Abstract
The mucosal immune network is a crucial barrier preventing pathogens from entering the body. The network of immune cells that mediates the defensive mechanisms in the mucosa is likely shaped by chemokines, which attract a wide range of immune cells to specific sites of the body. Chemokines have been divided into homeostatic or inflammatory depending upon their expression patterns. Additionally, several chemokines mediate direct killing of invading pathogens, as exemplified by CCL28, a mucosa-associated chemokine that exhibits antimicrobial activity against a range of pathogens. CXCL17 was the last chemokine ligand to be described, and the 17th member of the CXC chemokine family. Its expression pattern in 105 human tissues and cells indicates that CXCL17 is a homeostatic, mucosa-associated chemokine. Its strategic expression in mucosal tissues suggests that it is involved in innate immunity and/or sterility of the mucosa. To test the latter hypothesis, we tested CXCL17 for possible antibacterial activity against a panel of pathogenic and opportunistic bacteria. Our results indicate that CXCL17 has potent antimicrobial activities, and that its mechanism of antimicrobial action involves peptide-mediated bacterial membrane disruption. Since CXCL17 is strongly expressed in bronchi, we measured it in bronchoalveolar lavage fluids, and observed that it is strongly upregulated in idiopathic pulmonary fibrosis. We conclude that CXCL17 is an antimicrobial mucosal chemokine that may play a role in the pathogenesis of interstitial lung diseases.
Introduction
Humans interact with the outside world through external physical barriers, including the skin and mucosal sites. Exterior pathogens that gain access to these sites and successfully colonize them can cause local infections, which could give rise to systemic infections. Additionally, mucosal sites can harbor commensal bacteria that can, under certain conditions, become pathogenic. Therefore, the study of mucosal homeostasis is important to understand host defenses in these sites.
Chemokines are small chemotactic proteins that control the migration of leukocyte populations in the body (1). However, some chemokines exhibit other activities beyond the chemoattraction of immune cells. For example, the CXCL12/CXCR4 signaling axis is involved in brain development (2) and aids in wound healing following vascular injury (3).
Some chemokines are also found at several mucosal sites. Currently, the best known mucosal-associated chemokines are CCL25 and CCL28. CCL25 is known for its role in attracting CCR9+ α4β7+ T cells to the small intestine (4),(5). CCL28 is a chemokine expressed by epithelial cells whose expression is induced in the female mammary gland upon onset of lactation (6) where it plays a pivotal role in the recruitment of CCR10+ T and B cells to trigger IgA production (7). In addition CCL28 is known to be present in saliva and the vagina (7–8), and it also has been shown to have candidacidal activity (8).
Using a comprehensive human gene expression microarray database (9–11), which we call the Body Index of Gene Expression (BIGE), we undertook a systematic screen of chemokines in the human body by examining the expression profiles of all 48 known human chemokines (1). Since the BIGE database currently includes 105 normal human tissues or cell types, this analysis represents one of the most comprehensive chemokine expression analyses performed to date. This systematic screen yielded four chemokines whose expression is strongly associated with the mucosa: CCL28, CCL25 CXCL14 and CXCL17.
Two of these chemokines (CCL25 and CCL28) are known to be mucosal associated. CXCL14 was originally described to be expressed in breast and kidney (BRAK) (12) and subsequently in skin (13). It has also been linked to mucosal sites (14–15). However, little is known about CXCL17. It was the final chemokine ligand to be discovered (16), and therefore much of its biology remains uncharacterized although it is known to be expressed in trachea and stomach (16). Our screening results have expanded on these analyses and confirm that its expression is restricted to mucosal tissues. Therefore, we decided to explore the role of CXCL17 in the mucosa in more detail.
CCL28 (8, 17) has been shown to have broad spectrum antimicrobial activity against several microorganisms including Candida albicans (18). CXCL14 has also been shown to have antimicrobial activity (13, 19). Given the mucosal-associated expression of CXCL17, we hypothesized that it may also have antimicrobial activity. In the present study, we confirmed the expression of CXCL17 in various mucosal tissues. Our results indicate that CXCL17 has potent antimicrobial activity, and that this activity is mediated through a membrane disruptive mechanism.
Furthermore, our expression data indicates that CXCL17 is expressed in the lung airways. We therefore investigated a role for CXCL17 in idiopathic pulmonary fibrosis (IPF), a complex interstitial lung disease of unknown etiology. To this end, we measured CXCL17 in human bronchoaveolar lavage fluids (BALf) from patients with IPF. Our results indicate that CXCL17 is elevated in BALf from IPF patients, suggesting that CXCL17 may be involved in the pathogenesis of this disease.
Materials & Methods
Body Index of Gene Expression
The construction of the BIGE database has been described previously (9, 11). Briefly, tissues or cells corresponding to 105 different sites of the human body were obtained within 5 h post-mortem. RNA was prepared as described and used to prepare cDNA to be hybridized to U133 2.0 genearrays (Affymetrix, Santa Clara, CA). The resulting data were normalized and a probeset corresponding to CXCL17 (DMC) (226960_at) was used to determine the expression of this chemokine in the human body.
Real Time Quantitative-PCR Analysis
All Q-PCR data was generated using a Roche Lightcycler machine 480 using a Universal Probe Library (UPL) based system. Briefly, total RNA was extracted from each mouse tissue sample using TRIzol (Invitrogen Carlsbad, CA) followed by RNA purification and DNase digest using Qiagen’s RNEasy columns. Human RNA samples were purchased from Clontech (Mountain View, CA) and did not require any extra preparation. Equal concentrations of total RNA were used in a reverse transcription reaction to generate cDNA (Qiagen, Valencia, CA). 50ng of each cDNA was used per 40 cycle PCR run. Gene specific primers and corresponding UPL were used for each reaction to quantitatively detect the amount of CXCL17 and control genes transcripts in each tissue sample. The results were processed and analyzed using GraphPad Prism software (www.graphpad.com).
Immunohistochemistry
Selection of tissue sections: Tissue sections from formalin-fixed, paraffin embedded samples from well preserved areas of normal human tongue, colon mucosa and bronchial mucosa were selected from autopsy files and used to construct a tissue array. Two 5-mm tissue punches were obtained from the relevant tissue areas and subsequently included in a paraffin block. Each paraffin block contained four different samples from patients who died from natural causes.
Five micron tissue sections mounted on siliconized slides were deparaffinized in xylol and then rehydrated in ethanol solutions at progressively lower concentrations. Epitope retrieval was achieved by treatment in a pressure cooker for 15 minutes. The slides were then incubated with anti-CXCL17 antibody (R&D Systems, Minneapolis, MN) or isotype control. The primary antibody was incubated for 10 minutes using a polymer based visualization kit (DAKO, Carpinteria, CA) according to the manufacturer’s instructions; once the reaction was complete, it was revealed with diaminobenzidine and counterstained with hematoxylin. Any positivity in the tissue sections was evaluated by a pathologist and the sites and cells that stained positive were recorded.
Immunohistochemistry for CD68 and CD138: Epitope retrieval from tissue sections was achieved by heating at 95°C in citrate buffer for 3 minutes. The sections were stained with either anti-CD68 or anti-138 followed by polymer-based visualization kits (all from DAKO) according to manufacturer’s instructions. The sections were then developed with diaminobenzidine, counterstained with hematoxylin and examined under a light microscope.
Bacteriocidal Peptide Assays
A single bacterial colony was added to 5 ml Trypticase Soy Broth (TSB) and grown with shaking overnight at 37°C. 10 µl of each overnight culture growing in TSB was added to a fresh tube of TSB. The bacteria were shaken at 37°C until they reached mid log phase. The bacterial OD was measured at 600 nm to verify that the range was between 0.4–0.8. Mid-log bacteria were deposited by centrifugation at 10,000 × g for 3 min, washed three times with 10 mM PIPES (piperazine-1,4-bis(2-ethanesulfonic acid), pH 7.4, supplemented with 0.01 volume (1% v/v) trypticase soy broth (10 mM PIPES-TSB, pH 7.4). Bacteria, ~1–5 × 106 CFU/ml, were exposed to peptides at various concentrations in a total volume of 50 µl in a 96-well plate. The test samples are incubated at 37°C with shaking for 1 h, diluted 1:100 in 10 mM PIPES (pH 7.4), and plated on TSB agar plates using an Autoplate 4000 (Spiral Biotech Inc., Bethesda, MD, USA). After incubation overnight at 37°C, bacterial cell survival is determined by counting CFU. Each condition was performed in triplicate.
