Abstract
Background
The discovery of distinct subsets of non-small cell lung cancer (NSCLC) characterized by activation of driver oncogenes has greatly impacted personalized therapy. We hypothesized that the dominant oncogene in NSCLC would be associated with distinct patterns of metastatic spread in NSCLC at the time of diagnosis.
Methods
209 consecutive patients with stage IV non-squamous NSCLC with an EGFR mutation (N=39), KRAS mutation (N=49), ALK gene rearrangement (N=41), or wild-type for all three (triple negative, N=80) were included. The percentage of patients with metastatic disease at a given site was compared between each molecular cohort (EGFR, KRAS, or ALK) and the triple negative cohort.
Results
ALK gene rearrangement was significantly associated with pericardial disease (OR=4.61, 95% CI 1.30, 16.37, p=0.02) and pleural disease (OR=4.80, 95% CI 2.10, 10.97, p<0.001). Patients with ALK gene rearrangements (OR=5.50, 95% CI 1.76, 17.18, p= 0.003) and patients with EGFR mutations (OR=5.17, 95% CI 1.63, 16.43, p= 0.006) were predisposed to liver metastasis compared to the triple negative cohort. No molecular cohort had a predisposition to pulmonary nodules, adrenal, bone, or brain metastasis compared to the triple negative cohort. The mean number of metastatic disease sites in patients within the ALK rearranged cohort was significantly greater than the triple negative cohort (mean = 3.6 sites vs. 2.5 sites, p<0.0001).
Conclusion
The results support the hypothesis that the dominant molecular oncogenes in NSCLC are associated with different biological behaviors manifesting as distinct patterns of metastatic spread at the time of diagnosis.
Keywords: metastasis, Non-Small Cell Lung Cancer, EGFR, ALK tyrosine kinase receptor, KRAS
Introduction
For a long time NSCLC was treated as a single entity without regard to histology or molecular status. Over the last decade we have recognized that histology can predict both efficacy and safety of drugs used for the treatment of NSCLC.1, 2 Molecular analysis has provided an even more detailed classification of NSCLC. Activating mutations in the EGFR gene are both prognostic and predictive in that they are associated with improved survival irrespective of therapy and are associated with a significant response to EGFR tyrosine kinase inhibitors (TKIs) such as gefitinib or erlotinib.3, 4 Patients with EGFR mutations also show a significant improvement in progression free survival (PFS) compared to standard chemotherapy.5 More recently, fusions involving the anaplastic lymphoma kinase (ALK) gene were discovered in NSCLC.6 Patients with ALK gene rearrangements detected by fluorescence in situ hybridization (FISH) demonstrate significant objective response rates and PFS times to the oral TKI, crizotinib.7 The prognostic significance of ALK is somewhat unclear as studies in untreated, unselected populations are not yet available, although it was recently reported that ALK did not portend a favorable prognosis in NSCLC.8 Despite being the earliest recognized and the most frequently activated oncogene in lung cancer, KRAS mutations do not currently predict for benefit from any targeted or chemotherapeutic drugs and are associated with a worse survival.9–11
Sub-classification of patients with NSCLC using molecular diagnostics has permitted us to re-examine the characteristics and outcomes of patients with NSCLC. Indeed, evaluation of PFS for patients treated with pemetrexed, showed a significant benefit for ALK+ patients compared to patients without ALK gene rearrangement, EGFR mutation, or KRAS mutation (triple negative cohort).12 We initially made a clinical observation that a number of ALK+ patients had metastatic disease to the pericardium. We hypothesized that the biology of the tumor would, at least in part, regulate patterns of metastatic spread. We therefore formally analyzed patterns of metastatic spread by comparing different molecular cohorts based on current dominant oncogenes recognized in NSCLC. Here we provide the first study to examine variations in the patterns of metastatic spread in NSCLC by oncogenic driver status.
