Abstract
Inflammation is a hallmark of many important human diseases. Appropriate inflammation is critical for host defense. However, overactive response is detrimental to the host. Thus inflammation must be tightly regulated. The molecular mechanisms underlying the tight regulation of inflammation remain largely unknown. Ecotropic viral integration site 1 (EVI1), a proto-oncogene and zinc finger transcription factor, plays important roles in the normal development and leukemogenesis. However, its role in regulating NF-κB-dependent inflammation remains unknown. Here, we show that EVI1 negatively regulates nontypeable Haemophilus influenzae (NTHi)- and TNF-α-induced NF-κB-dependent inflammation in vitro and in vivo. EVI1 directly binds to the NF-κB p65 subunit and inhibits its acetylation at lysine 310, thereby inhibiting its DNA binding activity. Moreover, expression of EVI1 itself is induced by NTHi and TNF-α in a NF-κB-dependent manner, thereby unveiling a novel inducible negative feedback loop to tightly control NF-κB-dependent inflammation. Thus our study may not only provide important insights into the novel role of EVI1 in negatively regulating NF-κB-dependent inflammation, but may also shed light on the future development of novel anti-inflammatory strategies.
Keywords: EVI1, NF-κB, NTHi, p65 acetylation, inflammation
Introduction
Ecotropic viral integration site 1 (EVI1) was first identified as a common retroviral integration site in AKXD murine myeloid tumors with important roles in normal development and leukemogenesis (1). EVI1 is a proto-oncogene and transcriptionally activated by specific chromosomal rearrangement at 3q26 (2, 3). The aberrant expression of EVI1, as a result of either inv (3) (q21q26.2)/t (3;3) (q21;q26.2) or through other unknown mechanisms, is associated with human acute myelogenous leukemia (AML), myelodysplastic syndrome (MDS), and chronic myelogenous leukemia (CML) (2–5). It is also a zinc finger transcription factor localized to the nucleus. EVI1 encodes a nuclear protein of 145 kDa with domains characteristic of transcription regulators. It has 10 zinc fingers, seven of which are in the N-terminus and three of which are in the C-terminal part of the protein. A proline-rich region separates the two zinc finger domains, and a small acidic domain is located in the C-terminus of the second zinc finger domain (1). EVI1 binds to DNA through specific conserved sequences of GACAAGATA with the potential to interact with both co-repressors and co-activators (6–9). EVI1 has been shown to interact directly with the known transcriptional repressor C-terminal binding protein 2 (CtBP2) via two CtBP-binding consensus motifs at amino acids 544–607(10, 11). This binding has been suggested to recruit histone deacetylase complexes (HDACs) and lead to transcriptional repression via chromatin remodeling. In addition, the interaction of EVI1 with cyclic AMP response element binding protein (CBP) and p300/CBP-associated factor (P/CAF) was reported to result in the reversible acetylation of EVI1 and its co-localization in nuclear speckles (12). Based on our preliminary gene profiling study, we found that EVI1 is up-regulated by inflammatory stimuli including bacteria. However, its role in regulating inflammation remains largely unknown. Previous study has shown that mutation of the Evi1 gene in mice leads to chronic middle ear inflammatory disease (13), thereby implying a potential role of EVI1 in regulating inflammatory processes.
Inflammation is a hallmark and the root cause of many important human diseases, including airway inflammatory diseases (e.g., chronic obstructive pulmonary disease (COPD), asthma, and otitis media (OM)), atherosclerosis, infectious diseases, and cancer (14–17). Appropriate inflammation is a protective host defense response to remove the injurious stimuli as well as initiate tissue healing and repair process. However, overactive inflammation is clearly detrimental to the host, leading to inflammatory diseases. Thus, inflammation must be tightly regulated. The inflammatory response can be controlled at multiple levels (18), but the underlying molecular mechanisms still remain largely unknown, partly due to the complexity of the inflammatory response and the multitude of components involved. The transcription factor nuclear factor kappa-B (NF-κB) is activated by inflammatory stimuli such as bacteria and TNF-α and plays a critical role in mediating inflammatory responses by regulating the expression of pro-inflammatory mediators, including cytokines, chemokines, and adhesion molecules (19). NF-κB is activated via phosphorylation and degradation of IκB by the IκB kinase enzyme complex (IKKs) (20–23), which in turn leads to the nuclear translocation of NF-κB and the subsequent transcription of NF-κB-dependent genes, such as TNF-α, IL-1β and IL-8. Acetylation of p65, an important post-translational modification, plays a critical role in the regulation of the nuclear function of NF-κB, which leads to changes in its biological activity, such as alterations in DNA-binding activity and transcriptional activity (24–28). EVI1 acts as both transcriptional activator and repressor to recruit the HDACs and histone acetyltransferase (HAT). EVI1 itself can also be acetylated by P/CAF at lysine residues (12, 29). However, the role of EVI1 in controlling the acetylation of other molecules is still unknown. Moreover, the role of EVI1 in regulating the activation of NF-κB, a key regulator for proinflammatory responses, has yet to be determined.
Nontypeable Haemophilus influenzae (NTHi), a gram-negative bacterium, is an important human pathogen in both children and adults (30). In children, it causes OM, one of the most common childhood infections and the leading cause of conductive hearing loss in the United States (31, 32). In adults, it exacerbates COPD, the fourth leading cause of death in the United States (33, 34). Despite the need for prophylactic measures, the development of a vaccine to prevent NTHi infections has been difficult and still remains a great challenge. Moreover, inappropriate antibiotic treatment contributes to the worldwide emergence of antibiotic-resistant strains of NTHi. Therefore, there is an urgent need to develop alternative therapeutic strategies for the treatment of NTHi infections based on understanding the molecular pathogenesis of these infections. Like most other bacterial infections, NTHi infection is characterized by inflammation, which is mainly mediated by NF-κB-dependent up regulation of pro-inflammatory mediators (35–38).
Based on the essential involvement of NF-κB in NTHi-induced inflammatory responses and the up-regulation of EVI1 by inflammatory stimuli in our preliminary gene profiling studies, we hypothesized that EVI1 negatively regulates NTHi-induced inflammation via inhibition of NF-κB activity. Here, we show that EVI1 negatively regulates NTHi-induced NF-κB activation and the subsequent inflammatory response by regulating the acetylation of NF-κB p65 subunit at lysine 310, thereby inhibiting the DNA binding activity of NF-κB to κB sites. Given the important role of NF-κB in host immune and inflammatory response against bacterial infections, the current studies will not only provide novel insights into a previously unidentified role of EVI1 in the regulation of NF-κB, but may also lead to the development of novel therapeutic strategies for controlling overactive inflammatory response.
Material and methods
Reagents and antibodies
Recombinant mouse TNF-α was purchased from Roche (Mannheim, Germany). Anti-phospho-IκBα, anti-IκBα, anti-acetyl-p65 (Lys310), anti-phospho-p65 S536, anti-IKKβ, anti-EVI1, and anti-CtBP2 antibodies were purchased from Cell Signaling (MA, USA). Anti-actin, anti-p65, anti-EVI1, anti-tubulin, and anti-TFIID were purchased from Santa Cruz (CA, USA). Anti-Flag was purchased from Sigma-Aldrich (MO, USA). Anti-acetyl-Lysine was purchased from Upstate (NY, USA).
Mice and animal experiments
Junbo mutant mice were generated by ENU mutagenesis screen (39, 40) as previously reported (13). Genotyping was performed by Single-nucleotide polymorphism (SNP) genotyping assay on tail-derived genomic DNA.
