Abstract
A recent two-stage genome-wide association study (GWAS) identified five novel breast cancer susceptibility loci on chromosomes 9, 10 and 11. To provide more reliable estimates of the relative risk associated with these loci and investigate possible heterogeneity by subtype of breast cancer, we genotyped the variants rs2380205, rs1011970, rs704010, rs614367, rs10995190 in 39 studies from the Breast Cancer Association Consortium (BCAC), involving 49,608 cases and 48,772 controls of predominantly European ancestry. Four of the variants showed clear evidence of association (P ≤ 3 × 10−9) and weak evidence was observed for rs2380205 (P = 0.06). The strongest evidence was obtained for rs614367, located on 11q13 (per-allele odds ratio 1.21, P = 4 × 10−39). The association for rs614367 was specific to estrogen receptor (ER)-positive disease and strongest for ER plus progesterone receptor (PR)-positive breast cancer, whereas the associations for the other three loci did not differ by tumor subtype.
Keywords: breast cancer susceptibility, polymorphisms, genome wide association, risk factors, hormone receptor status, 11q13
Introduction
Recent genome-wide association studies (GWAS) have provided statistically robust evidence for the association of common genetic variants with breast cancer risk. In particular, variants in the gene regions of FGFR2, TOX3, MAP3K1, LSP1, SLC4A7, COX11, RAD51L1, and in chromosomal regions 8q24, 2q35, 5p12, 6q25, 1p11 and 9q21 (all MIM# 114480) were identified as susceptibility variants through GWAS [Ahmed et al., 2009; Antoniou et al., 2010; Broeks et al., 2011; Easton et al., 2007; Hunter et al., 2007; Milne et al., 2009; Stacey et al., 2007; Stacey et al., 2008; Thomas et al., 2009]. Typically, the variants in these loci occur commonly within the general population, but they confer only modest increases in risk with odds ratios (OR) ranging from 1.10 to 1.43 per allele. Together these variants explain approximately 5% of the familial risk for breast cancer. Despite these relatively small risk effects, the identification of new disease susceptibility loci using GWAS may contribute critically to our understanding of the mechanisms underlying breast cancer tumorigenesis. Furthermore, some loci are more strongly associated with specific tumor subtypes; for instance the FGFR2 rs2981582 variant is more strongly associated with estrogen receptor (ER)-positive than ER-negative disease [Broeks et al., 2011; Milne et al., 2009; Turnbull et al., 2010; Yang et al., 2011].
A recent two-stage GWAS conducted by Turnbull et al. [Turnbull et al., 2010] involving 3,659 cases with family history of breast cancer and 4,897 controls in the first stage, and 12,576 cases and 12,223 controls in the second stage, identified five novel susceptibility loci. The loci are on 11q13, 9p21, 10p15, 10q21 and 10q22 and are respectively close to the cyclin D1 (CCND1; MIM# 114500) and fibroblast growth factor genes (FGF3; MIM# 610706, FGF4; MIM# 104980, FGF19; MIM# 603891), the cyclin-dependent kinase inhibitors CDKN2A (MIM# 606719) and CDKN2B, (MIM# 600431), the zinc finger genes ZNF365 (MIM# 607818) and ZMIZ1 (MIM# 607159), and ANKRD16 [Turnbull et al., 2010]. Although the evidence for these associations was very strong, additional analyses, involving a much larger number of well-characterized breast cancer patients, are needed to independently confirm these associations and assess whether their risks vary with respect to tumor subtype. The Breast Cancer Association Consortium (BCAC), through its global collaborative approach, has gathered more than 96,000 breast cancer cases and controls for independent replication analysis, thereby providing a unique resource for this type of study [Breast Cancer Association Consortium, 2006; Easton et al., 2007].
Materials and Methods
Study Population
Ethics Statement: Written informed consent was obtained from all study participants and the analyses were approved by the institutional review boards at each study center.
Thirty-nine case-control studies from BCAC, that were not included previously in Turnbull et al. [Turnbull et al., 2010], participated in this pooled analysis. Of these, twenty-nine studies were conducted in Europe, five in North America, three in Asia and two in Australia. All studies provided information on disease status and age at diagnosis for cases and self-reported race/ethnicity for all subjects. All but five studies (BIGGS, HUBCS, KARBAC, ORIGO) also provided age at interview for controls. Family history of breast cancer among first degree relatives was provided by 13 studies (ABCF, BBCS, CECILE, CTS, ESTHER, GENICA, GESBC, KBCP, MARIE, MCBCS, SASBAC, SBCS, UCIBCS). ER and PR status as well as histology of the tumor were available for a subset of cases. This histopathology information was generally abstracted from medical reports. A total of 44,662 cases and 45,502 controls of European descent and 4,076 cases and 2,573 controls of Asian descent were included in this analysis. The description of study designs and final sample sizes per study are provided in the Supp. Table S1.
Genotyping and quality control
The rs1011970, rs2380205, rs10995190, rs704010, and rs614367 genetic variants were genotyped by MassARRAY® iPLEX Gold (Sequenom®, San Diego, CA, USA), TaqMan® (Applied Biosystems™, Foster City, CA, USA) and Fluidigm® technology (Fluidigm®, South San Francisco, CA, USA) (Supp. Table S1). The method used by each study is identified in Supp. Table S1. All studies included ≥2% duplicates and 93 CEPH DNAs (HAPMAPPT01, Coriell Institute for Medical Research, Cambden, NJ). The average genotype completion rate per variant was 99% and all genotype completion rates per study were greater than 95% for each variant. We used a χ2− test (1df) to verify that the genotype distributions for each SNP were consistent with those expected under Hardy- Weinberg equilibrium (HWE) within each study and separately among European and Asian control subjects. A Bonferroni correction for multiple tests was applied for the HWE test and gave a P value of 0.0002 as the cutoff for statistical significance, based on approximately 200 independent tests carried out. There was no evidence of departure from HWE for any SNP except rs614367 in one study (PBCS), which was therefore excluded from the analysis for this variant.
Statistical analysis
We used unconditional logistic regression to estimate OR and 95% CI. OR per allele or P values for trend were calculated by assuming a log-additive model. Pooled ORs were calculated using individual-level data. Logistic regression models were adjusted for study by including study specific indicator variables. Restricting the analysis to studies for which age at interview of controls was available, additional adjustment for age made no substantial difference in the results. Europeans and Asians were analyzed separately. Subgroup analyses were performed for breast cancer defined by hormone receptor status (ER and PR) and histological subtypes (ductal, lobular and other tumors) and by family history of breast cancer. For the analyses stratified by family history, we excluded studies with cases selected for family history of breast cancer (ABCS, CNIO-BCS, HEBCS, KARBAC, KConFab/AOCS, MBCSG, NC-BCFR; Supp. Table S1). Heterogeneity of OR across the studies or across the stratification groups was assessed using the Cochran Q test. All tests were two-sided. All analyses were performed using SAS (version 9.2; SAS Institute, Cary, NC).
Results
We analyzed single nucleotide polymorphisms (SNPs) rs1011970, rs2380205, rs10995190, rs704010 and rs614367 in 49,608 breast cancer cases and 48,772 controls from 39 studies participating in BCAC. Of these women, 93% were of European descent and 7% of Asian descent (Table 1). Genotype completion rates were on average >99% for each variant (at least 95% per study). Genotype frequencies for all SNPs were close to those expected under Hardy-Weinberg Equilibrium (HWE) with the exception of rs614367 in one study (PBCS), which was therefore excluded from the analysis of this variant.
Table 1.