Methods
Patient Selection
The University of Colorado Thoracic Oncology Program began to screen tumor biopsies from NSCLC patients for a series of different molecular drivers starting in 2008. In addition to routine EGFR and KRAS mutational testing ALK gene rearrangements were also assessed to identify patients for entry into defined molecular cohorts treated within the Phase I study of crizotinib (PF-02341066).5,8,11 Our initial ALK screening strategies intentionally enriched for those likely to be ALK positive, including not testing those who were previously proven EGFR or KRAS mutant, leading to a higher than expected prevalence of ALK+ patients at University of Colorado.13 However, in a desire to capture all ALK positive cases, we later adopted a policy of screening all NSCLC cases with available tissue. 14
An IRB-approved protocol permits clinical correlates to be made on all patients in whom molecular analyses have been conducted within the Colorado Molecular Correlates Lab (CMOCO). All NSCLC patients with stage IV cancer (TNM 7th edition) tested within CMOCO from June 2008 to May 2011 were eligible for assessment if there was either full triple testing results available (EGFR and KRAS mutation status and ALK gene FISH) or at least one positive result (ALK FISH+, KRAS mutation, or EGFR mutation) was found. Patients with more than one positive result in ALK, EGFR and KRAS were excluded. Data were collected by retrospective chart review and review of imaging for each patient, capturing NSCLC histology, age at metastatic diagnosis, sex, and smoking status and presence of metastatic disease in the predefined sites of brain, bone, adrenal, liver, pleura, pericardium, and intra- and extra-thoracic lymph nodes at the time of diagnosis or metastatic recurrence. Other sites of metastatic disease were noted but not formally analyzed due to the predicted low prevalence. Patients with squamous cell histology were excluded out of concern that histology may be significantly associated with sites of metastatic disease and the fact that the prevalence of the predominant gene abnormalities currently tested for would be significantly lower in this population thereby skewing the triple negative cohort. Patients were classified as either never smokers if they smoked less than 100 cigarettes, light smokers if they smoked ≤10 pack years, former smokers if they smoked > 10 pack years and quit more than one year prior to diagnosis, and current smokers if they smoked >10 pack years and were still smoking with one year of diagnosis. Patients were excluded if clinical information was incomplete.
Oncogene Testing
Mutational analyses were conducted as previously described in the Colorado Molecular Correlates laboratory (CMOCO).12 Briefly, EGFR exons 18–21 and KRAS exon 2 were amplified and sequenced using an ABI model 3730 capillary gel sequencer. Mutations were identified by visual inspection of the resulting chromatograms aligned using Mutation Surveyor, v3.24 (or higher). In some cases, mutation analysis in EGFR and KRAS was confirmed by SNaPshot analysis as previously described.15 The occurrence of an ALK gene rearrangement was assessed by FISH as previously described in the cytogenetics laboratory within CMOCO.13 Patients were deemed ALK FISH positive if >15% of tumor cells showed split red and green signals and/or single red (residual 3’) signals, otherwise the specimen was classified as ALK FISH negative.
Statistics
Descriptive analyses were performed on four molecularly defined groups: ALK FISH positive, EGFR mutant, KRAS mutant and triple negative and associated clinical data. Within each site of metastasis, Fisher's Exact Test was used to compare the proportion of patients falling in one molecular cohort to that for the triple negative cohort; and thus 27 comparisons were analyzed. One-way ANOVA was used to analyze the differences on number of metastatic sites among the four different molecular cohorts. Statistical analyses were performed by the University of Colorado Biostatistics and Bioinformatics Core using SAS/BASE and SAS/STAT software, Version 9.2 of the SAS System for Windows (SAS Institute Inc., Cary, NC, USA).
Results
209 consecutive patients with stage IV non-squamous NSCLC were evaluated in this study. One patient with an EGFR mutation and one patient with a KRAS mutation were excluded due to lack of records documenting sites of disease at the time of diagnosis of metastatic disease. One patient was excluded because of the presence of both an EGFR (HV773_774LM) and a KRAS mutation (G12V). One patient was excluded because of the presence of an ALK gene rearrangement and an EGFR mutation (S768I).
An ALK gene rearrangement was identified in 20% of patients, an EGFR mutation in 19% of patients, a KRAS mutation in 23% of patients, and 38% of patients had no abnormality in the three genes (Table 1). The vast majority of patients in this study displayed adenocarcinoma histology with only a few patients demonstrating large cell with or without neuroendocrine features or NSCLC not otherwise specified (NOS). The majority of patients analyzed here were categorized as stage IV at the time of diagnosis, but approximately 20% of patients had recurrent cancer.
Table 1.