For the NTHi-induced lung inflammation model in WT and Junbo (Jbo/+) mice, anaesthetized mice were intratracheally inoculated with NTHi, and saline was inoculated as control. The inoculated mice were then sacrificed by intraperitoneal inoculation of 100 mg/kg sodium pentobarbital at 9hours and 24hours after NTHi inoculation. For histological analysis, dissected lung was inflated and fixed with 10% buffered formaldehyde, embedded in paraffin, and sectioned at 5-µM thickness. Sections were then stained and inspected. For polymorphonuclear neutrophil (PMN) analysis, bronchoalveolar lavage (BAL) was performed by cannulating the trachea with sterilized phosphate-buffered saline (PBS). Cells from BAL fluid were stained with Hemacolor (EM Science) after cytocentrifugation (Thermo Electronic Co.). To assess the mRNA expression of pro-inflammatory mediators, total RNA was extracted from the lung of NTHi- and saline-inoculated mice at the time points indicated above and the real-time quantitative RT-PCR was performed as described previously (41). All animal experiments were approved by the Institutional Animal Care and Use Committee at University of Rochester and Georgia State University.
Bacteria strain and culture
Clinical isolate of NTHi strain 12 was used in in vitro cell culture experiments and in vivo animal experiments (42). Bacteria were grown on chocolate agar plate at 37°C in an atmosphere of 5% CO2 overnight and inoculated in brain heart infusion (BHI) broth supplemented with 3.5 µg of NAD per ml and Hemin. After overnight incubation, bacteria were sub cultured into 5 ml of fresh BHI, and the log phase NTHi, monitored by measurement of optical density (OD) value, was washed and suspended in PBS for in vitro cell experiments and in isotonic saline for in vivo animal experiments. For in vitro experiments, the cells were treated with NTHi at a multiplicity of infection (MOI) of 1:25 for various times as indicated.
Cell culture
Human airway epithelial A549 cell, human middle ear epithelial HMEEC-1 cell, and mouse macrophage RAW 264.7 cell were maintained as described previously (41, 43, 44). Both p65 knockout (p65−/−) MEFs and p65−/− MEFs reconstituted with p65 WT or p65 K310R constructs were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). All cells were cultured under standard conditions (5% CO2 in air in a humidified environment at 37°C)
Plasmids, transfections and luciferase reporter Assay
Flag-tagged full-length EVI1 (1–1052) was kindly provided by Dr. Giuseppina Nucifora (12). The N781I point mutation of EVI1 (EVI1 MT) was constructed using QuikChange™ Site-Directed Mutagenesis Kit (Stratagene). p65 WT, p65-K310R, a constitutively active form of IKKβ (IKKβ CA), a transdominant-negative mutant form of IκBα (IκBα S32/36A), and NF-κB-luciferase reporter were described previously (45). Cells were co-transfected with or without NF-κB-luciferase reporter plasmid and various expression plasmids as indicated in the figure legends. Empty vector was used as a control and was also added where necessary to ensure an equivalent amount of input DNA. All transient transfections were carried out in triplicate using a TransIT-LT1 reagent (Mirus Co.) following the manufacturer's instructions. At 40 hours after transfection, cells were inoculated with NTHi for 5hours before cell lysis for luciferase assay as described previously. Data from all experiments are presented as the relative luciferase activity (mean ± S.D.) from at least three independent sets of experiments, each with triplicate measurements.
RNA-mediated interference
Human and mouse EVI1 small interfering RNA (siRNA) oligonucleotides were purchased from Dharmacon and SantaCruz. The siRNA was transfected into A549 cells and MEF cells using Lipofectamine-2000 reagent (Invitrogen) following the manufacturer's instructions.
RNA isolation and real-time quantitative RT-PCR (Q-PCR)
Total RNA was isolated with TRIzol reagent (Invitrogen) by following the manufacturer’s instructions. For the reverse transcription reaction, TaqMan reverse transcription reagents (Applied Biosystems) were used. In brief, the reverse transcription reaction was performed for 60 min at 37 °C, followed by 60 min at 42 °C by using oligo (dT) and random hexamers. PCR amplifications were performed by using SYBR Green Universal Master Mix. In brief, reactions were performed in duplicate containing 2 X Universal Master Mix, 1 µl of template cDNA and 100nM primers in a final volume of 12.5 µl, and they were analyzed in a 96-well optical reaction plate (Applied Biosystems). The relative quantities of mRNAs were obtained by using the comparative Ct method and were normalized with mouse glyceraldehydes-3-phosphate dehydrogenase (GAPDH) or human cyclophilin as an endogenous control. The primers for human TNF-α, IL-1β, IL-8, MCP-1, ICAM-1, cyclophilin, and mouse TNF-α, IL-1β, MIP-2, and GAPDH were described previously (41). The primer sequences of human EVI1 and mouse EVI1 were as follows: human EVI1, forward: 5-AGGCATCCTGCTGGTCTTACCTTT-3, reverse: 5-TGGTACAAGCCGGAAGGAAACAGA-3’; mouse EVI1: forward: 5-TGTCAGTACACCAAGTGGCAGTGA-3, reverse: 5-AAGCCAGATTCTGCAGAGGGCTTA-3’.
Western blot (WB) analysis and immunoprecipitation (IP)
WB was performed as described previously (41, 46, 47). Briefly, cell lysates were prepared in a buffer containing 20 mM Tris-HCl (pH8.0), 0.5 M NaCl, 0.25% Triton X-100, 1 mM EDTA, 1 mM EGTA, 10 mM glycerophosphate, 10 mM NaF, 300 µM Na3VO4, 1 mM benzamidine, 2 µM PMSF, 1 mM DTT and protease inhibitor cocktail (Sigma, MO. USA) by scraping, incubation on ice for 30 min, and centrifugation at 12,000g for 15 min. Supernatant was collected and then subjected to SDS-PAGE, and transferred to polyvinylidine difluoride (PVDF) membranes. The membrane was blocked with 5% nonfat milk, incubated in a 1:2,000 dilution of a primary antibody, and then incubated with 1:2,000 dilution of the corresponding secondary antibody. The membrane was reacted with chemiluminescence reagent ECL to visualize the blots. For IP, cell lysates were immunoprecipitated with 2–3 µg of the appropriate antibodies overnight at 4°C and then conjugated to protein A/G-aga rose beads for 2 hours at 4°C.
Electrophoretic mobility shift assay (EMSA)
Five to seven µg of nuclear extracts were prepared and non-radioactive EMSA was performed using an EMSA kit according to the manufacturer’s instructions (Pierce) as described previously (45). Oligonucleotide (oligo) 5'-AGTTGAGGGGACTTTCCCAGGC-3' was used as the consensus κB site-containing probe. Oligo was obtained from Integrated DNA Technologies and end-labeled with biotin-11-UTP using Biotin 3’ End DNA labeling Kit according to the manufacturer’s instructions (Pierce).
Chromatin Immunoprecipitation (ChIP) assay
ChIP assays were performed with ChIP-IT™ Express Enzymatic Kit from Active Motif (Carlsbad, CA). Primers used for putative κB sites of proximal IL-8 promoter (chr4:74,606,178-74,606,231 hg19) were as follows: forward: 5’-CATCAGTTGCAAATCGTGGA-3’; reverse: 5’-TGCACCCTCATCTTTTCATT-3’ (48). After transfection with EVI1 siRNA or control siRNA, cells were stimulated with NTHi or TNF-α for the times indicated in the figure. Cross-linked chromatin was obtained according to the manufacturer’s protocol. Coprecipitated chromatin DNA with anti-p65 antibody and the corresponding input DNA was amplified by PCR with the primers specific for IL-8 promoter, which contain the κB binding site.
Measurement of myeloperoxidase activity
Myeloperoxidase (MPO) activity in homogenates of whole lung was determined as described previously (49, 50). All reagents were purchased from Sigma (St. Louis, MO). Briefly, equal weights (100 mg wet weight) of lung from various groups were suspended in 1 ml of buffer (0.5% hexadecyltrimethyl ammonium bromide in 50 mM phosphate buffer, pH 6.0) and sonicated twice at 30 cycles for 30 seconds on ice. Homogenates were cleared by centrifugation at 12,000 rpm at 4°C, and the supernatants were stored at −80°C. Protein content in the samples was determined with a Bio-Rad (Hercules, CA) assay kit. The samples were incubated with the substrate o-dianisidine dihydrochloride. This reaction was carried out in a 96-well plate by adding 290 µl of 50 mM phosphate buffer, 3 µl of substrate solution (containing 20 mg/ml o-dianisidine dihydrochloride), and 3 µl of H2O2 (20 mM). Sample (10 µl) was added to each well to start the reaction. Standard MPO (Sigma) was used in parallel to determine MPO activity in the sample. The reaction was stopped by adding 3 µl of sodium azide (30%). Light absorbance was measured at 460 nm. MPO activity was determined by using the curve obtained from the standard MPO.