Fungicidal Activity against Candida albicans
A single colony of Candida albicans was inoculated into 50 ml Sabouraud dextrose broth (SAB) overnight in a 37°C shaker. 100 µl of overnight culture was inoculated into 5 ml fresh SAB for 4 h in a 37°C shaker. Exponentially-growing C. albicans were deposited by centrifugation at 10,000 × g for 3 min, washed three times with 10 mM PIPES supplemented with 0.01 volume (1% v/v) SAB (10mM PIPES-SAB, pH 7.4). In triplicate, ~ 1–5 × 106 CFU/ml C. albicans were exposed to peptides at various concentrations in a total volume of 50 µl in a 96-well plate. Samples were incubated at 37°C with shaking for 2 h, diluted 1:100 in 10 mM PIPES (pH 7.4), and plated on SAB agar plates using an Autoplate 4000. After overnight growth at 37°C, C. albicans cell survival was determined by counting CFU.
Membrane Permeabilization Assays
The ability of CXCL17 to permeabilize live E. coli cells was assayed by measuring hydrolysis of o-nitrophenyl-β-d-galactopyranoside (ONPG) by cytoplasmic β-galactosidase and colorimetric detection of the ONPG hydrolysis product, o-nitrophenol. In lactose permease-deficient and β-galactosidase-constitutive E. coli ML35 cells, peptide-induced membrane disruption allows the ONPG substrate to diffuse into the bacterial cell for hydrolysis by cytoplasmic β-galactosidase. Log-phase E. coli ML35 cells were washed and resuspended in 10 mM PIPES-TSB. In triplicate, bacteria (5 × 106 CFU/ml) were exposed to peptides in the presence of 2.5 mM ONPG for 2 h at 37°C, and A405nm readings were taken every 30 s. The kinetics of ONPG hydrolysis was measured by determining the A405nm using a Spectra-Max plate spectrophotometer (Molecular Devices, Sunnyvale, CA). The resulting data was analyzed and compiled using GraphPad Prism.
BALf Samples
BALf samples were obtained from 12 patients with IPF (11 males and 1 female; 64.8 ± 5.5 years) and 5 healthy individuals (all males; 47.2 ± 12 years). Diagnosis of IPF was performed according to the American Thoracic Society/European Respiratory Society consensus (20). None of the patients had been treated with corticosteroids or immunosuppressive drugs at the time of the study. BAL was performed through flexible fiberoptic bronchoscopy under local anesthesia as described (21). Briefly, 300 ml of normal saline was instilled in 50-ml aliquots, with an average recovery of 60%-70%. The recovered BAL fluid was centrifuged at 250 g for 10 min at 4 °C and the supernatants were concentrated 10X and kept at −70 °C until use. The study was approved by the Bioethics committee at the National Institute of Respiratory Diseases, and informed consent was obtained from all subjects.
CXCL17 ELISA
BALf samples from healthy and diseased human subjects were analyzed for CXCL17 by sandwich ELISA by coating 96-well plates (NUNC, Rochester, NY) with primary monoclonal anti-human CXCL17 antibody (R&D Systems). Recombinant human CXCL17 (R&D Systems) was used as a standard. Bound standards and samples were detected by subsequent incubation with polyclonal anti-human CXCL17 antibody (R&D Systems) and horseradish peroxidase-conjugated mouse anti-human detection antibody (Abcam, Cambridge, MA). The binding was visualized using TMB (KPL, Gaithersburg MD). The reaction was stopped with 2N H2SO4 and absorbance was read at 450nm.
Results
The expression of CXCL17 is restricted to mucosal sites
We examined the expression profiles of each of 48 human chemokines in the BIGE database and selected chemokines that were highly and specifically expressed in mucosal sites. From this screen we identified several chemokines distinctly expressed in mucosal sites: CCL28, CCL25, CXCL14 and CXCL17. CCL28 is strongly expressed in salivary and lactating mammary gland (8, 22); CCL25 in thymus and intestine, confirming previous data (23); and CXCL14 is expressed in kidney, skin, and several mucosal tissues (15). Gene expression analyses using the BIGE database indicated that the expression of CXCL17 is restricted to mucosal sites including the digestive system, lung airways, the urethra and several sites of the female reproductive system (Figure 1). Given that CXCL17 is a relatively new member to the class of mucosal chemokines, we focused on the functional characterization of this chemokine.
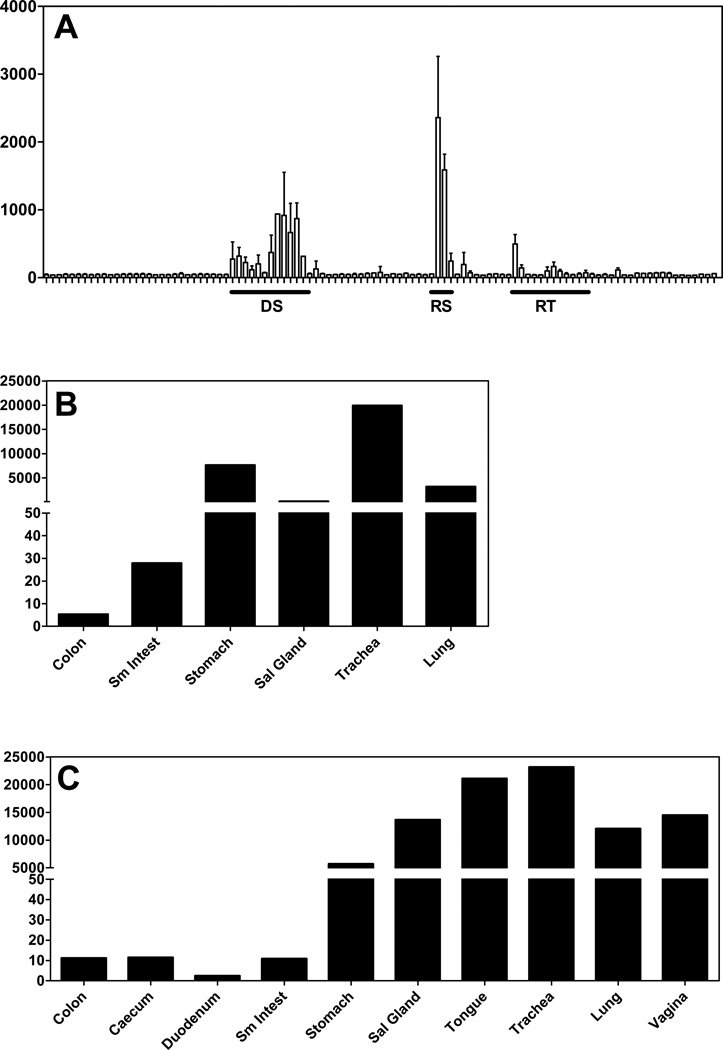
Figure 1. Microarray data indicates that CXCL17 is a mucosal chemokine.
(A) Mean expression values (y axis) from microarray data for 105 normal human tissues from the BIGE database (see Materials and Methods) (9, 11). Data are displayed across the x-axis grouped in organ systems. Y axis is the mean expression values. Highlighted organ systems, which have the highest expression of CXCL17, are: DS, digestive system; RS, respiratory system; RT, reproductive tract. The values for the tissues with the highest expression of CXCL17 are shown in Table 1. For a complete list of tissues included in the BIGE database consult (11). (B & C). Q-PCR in human and mouse mucosal tissue confirms that CXCL17 is highly expressed in mucosal tissues. Q-PCR with human (B) and mouse (C) CXCL17 specific primers was used to detect the expression of CXCL17 in each of the mucosal tissues (x axis). Expression Units of CXCL17 in mouse mucosal tissues (y axis) are based on Ct values averaged for multiple samples. Tissues were harvested from normal C57BL/6 mice. Expression for each tissue represents averaged expression for tissues collected from two different animals. Sm Intest, small intestine; Sal Gland, salivary gland.