Overall breast cancer risk effects in women of European descent and Asian descent of 5 GWAS identified loci [Turnbull et al., 2010]
| SNP | Position | Alleles | European women | Asian women | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | Turnbull et al. stage1 (3,659 ca/4,897 co) |
Turnbull et al. stage2 (12,576 ca/12,223 co) |
BCAC (44,662 ca/45,402 co) |
MAF | BCAC (4,076 ca/ 2,573 co) |
|||||||
| per-allele OR (95% CI) |
P value | per-allele OR (95% CI) |
P value | per-allele OR (95% CI) |
P value | per-allele OR (95% CI) |
P value | |||||
| rs1011970 | 9p21 | G>T | 0.16 | 1.20 (1.11–1.30) | 3×10−5 | 1.09 (1.04–1.14) | 0.00026 | 1.08 (1.05–1.11) | 3×10−9 | 0.08 | 1.13 (0.99–1.29) | 0.06 |
| rs2380205 | 10p15 | C>T | 0.44 | 0.86 (0.81–0.92) | 8×10−5 | 0.94 (0.91–0.98) | 0.0017 | 0.98 (0.96–1.00) | 0.06 | 0.14 | 1.00 (0.90–1.12) | 0.93 |
| rs10995190 | 10q21 | G>A | 0.16 | 0.76 (0.70–0.84) | 6×10−8 | 0.86 (0.82–0.91) | 1×10−8 | 0.88 (0.85–0.90) | 6×10−23 | 0.02 | 1.15 (0.89–1.48) | 0.28 |
| rs704010 | 10q22 | G>A | 0.37 | 1.15 (1.03–1.11) | 3×10−6 | 1.07 (1.03–1.11) | 0.00026 | 1.07 (1.05–1.10) | 4×10−13 | 0.34 | 1.09 (1.00–1.17) | 0.04 |
| rs614367 | 11q13 | C>T | 0.15 | 1.30 (1.20–1.41) | 4×10−8 | 1.15 (1.10–1.20) | 1×10−8 | 1.21 (1.17–1.24) | 4×10−39 | 0.02 | 1.01 (0.73–1.38) | 0.97 |
OR : Odds ratio
ca: cases
co: controls
Per-allele OR adjusted for study
All P values are two-sided
MAF: minor allele frequency (second listed)
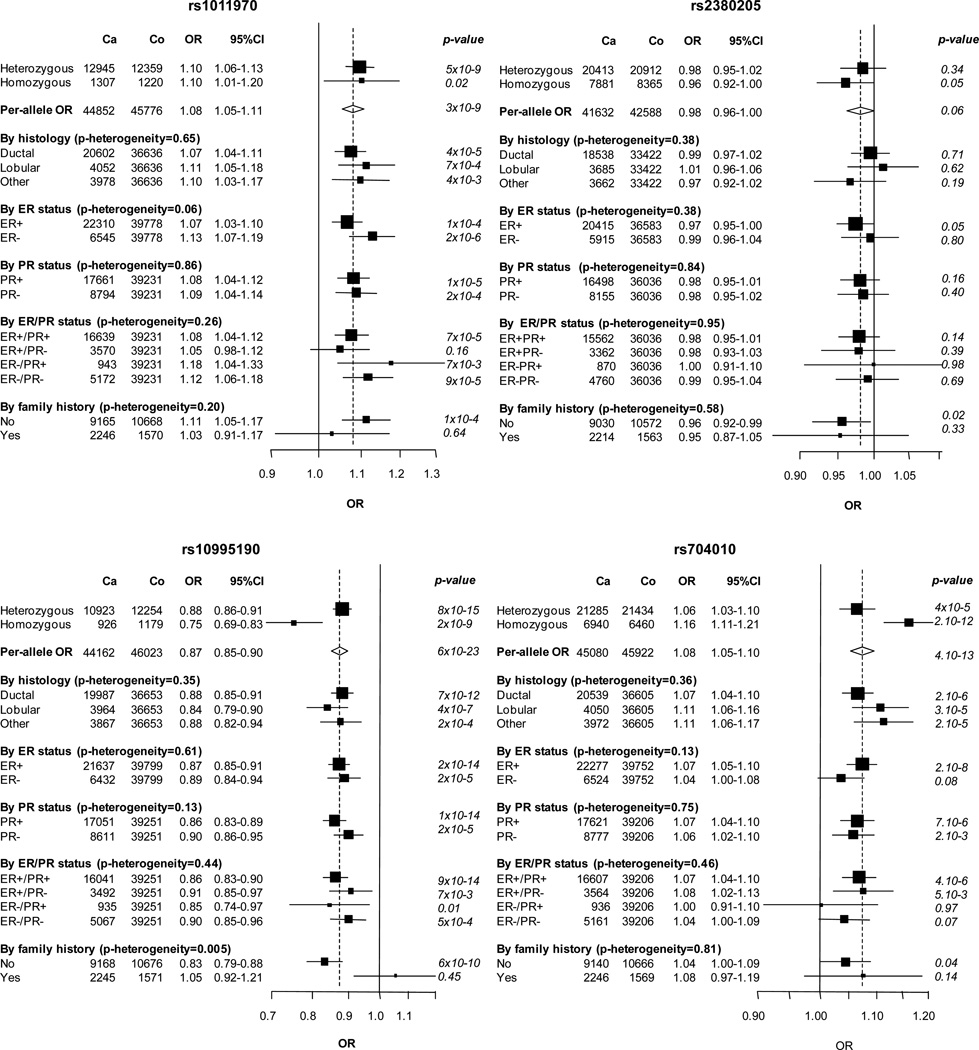
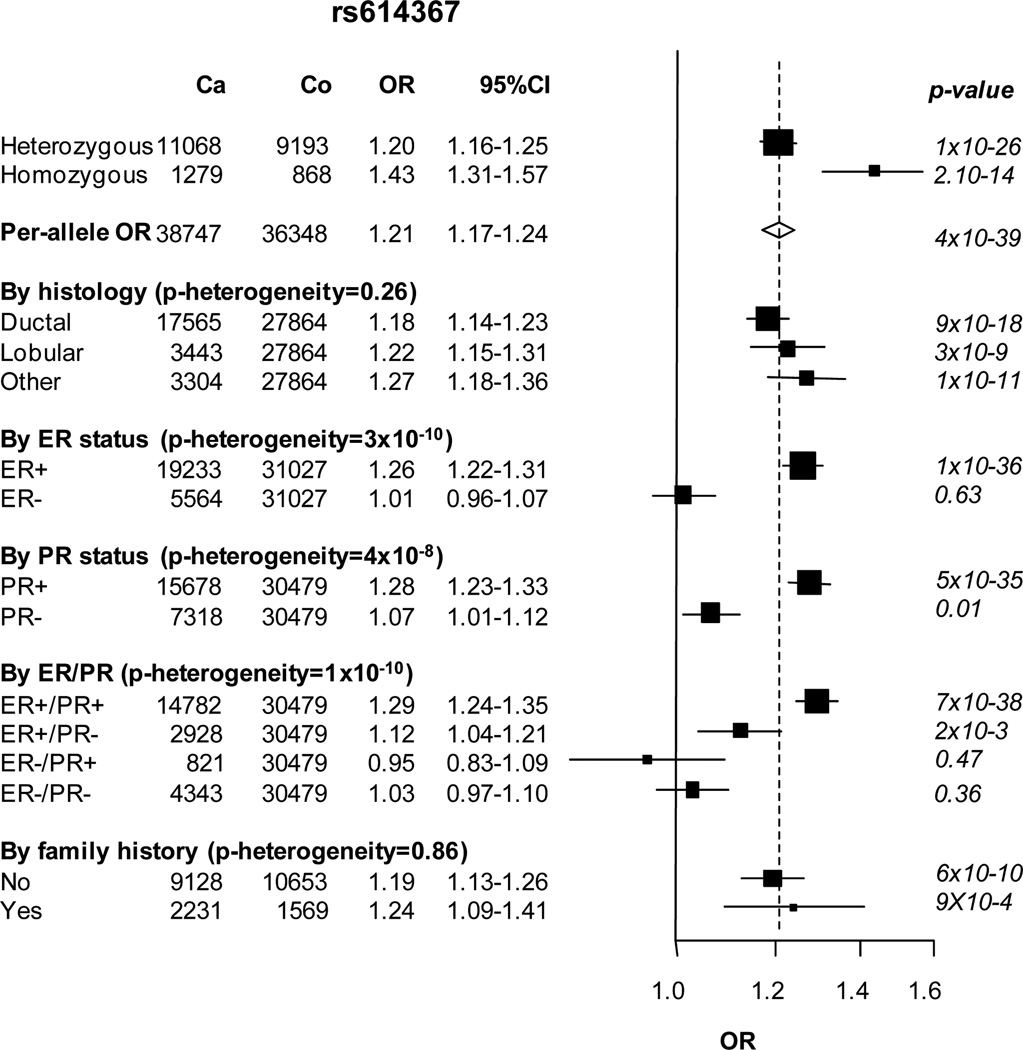
Four of the variants, rs1011970, rs10995190, rs704010 and rs614367, were associated with overall breast cancer risk in women of European descent (P < 1 × 10−8; Table 1, Figure 1 and Supp. Figure S1). Per-allele odds ratios (ORs) for these variants were very similar to those observed in the initial study by Turnbull et al. (Table 1) [Turnbull et al., 2010]. We estimated a lower OR for homozygotes at rs1011970 (OR=1.10 in our study vs. OR=1.29 in Turnbull et al) and rs10995190 (OR=0.75 in our study vs. OR=0.83). These differences, however, might be explained by the wide confidence intervals around the risk estimates due to low minor allele frequencies (MAF=0.16), respectively. Significant heterogeneity by study was only observed for the SNP rs1011970 (P heterogeneity = 0.01; Figure 1). This heterogeneity was due to the BSUCH study in which the per-allele OR was opposite directed to the overall estimated effect. After removing BSUCH from the analysis, heterogeneity between studies was not significant (P heterogeneity = 0.25), but the association of rs1011970 with breast cancer risk was similar (OR 1.08, P = 3 × 10−9 versus OR 1.09, P = 1 × 10−10, before and after exclusion of BSUCH, respectively). The SNP rs2380205 on 10p15 showed limited evidence for association with breast cancer risk (P = 0.06). The 95% confidence interval (CI) limits for the per-allele OR (0.98, 95% CI 0.96–1.00) excluded the OR estimate of 0.94 previously reported by Turnbull et al. [Turnbull et al., 2010], indicating either that the original association was false positive, or that the effect size is substantially smaller than previously reported.
Figure 1.
Forest plots of stratified analysis of the 5 variants in European women.