Statistical analysis
Data were shown as mean ± S.D. Statistical evaluation was done by unpaired Student's t test and p < 0.05 was considered as a significant difference.
Results
EVI1 negatively regulates bacteria-induced NF-κB-dependent inflammatory response in vitro
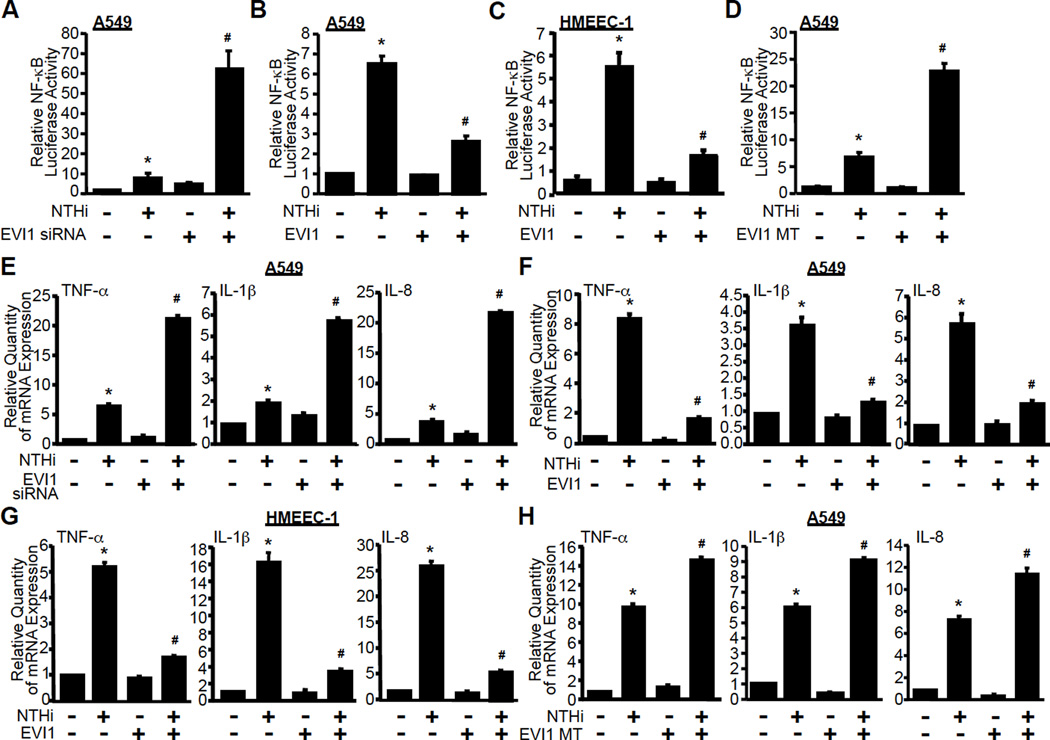
To test our hypothesis, we first investigated the role of EVI1 in NTHi-induced NF-κB luciferase promoter activity by using an EVI1 siRNA knockdown approach. Firstly, we confirmed the efficiency of EVI1-specific siRNA in reducing EVI1 expression in A549 cells. As expected, the expression of EVI1 at both mRNA and protein levels was markedly reduced by EVI1 siRNA (Supplementary Fig. 1A and 1B). Next, we determined if EVI1 knockdown affects NTHi-induced NF-κB activation in epithelial cells. As shown in Fig. 1A, NTHi-induced activation of NF-κB was greatly enhanced by EVI1 siRNA in human airway epithelial A549 cells, suggesting that EVI1 acts as a negative regulator of NTHi-induced NF-κB activation in airway epithelial cells. Next we sought to determine the generalizability of negative regulation of NF-κB activation by EVI1 by assessing its effect on NF-κB activation induced by TNF-α, a commonly used inflammation inducer. Similar to NTHi-induced NF-κB activation, TNF-α-induced NF-κB activation was also markedly enhanced by EVI1 siRNA in A549 cells (Supplementary Fig. 1C). We next confirmed the role of EVI1 in negatively regulating NTHi-induced NF-κB activation by overexpressing wild-type (WT) EVI1. Overexpression of EVI1 markedly reduced NTHi-induced NF-κB activation in A549 cells (Fig. 1B), human middle ear epithelial HMEEC-1 cells (Fig. 1C) and mouse macrophage RAW264.7 cells (Supplementary Fig. 1D). Similar results were also observed in TNF-α-induced NF-κB activation in A549 cells (Supplementary Fig. 1E). Together, these results demonstrate the negative role of EVI1 in regulating NTHi- and TNF-α–induced NF-κB activation.
Figure 1. EVI1 negatively regulates bacteria-induced NF-κB-dependent inflammatory response in airway epithelial and middle ear epithelial cells.
(A-D) A549 or HMEEC-1 cells were co-transfected with NF-κB-luciferase reporter plasmid together with either EVI1 siRNA (A), EVI1 WT (B & C), or EVI MT (D)construct for 40hours and then treated with or without NTHi for 5 hours. NF-κB-dependent promoter activity was measured by luciferase assay. (E-H) A549 or HMEEC-1 cells were transfected with either EVI1 siRNA (E), EVI1 WT (F & G), or EVI MT (H) construct for 40 hours and then treated with or without NTHi for 5 hours. Total RNA was extracted, and mRNA expression of TNF-α, IL-1β and IL-8 was measured by real-time quantitative RT-PCR (Q-PCR) assay. Data represent the mean ± SD of at least three independent experiments, and each experiment was performed in triplicate. *p < 0.05 vs. control; #p < 0.05 vs. NTHi alone.
Previously, a missense change in the C-terminal zinc finger region of the Evi1 gene was found to cause chronic middle ear inflammatory disease in mice (13). Thus, we generated EVI1 N782I mutation (EVI1 MT), a human homologue of mouse EVI1 N763I mutation (13), and determined the role of EVI1 N7821 mutation in NTHi-induced NF-κB activation by co-expressing theEVI1 MT. Consistent with our results obtained with EVI1 siRNA, overexpressing EVI1 MT also markedly enhanced NTHi-induced NF-κB activation in A549 cells (Fig. 1D) and RAW264.7 cells (Supplementary Fig. 1F), indicating that EVI1 MT indeed acts as a dominant-negative mutant. Similar results were also observed in TNF-α-induced NF-κB activation by co-expressing EVI1 MT in A549 cells (Supplementary Fig. 1G). Collectively, it is evident that EVI1 is a negative regulator of NF-κB activation induced not only by bacterial pathogen NTHi, but also by proinflammatory cytokine TNF-α.
Having identified EVI1 as a negative regulator of NF-κB activation, we next sought to determine if EVI1 also negatively regulates NF-κB-dependent transcription of several key proinflammatory mediators. As shown in Fig. 1E, Supplementary Fig. 1H, EVI1 siRNA greatly enhanced NTHi–induced mRNA expression of TNF-α, IL-1β, IL-8, MCP-1 and ICAM-1 in A549 cells. Similarly, TNF-α-induced mRNA expression of TNF-α, IL-1β, IL-8, MCP-1 and ICAM-1 was also potently enhanced by knockdown of EVI1 in A549 cells (Supplementary Fig. 1I and 1J). To further confirm the negative effect of EVI1 on NTHi- and TNF-α-induced up regulation of proinflammatory mediators, we evaluated the effects of overexpression of EVI1 WT and EVI1 MT expression plasmids on NTHi- and TNF-α-induced mRNA expression of TNF-α, IL-1β, and IL-8. As shown in Fig. 1F–H and Supplementary Fig. 1K, overexpressing EVI1 WT significantly reduced NTHi- and TNF-α-induced mRNA expression of TNF-α, IL-1β, and IL-8 in A549 and HMEEC-1 cells, whereas overexpressing EVI1 MT significantly increased NTHi-induced mRNA expression of TNF-α, IL-1β, and IL-8, similar to the findings with EVI1 siRNA. Taken together, these results confirmed our hypothesis that EVI1 is indeed a negative regulator of bacterial pathogen NTHi- and proinflammatory cytokine TNF-α-induced NF-κB-dependent inflammation in a variety of human cell types including respiratory epithelial cells and macrophage in vitro.