The initial report that described CXCL17 as a member of the chemokine superfamily demonstrated that this chemokine is expressed in several mucosal sites including the stomach, colon and trachea (16). Our data confirms and extends this observation. Given that the BIGE database contains 105 different tissues and cells, this allows us to conclude that CXCL17 is not only a mucosal chemokine but its expression in the human appears restricted to mucosal sites (Figure 1A; Table 1). We confirmed the microarray data using quantitative real time PCR (Q-PCR) in a panel of human and murine tissues (Figure 1B and 1C). The similar expression patterns between the genearrays and Q-PCR data confirmed the BIGE database expression data for CXCL17.
Table 1. The expression of CXCL17 is restricted to human mucosal tissues.
Shown are the tissues with the highest average signal intensity values of the probeset corresponding to CXCL17 (226960_at) from the BIGE database that includes 105 different human tissues and cells. A graphic representation of these data is shown in Figure 1A. The highest expression of CXCL17 in the BIGE human gene expression database corresponds to mucosal tissues. The mean intensity signal is the averaged microarray signal from the replicates for each tissue included in the BIGE database.
| Tissue | Mean intensity |
Tissue | Mean intensity |
|---|---|---|---|
| Trachea | 2359 | Lung | 242 |
| Bronchus | 1586 | Tongue | 224 |
| Stomach | 936 | Tongue_superior | 201 |
| Stomach_cardiac | 917 | Pancreas | 194 |
| Stomach_pyloric | 870 | Vagina | 165 |
| Stomach_fundus | 666 | Fallopian_tube | 143 |
| Urethra | 497 | Small_intestine | 130 |
| Esophagus | 371 | Tongue_main_corpus | 118 |
| Pharyngeal_mucosa | 317 | Tonsil | 113 |
| Small_intestine_duodenum | 315 | Cervix | 98 |
| Oral_mucosa | 271 | Vulva | 96 |
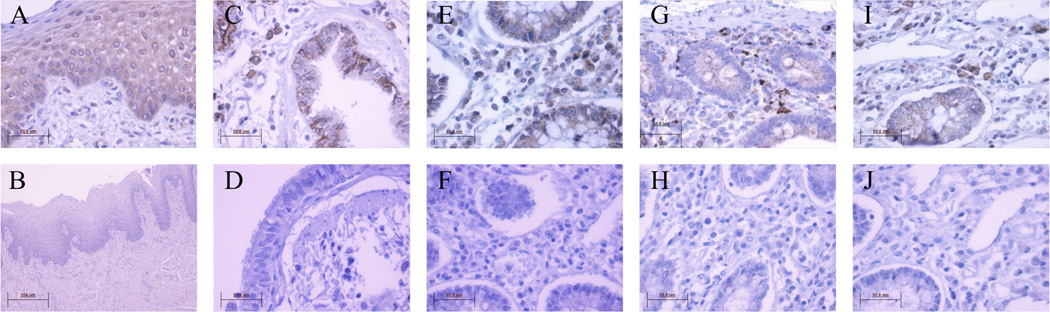
We next focused our study to provide a more in-depth analysis of the expression of CXCL17 in several mucosal sites (Figure 2). Using immunohistochemistry (IHC) the mRNA expression data was confirmed at the protein level for normal human bronchus, tongue and the gastrointestinal (GI) tract. In agreement with Pisabarro et al (16), we observed stronger CXCL17 expression near the epithelial layer exposed to the lumen of either the intestine or bronchus (data not shown), suggesting that CXCL17 is secreted into the lumen of these organs. In addition, within the mucosal tissues analyzed, there were distinct cells elsewhere that exhibit cytoplasmic staining for CXCL17 (Figure 2). Morphologically, these cells appeared to be macrophages and plasma cells. The macrophages showed indented nuclei and clear cytoplasm while the plasma cells have a peripheral nucleus with distinct chromatin staining (cartwheel) and abundant cytoplasm. Further evidence supporting the lineage of these CXCL17-producing cells was obtained by staining the same tissues with a macrophage marker (macrosialin; anti-CD68) and a plasma cell marker (syndecan 1; CD138). As shown in Supplementary Figure 1, cells with macrophage morphology stain with CD68 and cells with plasma cell morphology stain with CD138. Interestingly, we also observed CXCL17 staining in endothelial cells of some blood vessels present in these tissues (data not shown). This observation is a common feature of other chemokines, and suggests that CXCL17 may be involved in the extravasation of certain blood cells to mucosal sites.
Figure 2. CXCL17 exhibits specific expression patterns in several human mucosal tissues.
Positive staining for CXCL17 was detected in epithelial cells of the lingual tongue mucosa (A), the luminal facing respiratory mucosa (C), and the lamina propria (LP) colon mucosa (E). The respective isotype controls are shown in (B), (D), and (F). Additionally, discrete cells within each of these mucosal sites stained positive for CXCL17 (A, C, E). Staining with cell type specific markers, in addition to morphological analysis (supplementary figure 1), confirmed that these are CD68+ macrophages (G) and CD138+ plasma cells (I) within each of the mucosal sites analyzed (G and I are in the colon LP, other macrophage and plasma cell staining in tongue and respiratory mucosa are data not shown). The respective isotype controls are shown in (H) and (J). All images was captured at 40X magnification expect Figure B which was captured at 10X.
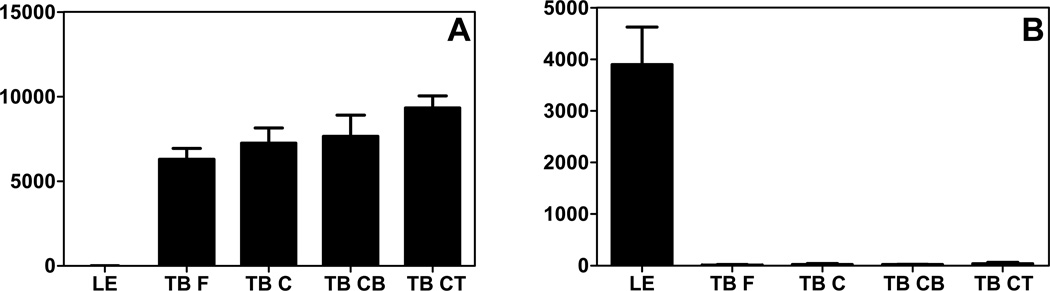
The expression of CXCL17 in mucosal tissues can exhibit surprising specificity. We previously constructed a gene expression database of primate tongue tissues which included taste buds (fungiform and circumvallate) and lingual epithelium collected by laser capture microdissection (10). The expression of CXCL17 in this database was compared to the expression of another chemokine strongly expressed in the tongue, CXCL14 (Figure 3). While CXCL14 is highly expressed in the taste buds, it is not expressed in the lingual epithelium (Figure 3 and (10, 24)). Conversely, both microarray and immunohistochemistry data indicates that CXCL17 is strongly expressed by the lingual epithelium but not in the taste buds (Figures 2 and 3). The functional significance of this highly specific chemokine expression in mucosal tissues is currently unknown.
Figure 3. CXCL14 and CXCL17 show mutually exclusive expression in the tongue.
CXCL14 and CXCL17 microarray gene expression data from either lingual epithelium or taste buds. This analysis was performed using a tongue gene expression database described previously (10). (A) CXCL14 is only expressed in the samples derived from taste buds (TB F, fungiform; TB C, circumvallate; TB CB, circumvallate base; TB CT, circumvallate top). (B) Conversely, CXCL17 is only expressed in lingual epithelium (LE). X axis: discrete tongue tissue; Y axis: mean expression values. All samples were obtained from a variable number of healthy donors depending on the tissue: lingual epithelium, 10 donors; fungiform taste buds, 6 donors; circumvallate, 4 donors; circumvallate base, 4 donors; circumvallate top, 4 donors. The data is shown as averages of data from all donors and error bars denote standard deviation of the mean.
CXCL17 is a Chemokine With Antimicrobial Properties
CXCL17 is structurally related to CCL28 and CXCL14 (1, 16), two other mucosal-expressed chemokines that have been reported to have antimicrobial activity against several pathogens including Candida albicans (8, 13). Given the structural similarity and the mucosal-specific expression of CXCL17 we hypothesized that CXCL17 also exhibits antimicrobial activity.