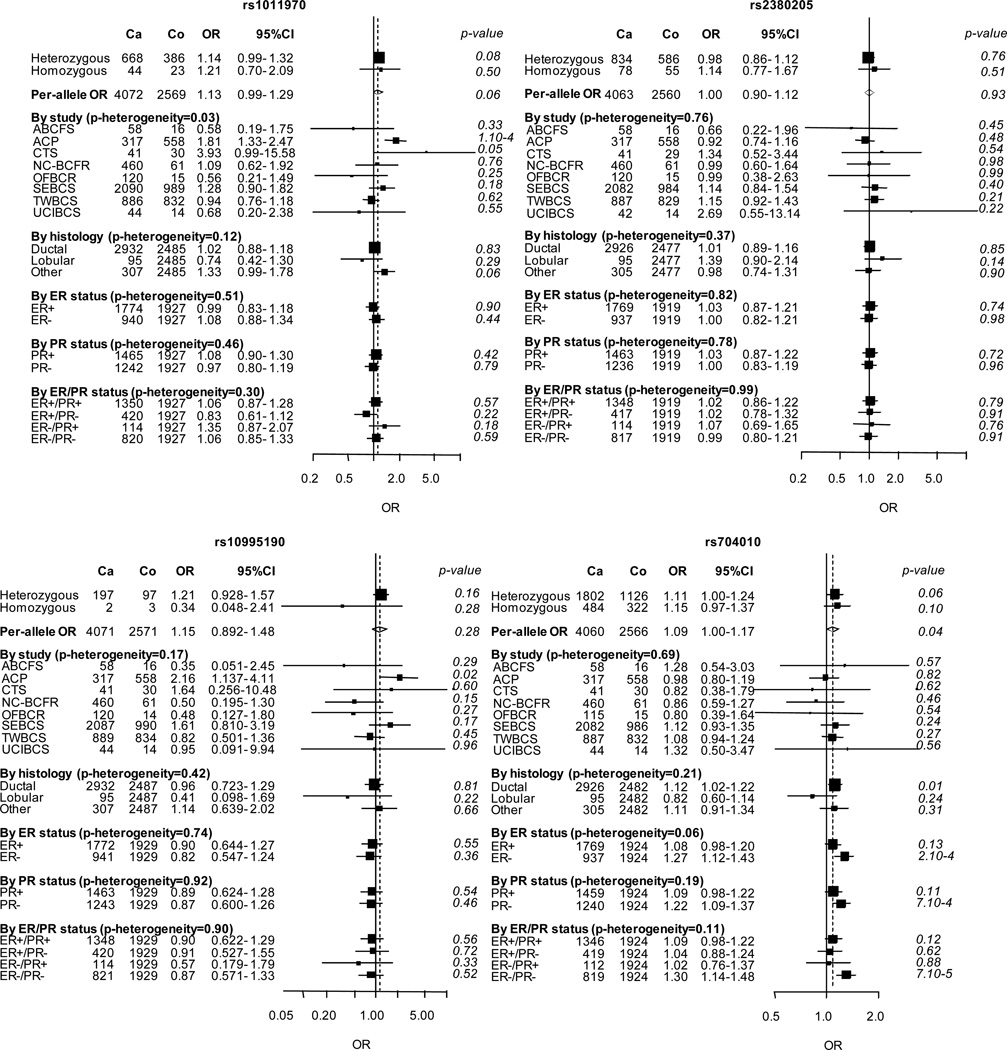
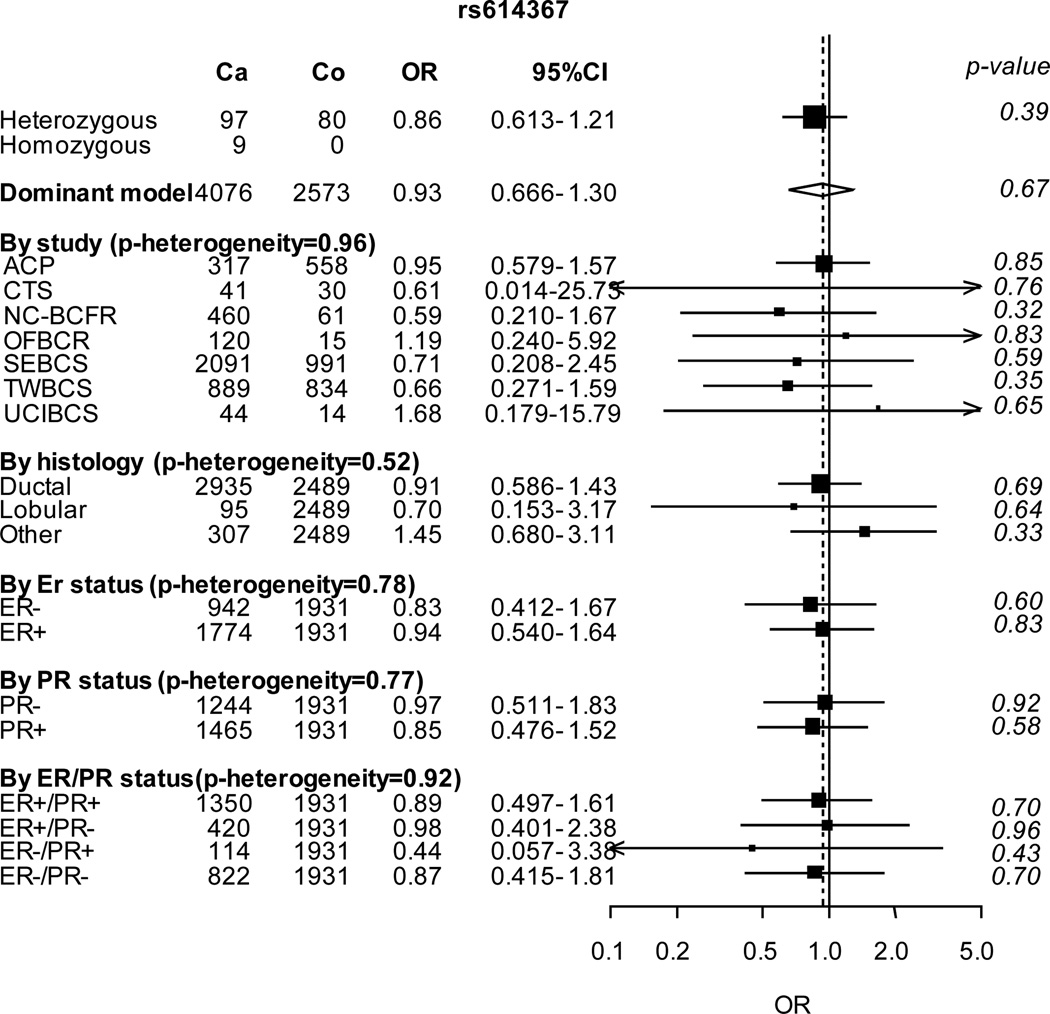
In women of Asian descent, none of the variants was significantly associated with breast cancer risk with the exception of a borderline association with rs704010 (Table 1). However, each of the variants exhibited much lower minor allele frequencies (MAF) in women of Asian descent (Table 1), and none of the estimated per-allele ORs differed significantly from those of European descent.
Next, subgroup analyses for breast cancer defined by hormone receptor status (ER and PR status), histopathological subtype (ductal, lobular and other tumors) and family history of breast cancer were performed separately in women of European and Asian descent. In Europeans, SNP rs614367 was significantly associated with ER-positive (OR 1.26; P = 1 × 10−36) but not with ER-negative breast cancer (OR 1.01; P = 0.63; P heterogeneity = 3 × 10−10; Figure 1). The association was stronger for ER-positive/PR-positive (OR 1.29; P = 7 × 10−38) than for ER-positive/PR-negative tumors (OR 1.12; P = 2 × 10−3; P heterogeneity = 9 × 10−4). The per-allele OR for rs1011970 was also slightly higher for ER-negative than for ER-positive breast cancer (OR 1.13; P = 2 × 10−6 versus OR 1.07; P = 1 × 10−4; Figure 1), but this difference was not significant (P heterogeneity = 0.06). The per-allele ORs for rs2380205, rs704010 and rs10995190 did not differ by tumor receptor status (Figure 1). There was no evidence for heterogeneity in the per-allele ORs by histopathological subtypes for any SNP. With respect to family history of breast cancer we observed that the OR of SNP rs10995190 was lower than 1 in women without family history (OR 0.83; P = 6 × 10−10), while it was greater than 1 in women with a family history of breast cancer (OR 1.05; P = 0.45; P heterogeneity = 5 × 10−3; Figure 1). No other SNP showed significant differences between women with and without family history of breast cancer (Figure 1).
Subgroup analyses in women of Asian descent showed that the association with rs704010 was stronger for ER-negative/PR-negative breast cancer (OR 1.30; P = 7 × 10−5; Figure 2). No heterogeneity by histopathological subtype was observed for any SNP. We did not perform analyses stratified by family history of breast cancer because the number of subjects was too small among Asian women.
Figure 2.
Forest plots of stratified analysis of the 5 variants in Asian women.
To examine potential associations between the breast cancer risk associated SNPs and gene expression we screened the publicly available Expression Quantitative Trait Locus (eQTL) database GENEVAR (www.sanger.ac.uk/resources/software/genevar). No associations with gene expression were observed.
Discussion
This is the largest association study in breast cancer to date and it provides independent and strong evidence for rs1011970, rs10995190, rs704010, and rs614367 being breast cancer susceptibility loci. These variants are located within the footprint of plausible candidate genes: CDKN2A/2B (rs1011970), ZMIZ1 (rs704010), ZNF365 (rs10995190), and CCND1 (rs614367) consistent with the critical role of cell cycle control, gene regulation and cell proliferation pathways in breast tumorigenesis. Each of these genes and one of the SNPs have been reported to be linked with other diseases or phenotypes. In particular, GWAS studies identified several SNPs in 9p21 near CDKN2 that have been associated with cutaneous nevi/melanoma [Falchi et al., 2009], glioma [Shete et al., 2009; Wrensch et al., 2009], type 2 diabetes [Zeggini et al., 2007] and coronary artery disease [Harismendy et al., 2011]. One SNP in the 3’ untranslated region of CDKN2A has been linked with pancreatic cancer [Chen et al., 2007]. All 9p21 SNPs differ from the breast cancer risk SNP rs1011970 described herein, yet this SNP is in linkage disequilibrium with the glioma SNP rs4977756 (r2 = 0.137; D’ = 1.0). Interestingly, the 9p21 interval is the second densest gene locus for predicted enhancers in the human genome and the one containing the most disease associated variants indicating that this chromosomal region has important regulatory function [Harismendy et al., 2011]. The ZMIZ1 is known to be a recombination partner to form an ABL1 fusion gene in B-cell acute lymphoblastic leukaemia [Soler et al., 2008] and a non-synonymous SNP of ZNF365 gene has been associated with Crohn’s disease [Haritunians et al., 2011]. Of note, the ZNF365 SNP rs10995190 now confirmed to be associated with breast cancer risk in this study has recently been associated with mammographic density which is considered one of the strongest risk factors for breast cancer [Lindstrom et al., 2011].