EVI1 negatively regulates bacteria-induced inflammation in mouse model in vivo
To further determine the in vivo biological role of EVI1 in negatively regulating inflammation, we explored the consequences of EVI1 mutations using the Junbo mutant mice (Jbo/+) generated from an ENU mutagenesis screen (13). Age- and sex-matched WT and Jbo/+ mice were intratracheally inoculated with NTHi, and the inflammatory response in the lung of infected mice was then monitored at 9hours or 24hours after NTHi inoculation. As shown in Fig. 2A, mRNA expression of NF-κB-regulated pro-inflammatory markers such as TNF-α, IL-1β and MIP-2 was markedly enhanced in lungs of Jbo/+ mice compared to that of WT littermate controls and was further enhanced with NTHi inoculation. Consistent with these results, histopathological analysis of the lung of NTHi-inoculated mice exhibited enhanced leukocyte infiltration in peribroncheal and interstitial areas in Jbo/+ mice compared with that in WT mice (Fig. 2B). Moreover, as shown in Fig. 2C, MPO activity, a key index of neutrophil activity, was also markedly enhanced in Jbo/+ mice compared with that in WT mice after NTHi inoculation. Similar results were also observed in the analysis of BAL fluids (Fig. 2D and 2E). Next, we confirmed if EVI1-mediated negative regulation of inflammatory response is dependent on the negative regulatory effect of EVI1 on NF-κB by assessing the effect of EVI1 siRNA on a transdominant-negative mutant of IκBα (IκBα S32/36A)-mediated inhibition of pro-inflammatory gene expression. As shown in Supplementary Fig. 1L, NTHi- and TNF-α-induced NF-κB-dependent promoter activity was enhanced by EVI1 siRNA and inhibited by overexpressing IκBα S32/36A. Moreover, EVI1 knockdown using EVI1 siRNA no longer enhanced NTHi- or TNF-α-induced NF-κB-dependent promoter activity in cells overexpressing IκBα S32/36A. Consistent with the findings from the luciferase assay, NTHi- and TNF-α-induced mRNA expression of TNF-α was enhanced by EVI1 knockdown and inhibited by overexpressing IκBα S32/36A. EVI1 siRNA no longer enhanced NTHi-induced mRNA expression of TNF-α and IL-8 in IκBα S32/36A overexpressing cells, suggesting that EVI1 negatively regulates NTHi-induced TNF-α mRNA expression via inhibition of NF-κB. (Supplementary Fig. 1M). Together, these results demonstrate that EVI1 is indeed a negative regulator of inflammatory response induced by bacterial pathogen NTHi and proinflammatory cytokine TNF-α in vitro and in vivo by negatively regulating NF-κB signaling.
Figure 2. EVI1 negatively regulates bacteria-induced inflammation in the lung of mice in vivo.
(A) Jbo/+ and wild-type littermate control (WT) mice were intratracheally inoculated with NTHi or saline as a control, and mRNA expression of TNF-α, IL-1β and MIP-2 was measured in the lung tissues of mice by Q-PCR assay. Data represent the mean ± SD of at least three independent experiments. (B)Representative slides of H&E staining with lung tissues from Jbo/+ and WT mice inoculated with NTHi or saline as a control (magnification ×200). (C) Lung tissues from Jbo/+ and WT mice inoculated with NTHi or saline were sonicated, and myeloperoxidase (MPO) activity was measured in the lung tissues homogenates. (D & E) Polymorphonuclear (PMN) cells were collected from the lung tissues of Jbo/+ and WT mice inoculated with NTHi or saline as a control by performing bronchoalveolar lavage (BAL). PMN numbers were counted (D) and stained with Hemacolor after cytocentrifugation (E). *p < 0.05 vs. saline; #p < 0.05 vs. NTHi in WT mice. All the Data are representative of at least three independent experiments.
EVI1 negatively regulates bacteria-induced activation of NF-κB by inhibiting its DNA-binding activity, likely independently of p65 nuclear translocation
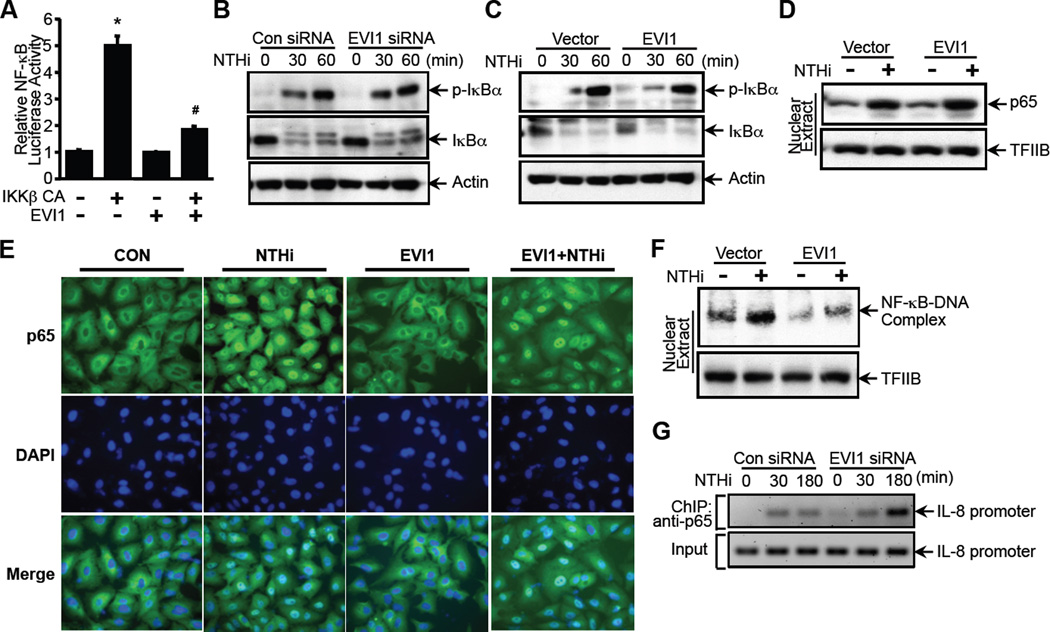
We next sought to determine how EVI1 inhibits NF-κB-dependent inflammation. The IκB kinase enzyme complex (IKKs) is part of the upstream NF-κB signal transduction cascade. The IκBα (inhibitor of kappa B) protein inactivate the NF-κB transcription factor by masking the nuclear localization signals (NLS) of NF-κB proteins and keeping them sequestered in an inactive state in the cytoplasm (20–22). Specifically, IKKs phosphorylate IκBα (23). This phosphorylation results in the degradation and dissociation of IκBα from NF-κB, which in turn leads to the nuclear translocation of NF-κB and the subsequent transcription of NF-κB-dependent genes. Among the three IKK isoforms IKKα, KKβ, and IKKγ, IKKβ plays a key role in NTHi-induced NF-κB activation by inducing phosphorylation of IκBα. Thus, we first determined if EVI1 negatively regulates NTHi-induced NF-κB activation by acting either upstream or downstream of IKKβ. To this end, A549 cells were co-transfected with a constitutively active form of IKKβ (IKKβ CA) alone or together with EVI1 WT construct. As shown in Fig. 3A, IKKβ CA significantly increased NF-κB luciferase activity, and overexpressing EVI1 markedly inhibited IKKβ CA-induced NF-κB luciferase activity. This result suggests that EVI1 negatively regulates NTHi-induced NF-κB activation by acting either at the level of or downstream of IKKβ.
Figure 3. EVI1 negatively regulates bacteria-induced activation of NF-κB by inhibiting its DNA-binding activity, likely independently of p65 nuclear translocation.