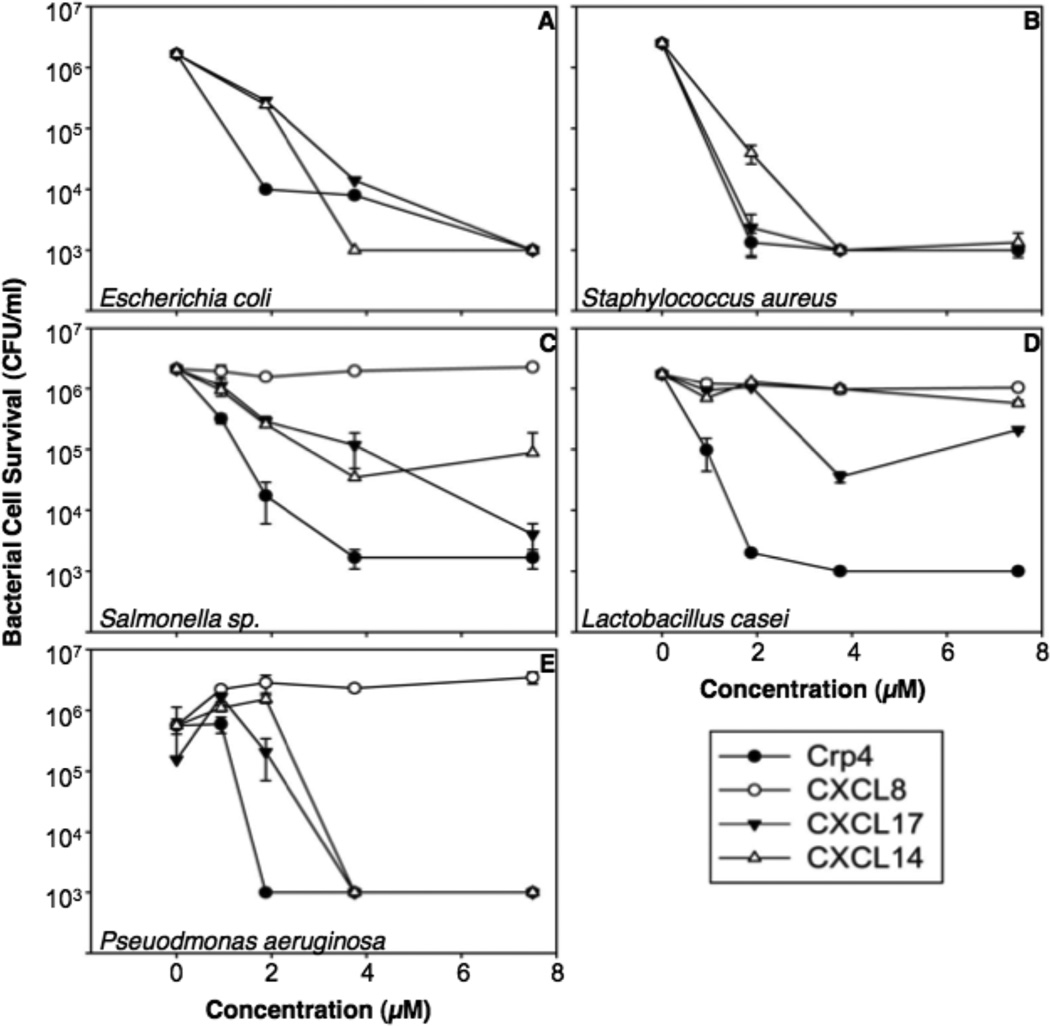
To test this hypothesis, we tested CXCL17’s ability to kill several bacterial strains as well as C. albicans in solution using microbicidal peptide assays. We compared CXCL17 to CXCL14 (another mucosal chemokine (15, 25)) and cryptdin-4 (Crp4). The latter is a well-characterized mouse Paneth cell α-defensin (26–27). We also compared CXCL17’s antimicrobial activity to CXCL8, a chemokine that lacks antimicrobial activity (28–29). The microorganisms were chosen for these studies because they can colonize the mucosa as either commensal or pathogenic microbes.
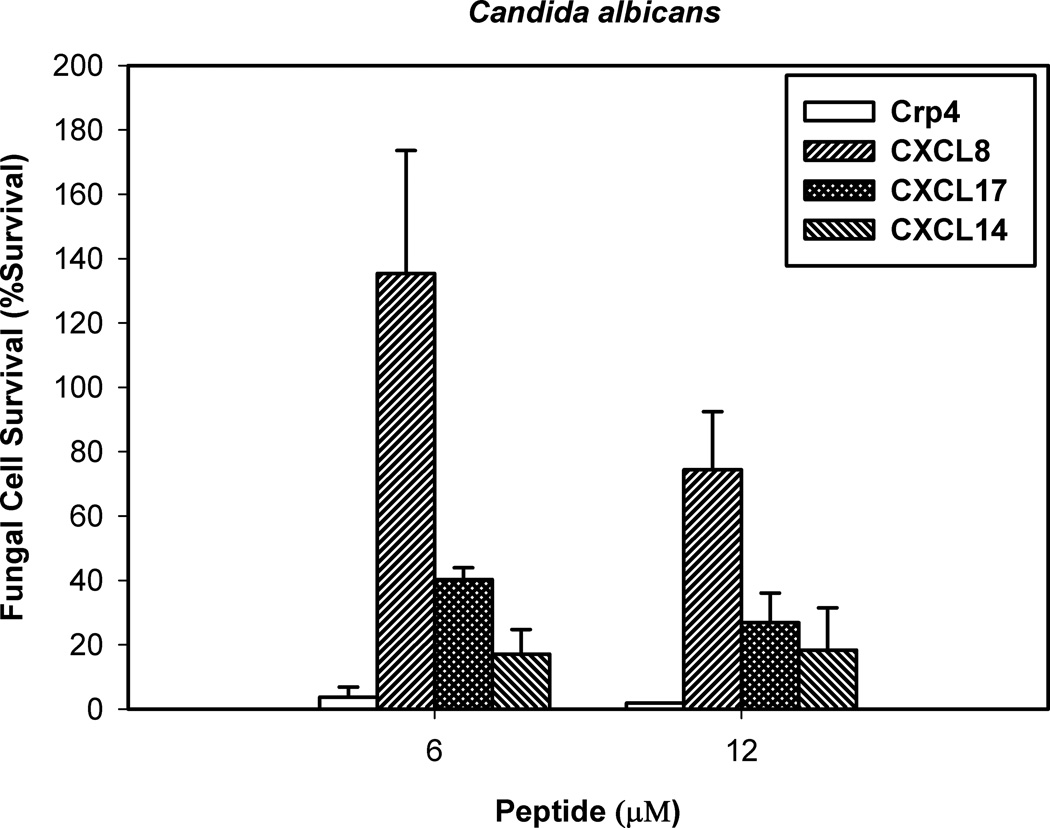
CXCL17 significantly reduced the survival of bacteria or fungi by 3–4 orders of magnitude in a dose-dependent manner (Figures 4 and 5). With the exception of Lactobacillus casei, CXCL17 exhibits microbicidal activity similar to that of Crp4, the most potent known mouse α-defensin (26), and is more active than CXCL14, a chemokine previously reported to have antimicrobial activity (13). Consistent with previous reports (28), CXCL8 failed to show antimicrobial activity against all bacterial species tested (Figure 4 and Supplementary Figure 2).
Figure 4. CXCL17 has broad spectrum bacteriocidal activity.
Exponentially growing Escherichia coli (A), Staphylococcus aureus (B), Salmonella enterica serovar Typhimurium 14028s (C), Lactobacillus casei (D), Pseudomonas aeruginosa (E), were exposed to chemokines at 37°C in 50 µl of PIPES-TSB buffer for 1 h (see Materials and Methods). Following peptide exposure, the bacteria were spread on TSB-agar plates and incubated overnight at 37°C. Surviving bacteria were counted as CFU/ml at each concentration and values at or below 1 × 103 CFU/ml mean that no detectable colonies were observed. Except for E. coli, assays were performed in triplicate and error bars denote standard deviation of the mean.
Figure 5. CXCL17 is active against the opportunistic fungal pathogen C. albicans.