The strongest association with breast cancer was for SNP rs614367 in European women. The estimated OR (1.21 overall, and 1.29 for ER-postive/PR-positive breast cancer) is comparable to that reported for the FGFR2 locus, the most strongly associated known common susceptibility variant for breast cancer. SNP rs614367 is located in an LD block of ~170kb on 11q13 that contains no known genes. This polymorphism lies ~130kb upstream of CCND1, encoding cyclin D1, which is known to be mutated, amplified or overexpressed in various cancers, including breast cancer [Dickson et al., 1995; Kim and Diehl, 2009]. Cyclin D1 together with cyclin-dependent kinases CDK4 and CDK6 mediate phosphorylation of the retinoblastoma protein (Rb) in the cell cycle G1 phase, leading to inactivation of pRb and commitment of mammalian cells to proceed to cell division in response to multiple signaling pathways, including tyrosine kinase and ER signaling [Lange and Yee, 2011]. If the association with rs614367 proves to be functionally related to CCND1, the stronger association of rs614367 with ER-positive disease would be consistent with the role of CCND1 as a mediator of estrogen induced cell proliferation. There is evidence from cell line models that cyclin D1 expression together with inactivation of pRb are features of poor response to endocrine therapies [Lange and Yee, 2011]. However, it is not certain at this stage whether or not the association between rs614367 and breast cancer risk is mediated through CCNC1. Whether 11q13 genetic variation affects the role of cyclin D1 as an oncogenic driver remains to be determined, as other plausible candidates, including FGF4 and FGF19 located at distances of 180kb and 270kb from rs614367, respectively, might also be involved.
The absence of any general breast cancer risk effects in women of Asian descent may be attributed to much lower MAFs of the SNPs tested in this present study and therefore lack of statistical power. Yet, the finding of an association of rs704010 with ER-negative/PR-negative breast cancer suggests a potential relevance in this ethnic group, but much larger sample sizes will be needed for the identification of SNP associations with breast cancer risk as well as patient and tumor characteristics
In conclusion, we confirm the association of four new breast cancer susceptibility loci, provide precise estimates of the associated risks, and provide evidence of variation in the strength of associations by hormone receptor status. We are currently following up these findings through fine-mapping approaches to identify the causal SNPs and genes. This should in turn allow further studies on the impact of the risk causing variants on gene function, and hence explain the observed associations at the molecular level.
Supplementary Material
Acknowledgments
We thank: Maggie Angelakos, Judi Maskiell, Gillian Dite (ABCFS); Laura van 't Veer, Linde Braaf, Senno Verhoef, Frans Hogervorst, Bas Bueno-de-Mesquita (ABCS); Eileen Williams, Elaine Ryder-Mills, Kara Sargus (BBCS); Niall McInerney, Gabrielle Colleran, Andrew Rowan, Angela Jones (BIGGS); Charo Alonso, Tais Moreno, Guillermo Pita, Primitiva Menendez, Anna González-Neira (CNIO-BCS); Sylvia Rabstein, Anne Spickenheuer, Hans-Peter Fischer, Beate Pesch, Volker Harth, Christian Baisch (GENICA); Ursula Eilber, Tanya Koehler (GESBC); Eija Myöhänen, Helena Kemiläinen (KBCP); Heather Thorne, Eveline Niedermayr (kConFab/AOCS); D Bowtell, A deFazio, D Gertig, A Green, P Webb (the AOCS Management Group); A Green, P Parsons, N Hayward, P Webb, D Whiteman (the ACS Management Group); Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel, Kathleen Corthouts (LMBC); Tracy Slanger, Elke Mutschelknauss, S. Behrens, R. Birr, W. Busch, U. Eilber, B. Kaspereit, N. Knese, K. Smit (MARIE); Meera Sangaramoorthy (NC-BCFR); Meeri Otsukka, Kari Mononen (OBCS); Julia Knight, Nayana Weerasooriya (OFBCR); E. Krol-Warmerdam, J. Blom (ORIGO); Louise Brinton, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner (PBCS); Sue Higham, Helen Cramp, Dan Connley (SBCS); Irene Masunaka (UCIBCS); Bernard Peissel, Nadia Zaffaroni, Marco A. Pierotti, Monica Barile, Bernardo Bonanni and the personnel of the Cancer Genetics Testing laboratory (MBCSG); The Breakthrough Breast Cancer and the Institute of Cancer Research, the Study participants, Study staff, and the doctors, nurses and other health care staff and data providers who have contributed to the Study (UKBGS).
Grant Number/ Funding Information
Part of this work was supported by the European Community´s Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175) (COGS). The BCAC is funded by Cancer Research UK (C1287/A10118 and C1287/A12014). Meetings of the BCAC have been funded by the European Union COST program (BM0606). D.F.E. is a Principal Research Fellow of Cancer Research UK. The ABCFS, NC-BCFR and OFBCR work was supported by the United States National Cancer Institute, National Institutes of Health (NIH) under RFA-CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario (U01 CA69467), Cancer Prevention Institute of California (U01 CA69417), and University of Melbourne (U01 CA69638). Samples from the NC-BCFR were processed and distributed by the Coriell Institute for Medical Research. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Australia Fellow and a Victorian Breast Cancer Research Consortium Group Leader. M.C.S. is a NHMRC Senior Research Fellow and a Victorian Breast Cancer Research Consortium Group Leader. The ABCS study was supported by the Dutch Cancer Society (grants NKI DCS 2007-3839 and 2009-4363) and the Dutch National Genomics Initiative. The ACP is funded by the Breast Cancer Research Trust. The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. The BBCS is funded by Cancer Research UK and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). BIGGS: ES is supported by NIHR Comprehensive Biomedical Research Centre, Guy's and St. Thomas' NHS Foundation Trust in partnership with King's College London. IT is supported by the Oxford Biomedical Research Centre. The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ). CECILE study was funded by Fondation de France (grant 2004012618; 2007005156), Institut National du Cancer (INCa grant 2007-1 SPC2; 2008-1-CP-4; 2009-1-SHS SP-04), Association pour la Recherche contre le Cancer (ARC grant 2008-1-CP-4). The CNIO-BCS was supported by the Genome Spain Foundation, the Red Temática de Investigacion Cooperativa en Cancer and grants from the Asociación Espanola Contra el Cancer and the Fondo de Investigacion Sanitario (PI081583 and PI081120). The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council and Herlev Hospital. The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The GENICA Network was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, Germany; The LMBC was supported by the ‘Stichting tegen Kanker’ (grants 232-2008 and 196-2010). The GC-HBOC was supported by Deutsche Krebshilfe (107054), the Dietmar-Hopp Foundation, the Helmholtz society and the German Cancer Research Centre (DKFZ). The HABCS study was supported by the Rudolf Bartling Foundation and by an intramural grant from Hannover Medical School. The HEBCS study has been financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society, and the Sigrid Juselius Foundation. The HMBCS was supported by short-term fellowships from the German Academic Exchange Program (to N.B), and the Friends of Hannover Medical School (to N.B.). The HUBCS was supported by a grant from the German Federal Ministry of Research and Education (RUS08/017). The KARBAC work was supported by the Swedish Cancer Society and the Gustav V Jubilee Foundation. kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC, the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia and the Cancer Foundation of Western Australia. The kConFab Clinical Follow Up Study was funded by the NHMRC (145684, 288704, 454508). Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command (DAMD17-01-1-0729), the Cancer Council of Tasmania and Cancer Foundation of Western Australia and the NHMRC (199600). G.C.T. and P.W. are supported by the NHMRC. The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, the Academy of Finland and by the strategic funding of the University of Eastern Finland. LMBC is supported by the 'Stichting tegen Kanker' (232-2008 and 196-2010). The MARIE study was supported by the Deutsche Krebshilfe e.V. (70-2892-BR I), the Hamburg Cancer Society, the German Cancer Research Center and the genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany (01KH0402). MBCSG is supported by grants from Ministero della Salute (Extraordinary National Cancer Program 2006 “Alleanza contro il Cancro”, and “Progetto Tumori Femminili” to PR), Ministero dell’Universita’ e Ricerca (RBLAO3-BETH to PR), Fondazione Italiana per la Ricerca sul Cancro (Special Project “Hereditary tumors”), Associazione Italiana per la Ricerca sul Cancro (4017 and by funds from Italian citizens who allocated the 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects “5×1000”). The MCBCS was supported by the NIH grants [CA122340, CA128978] and a Specialized Program of Research Excellence (SPORE) in Breast Cancer [CA116201]. MCCS is supported by Cancer Council Victoria and by NHMRC (grants 209057, 251533, 396414, 504711, 504715). The NBCS was supported by grants from the Norwegian Research council, 155218/V40, 175240/S10 to ALBD, FUGE-NFR 181600/V11 to VNK and a Swizz Bridge Award to ALBD. The OBCS was supported by research grants from the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the Academy of Finland, the University of Oulu, and the Oulu University Hospital. The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The SBCS was supported by Yorkshire Cancer Research and the Breast Cancer Campaign. The SEBCS was supported by the Korea Health 21 R&D Project [AO30001], Ministry of Health and Welfare, Republic of Korea. The SZBCS was supported by Grant PBZ_KBN_122/P05/2004; Katarzyna Jaworska is a fellow of International PhD program, Postgraduate School of Molecular Medicine, Warsaw Medical University, supported by the Polish Foundation of Science. The TWBCS is supported by the Taiwan Biobank project of the Institute of Biomedical Sciences, Academia Sinica, Taiwan. The UCIBCS component of this research was supported by the NIH [CA58860, CA92044] and the Lon V Smith Foundation (LVS39420). ORIGO were funded by grants from the Dutch Cancer Society (UL1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The UKBGS thank Breakthrough Breast Cancer and the Institute of Cancer Research for funding. The ICR acknowledge NHS funding to the NIHR Biomedical Research Centre.