(A) A549 cells were co-transfected with NF-κB-luciferase reporter plasmid together with a constitutively active form of IKKβ (IKKβ CA), and NF-κB-dependent promoter activity was measured by luciferase assay. (B & C) A549 cells transfected either with EVI1 siRNA (B) or EVI1 WT construct (C) were treated with NTHi for the time indicated in the figure, and analyzed by western blot analysis with the indicated antibodies. (D) A549 cells co-transfected with p65 and EVI1 WT or control vector were treated with NTHi, and nuclear extracts were analyzed by western blot analysis with the indicated antibodies. (E) A549 cells co-transfected with EVI1 and p65 were treated with NTHi and immunostained with DAPI and anti-p65 antibodies. Cells were visualized by fluorescence microscopy. (F) A549 cells co-transfected with p65 and EVI1 WT or control vector were treated with NTHi, and DNA-binding activity of NF-κB was assessed by electrophoretic mobility shift assay (EMSA). (G) A549 cells transfected with EVI1 siRNA or control siRNA were treated with NTHi for the times indicated in the figure, and chromatin immunoprecipitation (ChIP) assay was conducted using antibody against p65 and analyzed by PCR using specific primers for the IL-8 promoter sequences spanning the κB binding sites. *p < 0.05 vs. control; #p < 0.05 vs. IKKβ CA alone. Data are representative of at least three or more independent experiments.
Because phosphorylation and degradation of IκBα and subsequent nuclear translocation of NF-κB are critical for NF-κB activation, we next determined if EVI1 negatively regulates NTHi-induced NF-κB activation by altering phosphorylation and degradation of IκBα. As shown in Fig. 3B and 3C, neither knockdown of EVI1 expression by siRNA nor overexpressing EVI1 exhibited any significant inhibitory effect on NTHi-induced IκBα phosphorylation and degradation. These results thus led us to determine if EVI1 inhibits NTHi-induced NF-κB activation by regulating nuclear translocation of p65 by performing western blot analysis of the nuclear extract. As shown in Fig. 3D, EVI1 exhibited no inhibitory effect on NTHi-induced p65 translocation, which was further confirmed by p65 immunofluorescence staining (Fig. 3E). Similarly, EVI1 also exhibited no inhibitory effect on TNF-α-induced p65 translocation (Supplementary Fig. 2A and 2B). These results thus led us conclude that the negative regulation of NF-κB activation by EVI1 may occur at the level further downstream of p65 nuclear translocation.
Because the DNA-binding activity of the NF-κB complex is critical for NF-κB to exert its transcriptional regulatory activity, we next investigated the effect of EVI1 on NTHi-induced DNA-binding activity of NF-κB by performing electrophoretic mobility shift assay (EMSA). As shown in Fig. 3F, overexpressing EVI1 significantly deceased NTHi-induced DNA-binding activity of NF-κB. Moreover, NTHi- and TNF-α-induced DNA-binding activity of NF-κB was further enhanced and remained sustained with EVI1 knockdown using EVI1 siRNA as assessed by performing ChIP assays (Fig. 3G, Supplementary Fig. 2C). Taken together, these data suggest that EVI1 negatively regulates NTHi-induced NF-κB activation by inhibiting its DNA-binding activity, likely independent of p65 nuclear translocation.
NTHi induces direct interaction of EVI1 with p65
We have found that EVI1 negatively regulates NTHi-induced NF-κB activation and subsequent inflammatory responses by inhibiting DNA-binding activity of the NF-κB complex. Next, we sought to determine if NTHi induces interaction between EVI1 and p65, the major subunit of NF-κB complex, by performing co-immunoprecipitation analysis of interaction between EVI1 and p65. As shown in Fig. 4A, NTHi markedly increased the interaction of EVI1 with p65 in A549 cells. This interaction was confirmed by reverse immunoprecipitation as indicated in Fig. 4B. To further confirm the interaction of EVI1 with p65, triple immunostaining with antibodies against DAPI, EVI1 and p65 was performed. As shown in Fig. 4C and Supplementary Fig. 3A, the majority of EVI1 is mainly localized in the nucleus, whereas p65 is mainly localized in the cytoplasm in the absence of NTHi but is translocated to the nucleus and co-localized with EVI1 in the nucleus in response to NTHi. We next determined if endogenous EVI1 directly interacts with endogenous p65 and if such a direct interaction is further enhanced upon NTHi and TNF-α treatment by performing co-immunoprecipitation analysis. As shown in Fig. 4D, endogenous EVI1 indeed directly interacts with endogenous p65, and both NTHi and TNF-α treatment markedly enhanced their direct interaction. Since EVI1 has been shown to interact directly with the known transcriptional repressor CtBP2 via two CtBP-binding consensus motifs (10, 11), we determined if NTHi induces the formation of p65/CtBP2 repressor complexes and EVI1 knockdown using EVI1 siRNA inhibits their interaction, thereby resulting in enhanced activation of NF-κB. As shown in Supplementary Fig. 3B, protein-protein interaction between p65 and CtBP2 was observed in cells treated with NTHi, and EVI1 knockdown using EVI1 siRNA inhibited their interaction. Similar results were also observed in cells treated with TNF-α (Data not shown). However, the data with TNF-α was not as definitive as that with NTHi. These results thus suggest that NTHi or TNF-α induces direct physical interaction of EVI1 with p65 in the nucleus, and EVI1 promotes the association of CtBP2 with p65, which may in turn lead to the inhibition of NF-κB activation.
Figure 4. EVI1 negatively regulates bacteria-induced NF-κB activation via direct interaction with p65.
(A & B) A549 cells co-transfected with Flag-EVI1 and p65 were treated with NTHi for the times indicated in the figure, and the proteins were then immunoprecipitated with anti-Flag antibody (A) or anti-p65 antibody (B). Immunoprecipitations were then analyzed by western blot analysis with the indicated antibodies. (C) A549 cells co-transfected with Flag-EVI1 and p65 were treated with NTHi and then triple-immunostained with DAPI, anti-p65 and anti-Flag antibodies. Slides were visualized by fluorescence microscopy. (D) A549 cells were treated with NTHi or TNF-α for the times indicated in the figure, and the proteins were then immunoprecipitated with anti-p65 antibody and analyzed by western blot analysis with the indicated antibodies. Data are representative of at least three independent experiments.