Log-phase C. albicans were exposed to chemokines at 6 and 12 µM protein concentrations at 37°C in 50 µl of PIPES-SAB buffer for 1 h. Following peptide exposure, the bacteria were spread on SAB-agar plates and incubated overnight at 37°C. Surviving Candida colonies were counted as CFU/ml at each concentration and values at or below 1 × 103 CFU/ml mean no detectable colonies were observed. Assays were performed in triplicate and error bars denote standard deviation of the mean.
CXCL17 Induces E. coli Cell Permeabilization
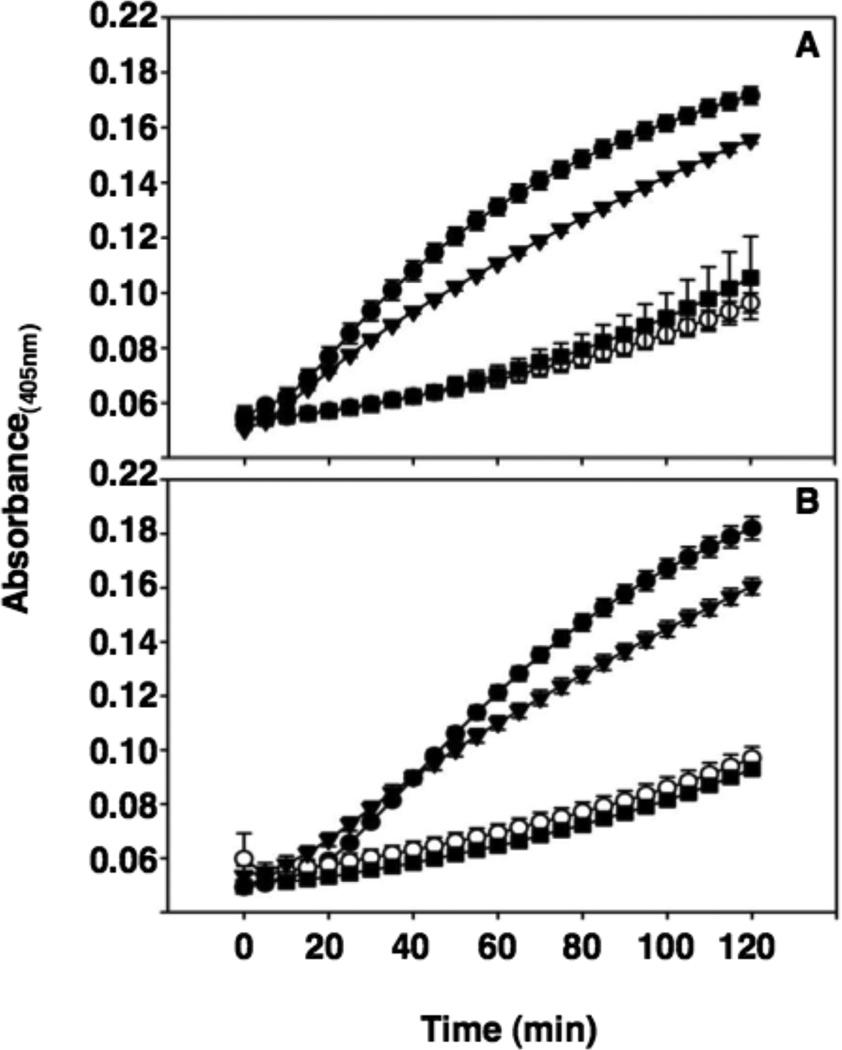
We then sought to explore the antimicrobial mechanism of CXCL17. Given the similarities between the bacteriocidal effects of CXCL17 and Crp4, we hypothesized that CXCL17 could induce permeabilization of live E. coli similar to Crp4 (24, 30). To investigate this, we tested CXCL17 in a membrane permeabilization assay. This is a common method to gain insight into the antimicrobial mechanism of peptides, such as Crp-4 (31).
In the permeabilization assay, CXCL17 disturbed cellular homeostasis enough to enable diffusion of the ONPG substrate into the cells for conversion to ONP (Figure 6), an activity that results from disturbing the integrity of bacterial membranes transiently or by formation of stable pores (24, 32). These results indicate that CXCL17 induces permeabilization of bacterial membranes. As a control, we tested CXCL8 in the membrane permeabilization assay, a chemokine that does not exhibit antimicrobial activity (Figure 4 and Supplementary Figure 2). CXCL8 does not induce diffusion of ONPG, indicating that it does not disturb bacterial membranes (Figure 6).
Figure 6. CXCL17 induces permeabilization of live E. coli membranes as measured by ONPG conversion.
Log-phase E. coli ML35 cells were exposed to either 3.75 µM (A) or 1.625 µM (B) Crp4, CXCL17, CXCL8, and ProCryptdin4 in the presence of 2-nitrophenyl β-D-galactopyranoside (ONPG) for 2 h at 37°C. β-galactosidase mediated hydrolysis of ONPG was measured at A405. Symbols: Crp4, -●-; CXCL8, -○-; CXCL17, - ▼-; and, ProCrp4, -■-.
CXCL17 Is Elevated In Idiopathic Pulmonary Fibrosis
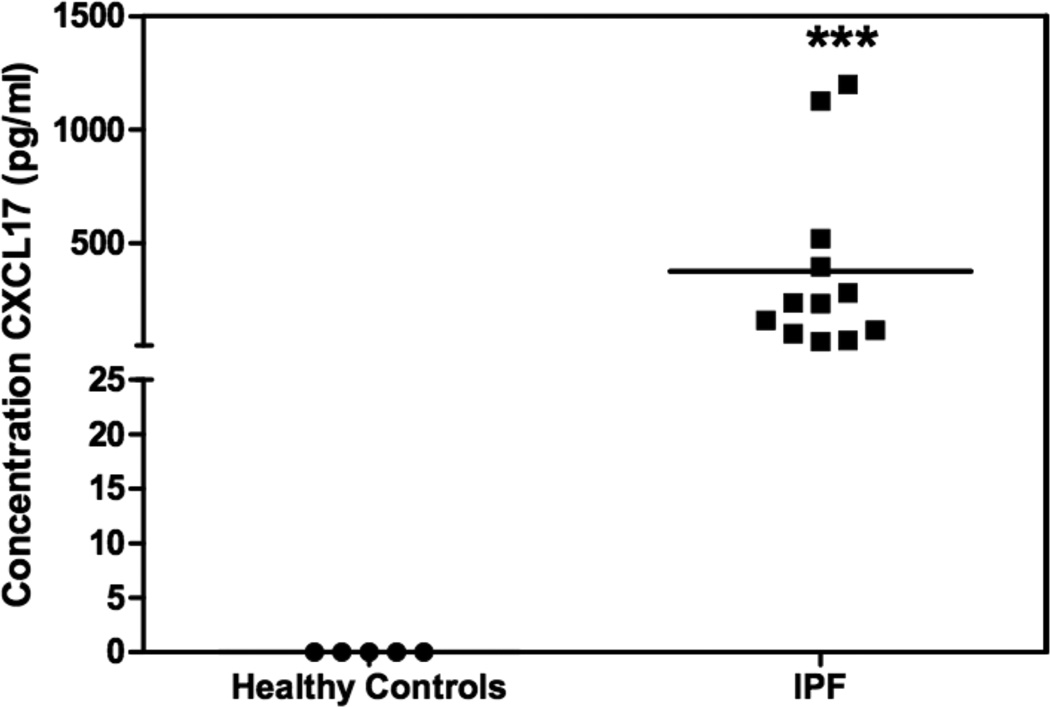
Given the high CXCL17 expression in the airways (Figure 1, 2 and (16)), we next asked whether CXCL17 could be associated with human lung disease. To approach this question, we developed a CXCL17 ELISA, and used it to measure the levels of this chemokine in bronchoalveolar lavage fluids obtained from patients with IPF or healthy donors. Our results indicate that CXCL17 levels are significantly elevated in BALf from patients with IPF (376.6 pg/ml versus undetectable (<62.5 pg/ml); Figure 7).
Figure 7. CXCL17 is elevated in BALf samples of patients diagnosed with IPF compared to healthy controls.