Footnotes
Due to length, grant number and funding information are listed in the Acknowledgments
References
- Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, Morrison J, Maranian M, Pooley KA, Luben R, Eccles D, Evans DG, Fletcher O, Johnson N, Dos SS I, Peto J, Stratton MR, Rahman N, Jacobs K, Prentice R, Anderson GL, Rajkovic A, Curb JD, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Diver WR, Bojesen S, Nordestgaard BG, Flyger H, Dork T, Schurmann P, Hillemanns P, Karstens JH, Bogdanova NV, Antonenkova NN, Zalutsky IV, Bermisheva M, Fedorova S, Khusnutdinova E, Kang D, Yoo KY, Noh DY, Ahn SH, Devilee P, van Asperen CJ, Tollenaar RA, Seynaeve C, Garcia-Closas M, Lissowska J, Brinton L, Peplonska B, Nevanlinna H, Heikkinen T, Aittomaki K, Blomqvist C, Hopper JL, Southey MC, Smith L, Spurdle AB, Schmidt MK, Broeks A, van Hien RR, Cornelissen S, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Schmutzler RK, Burwinkel B, Bartram CR, Meindl A, Brauch H, Justenhoven C, Hamann U, Chang-Claude J, Hein R, Wang-Gohrke S, Lindblom A, Margolin S, Mannermaa A, Kosma VM, Kataja V, Olson JE, Wang X, Fredericksen Z, Giles GG, Severi G, Baglietto L, English DR, Hankinson SE, Cox DG, Kraft P, Vatten LJ, Hveem K, Kumle M, Sigurdson A, Doody M, Bhatti P, Alexander BH, Hooning MJ, van den Ouweland AM, Oldenburg RA, Schutte M, Hall P, Czene K, Liu J, Li Y, Cox A, Elliott G, Brock I, Reed MW, Shen CY, Yu JC, Hsu GC, Chen ST, Anton-Culver H, Ziogas A, Andrulis IL, Knight JA, Beesley J, Goode EL, Couch F, Chenevix-Trench G, Hoover RN, Ponder BA, Hunter DJ, Pharoah PD, Dunning AM, Chanock SJ, Easton DF. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton-Culver H, Cohen PF, Gildea ME, Ziogas A. Characteristics of BRCA1 mutations in a population-based case series of breast and ovarian cancer. Eur J Cancer. 2000;36:1200–1208. doi: 10.1016/s0959-8049(00)00110-6. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, Healey S, Morrison J, Kartsonaki C, Lesnick T, Ghoussaini M, Barrowdale D, Peock S, Cook M, Oliver C, Frost D, Eccles D, Evans DG, Eeles R, Izatt L, Chu C, Douglas F, Paterson J, Stoppa-Lyonnet D, Houdayer C, Mazoyer S, Giraud S, Lasset C, Remenieras A, Caron O, Hardouin A, Berthet P, Hogervorst FB, Rookus MA, Jager A, van den OA, Hoogerbrugge N, van der Luijt RB, Meijers-Heijboer H, Gomez Garcia EB, Devilee P, Vreeswijk MP, Lubinski J, Jakubowska A, Gronwald J, Huzarski T, Byrski T, Gorski B, Cybulski C, Spurdle AB, Holland H, Goldgar DE, John EM, Hopper JL, Southey M, Buys SS, Daly MB, Terry MB, Schmutzler RK, Wappenschmidt B, Engel C, Meindl A, Preisler-Adams S, Arnold N, Niederacher D, Sutter C, Domchek SM, Nathanson KL, Rebbeck T, Blum JL, Piedmonte M, Rodriguez GC, Wakeley K, Boggess JF, Basil J, Blank SV, Friedman E, Kaufman B, Laitman Y, Milgrom R, Andrulis IL, Glendon G, Ozcelik H, Kirchhoff T, Vijai J, Gaudet MM, Altshuler D, Guiducci C, Loman N, Harbst K, Rantala J, Ehrencrona H, Gerdes AM, Thomassen M, Sunde L, Peterlongo P, Manoukian S, Bonanni B, Viel A, Radice P, Caldes T, de la HM, Singer CF, Fink-Retter A, Greene MH, Mai PL, Loud JT, Guidugli L, Lindor NM, Hansen TV, Nielsen FC, Blanco I, Lazaro C, Garber J, Ramus SJ, Gayther SA, Phelan C, Narod S, Szabo CI, Benitez J, Osorio A, Nevanlinna H, Heikkinen T, Caligo MA, Beattie MS, Hamann U, Godwin AK, Montagna M, Casella C, Neuhausen SL, Karlan BY, Tung N, Toland AE, Weitzel J, Olopade O, Simard J, Soucy P, Rubinstein WS, Arason A, Rennert G, Martin NG, Montgomery GW, Chang-Claude J, Flesch-Janys D, Brauch H, Severi G, Baglietto L, Cox A, Cross SS, Miron P, Gerty SM, Tapper W, Yannoukakos D, Fountzilas G, Fasching PA, Beckmann MW, Dos SS I, Peto J, Lambrechts D, Paridaens R, Rudiger T, Forsti A, Winqvist R, Pylkas K, Diasio RB, Lee AM, Eckel-Passow J, Vachon C, Blows F, Driver K, Dunning A, Pharoah PP, Offit K, Pankratz VS, Hakonarson H, Chenevix-Trench G, Easton DF, Couch FJ. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beesley J, Jordan SJ, Spurdle AB, Song H, Ramus SJ, Kjaer SK, Hogdall E, DiCioccio RA, McGuire V, Whittemore AS, Gayther SA, Pharoah PD, Webb PM, Chenevix-Trench G. Association between single-nucleotide polymorphisms in hormone metabolism and DNA repair genes and epithelial ovarian cancer: results from two Australian studies and an additional validation set. Cancer Epidemiol Biomarkers Prev. 2007;16:2557–2565. doi: 10.1158/1055-9965.EPI-07-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanova N, Cybulski C, Bermisheva M, Datsyuk I, Yamini P, Hillemanns P, Antonenkova NN, Khusnutdinova E, Lubinski J, Dork T. A nonsense mutation (E1978X) in the ATM gene is associated with breast cancer. Breast Cancer Res Treat. 2009;118:207–211. doi: 10.1007/s10549-008-0189-9. [DOI] [PubMed] [Google Scholar]
- Breast Cancer Association Consortium. Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. J Natl Cancer Inst. 2006;98:1382–1396. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, Apicella C, Smith LD, Hammet F, Southey MC, van ' V, de GR, Smit VT, Fasching PA, Beckmann MW, Jud S, Ekici AB, Hartmann A, Hein A, Schulz-Wendtland R, Burwinkel B, Marme F, Schneeweiss A, Sinn HP, Sohn C, Tchatchou S, Bojesen SE, Nordestgaard BG, Flyger H, Orsted DD, Kaur-Knudsen D, Milne RL, Perez JI, Zamora P, Rodriguez PM, Benitez J, Brauch H, Justenhoven C, Ko YD, Hamann U, Fischer HP, Bruning T, Pesch B, Chang-Claude J, Wang-Gohrke S, Bremer M, Karstens JH, Hillemanns P, Dork T, Nevanlinna HA, Heikkinen T, Heikkila P, Blomqvist C, Aittomaki K, Aaltonen K, Lindblom A, Margolin S, Mannermaa A, Kosma VM, Kauppinen JM, Kataja V, Auvinen P, Eskelinen M, Soini Y, Chenevix-Trench G, Spurdle AB, Beesley J, Chen X, Holland H, Lambrechts D, Claes B, Vandorpe T, Neven P, Wildiers H, Flesch-Janys D, Hein R, Loning T, Kosel M, Fredericksen ZS, Wang X, Giles GG, Baglietto L, Severi G, McLean C, Haiman CA, Henderson BE, Le ML, Kolonel LN, Grenaker AG, Kristensen V, Borresen-Dale AL, Hunter DJ, Hankinson SE, Andrulis IL, Marie MA, O'Malley FP, Devilee P, Huijts PE, Tollenaar RA, van Asperen CJ, Seynaeve CS, Chanock SJ, Lissowska J, Brinton L, Peplonska B, Figueroa J, Yang XR, Hooning MJ, Hollestelle A, Oldenburg RA, Jager A, Kriege M, Ozturk B, van Leenders GJ, Hall P, Czene K, Humphreys K, Liu J, Cox A, Connley D, Cramp HE, Cross SS, Balasubramanian SP, Reed MW, Dunning AM, Easton DF, Humphreys MK, Caldas C, Blows F, Driver K, Provenzano E, Lubinski J, Jakubowska A, Huzarski T, Byrski T, Cybulski C, Gorski B, Gronwald J, Brennan P, Sangrajrang S, Gaborieau V, Shen CY, Hsiung CN, Yu JC, Chen ST, Hsu GC, Hou MF, Huang CS, Anton-Culver H, Ziogas A, Pharoah PD, Garcia-Closas M. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20:3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catucci I, Verderio P, Pizzamiglio S, Manoukian S, Peissel B, Barile M, Tizzoni L, Bernard L, Ravagnani F, Galastri L, Pierotti MA, Radice P, Peterlongo P. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis. 2009;30:544–545. doi: 10.1093/carcin/bgn289. [DOI] [PubMed] [Google Scholar]
- Chang-Claude J, Eby N, Kiechle M, Bastert G, Becher H. Breastfeeding and breast cancer risk by age 50 among women in Germany. Cancer Causes Control. 2000;11:687–695. doi: 10.1023/a:1008907901087. [DOI] [PubMed] [Google Scholar]
- Chen J, Li D, Wei C, Sen S, Killary AM, Amos CI, Evans DB, Abbruzzese JL, Frazier ML. Aurora-A and p16 polymorphisms contribute to an earlier age at diagnosis of pancreatic cancer in Caucasians. Clin Cancer Res. 2007;13:3100–3104. doi: 10.1158/1078-0432.CCR-06-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleran G, McInerney N, Rowan A, Barclay E, Jones AM, Curran C, Miller N, Kerin M, Tomlinson I, Sawyer E. The TGFBR1*6A/9A polymorphism is not associated with differential risk of breast cancer. Breast Cancer Res Treat. 2010;119:437–442. doi: 10.1007/s10549-009-0395-0. [DOI] [PubMed] [Google Scholar]