Characteristics of patients
| Molecular Cohort | |||||
|---|---|---|---|---|---|
| Characteristic | ALK+ | EGFR+ | KRAS+ | Triple Negative |
Total |
| Total | 41 (20) | 39 (19) | 49 (23) | 80 (38) | 209 |
| Histology | |||||
| Adenocarcinoma | 38 (93) | 37 (95) | 47 (96) | 78 (98) | 200 (96) |
| Large cell | 1 (2) | 0 | 2 (4) | 0 | 5 (2) |
| NOS | 2 (5) | 2 (5) | 0 | 2 (3) | 6 (3) |
| Staging | |||||
| Stage IV at diagnosis | 35 (85) | 28 (72) | 39 (80) | 63 (79) | 165 (79) |
| Recurrent disease | 6 (15) | 11 (28) | 10 (20) | 17 (21) | 44 (21) |
| Smoking history | |||||
| Never (<100 cigarettes) | 31 (76) | 22 (56) | 6* (12) | 25 (31) | 84 (40) |
| Light (≤10 py) | 5 (12) | 5 (13) | 1 (2) | 9 (11) | 20 (10) |
| Current/former | 5 (12) | 12 (31) | 42 (86) | 46 (58) | 105 (50) |
| Mean Pack Years | 22.6 | 43.9 | 46.2 | 39.8 | 42 |
| Sex | |||||
| Male | 21 (51) | 10 (26) | 14 (29) | 38 (48) | 83 (40) |
| Female | 20 (49) | 29 (74) | 35 (71) | 42 (52) | 126 (60) |
| Median age, years (range) † | 51 (21–78) | 62 (45–78) | 59.5 (32–82) | 62 (41–82) | 59 (21–82) |
includes one pipe smoker,
age at diagnosis of metastatic disease
Baseline characteristics of evaluable patients are show in Table 1. The proportion of heavy smokers significantly differed across the molecular cohorts, with the triple negative and KRAS mutation groups showing the highest proportions and the EGFR and ALK+ groups showing the lowest (p<0.0001). Patients with EGFR and KRAS gene mutations were more often female than the triple negative cohort, whereas patients with ALK gene rearrangements exhibited a similar sex distribution; overall the distribution of molecular cohorts differed significantly between males and females (p = 0.03). EGFR and KRAS mutation positive patients were diagnosed with metastatic disease at a similar age as the triple negative cohort and ALK positive patients (p<0.0001) tended to be younger consistent with previous reports.16
The majority of patients underwent testing for all three molecular markers (80%). A smaller number underwent testing for only two of the three biomarkers (18%) (Table 2). Only three patients, all with an ALK gene rearrangement, had only one test performed. Biopsy material from the primary tumor was most often used for molecular testing, followed by metastatic sites then lymph nodes (Table 3). Only 11 patients had more than one site biopsied. Documentation of imaging modalities across the molecular cohorts did not differ significantly (Table 4). PET/CT and brain MR, for example, were performed and documented in the majority of cases across all molecular cohorts.
Table 2.
Molecular testing performed on patients
| Molecular Cohort | |||||
|---|---|---|---|---|---|
| Test(s) Performed | ALK+ | EGFR+ | KRAS+ | Triple Negative |
Total Number |
| EGFR, KRAS, ALK | 32 (78) | 29 (74) | 27 (55) | 80 (100) | 168 (80) |
| EGFR and KRAS only | 0 | 10 (26) | 22 (45) | 0 | 32 (15) |
| EGFR and ALK only | 6 (15) | 0 | 0 | 0 | 6 (3) |
| ALK only | 3 (7) | 0 | 0 | 0 | 3 (1) |
| Total | 41 | 39 | 49 | 80 | 209 |
Table 3.
Tumor sites used for molecular testing
| Molecular Cohort | |||||
|---|---|---|---|---|---|
| Test(s) Performed | ALK+ | EGFR+ | KRAS+ | Triple Negative |
Total Number |
| Primary | 18 | 17 | 24 | 45 | 104 |
| Lymph Node | 10 | 4 | 3 | 11 | 28 |
| Metastatic site | 10 | 13 | 20 | 22 | 65 |
| Primary + other site | 0 | 1 | 1 | 1 | 3 |
| Primary with multiple biopsies | 0 | 1 | 0 | 0 | 1 |
| Multiple metastatic site biopsied | 3 | 3 | 1 | 1 | 8 |
| Total | 41 | 39 | 49 | 80 | 209 |
Table 4.