CXCL17 was detected in human BALf samples using a CXCL17-specific sandwich ELISA. The BALf samples were concentrated to 10X prior to ELISA analysis. Recombinant human CXCL17 was used as the standard. CXCL17 was below the level of detection in healthy samples, but was detected at robust levels in samples from patients previously diagnosed with IPF. The CXCL17 levels in the IPF samples were statistically significant (indicated by ***) compared to healthy samples (p=0.01). Assays were performed in duplicate. X axis: samples; Y axis: concentration of CXCL17 (pg/ml).
Discussion
CXCL17 was the last chemokine ligand to be discovered (16) and consequently, there are few studies on this chemokine (16, 33–34). Pisabarro et al (16) first reported this chemokine and it still represents it most detailed characterization. Weinstein et al (33) called this chemokine Vascular Endothelial Growth Factor (VEGF)-correlated chemokine 1 (VCC-1) because they found it expressed in several cancers and found that it induces VEGF expression. They observed that expression of CXCL17 in a NIH3T3 cells favored tumor progression in vivo, and they ascribed this effect to the ability of CXCL17 to promote angiogenesis. Mu et al (34) found CXCL17 expressed in hepatocellular carcinoma and concluded that it favors tumor progression in vivo.
Pisabarro et al (16) analyzed the expression of CXCL17 mRNA in human adult stomach, thyroid, spinal cord, lymph node, trachea, adrenal gland and peripheral blood leukocytes. They concluded that it is expressed in stomach and trachea. In addition, they detected CXCL17 in adult lung and intestine by immunohistochemistry. In our present study, we present microarray data on 105 tissues and cells of the human body (Figure 1). Therefore, our studies both confirm and extend the results of Pisabarro et al (16) and allow us to conclude that CXCL17 is a chemokine that exhibits restricted expression in mucosal tissues. In fact, of the four mucosal-associated chemokines (CCL25, CCL28, CXCL14 and CXCL17), only CCL28 and CXCL17 are mucosal-restricted, since CCL25 is also expressed in the thymus (23) and CXCL14 is also expressed in the skin and kidney (12–13). CCL28 has a very important function in the mucosa, namely, the recruitment of cells that mediate IgA production in the mammary gland (7). We therefore hypothesize that CXCL17 must also mediate important mucosal-specific functions. The results of our present study, along with the pioneering Pisabarro report (16), indicate that CXCL17 should be recognized as an important mucosal chemokine.
The discrete mucosal expression pattern of CXCL17 in normal human tissues strongly suggests that it is a homeostatic chemokine (Figures 1, 2 and 3). It has been reported to chemoattract dendritic cells and monocytes (16, 33–34). Importantly, its receptor has not been described yet. Homeostatic chemokines tend to have specific receptors (1). We predict that the CXCL17 receptor should be expressed in monocytes, dendritic cells, as well as discrete cell populations present in mucosal tissues that may be recruited by CXCL17.
Both CCL28 and CXCL14 have broad antimicrobial activity (8, 13, 18); several other chemokines have antibacterial activity against a narrow range of species (35–37). Other chemokines, like CXCL8, do not have antimicrobial properties ((28–29), Figure 4 and Supplemental Figure 2). It is noteworthy that 3 chemokines that exhibit broad antimicrobial properties (CCL28, CXCL14 and CXCL17) are both homeostatic and expressed in the mucosa, suggesting that this activity is associated with their mucosal expression pattern.
A surprising observation is the high level of specificity that mucosal chemokines can exhibit in their expression patterns within mucosal tissues (Figure 3). Both CXCL14 and CXCL17 are expressed in the tongue. However, CXCL14 is only expressed in the taste buds while CXCL17 is only expressed in the lingual epithelium (Figure 3). This suggests highly specific functions for each chemokine within these tissues. In fact, CXCL14 is the highest expressed gene in primate taste buds (10), although at present its function there remains obscure. In the gut mucosa, CXCL17 is preferentially expressed by epithelial cells adjacent to the lumen versus cells adjoining the lamina propria (LP) (16) and (data not shown). However, within the colon LP we also detected discrete cells that express CXCL17. Their morphology as well as IHC staining with cell specific markers, indicates that the CXCL17-expressing cells include macrophages (expressing CD68) and plasma cells (expressing CD138) (Figure 2 and supplementary Figure 1). These cells were also observed in the bronchus and tongue (Figure 2); as in the colon LP, they were identified as macrophage and plasma cells through their expression of CD68 and CD138, respectively (data not shown). Their ability to express CXCL17 suggests that they may use this chemokine to attract other cells for as yet unknown functions within the mucosa.
Our data shows that CXCL17 can have direct effects on the microbial populations at mucosal sites (Figures 4 and 5). We tested commensal and pathogenic microbes, including some that are known to cause infection at mucosal sites. Our results strongly suggest that CXCL17 could play a direct role in shaping the microbiome. This conclusion is supported by immunohistochemistry data, which indicates that CXCL17 is preferentially expressed in mucosal sites adjacent to the lumen ((16) and data not shown). Additionally, chemokines bind to the heparin sulfate, which is part of the extracellular matrix of epithelial cells (38). This binding helps localize chemokines within a specific region and aids the development of a gradient within the tissue (39). This phenomenon could help increase the concentration of CXCL17 in mucosal surfaces.
Prior to the outset of these studies, we analyzed the primary protein structure of CXCL17 to determine if it shared properties with defensins, a well-characterized antimicrobial family. However, helical wheel projection analyses of CXCL17 do not predict amphipathic helices (data not shown), evidence that the CXCL17 antimicrobial mechanism of action differs from that of other linear antimicrobial peptides. Given the constraints to the general chemokine structure imposed by the disulfide bonds, CXCL17 may have a different distribution of charge and hydrophobicity, which contribute to its antimicrobial effects. For example, dimeric human neutrophil defensin-3 (HNP-2) forms stable, 20 Å multimeric pores after insertion in model phospholipid bilayers (31), but rabbit NP-1 monomers create short-lived bacterial membrane defects of widely varied magnitude (40). Clearly, CXCL17 induces membrane permeabilization in E. coli cells (Figure 6), but whether it does so by formation of stable pores or transient membrane defects remains a topic for future study.
Lastly we explored whether CXCL17 could be associated with human disease. Pisabarro et al (16) analyzed the expression of CXCL17 in asthma or chronic obstructive pulmonary disease tissue sections by immunohistochemistry but could not conclude whether there was increased CXCL17 expression. The development of a human CXCL17 ELISA allowed us to quantitate the levels of this chemokine in BALf of patients with idiopathic pulmonary fibrosis (IPF). Interestingly, IPF has been recently associated with a Single Nucleotide Polymorphism (SNP) in the promoter region of the airway mucin gene (MUC5B) that strongly increases the production of this protein (41). In that study, MUC5B was detected in the cytoplasm of the secretory columnar cells of the bronchi and in larger proximal bronchioles where CXCL17 is also expressed ((16) and data not shown). Our data demonstrates that CXCL17 is elevated in IPF (Figure 7). This is the first observation that CXCL17 is over-expressed in IPF, the most aggressive human interstitial lung disease. IPF is a progressive and usually lethal lung disease characterized by an aberrant activation of bronchiolar/alveolar epithelial cells, expansion of the myofibroblasts population followed by the abnormal remodeling of the lung parenchyma (42). However, CXCL17’s putative role in this disease remains unclear. The increase in CXCL17 may be related to its antimicrobial mechanisms since infections are common in these advanced stage IPF patients. Actually, a characteristic finding in IPF lungs is the presence of areas of cystic fibrotic airspaces (honeycombing) usually filled with mucinous exudate. Chronic and even acute inflammation is usually detected in and around these honeycomb areas (43). In addition, it has been suggested that CXCL17 facilitates the antitumor host immune response by inducing the infiltration and accumulation of immature myeloid Dendritic Cells (DCs), and these cells have been found increased in IPF lungs, although its role in this disease has not been elucidated (44–45). Finally, a substantial vascular remodeling occurs in the fibrotic lung disorders and increased staining for VEGF, a potent angiogenic factor and a putative CXCL17-inducing factor, has been observed the IPF lungs (46). Further studies will be necessary to evaluate the cellular source of this chemokine in IPF lungs and to understand its contribution to the complex mechanisms involved in this disease.