- de Bock GH, Schutte M, Krol-Warmerdam EM, Seynaeve C, Blom J, Brekelmans CT, Meijers-Heijboer H, van Asperen CJ, Cornelisse CJ, Devilee P, Tollenaar RA, Klijn JG. Tumour characteristics and prognosis of breast cancer patients carrying the germline CHEK2*1100delC variant. J Med Genet. 2004;41:731–735. doi: 10.1136/jmg.2004.019737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De ML, Van LE, De NK, Moerman P, Pochet N, Hendrickx W, Wildiers H, Paridaens R, Smeets A, Christiaens MR, Vergote I, Leunen K, Amant F, Neven P. Does estrogen receptor negative/progesterone receptor positive breast carcinoma exist? J Clin Oncol. 2008:335–336. doi: 10.1200/JCO.2007.14.8411. [DOI] [PubMed] [Google Scholar]
- De VG, Verderio P, Pizzamiglio S, Manoukian S, Barile M, Fortuzzi S, Ravagnani F, Pierotti MA, Radice P, Peterlongo P. Evidences for association of the CASP8 -652 6N del promoter polymorphism with age at diagnosis in familial breast cancer cases. Breast Cancer Res Treat. 2009;113:607–608. doi: 10.1007/s10549-008-9963-y. [DOI] [PubMed] [Google Scholar]
- Dickson C, Fantl V, Gillett C, Brookes S, Bartek J, Smith R, Fisher C, Barnes D, Peters G. Amplification of chromosome band 11q13 and a role for cyclin D1 in human breast cancer. Cancer Lett. 1995;90:43–50. doi: 10.1016/0304-3835(94)03676-a. [DOI] [PubMed] [Google Scholar]
- Ding SL, Yu JC, Chen ST, Hsu GC, Kuo SJ, Lin YH, Wu PE, Shen CY. Genetic variants of BLM interact with RAD51 to increase breast cancer susceptibility. Carcinogenesis. 2009;30:43–49. doi: 10.1093/carcin/bgn233. [DOI] [PubMed] [Google Scholar]
- Dite GS, Jenkins MA, Southey MC, Hocking JS, Giles GG, McCredie MR, Venter DJ, Hopper JL. Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 germline mutations. J Natl Cancer Inst. 2003;95:448–457. doi: 10.1093/jnci/95.6.448. [DOI] [PubMed] [Google Scholar]
- Dork T, Bendix R, Bremer M, Rades D, Klopper K, Nicke M, Skawran B, Hector A, Yamini P, Steinmann D, Weise S, Stuhrmann M, Karstens JH. Spectrum of ATM gene mutations in a hospital-based series of unselected breast cancer patients. Cancer Res. 2001;61:7608–7615. [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowman R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le ML, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Brinton L, Chanock S, Lissowska J, Peplonska B, Nevanlinna H, Fagerholm R, Eerola H, Kang D, Yoo KY, Noh DY, Ahn SH, Hunter DJ, Hankinson SE, Cox DG, Hall P, Wedren S, Liu J, Low YL, Bogdanova N, Schurmann P, Dork T, Tollenaar RA, Jacobi CE, Devilee P, Klijn JG, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MW, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den OA, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X, Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BA. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkko H, Xia B, Nikkila J, Schleutker J, Syrjakoski K, Mannermaa A, Kallioniemi A, Pylkas K, Karppinen SM, Rapakko K, Miron A, Sheng Q, Li G, Mattila H, Bell DW, Haber DA, Grip M, Reiman M, Jukkola-Vuorinen A, Mustonen A, Kere J, Aaltonen LA, Kosma VM, Kataja V, Soini Y, Drapkin RI, Livingston DM, Winqvist R. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- Falchi M, Bataille V, Hayward NK, Duffy DL, Bishop JA, Pastinen T, Cervino A, Zhao ZZ, Deloukas P, Soranzo N, Elder DE, Barrett JH, Martin NG, Bishop DT, Montgomery GW, Spector TD. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching PA, Loehberg CR, Strissel PL, Lux MP, Bani MR, Schrauder M, Geiler S, Ringleff K, Oeser S, Weihbrecht S, Schulz-Wendtland R, Hartmann A, Beckmann MW, Strick R. Single nucleotide polymorphisms of the aromatase gene (CYP19A1), HER2/neu status, and prognosis in breast cancer patients. Breast Cancer Res Treat. 2008;112:89–98. doi: 10.1007/s10549-007-9822-2. [DOI] [PubMed] [Google Scholar]
- Flesch-Janys D, Slanger T, Mutschelknauss E, Kropp S, Obi N, Vettorazzi E, Braendle W, Bastert G, Hentschel S, Berger J, Chang-Claude J. Risk of different histological types of postmenopausal breast cancer by type and regimen of menopausal hormone therapy. Int J Cancer. 2008;123:933–941. doi: 10.1002/ijc.23655. [DOI] [PubMed] [Google Scholar]
- Fletcher O, Johnson N, Palles C, Dos SS I, McCormack V, Whittaker J, Ashworth A, Peto J. Inconsistent association between the STK15 F31I genetic polymorphism and breast cancer risk. J Natl Cancer Inst. 2006;98:1014–1018. doi: 10.1093/jnci/djj268. [DOI] [PubMed] [Google Scholar]
- Frank B, Hemminki K, Wappenschmidt B, Meindl A, Klaes R, Schmutzler RK, Bugert P, Untch M, Bartram CR, Burwinkel B. Association of the CASP10 V410I variant with reduced familial breast cancer risk and interaction with the CASP8 D302H variant. Carcinogenesis. 2006;27:606–609. doi: 10.1093/carcin/bgi248. [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S, Welch R, Lissowska J, Peplonska B, Szeszenia-Dabrowska N, Zatonski W, Bardin-Mikolajczak A, Struewing JP. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet. 2006;119:376–388. doi: 10.1007/s00439-006-0135-z. [DOI] [PubMed] [Google Scholar]
- Giles GG, English DR. The Melbourne Collaborative Cohort Study. IARC Sci Publ. 2002;156:69–70. [PubMed] [Google Scholar]
- Han S, Lee KM, Choi JY, Park SK, Lee JY, Lee JE, Noh DY, Ahn SH, Han W, Kim DH, Hong YC, Ha E, Yoo KY, Kang D. CASP8 polymorphisms, estrogen and progesterone receptor status, and breast cancer risk. Breast Cancer Res Treat. 2008;110:387–393. doi: 10.1007/s10549-007-9730-5. [DOI] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritunians T, Jones MR, McGovern DP, Shih DQ, Barrett RJ, Derkowski C, Dubinsky MC, Dutridge D, Fleshner PR, Ippoliti A, King L, Leshinsky-Silver E, Levine A, Melmed GY, Mengesha E, Vasilauskas EA, Ziaee S, Rotter JI, Targan SR, Taylor KD. Variants in ZNF365 isoform D are associated with Crohn's disease. Gut. 2011 doi: 10.1136/gut.2010.227256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartikainen JM, Tuhkanen H, Kataja V, Dunning AM, Antoniou A, Smith P, Arffman A, Pirskanen M, Easton DF, Eskelinen M, Uusitupa M, Kosma VM, Mannermaa A. An autosome-wide scan for linkage disequilibrium-based association in sporadic breast cancer cases in eastern Finland: three candidate regions found. Cancer Epidemiol Biomarkers Prev. 2005;14:75–80. [PubMed] [Google Scholar]
- Hartikainen JM, Tuhkanen H, Kataja V, Eskelinen M, Uusitupa M, Kosma VM, Mannermaa A. Refinement of the 22q12-q13 breast cancer--associated region: evidence of TMPRSS6 as a candidate gene in an eastern Finnish population. Clin Cancer Res. 2006;12:1454–1462. doi: 10.1158/1078-0432.CCR-05-1417. [DOI] [PubMed] [Google Scholar]
- Hsu HM, Wang HC, Chen ST, Hsu GC, Shen CY, Yu JC. Breast cancer risk is associated with the genes encoding the DNA double-strand break repair Mre11/Rad50/Nbs1 complex. Cancer Epidemiol Biomarkers Prev. 2007;16:2024–2032. doi: 10.1158/1055-9965.EPI-07-0116. [DOI] [PubMed] [Google Scholar]
- Huijts PE, Vreeswijk MP, Kroeze-Jansema KH, Jacobi CE, Seynaeve C, Krol-Warmerdam EM, Wijers-Koster PM, Blom JC, Pooley KA, Klijn JG, Tollenaar RA, Devilee P, van Asperen CJ. Clinical correlates of low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch cohort of incident breast cancer cases. Breast Cancer Res. 2007;9:R78. doi: 10.1186/bcr1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowska A, Jaworska K, Cybulski C, Janicka A, Szymanska-Pasternak J, Lener M, Narod SA, Lubinski J. Do BRCA1 modifiers also affect the risk of breast cancer in non-carriers? Eur J Cancer. 2009;45:837–842. doi: 10.1016/j.ejca.2008.10.021. [DOI] [PubMed] [Google Scholar]