Imaging data collection patients
| Molecular Cohort | |||||
|---|---|---|---|---|---|
| Characteristic | ALK+ | EGFR+ | KRAS+ | Triple Negative |
Total Number |
| Total | 41 | 39 | 49 | 80 | 209 |
| Brain Imaging | |||||
| MRI | 31 (76) | 25 (64) | 35 (71) | 48 (60) | 139 (67) |
| CT | 2 (5) | 2 (5) | 6 (12) | 5 (6) | 15 (7) |
| No imaging | 8 (20) | 12 (31) | 8 (16) | 27 (34) | 55 (27) |
| Body Imaging | |||||
| PET/CT | 30 (73) | 35 (90) | 43 (88) | 72 (90) | 180 (86) |
| Bone Scan | 3 (7) | 5 (13) | 5 (10) | 5 (6) | 18 (8) |
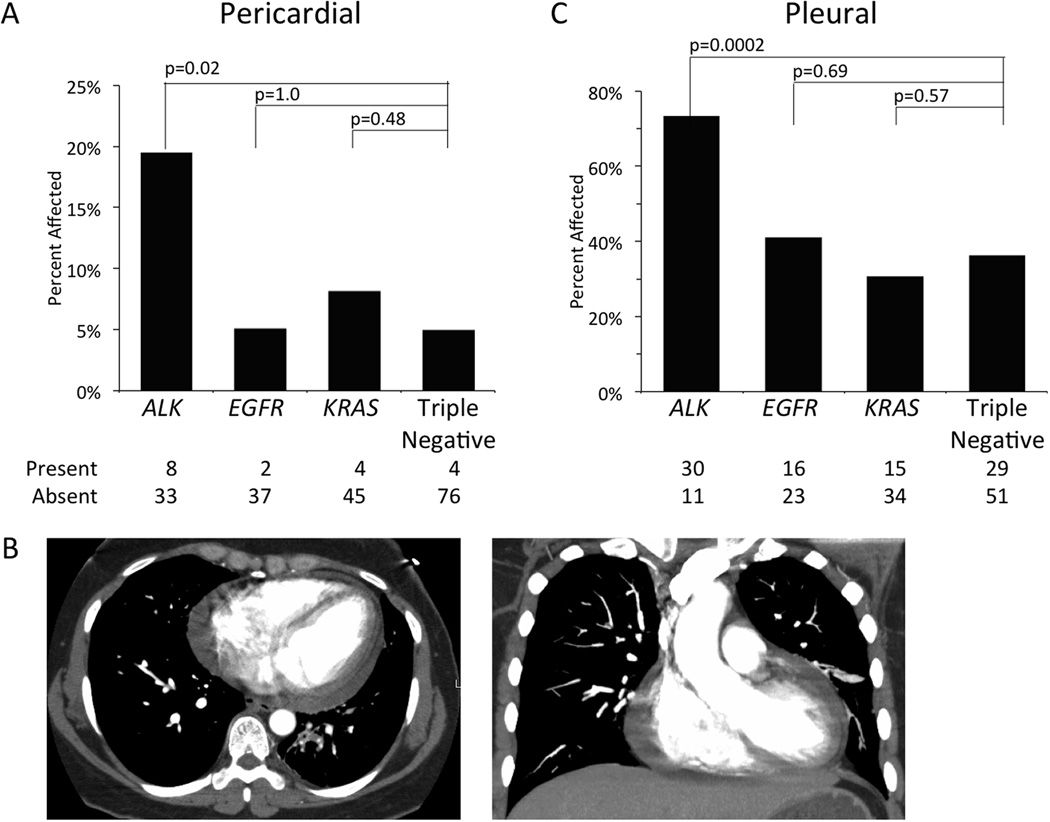
Only 18 of 209 patients (9%) exhibited evidence of pericardial spread at the time of diagnosis, but 8 of 41 (20%) patients in the ALK+ cohort had pericardial spread (Figure 1A). Patients with an ALK gene rearrangement were significantly more likely to have metastatic spread to the pericardium than the triple negative cohort (OR=4.61, 95% CI 1.30, 16.37, p=0.02). EGFR (OR=1.03, 95% CI 0.18, 5.87, p=1.0) and KRAS (OR=1.69 95% CI 0.40, 7.09, p=0.48) mutant patients were similar to the triple negative with respect to metastatic spread to the pericardium.
Figure 1. Metastases to Serous Membranes.
The percentage of patients with the presence of metastatic disease in the pericardium (A) or pleura (C) is shown by molecular cohort. Absolute numbers of patients with or without metastases in each cohort is provided below. Panel B depicts axial (left) or coronal (right) CT scan slices showing a pericardial effusion in a 42 year old female patient with an ALK gene rearrangement.
A large proportion of patients, 90 of 209 (43%), exhibited pleural disease (Figure 1C). ALK positive patients were also more likely to have spread to the pleura than patients in the triple negative cohort (OR=4.80, 95% CI 2.10, 10.97, p<0.001). There was no difference in the EGFR (OR=1.22, 95% CI 0.56, 2.68, p=0.69) and KRAS cohorts (OR=0.78, 95% CI 0.36, 1.66, p=0.57) with respect to pleural spread compared to the triple negative cohort.