The levels of CXCL17 in the healthy BALf samples were below the limits of detection for our ELISA assay (below 62.5pg/ml). This finding may be partially due to the large volumes of saline solution used to collect the BALf samples (300ml). Therefore, the inherent dilution of CXCL17 during the collection of the BALf may account for the failure to detect CXCL17 in the healthy BALf samples.
IPF is one of several interstitial lung diseases, and its differential diagnosis may be difficult. We have profiled several of these diseases using genearrays and found that they exhibit a distinct gene expression signature (47). Nevertheless, their diagnosis is still challenging; our finding of elevated levels of CXCL17 in IPF raises the possibility that it may be a candidate biomarker to assist in the diagnosis of this disease. However, in order to confirm this, it is necessary to study CXCL17 levels in other lung pathologies. We conclude that these observations open a new field of research. Besides its diagnostic potential, our observations also raise the possibility that CXCL17 may be involved in the pathogenesis of IPF. Future experiments will aim to answer the questions raised by the present study.
Supplementary Material
Acknowledgments
The authors wish to thank Tracy Handel for helpful discussions and Carolina Garcia de Alba for her assistance with the CXCL17 ELISAs.
Abbreviations used in this paper
- BALf
bronchoalveolar lavage fluid
- BIGE
Body Index of Gene Expression
- Crp4
cryptdin-4
- DCs
dendritic cells
- ELISA
enzyme linked immunosorbent assay
- GI
gastrointestinal tract
- IHC
immunohistochemistry
- IPF
idiopathic pulmonary fibrosis
- LP
lamina propria
- ONPG
o-nitrophenyl-β-d-galactopyranoside
- ONP
o-nitrophenol
- Q-PCR
quantitative real-time polymerase chain reaction
- VEGF
vascular endothelial growth factor
Footnotes
This study was supported by National Institutes of Health grants (NIAID) R21AI083540-01 to AZ and DK044632, AI059346 grants to AJO. AJO is also recipient of USC Norris Cancer Center Support Grant P30 CA014089. AMB is supported by NIH Immunology Research Training Program Grant T32AI60573 and a UCMEXUS grant (51934).
References
- 1.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7:243. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Der Meer P, Goldberg SH, Fung KM, Sharer LR, Gonzalez-Scarano F, Lavi E. Expression pattern of CXCR3, CXCR4, and CCR3 chemokine receptors in the developing human brain. J Neuropathol Exp Neurol. 2001;60:25–32. doi: 10.1093/jnen/60.1.25. [DOI] [PubMed] [Google Scholar]
- 3.Li M, Yu J, Li Y, Li D, Yan D, Ruan Q. CXCR4+ progenitors derived from bone mesenchymal stem cells differentiate into endothelial cells capable of vascular repair after arterial injury. Cell Reprogram. 2010;12:405–415. doi: 10.1089/cell.2009.0088. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel EJ, Campbell JJ, Haraldsen G, Pan J, Boisvert J, Roberts AI, Ebert EC, Vierra MA, Goodman SB, Genovese MC, Wardlaw AJ, Greenberg HB, Parker CM, Butcher EC, Andrew DP, Agace WW. Lymphocyte CC chemokine receptor 9 and epithelial thymus-expressed chemokine (TECK) expression distinguish the small intestinal immune compartment: Epithelial expression of tissue-specific chemokines as an organizing principle in regional immunity. J Exp Med. 2000;192:761–768. doi: 10.1084/jem.192.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onai N, Kitabatake M, Zhang YY, Ishikawa H, Ishikawa S, Matsushima K. Pivotal role of CCL25 (TECK)-CCR9 in the formation of gut cryptopatches and consequent appearance of intestinal intraepithelial T lymphocytes. Int Immunol. 2002;14:687–694. doi: 10.1093/intimm/dxf035. [DOI] [PubMed] [Google Scholar]
- 6.Morteau O, Gerard C, Lu B, Ghiran S, Rits M, Fujiwara Y, Law Y, Distelhorst K, Nielsen EM, Hill ED, Kwan R, Lazarus NH, Butcher EC, Wilson E. An indispensable role for the chemokine receptor CCR10 in IgA antibody-secreting cell accumulation. J Immunol. 2008;181:6309–6315. doi: 10.4049/jimmunol.181.9.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson E, Butcher EC. CCL28 controls immunoglobulin (Ig)A plasma cell accumulation in the lactating mammary gland and IgA antibody transfer to the neonate. J Exp Med. 2004;200:805–809. doi: 10.1084/jem.20041069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hieshima K, Ohtani H, Shibano M, Izawa D, Nakayama T, Kawasaki Y, Shiba F, Shiota M, Katou F, Saito T, Yoshie O. CCL28 has dual roles in mucosal immunity as a chemokine with broad-spectrum antimicrobial activity. J Immunol. 2003;170:1452–1461. doi: 10.4049/jimmunol.170.3.1452. [DOI] [PubMed] [Google Scholar]
- 9.Roth RB, Hevezi P, Lee J, Willhite D, Lechner SM, Foster AC, Zlotnik A. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- 10.Hevezi P, Moyer BD, Lu M, Gao N, White E, Echeverri F, Kalabat D, Soto H, Laita B, Li C, Yeh SA, Zoller M, Zlotnik A. Genome-wide analysis of gene expression in primate taste buds reveals links to diverse processes. PLoS One. 2009;4:e6395. doi: 10.1371/journal.pone.0006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J, Hever A, Willhite D, Zlotnik A, Hevezi P. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J. 2005;19:1356–1358. doi: 10.1096/fj.04-3552fje. [DOI] [PubMed] [Google Scholar]
- 12.Hromas R, Broxmeyer HE, Kim C, Nakshatri H, Christopherson K, 2nd, Azam M, Hou YH. Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem Biophys Res Commun. 1999;255:703–706. doi: 10.1006/bbrc.1999.0257. [DOI] [PubMed] [Google Scholar]
- 13.Maerki C, Meuter S, Liebi M, Muhlemann K, Frederick MJ, Yawalkar N, Moser B, Wolf M. Potent and broad-spectrum antimicrobial activity of CXCL14 suggests an immediate role in skin infections. J Immunol. 2009;182:507–514. doi: 10.4049/jimmunol.182.1.507. [DOI] [PubMed] [Google Scholar]
- 14.Shellenberger TD, Wang M, Gujrati M, Jayakumar A, Strieter RM, Burdick MD, Ioannides CG, Efferson CL, El-Naggar AK, Roberts D, Clayman GL, Frederick MJ. BRAK/CXCL14 is a potent inhibitor of angiogenesis and a chemotactic factor for immature dendritic cells. Cancer Res. 2004;64:8262–8270. doi: 10.1158/0008-5472.CAN-04-2056. [DOI] [PubMed] [Google Scholar]
- 15.Meuter S, Moser B. Constitutive expression of CXCL14 in healthy human and murine epithelial tissues. Cytokine. 2008;44:248–255. doi: 10.1016/j.cyto.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Pisabarro MT, Leung B, Kwong M, Corpuz R, Frantz GD, Chiang N, Vandlen R, Diehl LJ, Skelton N, Kim HS, Eaton D, Schmidt KN. Cutting edge: novel human dendritic cell- and monocyte-attracting chemokine-like protein identified by fold recognition methods. J Immunol. 2006;176:2069–2073. doi: 10.4049/jimmunol.176.4.2069. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Soto H, Oldham ER, Buchanan ME, Homey B, Catron D, Jenkins N, Copeland NG, Gilbert DJ, Nguyen N, Abrams J, Kershenovich D, Smith K, McClanahan T, Vicari AP, Zlotnik A. Identification of a novel chemokine (CCL28), which binds CCR10 (GPR2) J Biol Chem. 2000;275:22313–22323. doi: 10.1074/jbc.M001461200. [DOI] [PubMed] [Google Scholar]
- 18.Liu B, Wilson E. The antimicrobial activity of CCL28 is dependent onC-terminal positively-charged amino acids. Eur J Immunol. 2010;40:186–196. doi: 10.1002/eji.200939819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frick IM, Nordin SL, Baumgarten M, Morgelin M, Sorensen OE, Olin AI, Egesten A. Constitutive and inflammation-dependent antimicrobial peptides produced by epithelium are differentially processed and inactivated by the commensal Finegoldia magna and the pathogen Streptococcus pyogenes. J Immunol. 2011;187:4300–4309. doi: 10.4049/jimmunol.1004179. [DOI] [PubMed] [Google Scholar]
