate expression of IL-13 receptor alpha2 on human fibroblasts. Biochem Biophys Res Commun. 2003;312:1248–1255. doi: 10.1016/j.bbrc.2003.11.077. [DOI] [PubMed] [Google Scholar]
- 44.David M, Ford D, Bertoglio J, Maizel AL, Pierre J. Induction of the IL-13 receptor alpha2-chain by IL-4 and IL-13 in human keratinocytes: involvement of STAT6, ERK and p38 MAPK pathways. Oncogene. 2001;20:6660–6668. doi: 10.1038/sj.onc.1204629. [DOI] [PubMed] [Google Scholar]
- 45.Moynihan BJ, Tolloczko B, El Bassam S, Ferraro P, Michoud MC, Martin JG, Laberge S. IFN-gamma, IL-4 and IL-13 modulate responsiveness of human airway smooth muscle cells to IL-13. Respir Res. 2008;9:84. doi: 10.1186/1465-9921-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee MK, Pardoux C, Hall MC, Lee PS, Warburton D, Qing J, Smith SM, Derynck R. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007;26:3957–3967. doi: 10.1038/sj.emboj.7601818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moore PE, Church TL, Chism DD, Panettieri RA, Jr., Shore SA. IL-13 and IL-4 cause eotaxin release in human airway smooth muscle cells: a role for ERK. Am J Physiol Lung Cell Mol Physiol. 2002;282:L847–853. doi: 10.1152/ajplung.00245.2001. [DOI] [PubMed] [Google Scholar]
- 48.Scherf W, Burdach S, Hansen G. Reduced expression of transforming growth factor beta 1 exacerbates pathology in an experimental asthma model. Eur J Immunol. 2005;35:198–206. doi: 10.1002/eji.200425209. [DOI] [PubMed] [Google Scholar]
- 49.Ray A, Khare A, Krishnamoorthy N, Qi Z, Ray P. Regulatory T cells in many flavors control asthma. Mucosal Immunol. 2010;3:216–229. doi: 10.1038/mi.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han Y, Meng T, Murray NR, Fields AP, Brasier AR. Interleukin-1-induced nuclear factor-kappaB-IkappaBalpha autoregulatory feedback loop in hepatocytes. A role for protein kinase calpha in post-transcriptional regulation of ikappabalpha resynthesis. J Biol Chem. 1999;274:939–947. doi: 10.1074/jbc.274.2.939. [DOI] [PubMed] [Google Scholar]
- 51.de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE. Interleukin 10(IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med. 1991;174:1209–1220. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franchimont N, Rydziel S, Canalis E. Interleukin 6 is autoregulated by transcriptional mechanisms in cultures of rat osteoblastic cells. J Clin Invest. 1997;100:1797–1803. doi: 10.1172/JCI119707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.