- John EM, Hopper JL, Beck JC, Knight JA, Neuhausen SL, Senie RT, Ziogas A, Andrulis IL, Anton-Culver H, Boyd N, Buys SS, Daly MB, O'Malley FP, Santella RM, Southey MC, Venne VL, Venter DJ, West DW, Whittemore AS, Seminara D. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justenhoven C, Pierl CB, Haas S, Fischer HP, Baisch C, Hamann U, Harth V, Pesch B, Bruning T, Vollmert C, Illig T, Dippon J, Ko YD, Brauch H. The CYP1B1_1358_GG genotype is associated with estrogen receptor-negative breast cancer. Breast Cancer Res Treat. 2008;111:171–177. doi: 10.1007/s10549-007-9762-x. [DOI] [PubMed] [Google Scholar]
- Kilpivaara O, Bartkova J, Eerola H, Syrjakoski K, Vahteristo P, Lukas J, Blomqvist C, Holli K, Heikkila P, Sauter G, Kallioniemi OP, Bartek J, Nevanlinna H. Correlation of CHEK2 protein expression and c.1100delC mutation status with tumor characteristics among unselected breast cancer patients. Int J Cancer. 2005;113:575–580. doi: 10.1002/ijc.20638. [DOI] [PubMed] [Google Scholar]
- Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. 2009;220:292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange CA, Yee D. Killing the second messenger: targeting loss of cell cycle control in endocrine-resistant breast cancer. Endocr Relat Cancer. 2011;18:C19–C24. doi: 10.1530/ERC-11-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Choi JY, Park SK, Chung HW, Ahn B, Yoo KY, Han W, Noh DY, Ahn SH, Kim H, Wei Q, Kang D. Genetic polymorphisms of ataxia telangiectasia mutated and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:821–825. doi: 10.1158/1055-9965.EPI-04-0330. [DOI] [PubMed] [Google Scholar]
- Lindblom A, Rotstein S, Larsson C, Nordenskjold M, Iselius L. Hereditary breast cancer in Sweden: a predominance of maternally inherited cases. Breast Cancer Res Treat. 1992;24:159–165. doi: 10.1007/BF01961248. [DOI] [PubMed] [Google Scholar]
- Lindstrom S, Vachon CM, Li J, Varghese J, Thompson D, Warren R, Brown J, Leyland J, Audley T, Wareham NJ, Loos RJ, Paterson AD, Rommens J, Waggott D, Martin LJ, Scott CG, Pankratz VS, Hankinson SE, Hazra A, Hunter DJ, Hopper JL, Southey MC, Chanock SJ, Silva IS, Liu J, Eriksson L, Couch FJ, Stone J, Apicella C, Czene K, Kraft P, Hall P, Easton DF, Boyd NF, Tamimi RM. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat Genet. 2011;43:185–187. doi: 10.1038/ng.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski J, Korzen M, Gorski B, Cybulski C, Debniak T, Jakubowska A, Jaworska K, Wokolorczyk D, Medrek K, Matyjasik J, Huzarski T, Byrski T, Gronwald J, Masojc B, Lener M, Szymanska A, Szymanska-Pasternak J, Serrano-Fernandez P, Piegat A, Ucinski R, Domagala P, Domagala W, Chosia M, Kladny J, Gorecka B, Narod S, Scott R. Genetic contribution to all cancers: the first demonstration using the model of breast cancers from Poland stratified by age at diagnosis and tumour pathology. Breast Cancer Res Treat. 2009;114:121–126. doi: 10.1007/s10549-008-9974-8. [DOI] [PubMed] [Google Scholar]
- MacPherson G, Healey CS, Teare MD, Balasubramanian SP, Reed MW, Pharoah PD, Ponder BA, Meuth M, Bhattacharyya NP, Cox A. Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst. 2004;96:1866–1869. doi: 10.1093/jnci/dji001. [DOI] [PubMed] [Google Scholar]
- Mann GJ, Thorne H, Balleine RL, Butow PN, Clarke CL, Edkins E, Evans GM, Fereday S, Haan E, Gattas M, Giles GG, Goldblatt J, Hopper JL, Kirk J, Leary JA, Lindeman G, Niedermayr E, Phillips KA, Picken S, Pupo GM, Saunders C, Scott CL, Spurdle AB, Suthers G, Tucker K, Chenevix-Trench G. Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res. 2006;8:R12. doi: 10.1186/bcr1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin S, Werelius B, Fornander T, Lindblom A. BRCA1 mutations in a population-based study of breast cancer in Stockholm County. Genet Test. 2004;8:127–132. doi: 10.1089/gte.2004.8.127. [DOI] [PubMed] [Google Scholar]
- McInerney N, Colleran G, Rowan A, Walther A, Barclay E, Spain S, Jones AM, Tuohy S, Curran C, Miller N, Kerin M, Tomlinson I, Sawyer E. Low penetrance breast cancer predisposition SNPs are site specific. Breast Cancer Res Treat. 2009;117:151–159. doi: 10.1007/s10549-008-0235-7. [DOI] [PubMed] [Google Scholar]
- Milne RL, Benitez J, Nevanlinna H, Heikkinen T, Aittomaki K, Blomqvist C, Arias JI, Zamora MP, Burwinkel B, Bartram CR, Meindl A, Schmutzler RK, Cox A, Brock I, Elliott G, Reed MW, Southey MC, Smith L, Spurdle AB, Hopper JL, Couch FJ, Olson JE, Wang X, Fredericksen Z, Schurmann P, Bremer M, Hillemanns P, Dork T, Devilee P, van Asperen CJ, Tollenaar RA, Seynaeve C, Hall P, Czene K, Liu J, Li Y, Ahmed S, Dunning AM, Maranian M, Pharoah PD, Chenevix-Trench G, Beesley J, Bogdanova NV, Antonenkova NN, Zalutsky IV, Anton-Culver H, Ziogas A, Brauch H, Justenhoven C, Ko YD, Haas S, Fasching PA, Strick R, Ekici AB, Beckmann MW, Giles GG, Severi G, Baglietto L, English DR, Fletcher O, Johnson N, Dos SS I, Peto J, Turnbull C, Hines S, Renwick A, Rahman N, Nordestgaard BG, Bojesen SE, Flyger H, Kang D, Yoo KY, Noh DY, Mannermaa A, Kataja V, Kosma VM, Garcia-Closas M, Chanock S, Lissowska J, Brinton LA, Chang-Claude J, Wang-Gohrke S, Shen CY, Wang HC, Yu JC, Chen ST, Bermisheva M, Nikolaeva T, Khusnutdinova E, Humphreys MK, Morrison J, Platte R, Easton DF. Risk of estrogen receptor-positive and -negative breast cancer and single-nucleotide polymorphism 2q35-rs13387042. J Natl Cancer Inst. 2009;101:1012–1018. doi: 10.1093/jnci/djp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RL, Ribas G, Gonzalez-Neira A, Fagerholm R, Salas A, Gonzalez E, Dopazo J, Nevanlinna H, Robledo M, Benitez J. ERCC4 associated with breast cancer risk: a two-stage case-control study using high-throughput genotyping. Cancer Res. 2006;66:9420–9427. doi: 10.1158/0008-5472.CAN-06-1418. [DOI] [PubMed] [Google Scholar]
- Neven P, Brouckaert O, Van BV, Vanden BI, Hendrickx W, Cho H, Deraedt K, Van CB, Van HS, Moerman P, Amant F, Leunen K, Smeets A, Wildiers H, Paridaens R, Vergote I, Christiaens MR. In early-stage breast cancer, the estrogen receptor interacts with correlation between human epidermal growth factor receptor 2 status and age at diagnosis, tumor grade, and lymph node involvement. J Clin Oncol. 2008;26:1768–1769. doi: 10.1200/JCO.2007.15.6141. [DOI] [PubMed] [Google Scholar]
- Nordgard SH, Johansen FE, Alnaes GI, Bucher E, Syvanen AC, Naume B, Borresen-Dale AL, Kristensen VN. Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer. 2008;47:680–696. doi: 10.1002/gcc.20569. [DOI] [PubMed] [Google Scholar]
- Olson JE, Ma CX, Pelleymounter LL, Schaid DJ, Pankratz VS, Vierkant RA, Fredericksen ZS, Ingle JN, Wu Y, Couch F, Sellers TA, Weinshilboum RM, Vachon CM. A comprehensive examination of CYP19 variation and breast density. Cancer Epidemiol Biomarkers Prev. 2007;16:623–625. doi: 10.1158/1055-9965.EPI-06-0781. [DOI] [PubMed] [Google Scholar]
- Pesch B, Ko Y, Brauch H, Hamann U, Harth V, Rabstein S, Pierl C, Fischer HP, Baisch C, Justenhoven C, Ranft U, Bruning T. Factors modifying the association between hormone-replacement therapy and breast cancer risk. Eur J Epidemiol. 2005;20:699–711. doi: 10.1007/s10654-005-0032-0. [DOI] [PubMed] [Google Scholar]
- Rafii S, O'Regan P, Xinarianos G, Azmy I, Stephenson T, Reed M, Meuth M, Thacker J, Cox A. A potential role for the XRCC2 R188H polymorphic site in DNA-damage repair and breast cancer. Hum Mol Genet. 2002;11:1433–1438. doi: 10.1093/hmg/11.12.1433. [DOI] [PubMed] [Google Scholar]