As expected, the majority of patients, 144 of 209 (69%), displayed spread to intrathoracic lymph nodes, whereas only 37 of 209 (18%) had involvement of extrathoracic lymph nodes (Figure 2). Patients with ALK gene rearrangements demonstrated a numerically higher incidence of spread to intrathoracic lymph nodes (OR=2.34, 95% CI 0.92–5.98, p=0.09) and extrathoracic lymph nodes (OR=2.25, 95% CI 0.96, 5.29, p=0.07) compared to the triple negative cohort, however this did not reach statistical significance.
Figure 2. Metastases to Lymph Nodes.
The percentage of patients with the presence of metastatic disease in the intrathoracic (A) or extrathoracic (B) lymph nodes is shown by molecular cohort. Absolute numbers of patients with or without metastases in each cohort is provided below.
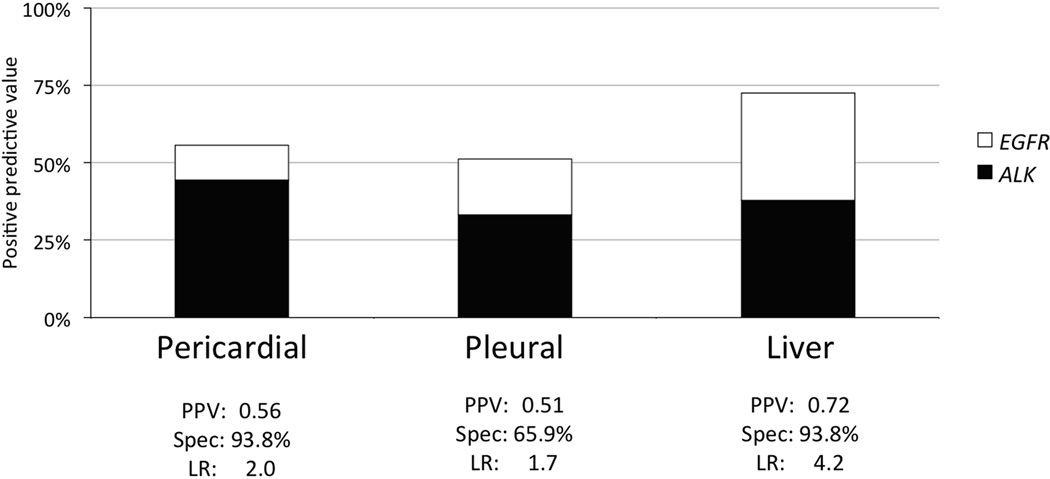
Patients with ALK gene rearrangements (OR=5.50, 95% CI 1.76, 17.18, p= 0.003) and patients with EGFR mutations (OR=5.17, 95% CI 1.63, 16.43, p= 0.006) were predisposed to liver metastasis compared to the triple negative cohort (Figure 3). Out of the 29 patients in this study with liver metastasis, 21 (72%) patients were ALK or EGFR+. Given the high proportion of patients with “actionable” genetic changes (i.e. targeted by a highly active drug) found to have liver metastases, we determined the likelihood ratios for an EGFR mutation or ALK gene rearrangement in the presence of liver, pleural, or pericardial metastases and the predictive values of these sites of disease (Figure 4).
Figure 3. Metastases to the Liver.
The percentage of patients with the presence of metastatic disease in the liver is shown by molecular cohort. Absolute numbers of patients with or without metastases in each cohort is provided below.
Figure 4.
Positive predictive value (PPV) and the contribution of EGFR (open) and ALK (filled) are shown in the bar graph. Also displayed are the specificity and likelihood ratio (LR) for the presence of an actionable gene alteration in EGFR or ALK given known metastases to the pericardium, pleura or liver.
No molecular cohort demonstrated a significant predilection to pulmonary nodules, adrenal, bone, or brain metastases compared to the triple negative cohort (Figure 5). Brain imaging was not documented in 27% of patients, although the lack of imaging in each cohort was similar (Table 4). To determine whether this factor influenced our findings, we re-analyzed the data using only patients with brain imaging and found similar results (data not shown). KRAS mutation was not associated with increased risk of any metastatic site compared to the triple negative cohort.
Figure 5. Metastases to other Distant Sites.
The percentage of patients with the presence of metastatic disease in the bone (A), pulmonary nodules (B), adrenal gland(s) (C), and brain (D) is shown by molecular cohort. Absolute numbers of patients with or without metastases in each cohort is provided below.