EVI1 negatively regulates NTHi-induced NF-κB activation via inhibition of p65 acetylation at lysine 310
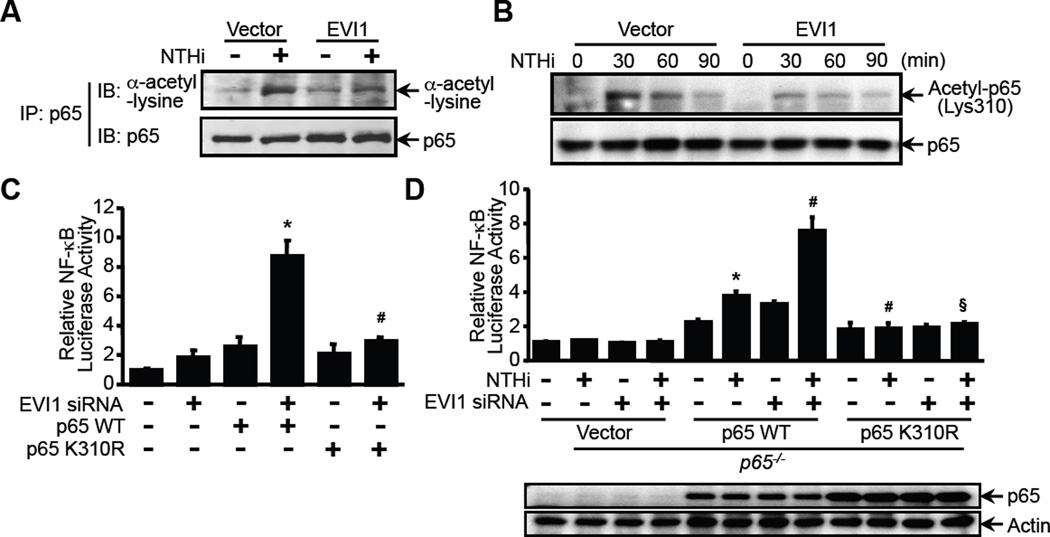
Having demonstrated that EVI1 directly interacts with p65 and inhibits NTHi-induced DNA-binding activity, the molecular mechanism underlying the inhibition of DNA-binding activity of p65 by EVI1 is still unknown. Post-translational modifications, particularly acetylation, have been shown to play a critical role in NF-κB activation by enhancing the DNA-binding activity of p65 to the κB site (45). Because EVI1 was known to potentially recruit both HDACs and p300/pCAF (12), it is logical to hypothesize that EVI1 may negatively regulate NTHi-induced DNA-binding activity of NF-κB by inhibiting acetylation of p65. We thus determined if EVI1 inhibits NTHi-induced p65 acetylation by assessing the effect of EVI1 overexpression in p65-overexpressing cells. Cells were first transfected with p65 WT with or without EVI1, and NTHi-induced p65 acetylation was then measured (Fig. 5A and 5B). Interestingly, as shown in Fig. 5A, overexpressing EVI1 markedly inhibited NTHi-induced p65 acetylation. Because acetylation of p65 at lysine 310 (p65 K310) is required for full transactivation activity of the NF-κB complex, we next determined if EVI1 inhibits the acetylation of p65 at K310(24). As shown in Fig. 5B, the acetylation of p65 at K310 induced by NTHi is markedly reduced by overexpressing EVI1. To further confirm the functional involvement of p65 K310, A549 cells were transfected with EVI1 siRNA alone or together with p65 WT or p65 K310R (a p65 mutant at K310) and NF-κB luciferase activity was assayed. As shown in Fig. 5C, knockdown of EVI1 using EVI1 siRNA markedly enhanced NF-κB activation in cells co-transfected with WT p65 but not with p65 K310R. To further confirm the functional involvement of p65 K310 in inhibition of NF-κB activation by EVI1, we assessed the effect of EVI1 knockdown on NTHi-induced NF-κB activity in p65−/− MEF cells that were reconstituted with either WT p65 expression plasmid or p65-K310R mutant. The expression of p65 WT and p65 K310R in p65−/− cells was first confirmed by WB analysis (Fig. 5D, bottom panel). As shown in Fig. 5D (upper panel), EVI1 knockdown markedly enhanced NTHi-induced NF-κB activation inp65−/− MEF cells that were reconstituted with WT p65 expression plasmid but not with p65-K310R mutant.
Figure 5. EVI1 negatively regulates bacteria-induced NF-κB activation via inhibition of p65 acetylation at lysine 310.
(A & B) A549 cells co-transfected with EVI1 and p65 were treated with NTHi, and the cell lysates were immnoprecipitated with anti-p65 antibody and analyzed by western blot analysis with the indicated antibodies.(C) A549 cells were co-transfected with NF-κB-luciferase reporter plasmid together with EVI1 siRNA and p65 WT or p65 K310R constructs, and NF-κB-dependent promoter activity was measured by luciferase assay.*p < 0.05 vs. p65 WT alone; #p < 0.05 vs. p65 WT with EVI1 siRNA.(D) p65−/− MEF cells reconstituted with vector, p65 WT, or p65 K310R were co-transfected with NF-κB-luciferase reporter plasmid together with EVI1 siRNA or control vector. Cells were treated with NTHi, and cell lysate were analyzed by western blot analysis with anti-p65 and tubulin antibodies (bottom panel). NF-κB-dependent promoter activity was measured by luciferase assay (upper panel). *p < 0.05 vs. NTHi in p65−/− MEF cells; #p < 0.05 vs. NTHi in p65−/− MEF cells reconstituted with p65 WT; §p < 0.05 vs. NTHi treatment with EVI1 siRNA in p65−/− MEF cells reconstituted with p65 WT. For western blot, data are representative of three independent experiments. For luciferase assay, data represent the mean ± SD of at least three independent experiments, and each experiment was performed in triplicate.
Next, we determined if EVI1 regulates endogenous acetylation of p65 by assessing the effect of EVI1 siRNA on NTHi- and TNF-α-induced acetylation of endogenous p65. As shown in Supplementary Fig. 4, NTHi-induced acetylation of endogenous p65, and knockdown of EVI1 using EVI1 siRNA further enhanced and led to the sustained acetylation of p65. It has been previously reported that acetylation of p65 is dependent on p65 phosphorylation (51). Thus we further determined if EVI1 regulates acetylation of p65 by regulating p65 phosphorylation. As shown in Supplementary Fig. 4A, NTHi-induced p65 acetylation was further enhanced by EVI1 siRNA, whereas phosphorylation of p65 at Ser 536 remained unaffected by EVI1 siRNA. Similar results were also found in TNF-α treated cells (Supplementary Fig. 4B). Taken together, our data demonstrate that EVI1 acts as a negative regulator of NTHi-induced DNA-binding activity of NF-κB by inhibiting acetylation of p65 at the K310 site, thereby leading to the inhibition of NF-κB-dependent inflammation.
Expression of EVI1 itself is also induced by NTHi both in vitro and in vivo
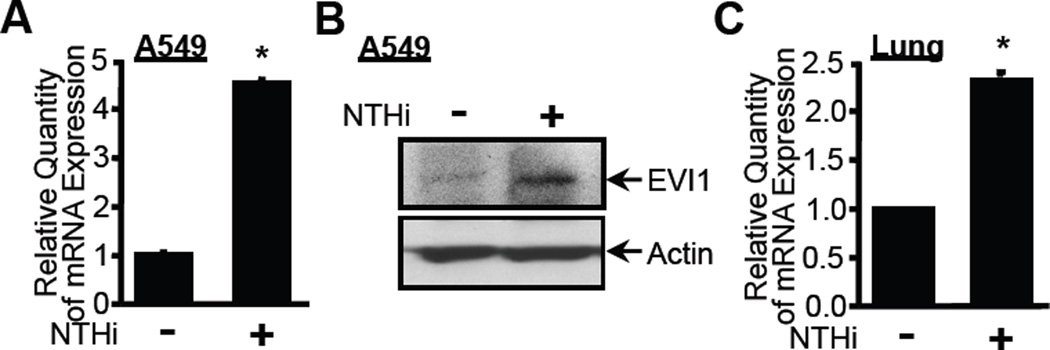
Negative feedback regulation plays a critical role in preventing overactive and detrimental inflammatory response in a variety of human inflammatory diseases including infectious diseases (52). It is well known that many genes involved in inflammatory response undergo changes in expression pattern in response to inflammatory stimuli such as bacteria. Because EVI1 negatively regulates TNF-α- and NTHi-induced NF-κB-dependent inflammatory response, we hypothesized that EVI1 itself may also be induced by NTHi, which may in turn lead to the inhibition of NTHi-induced inflammatory response, thereby preventing an overactive inflammatory response. To test our hypothesis, we determined if NTHi induces EVI1 expression. As shown in Fig. 6A, EVI1 expression at the mRNA level was markedly up-regulated by NTHi in A549 cells. The induction of EVI1 by NTHi was also confirmed at the protein level by performing western blot analysis (Fig. 6B). Moreover, up-regulation of EVI1 by NTHi was also observed in the lung of WT mice inoculated with NTHi (Fig. 6C). Together, these data suggest that EVI1 itself is also induced by NTHi, which in turn leads to the inhibition of NTHi-induced inflammatory response.
Figure 6. Expression of EVI1 itself is induced by NTHi both in vitro and in vivo.
(A) A549 cells were treated with NTHi, and mRNA expression of EVI1 was measured by Q-PCR analysis. (B) A549 cells were treated with NTHi, and expression of EVI1 at protein level was assessed by western blot analysis with anti-EVI1 antibody. (C) WT mice were intratracheally inoculated with NTHi, and mRNA expression of Evi1 was measured in the lung tissues of mice by Q-PCR analysis. Data represent the mean ± S.D (n = 3). *p < 0.05 vs. control.