- 20.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, Lynch DA, Ryu JH, Swigris JJ, Wells AU, Ancochea J, Bouros D, Carvalho C, Costabel U, Ebina M, Hansell DM, Johkoh T, Kim DS, King TE, Jr, Kondoh Y, Myers J, Muller NL, Nicholson AG, Richeldi L, Selman M, Dudden RF, Griss BS, Protzko SL, Schunemann HJ. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, Kaminski N. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med. 2005;2:e251. doi: 10.1371/journal.pmed.0020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourges D, Meurens F, Berri M, Chevaleyre C, Zanello G, Levast B, Melo S, Gerdts V, Salmon H. New insights into the dual recruitment of IgA+ B cells in the developing mammary gland. Mol Immunol. 2008;45:3354–3362. doi: 10.1016/j.molimm.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 23.Vicari AP, Figueroa DJ, Hedrick JA, Foster JS, Singh KP, Menon S, Copeland NG, Gilbert DJ, Jenkins NA, Bacon KB, Zlotnik A. TECK: a novel CC chemokine specifically expressed by thymic dendritic cells and potentially involved in T cell development. Immunity. 1997;7:291–301. doi: 10.1016/s1074-7613(00)80531-2. [DOI] [PubMed] [Google Scholar]
- 24.Hadjicharalambous C, Sheynis T, Jelinek R, Shanahan MT, Ouellette AJ, Gizeli E. Mechanisms of alpha-defensin bactericidal action: comparative membrane disruption by Cryptdin-4 and its disulfide-null analogue. Biochemistry. 2008;47:12626–12634. doi: 10.1021/bi800335e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurth I, Willimann K, Schaerli P, Hunziker T, Clark-Lewis I, Moser B. Monocyte selectivity and tissue localization suggests a role for breast and kidney-expressed chemokine (BRAK) in macrophage development. J Exp Med. 2001;194:855–861. doi: 10.1084/jem.194.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mastroianni JR, Ouellette AJ. Alpha-defensins in enteric innate immunity: functional Paneth cell alpha-defensins in mouse colonic lumen. J Biol Chem. 2009;284:27848–27856. doi: 10.1074/jbc.M109.050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda K, Nakamura K, Yoshioka S, Fukaya R, Sakai N, Ayabe T. Regulation of microbiota by antimicrobial peptides in the gut. Adv Otorhinolaryngol. 2011;72:97–99. doi: 10.1159/000324625. [DOI] [PubMed] [Google Scholar]
- 28.Bjorstad A, Fu H, Karlsson A, Dahlgren C, Bylund J. Interleukin-8-derived peptide has antibacterial activity. Antimicrob Agents Chemother. 2005;49:3889–3895. doi: 10.1128/AAC.49.9.3889-3895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linge HM, Collin M, Giwercman A, Malm J, Bjartell A, Egesten A. The antibacterial chemokine MIG/CXCL9 is constitutively expressed in epithelial cells of the male urogenital tract and is present in seminal plasma. J Interferon Cytokine Res. 2008;28:191–196. doi: 10.1089/jir.2007.0100. [DOI] [PubMed] [Google Scholar]
- 30.Satchell DP, Sheynis T, Shirafuji Y, Kolusheva S, Ouellette AJ, Jelinek R. Interactions of mouse Paneth cell alpha-defensins and alpha-defensin precursors with membranes. Prosegment inhibition of peptide association with biomimetic membranes. J Biol Chem. 2003;278:13838–13846. doi: 10.1074/jbc.M212115200. [DOI] [PubMed] [Google Scholar]
- 31.Hristova K, Selsted ME, White SH. Interactions of monomeric rabbit neutrophil defensins with bilayers: comparison with dimeric human defensin HNP-2. Biochemistry. 1996;35:11888–11894. doi: 10.1021/bi961100d. [DOI] [PubMed] [Google Scholar]
- 32.Figueredo SM, Weeks CS, Young SK, Ouellette AJ. Anionic amino acids near the pro-alpha-defensin N terminus mediate inhibition of bactericidal activity in mouse pro-cryptdin-4. J Biol Chem. 2009;284:6826–6831. doi: 10.1074/jbc.M807024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinstein EJ, Head R, Griggs DW, Sun D, Evans RJ, Swearingen ML, Westlin MM, Mazzarella R. VCC-1, a novel chemokine, promotes tumor growth. Biochem Biophys Res Commun. 2006;350:74–81. doi: 10.1016/j.bbrc.2006.08.194. [DOI] [PubMed] [Google Scholar]
- 34.Mu X, Chen Y, Wang S, Huang X, Pan H, Li M. Overexpression of VCC-1 gene in human hepatocellular carcinoma cells promotes cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2009;41:631–637. doi: 10.1093/abbs/gmp051. [DOI] [PubMed] [Google Scholar]
- 35.Yang D, Chen Q, Hoover DM, Staley P, Tucker KD, Lubkowski J, Oppenheim JJ. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J Leukoc Biol. 2003;74:448–455. doi: 10.1189/jlb.0103024. [DOI] [PubMed] [Google Scholar]
- 36.Yung SC, Parenti D, Murphy PM. Host chemokines bind to Staphylococcus aureus and stimulate protein A release. J Biol Chem. 2011;286:5069–5077. doi: 10.1074/jbc.M110.195180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eliasson M, Egesten A. Antibacterial chemokines--actors in both innate and adaptive immunity. Contrib Microbiol. 2008;15:101–117. doi: 10.1159/000136317. [DOI] [PubMed] [Google Scholar]
- 38.Lortat-Jacob H, Grosdidier A, Imberty A. Structural diversity of heparan sulfate binding domains in chemokines. Proc Natl Acad Sci U S A. 2002;99:1229–1234. doi: 10.1073/pnas.032497699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshie O, Imai T, Nomiyama H. Chemokines in immunity. Adv Immunol. 2001;78:57–110. doi: 10.1016/s0065-2776(01)78002-9. [DOI] [PubMed] [Google Scholar]
- 40.Hristova K, Selsted ME, White SH. Critical role of lipid composition in membrane permeabilization by rabbit neutrophil defensins. J Biol Chem. 1997;272:24224–24233. doi: 10.1074/jbc.272.39.24224. [DOI] [PubMed] [Google Scholar]
- 41.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, Fingerlin TE, Zhang W, Gudmundsson G, Groshong SD, Evans CM, Garantziotis S, Adler KB, Dickey BF, du Bois RM, Yang IV, Herron A, Kervitsky D, Talbert JL, Markin C, Park J, Crews AL, Slifer SH, Auerbach S, Roy MG, Lin J, Hennessy CE, Schwarz MI, Schwartz DA. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King TE, Jr, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378:1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 43.Katzenstein AL, Mukhopadhyay S, Myers JL. Diagnosis of usual interstitial pneumonia and distinction from other fibrosing interstitial lung diseases. Hum Pathol. 2008;39:1275–1294. doi: 10.1016/j.humpath.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Hiraoka N, Yamazaki-Itoh R, Ino Y, Mizuguchi Y, Yamada T, Hirohashi S, Kanai Y. CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology. 2011;140:310–321. doi: 10.1053/j.gastro.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 45.Tsoumakidou M, Karagiannis KP, Bouloukaki I, Zakynthinos S, Tzanakis N, Siafakas NM. Increased Bronchoalveolar Lavage Fluid CD1c Expressing Dendritic Cells in Idiopathic Pulmonary Fibrosis. Respiration. 2009;78:446–452. doi: 10.1159/000226244. [DOI] [PubMed] [Google Scholar]
- 46.Ebina M, Shimizukawa M, Shibata N, Kimura Y, Suzuki T, Endo M, Sasano H, Kondo T, Nukiwa T. Heterogeneous increase in CD34-positive alveolar capillaries in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2004;169:1203–1208. doi: 10.1164/rccm.200308-1111OC. [DOI] [PubMed] [Google Scholar]
- 47.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, Aziz N, Kaminski N, Zlotnik A. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med. 2006;173:188–198. doi: 10.1164/rccm.200504-644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.