The majority of the patients in this study had metastatic disease at diagnosis, however 21% of patients included in this analysis had metastatic disease at recurrence (i.e., patients initially with a stage I–III NSCLC that later recurred with sites of metastatic disease) (Table 1). To determine whether the inclusion of “recurrent” metastatic patients could have influenced these results presented here, we re-analyzed the data with only patients who had metastatic disease at first diagnosis. A similar trend for increased prevalence of pleural and pericardial effusion in ALK+ patients and for liver metastases in patients with ALK+ or EGFR mutant NSCLC was observed (Supplemental Figure 1). For all other sites of disease, the relative prevalence amongst the molecular cohorts at each organ site was similar (data not shown).
We also evaluated the number of metastatic disease sites for each cohort, limiting our analysis to the disease sites described above (Figure 6). The EGFR (mean=2.5 sites, p=0.79) and KRAS mutation cohorts (mean=2.4 sites, p=0.72) exhibited a similar number of metastatic sites to the triple negative cohort (mean=2.5 sites), whereas the ALK+ cohort exhibited more sites of metastatic disease than the triple negative cohort (mean=3.6 sites, p<0.0001).
Figure 6. Number of Metastatic Sites at Presentation.
The mean numbers of metastatic sites of disease, including intra- and extra-thoracic lymph node(s), for each molecular cohort is displayed. One-way ANOVA was used to compare each cohort to the triple negative cohort. Error bars represent standard error.
Less common sites of metastatic spread were also recorded for each patient in this study. No formal analyses were performed given the small number of patients displaying each of these rare sites. Interestingly 3 patients were found to have retinal metastases, 2 in the ALK+ cohort and 1 in the EGFR mutation cohort, whereas no patients in any other molecular cohort had this finding. Retinal metastases have been reported previously in lung cancer, but before molecular testing was routinely performed.17
Discussion
Here we report analysis of the association between molecular oncogene status and patterns of metastatic spread in treatment naÔve NSCLC. We observed a higher incidence of pericardial, pleural, and liver metastasis in ALK+ patients compared to patients without an EGFR, KRAS or ALK oncogene abnormality. Patients with an EGFR mutation also had a higher rate of liver metastases compare to the triple negative cohort.
A much higher than expected number of ALK+ patients were present in this study partly because of our role as a referral site for the phase I study of crizotinib and our initial screening strategy which enriched the detection of these patients. Undoubtedly, these elevated numbers have facilitated the identification of oncogene specific patterns of spread for ALK that might otherwise have been missed given its relative rarity as a molecular subtype of NSCLC. An expected percentage of EGFR and KRAS mutant patients were identified in our study.3, 18
We recognize that the triple negative cohort is a heterogeneous cohort and a number of patients evaluated in this study underwent evaluation for mutations in other molecular oncogenes such as BRAF, MET, and HER2. Currently, KRAS, EGFR, and ALK are the most established and commonly tested oncogenes in NSCLC and our testing patterns dictated the categorization used in this study. As we collect more data on other oncogenes in NSCLC we expect to refine the model described here. The vast majority of patients underwent triple testing for all three molecular markers analyzed in this study, however 20% of patients had only one or two tests performed. As necessitated by the entry criteria for this study, all of the patients with incomplete testing for all three biomarkers demonstrated a positive result for one of the biomarkers. We believe that this criterion is justified given the low likelihood of a patient harboring more than one positive biomarker result within this subset of analytes.19, 20 Indeed, data from this study show that of the 211 patients with non-squamous NSCLC that underwent double or triple testing, only two patients (~1%) were found to have more than one positive biomarker result, and both were associated with less common forms of mutation in EGFR.
We chose to collect data only at the time of diagnosis as a longitudinal study might be skewed by survival time. We included patients with recurrent metastatic disease in this study, however analysis of only patients with metastatic disease at first diagnosis yielded remarkably similar results, which is consistent with our hypothesis that biology is a critical factor in determining patterns of metastatic spread. Furthermore, treatment with either chemotherapy or targeted therapies may induce patterns of spread that do not reflect the natural history of the disease. As an example, patients treated over a significant time period may develop more brain metastases because of poor CNS penetration by many of the drugs used to treat NSCLC.21, 22
We used clinical staging (TNM 7th edition) to classify sites of metastatic disease based on imaging studies, as it would not be feasible to perform pathologic confirmation for each metastatic sites documented. The rates of site-specific metastases are difficult to compare with previously published data given the recent shift of pleural and pericardial disease from a T4 classification in the 6th edition TNM to an M1a classification based on the 7th edition TNM.23 A higher incidence of liver metastases in ALK+ patients was recently reported.24, 25 While this is the first study to examine patterns of spread by different molecular oncogenes, a pattern of miliary spread of pulmonary metastases has previously been reported in association with EGFR in-frame deletions of exon 19 consistent with oncogene status driving the specific clinical presentation of the disease.26
In addition to the organ-specific patterns observed here, patients with ALK gene rearrangements were found to have more involved sites of metastasis at the time of diagnosis compared to patients in the triple negative cohort. On the surface, this finding might be considered an indicator of poor prognosis. Currently, the prognosis of ALK positive patients compared to other molecular cohorts has not been determined definitively.8, 19, 27 Analysis of prognosis in this group is hindered by the fact that most patients who have been identified as ALK positive have been treated with crizotinib.8 Alternatively, this finding could be explained by a lag in time to diagnosis in this population of patients who are mostly younger and tend to be never or light smokers. Finally, it is possible that a slower proliferating cancer with similar metastatic potential might take longer to present with symptoms and explain this result.