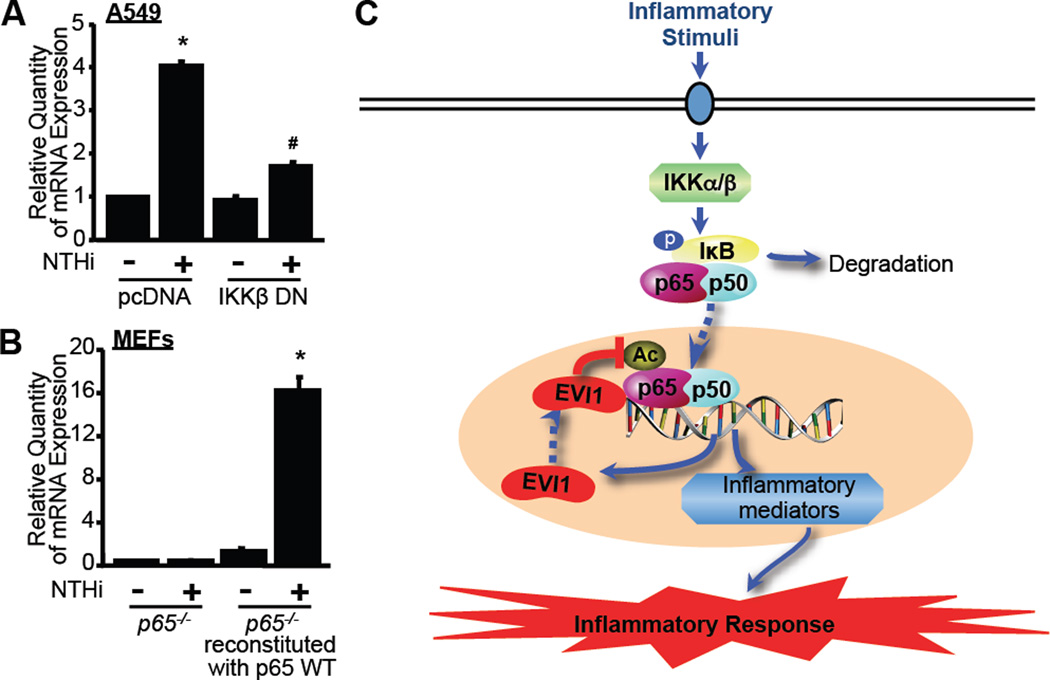
EVI1 is induced by NTHi via an IKKβ-p65-dependent mechanism, thereby unveiling a novel negative feedback loop of NF-κB-dependent inflammation
Because our data indicate that EVI1 is induced by NTHi, a potent inducer for inflammation, we sought to determine if NTHi-induced EVI1 expression is also controlled by IKKβ-NF-κB pathway. Interestingly, as shown in Fig. 7A, NTHi-induced EVI1 expression was markedly inhibited by overexpressing a dominant-negative mutant form of IKKβ (IKKβ DN) in A549 cells. We next investigated the role of p65 in NTHi-induced EVI1 expression. As shown in Fig. 7B, no induction of EVI1 expression by NTHi was observed in p65−/− MEF cells. In contrast, NTHi markedly induced EVI1 expression in p65−/− MEF cells that were reconstituted with p65 WT construct (Fig. 7B). These data thus unveils a novel EVI1-dependent negative feedback loop controlling NF-κB-dependent inflammation.
Figure 7. EVI1 induction by NTHi is IKKβ- and p65-dependent.
(A) A549 cells transfected with a dominant-negative mutant form of IKKβ (IKKβ DN) or pcDNA as a control vector were treated with NTHi, and mRNA expression of EVI1 was measured by Q-PCR analysis. (B) p65−/− MEF cells or p65−/− MEF cells reconstituted with p65 WT were treated with NTHi, and mRNA expression of Evi1 was measured by Q-PCR analysis. Data represent the mean ± SD of at least three independent experiments. *p < 0.05 vs. control in pcDNA (A) and control in p65−/− MEF cells reconstituted with p65 WT (B); #p < 0.05 vs. NTHi in pcDNA. (C) Schematic representation describing how EVI1 acts as an inducible negative regulator for inflammation induced either by bacterial pathogen NTHi or cytokine TNF-α. As indicated, EVI1 expression is induced by inflammatory stimuli and, in turn, negatively regulates NTHi- or TNF-α-induced NF-κB activation and subsequent inflammation by directly binding to p65 and inhibiting acetylation of p65 at the lysine 310 residue. Abbreviations used: IKKα/β: IκB kinase α/β; p: phosphorylation; Ac: acetylation; EVI1: Ecotropic Viral Integration Site 1.
Discussion
In the present study, we provide direct evidence that EVI1 acts as a negative feedback regulator of NTHi-induced NF-κB-dependent inflammation in human respiratory epithelial cell in vitro and in mouse model of lung inflammation in vivo. Interestingly, expression of EVI1 itself is also induced by NTHi in an NF-κB-dependent manner, which, in turn, leads to the inhibition of acetylation of p65 at K310 site, thereby leading to the decreased DNA-binding activity of NF-κB and subsequent inhibition of inflammatory response (Fig. 7C). This study will not only provide new insights into the novel role of EVI1 in the regulation of NF-κB-dependent inflammatory response through a negative feedback loop, but may also lead to the development of new therapeutic interventions for controlling inflammation.
Of particular interest in this study is the identification of EVI1 as a novel negative feedback regulator of NF-κB-dependent inflammatory response in vitro and in vivo. EVI1 was originally identified as a proto-oncogene and zinc finger transcription factor with important roles in normal development and leukemogenesis. As a proto-oncogene, the aberrant expression of EVI1is associated with AML (2, 4), MDS (5), and CML (3). High expression of EVI1 is detectable in around 8% of AML cases and is a poor prognostic indicator (4, 53). As a transcription factor, EVI1 has the potential to interact with both co-repressors and co-activators and is involved in many signaling pathways for both co-repression and co-activation of cell cycle genes (6–9). It has also been demonstrated that Smad3 interacts with the first zinc finger domain of EVI1. As a consequence, EVI1 disturbs the TGF-β signaling pathway, which is known to be a negative regulator for cellular growth and differentiation (54). However, there has been no report demonstrating a role of EVI1 in regulating NF-κB activity and the subsequent inflammatory response. In the present study, we show that EVI1 negatively regulates NTHi- and TNF-α-induced NF-κB activation and inflammatory response in vitro and in vivo. This finding is in line with the previous study demonstrating that the loss of function mutation in the second zinc finger domain of Evi1 in the Junbo mouse is associated with the predisposition to chronic OM (13). Interestingly, human EVI1 MT that is homologous to the point mutation of Junbo mouse indeed acts as a dominant-negative mutant in our in vitro studies. Thus, our results unveil a previously unrecognized role of EVI1 as a negative feedback regulator of NF-κB-dependent inflammation in upper respiratory inflammatory diseases.
In the current study, the generalizability of our finding that EVI1 acts as a negative regulator of NF-κB-dependent inflammation was further confirmed by evaluating the negative regulation of TNF-α-induced NF-κB-dependent inflammation by EVI1. Since we found that EVI1 inhibits inflammation by directly targeting NF-κB, it seems reasonable to postulate that EVI1 may act as a negative regulator of NF-κB-dependent inflammation induced by other inflammatory stimuli as well.
Another major interesting finding in this study is that EVI1 inhibits NTHi-induced DNA-binding activity of NF-κB by directly interacting with p65 and also inhibiting the acetylation of p65. The NF-κB family of transcription factors consists of five members in mammalian cells: RelA (p65), RelB, c-Rel, p50/p105, and p52/p100. NF-κB is a dimeric transcription factor consisting of homo- or heterodimers of Rel-related proteins (55). The most important heterodimer consisting of two subunits, RelA/p65 and p50, is involved in the regulation of a variety of physiologic and pathologic processes, including proliferation, differentiation, survival, tumorigenesis and inflammation (55). In the inactive state, NF-κB resides in the cytoplasm and forms a multi-protein complex with an inhibitory subunit, inhibitor of NF-κB (IκB). Upon activation by external stimuli, the inflammatory signal converges on and activates a set of IκB kinases known as the IκB kinase (IKK) complex, which phosphorylate NF-κB, targeting it for proteasomal degradation. Once released from the IκBα complex, NF-κB translocates to the nucleus, where it binds to DNA and promotes the transcription of pro-inflammatory mediators, including cytokines, chemokines, and adhesion molecules, thereby playing a critical role in mediating inflammatory response (19). Our studies indicate that EVI1 negatively regulates NTHi-induced NF-κB activation via a mechanism independent of IκBα phosphorylation or degradation (Fig. 3B and 3C), and p65 nuclear translocation (Fig. 3D and 3E), but dependent on the inhibition of DNA-binding activity of the NF-κB complex (Fig. 3F and 3G).