- Schmidt MK, Tollenaar RA, de Kemp SR, Broeks A, Cornelisse CJ, Smit VT, Peterse JL, van Leeuwen FE, van't Veer LJ. Breast cancer survival and tumor characteristics in premenopausal women carrying the CHEK2*1100delC germline mutation. J Clin Oncol. 2007;25:64–69. doi: 10.1200/JCO.2006.06.3024. [DOI] [PubMed] [Google Scholar]
- Schrauder M, Frank S, Strissel PL, Lux MP, Bani MR, Rauh C, Sieber CC, Heusinger K, Hartmann A, Schulz-Wendtland R, Strick R, Beckmann MW, Fasching PA. Single nucleotide polymorphism D1853N of the ATM gene may alter the risk for breast cancer. J Cancer Res Clin Oncol. 2008;134:873–882. doi: 10.1007/s00432-008-0355-9. [DOI] [PubMed] [Google Scholar]
- Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, Simon M, Marie Y, Boisselier B, Delattre JY, Hoang-Xuan K, El HS, Idbaih A, Zelenika D, Andersson U, Henriksson R, Bergenheim AT, Feychting M, Lonn S, Ahlbom A, Schramm J, Linnebank M, Hemminki K, Kumar R, Hepworth SJ, Price A, Armstrong G, Liu Y, Gu X, Yu R, Lau C, Schoemaker M, Muir K, Swerdlow A, Lathrop M, Bondy M, Houlston RS. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler G, Radford-Weiss I, Ben-Abdelali R, Mahlaoui N, Ponceau JF, Macintyre EA, Vekemans M, Bernard OA, Romana SP. Fusion of ZMIZ1 to ABL1 in a B-cell acute lymphoblastic leukaemia with a t(9;10)(q34;q22.3) translocation. Leukemia. 2008;22:1278–1280. doi: 10.1038/sj.leu.2405033. [DOI] [PubMed] [Google Scholar]
- Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, Masson G, Jakobsdottir M, Thorlacius S, Helgason A, Aben KK, Strobbe LJ, bers-Akkers MT, Swinkels DW, Henderson BE, Kolonel LN, Le ML, Millastre E, Andres R, Godino J, Garcia-Prats MD, Polo E, Tres A, Mouy M, Saemundsdottir J, Backman VM, Gudmundsson L, Kristjansson K, Bergthorsson JT, Kostic J, Frigge ML, Geller F, Gudbjartsson D, Sigurdsson H, Jonsdottir T, Hrafnkelsson J, Johannsson J, Sveinsson T, Myrdal G, Grimsson HN, Jonsson T, von HS, Werelius B, Margolin S, Lindblom A, Mayordomo JI, Haiman CA, Kiemeney LA, Johannsson OT, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, Jakobsdottir M, Bergthorsson JT, Gudmundsson J, Aben KK, Strobbe LJ, Swinkels DW, van Engelenburg KC, Henderson BE, Kolonel LN, Le ML, Millastre E, Andres R, Saez B, Lambea J, Godino J, Polo E, Tres A, Picelli S, Rantala J, Margolin S, Jonsson T, Sigurdsson H, Jonsdottir T, Hrafnkelsson J, Johannsson J, Sveinsson T, Myrdal G, Grimsson HN, Sveinsdottir SG, Alexiusdottir K, Saemundsdottir J, Sigurdsson A, Kostic J, Gudmundsson L, Kristjansson K, Masson G, Fackenthal JD, Adebamowo C, Ogundiran T, Olopade OI, Haiman CA, Lindblom A, Mayordomo JI, Kiemeney LA, Gulcher JR, Rafnar T, Thorsteinsdottir U, Johannsson OT, Kong A, Stefansson K. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- Syrjakoski K, Vahteristo P, Eerola H, Tamminen A, Kivinummi K, Sarantaus L, Holli K, Blomqvist C, Kallioniemi OP, Kainu T, Nevanlinna H. Population-based study of BRCA1 and BRCA2 mutations in 1035 unselected Finnish breast cancer patients. J Natl Cancer Inst. 2000;92:1529–1531. doi: 10.1093/jnci/92.18.1529. [DOI] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, Hankinson SE, Hutchinson A, Wang Z, Yu K, Chatterjee N, Garcia-Closas M, Gonzalez-Bosquet J, Prokunina-Olsson L, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Diver R, Prentice R, Jackson R, Kooperberg C, Chlebowski R, Lissowska J, Peplonska B, Brinton LA, Sigurdson A, Doody M, Bhatti P, Alexander BH, Buring J, Lee IM, Vatten LJ, Hveem K, Kumle M, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover RN, Chanock SJ, Hunter DJ. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, Seal S, Ghoussaini M, Hines S, Healey CS, Hughes D, Warren-Perry M, Tapper W, Eccles D, Evans DG, Hooning M, Schutte M, van den OA, Houlston R, Ross G, Langford C, Pharoah PD, Stratton MR, Dunning AM, Rahman N, Easton DF. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve S, Fevotte J, Anger A, Truong T, Lamkarkach F, Gaye O, Kerbrat P, Arveux P, Miglianico L, Imbernon E, Guenel P. Breast cancer risk by occupation and industry: analysis of the CECILE study, a population-based case-control study in France. Am J Ind Med. 2011;54:499–509. doi: 10.1002/ajim.20952. [DOI] [PubMed] [Google Scholar]
- Wedren S, Lovmar L, Humphreys K, Magnusson C, Melhus H, Syvanen AC, Kindmark A, Landegren U, Fermer ML, Stiger F, Persson I, Baron J, Weiderpass E. Oestrogen receptor alpha gene haplotype and postmenopausal breast cancer risk: a case control study. Breast Cancer Res. 2004;6:R437–R449. doi: 10.1186/bcr811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischer M, Bojesen SE, Tybjaerg-Hansen A, Axelsson CK, Nordestgaard BG. Increased risk of breast cancer associated with CHEK2*1100delC. J Clin Oncol. 2007;25:57–63. doi: 10.1200/JCO.2005.05.5160. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Apostolidou S, Raum E, Rothenbacher D, Fiegl H, Menon U, Stegmaier C, Jacobs IJ, Brenner H. Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PLoS One. 2008;3:e2656. doi: 10.1371/journal.pone.0002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, Kosel ML, LaChance DH, McCoy L, O'Neill BP, Patoka J, Pico AR, Prados M, Quesenberry C, Rice T, Rynearson AL, Smirnov I, Tihan T, Wiemels J, Yang P, Wiencke JK. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, Gaudet M, Schmidt MK, Broeks A, Cox A, Fasching PA, Hein R, Spurdle AB, Blows F, Driver K, Flesch-Janys D, Heinz J, Sinn P, Vrieling A, Heikkinen T, Aittomaki K, Heikkila P, Blomqvist C, Lissowska J, Peplonska B, Chanock S, Figueroa J, Brinton L, Hall P, Czene K, Humphreys K, Darabi H, Liu J, van ' V, van Leeuwen FE, Andrulis IL, Glendon G, Knight JA, Mulligan AM, O'Malley FP, Weerasooriya N, John EM, Beckmann MW, Hartmann A, Weihbrecht SB, Wachter DL, Jud SM, Loehberg CR, Baglietto L, English DR, Giles GG, McLean CA, Severi G, Lambrechts D, Vandorpe T, Weltens C, Paridaens R, Smeets A, Neven P, Wildiers H, Wang X, Olson JE, Cafourek V, Fredericksen Z, Kosel M, Vachon C, Cramp HE, Connley D, Cross SS, Balasubramanian SP, Reed MW, Dork T, Bremer M, Meyer A, Karstens JH, Ay A, Park-Simon TW, Hillemanns P, rias Perez JI, Menendez RP, Zamora P, Benitez J, Ko YD, Fischer HP, Hamann U, Pesch B, Bruning T, Justenhoven C, Brauch H, Eccles DM, Tapper WJ, Gerty SM, Sawyer EJ, Tomlinson IP, Jones A, Kerin M, Miller N, McInerney N, Anton-Culver H, Ziogas A, Shen CY, Hsiung CN, Wu PE, Yang SL, Yu JC, Chen ST, Hsu GC, Haiman CA, Henderson BE, Le ML, Kolonel LN, Lindblom A, Margolin S, Jakubowska A, Lubinski J, Huzarski T, Byrski T, Gorski B, Gronwald J, Hooning MJ, Hollestelle A, van den Ouweland AM, Jager A, Kriege M, Tilanus-Linthorst MM, Collee M, Wang-Gohrke S, Pylkas K, Jukkola-Vuorinen A, Mononen K, Grip M, Hirvikoski P, Winqvist R, Mannermaa A, Kosma VM, Kauppinen J, Kataja V, Auvinen P, Soini Y, Sironen R, Bojesen SE, Orsted DD, Kaur-Knudsen D, Flyger H, Nordestgaard BG, Holland H, Chenevix-Trench G, Manoukian S, Barile M, Radice P, Hankinson SE, Hunter DJ, Tamimi R, Sangrajrang S, Brennan P, McKay J, Odefrey F, Gaborieau V, Devilee P, Huijts PE, Tollenaar RA, Seynaeve C, Dite GS, Apicella C, Hopper JL, Hammet F, Tsimiklis H, Smith LD, Southey MC, Humphreys MK, Easton D, Pharoah P, Sherman ME, Garcia-Closas M. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103:250–263. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, McCarthy MI, Hattersley AT. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziogas A, Gildea M, Cohen P, Bringman D, Taylor TH, Seminara D, Barker D, Casey G, Haile R, Liao SY, Thomas D, Noble B, Kurosaki T, Anton-Culver H. Cancer risk estimates for family members of a population-based family registry for breast and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:103–111. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.