The hypothesis presented here is that biology drives metastasis, but one limitation of this study is that patients were categorized patients based on a limited set of oncogene abnormalities. Additionally, there is clearly significant genetic heterogeneity within all of the molecular cohorts, such as specific type of EGFR mutations (e.g., L858R vs. exon 19 deletions) or KRAS mutations (e.g., G12C vs. G12D). However, the number of patients in the study did not allow characterization based on the specific type of EGFR or KRAS mutations. The fact that KRAS behaved similarly to the triple negative cohort with respect to sites of metastatic spread could reflect greater biological heterogeneity in the KRAS cohort compared to the EGFR and ALK cohorts. Alternatively, these results may reflect the role of KRAS in tumor initiation rather than always being a dominant oncogene to which tumors are “addicted.”28 EGFR and ALK+ tumors may be more uniform based on their stricter dependence on these dominant signaling pathways.6, 29
The clinical results reported here are supported by preclinical models showing that specific genetic pathways mediate the sites of cancer metastases.30–32 Gene expression signatures derived from cell lines that repeatedly metastasize to a given organ site suggest that this process is programmed rather than just a stochastic result.33 Animal studies using tail vein injections of NSCLC cell lines with different oncogene drivers could be used to further study the genetic programs underlying site-specific organ metastatic patterns in lung cancer. The number of patients in each molecular cohort is relatively small and none of the significant associations identified are absolute, thus this data should not be used as the only criteria for deciding which patients should undergo molecular testing. While we would advocate testing all NSCLC patients for relevant molecular markers, the clear association of distinct patterns of metastatic spread with certain highly targetable oncogenes, could provide additional clinical prompts for molecular testing among those oncologists not conducting widespread screening in their lung cancer patients at the present time.
Supplementary Material
Supplemental Figure 1. Patients with metastatic disease at first diagnosis (i.e., excluding patients with recurrent disease) were evaluated for the presence or absence of disease at selected metastatic sites. The percentages of patients with the presence of metastatic disease in the pericardium (A), pleura (B), and liver (C) are shown by molecular cohort. Absolute numbers of patients with or without metastases in each cohort is provided below each bar graph.
Acknowledgements
This work was supported by the University of Colorado Lung SPORE grant with a Career Development Award to RCD, support to MVG, and support to the Lung SPORE Biostatistics, Informatics and Bioinformatics Core (P50CA058187). The University of Colorado Cytogenetics Core also provided technical support for this work (P30CA046934). Dr. Peter Sachs in Department of Radiology assisted with radiology images.
Footnotes
Financial Disclosures: None.
References
- 1.Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(21):3543–3551. doi: 10.1200/JCO.2007.15.0375. [DOI] [PubMed] [Google Scholar]
- 2.Sandler A, Gray R, Perry MC, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. The New England journal of medicine. 2006;355(24):2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 3.Douillard JY, Shepherd FA, Hirsh V, et al. Molecular predictors of outcome with gefitinib and docetaxel in previously treated non-small-cell lung cancer: data from the randomized phase III INTEREST trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(5):744–752. doi: 10.1200/JCO.2009.24.3030. [DOI] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England journal of medicine. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 6.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 7.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. The New England journal of medicine. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw A. Impact of crizotinib on survival in patients with advanced, ALK-positive NSCLC compared with historical controls. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(15S):4773. [Google Scholar]
- 9.Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(11):3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. British journal of cancer. 2005;92(1):131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camidge DR, Kono SA, Lu X, et al. Anaplastic Lymphoma Kinase Gene Rearrangements in Non-small Cell Lung Cancer are Associated with Prolonged Progression-Free Survival on Pemetrexed. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2011;6(4):774–780. doi: 10.1097/JTO.0b013e31820cf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camidge DR, Kono SA, Flacco A, et al. Optimizing the Detection of Lung Cancer Patients Harboring Anaplastic Lymphoma Kinase (ALK) Gene Rearrangements Potentially Suitable for ALK Inhibitor Treatment. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varella-Garcia M, Cho Y, Lu X, et al. ALK gene rearrangements in unselected caucasians with non-small cell lung carcinoma (NSCLC) J Clin Oncol. 2010;28(15S) Abstract #10533. [Google Scholar]
- 15.Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. The Journal of molecular diagnostics : JMD. 2011;13(1):74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2009;22(4):508–515. doi: 10.1038/modpathol.2009.2. [DOI] [PubMed] [Google Scholar]
- 17.Rossi A, Manto A, Maione P, Gridelli C. Synchronous bilateral retinal metastases from lung adenocarcinoma. Tumori. 2005;91(3):287–289. doi: 10.1177/030089160509100318. [DOI] [PubMed] [Google Scholar]
- 18.Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(18):5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varella-Garcia M, Cho Y, Lu X, et al. ALK gene rearrangements in unselected caucasians with non-small cell lung carcinoma (NSCLC) Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(15S):10533. [Google Scholar]
- 20.Camidge DR, Kono SA, Flacco A, et al. Optimizing the detection of lung cancer patients harboring anaplastic lymphoma kinase (ALK) gene rearrangements potentially suitable for ALK inhibitor treatment. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(22):5581–5590. doi: 10.1158/1078-0432.CCR-10-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai H, Chen Y, Elmquist WF. Distribution of the novel antifolate pemetrexed to the brain. The Journal of pharmacology and experimental therapeutics. 2005;315(1):222–229. doi: 10.1124/jpet.105.090043. [DOI] [PubMed] [Google Scholar]
- 22.Lee YJ, Choi HJ, Kim SK, et al. Frequent central nervous system failure after clinical benefit with epidermal growth factor receptor tyrosine kinase inhibitors in Korean patients with nonsmall-cell lung cancer. Cancer. 2010;116(5):1336–1343. doi: 10.1002/cncr.24877. [DOI] [PubMed] [Google Scholar]
- 23.Ou SH, Zell JA. Validation study of the proposed IASLC staging revisions of the T4 and M non-small cell lung cancer descriptors using data from 23,583 patients in the California Cancer Registry. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2008;3(3):216–227. doi: 10.1097/JTO.0b013e318164545d. [DOI] [PubMed] [Google Scholar]
- 24.Varella-Garcia M, Iafrate JA, Pao W, et al. ALK Fusion and MET Amplification as Molecular Biomarkers and Therapeutic Targets in Advanced Lung Adenocarcinomas in hte Lung Cancer Mutation Consortium. Journal of Thoracic Oncology. 2011;6(6):S291. [Google Scholar]
- 25.Yang P. Lung Cancer in Never-Smokers: prognostic Implications. Journal of Thoracic Oncology. 2011;6(6):S50. [Google Scholar]
- 26.Laack E, Simon R, Regier M, et al. Miliary never-smoking adenocarcinoma of the lung: strong association with epidermal growth factor receptor exon 19 deletion. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer. 2011;6(1):199–202. doi: 10.1097/JTO.0b013e3181fb7cf1. [DOI] [PubMed] [Google Scholar]
- 27.Kim D, Lee JK, Park HS, et al. Comparative analyses of overall survival of anaplastic lymphoma kinase (ALK)–positive advanced non-small cell lung cancer (NSCLC) patients who did not receive ALK inhibitors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(suppl) abstract 7515. [Google Scholar]
- 28.Singh A, Greninger P, Rhodes D, et al. A gene expression signature associated with "K-Ras addiction" reveals regulators of EMT and tumor cell survival. Cancer cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothenberg SM, Engelman JA, Le S, Riese DJ, 2nd, Haber DA, Settleman J. Modeling oncogene addiction using RNA interference. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(34):12480–12484. doi: 10.1073/pnas.0803217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer cell. 2003;3(6):537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 31.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang AC, Massague J. Molecular basis of metastasis. The New England journal of medicine. 2008;359(26):2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta GP, Nguyen DX, Chiang AC, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446(7137):765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Patients with metastatic disease at first diagnosis (i.e., excluding patients with recurrent disease) were evaluated for the presence or absence of disease at selected metastatic sites. The percentages of patients with the presence of metastatic disease in the pericardium (A), pleura (B), and liver (C) are shown by molecular cohort. Absolute numbers of patients with or without metastases in each cohort is provided below each bar graph.