Recent studies have suggested that degradation of IκBα and nuclear translocation of NF-κB are insufficient to induce a maximal NF-κB-dependent transcriptional activity. Rather, the NF-κB complex must undergo additional post-translational modifications. Among all known post-translational modifications of p65, acetylation of p65 has been shown to play a critical role in controlling the duration and strength of NF-κB signaling and regulating various biological functions of NF-κB, including DNA binding, transactivation and association with the inhibitor IκBα (38, 51, 56). The p300 and CBP have been shown to play major roles in the acetylation of p65, while HDAC3 appears to be critical for the deacetylation of p65. Endogenous p65 is acetylated in a stimulus-coupled manner after activation of cells with NTHi or other stimuli (45). Interestingly, our data demonstrate that EVI1 directly binds to p65 and inhibits p65 acetylation (Fig. 4 and 5). Site-specific acetylation of p65 regulates discrete biological activity of the NF-κB complex. It has been previously reported that acetylation of p65 at the K310 residue is important for the transcriptional activity of NF-κB because acetylation of K310 is required for optimal p65 transcriptional activity by enhancing NF-κB/DNA binding and attenuating its interaction with IκBα (24). In our study, we found that overexpression of EVI1 significantly inhibits NTHi-induced acetylation of p65 at the K310 site. Further analysis using a site-specific mutant of 310 and p65−/− MEFs reconstituted with either WT p65 or p65-K310R mutant confirmed that the lysine 310 residue is critical for EVI1-regulated acetylation of p65 by NTHi. Thus, our studies provide evidence that EVI1, a proto-oncogene, negatively regulates NF-κB-dependent inflammation by inhibiting p65 acetylation.
Of additional interest in this study is that EVI1 itself is markedly induced by NTHi in an NF-κB-dependent manner, which in turn leads to the inhibition of the NTHi-induced inflammatory response, thereby preventing an overactive inflammatory response. The inflammatory response triggered by bacteria is a protective attempt by the host to remove the injurious stimuli and to initiate the healing process, but excessive inflammatory response is detrimental to the host, due to severe tissue damage (57, 58). To avoid overactive and detrimental inflammatory response in infectious disease, bacteria-induced inflammatory response must be tightly regulated. The host has evolved a variety of strategies to prevent detrimental inflammatory response during bacterial infections. Bacteria-induced negative feedback regulation is thought to play a critical role in preventing overactive inflammatory response by bacterial pathogens. However, despite the importance of tight regulation in preventing overactive inflammatory response, the molecular mechanisms underlying the negative feedback regulation of inflammation in the pathogenesis of NTHi infection remain largely unknown. In this study, we found that EVI1 acts as an inducible negative feedback regulator of NTHi-induced NF-κB-dependent inflammatory response in vitro and in vivo. Thus, up regulating EVI1 may represent an interesting therapeutic strategy for controlling overactive inflammation in bacterial infections.
Supplementary Material
Acknowledgement
We are grateful to Drs. G. Nucifora and L.F. Chen for kindly providing plasmid constructs and cells.
Abbreviations used in this manuscript
- EVI1
Ecotropic viral integration site 1
- HDAC
histone deacetylase complex
- NTHi
Nontypeable Haemophilus influenzae
Footnotes
Disclosures
The authors have no financial conflict of interest.
This work was supported by grants from National Institute of Health DC005843 andDC004562 (to J.-D.L), DC010048 (to X.X.), by American Heart Association 10SDG2630077 (to J.H.L.), by Basic Science Research Program from the National Research Foundation of Korea 2011-0026075(to C.H.W.), and by funding from the Medical Research Council, UK (to M.T.C and S.D.M.B).
References
- 1.Morishita K, Parker DS, Mucenski ML, Jenkins NA, Copeland NG, Ihle JN. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- 2.Morishita K, Parganas E, William CL, Whittaker MH, Drabkin H, Oval J, Taetle R, Valentine MB, Ihle JN. Activation of EVI1 gene expression in human acute myelogenous leukemias by translocations spanning 300–400 kilobases on chromosome band 3q26. Proc Natl Acad Sci U S A. 1992;89:3937–3941. doi: 10.1073/pnas.89.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitani K, Ogawa S, Tanaka T, Miyoshi H, Kurokawa M, Mano H, Yazaki Y, Ohki M, Hirai H. Generation of the AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. EMBO J. 1994;13:504–510. doi: 10.1002/j.1460-2075.1994.tb06288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lugthart S, van Drunen E, van Norden Y, van Hoven A, Erpelinck CA, Valk PJ, Beverloo HB, Lowenberg B, Delwel R. High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008;111:4329–4337. doi: 10.1182/blood-2007-10-119230. [DOI] [PubMed] [Google Scholar]
- 5.Alessandrino EP, Amadori S, Cazzola M, Locatelli F, Mecucci C, Morra E, Saglio G, Visani G, Tura S. Myelodysplastic syndromes: recent advances. Haematologica. 2001;86:1124–1157. [PubMed] [Google Scholar]
- 6.Yatsula B, Lin S, Read AJ, Poholek A, Yates K, Yue D, Hui P, Perkins AS. Identification of binding sites of EVI1 in mammalian cells. J Biol Chem. 2005;280:30712–30722. doi: 10.1074/jbc.M504293200. [DOI] [PubMed] [Google Scholar]
- 7.Morishita K, Suzukawa K, Taki T, Ihle JN, Yokota J. EVI-1 zinc finger protein works as a transcriptional activator via binding to a consensus sequence of GACAAGATAAGATAAN1–28 CTCATCTTC. Oncogene. 1995;10:1961–1967. [PubMed] [Google Scholar]
- 8.Delwel R, Funabiki T, Kreider BL, Morishita K, Ihle JN. Four of the seven zinc fingers of the Evi-1 myeloid-transforming gene are required for sequence-specific binding to GA(C/T)AAGA(T/C)AAGATAA. Mol Cell Biol. 1993;13:4291–4300. doi: 10.1128/mcb.13.7.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins AS, Fishel R, Jenkins NA, Copeland NG. Evi-1, a murine zinc finger proto-oncogene, encodes a sequence-specific DNA-binding protein. Mol Cell Biol. 1991;11:2665–2674. doi: 10.1128/mcb.11.5.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer S, Brouillet JP, Kilbey A, Fulton R, Walker M, Crossley M, Bartholomew C. Evi-1 transforming and repressor activities are mediated by CtBP co-repressor proteins. J Biol Chem. 2001;276:25834–25840. doi: 10.1074/jbc.M102343200. [DOI] [PubMed] [Google Scholar]
- 11.Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty S, Senyuk V, Sitailo S, Chi Y, Nucifora G. Interaction of EVI1 with cAMP-responsive element-binding protein-binding protein (CBP) and p300/CBP-associated factor (P/CAF) results in reversible acetylation of EVI1 and in co-localization in nuclear speckles. J Biol Chem. 2001;276:44936–44943. doi: 10.1074/jbc.M106733200. [DOI] [PubMed] [Google Scholar]
- 13.Parkinson N, Hardisty-Hughes RE, Tateossian H, Tsai HT, Brooker D, Morse S, Lalane Z, MacKenzie F, Fray M, Glenister P, Woodward AM, Polley S, Barbaric I, Dear N, Hough TA, Hunter AJ, Cheeseman MT, Brown SD. Mutation at the Evi1 locus in Junbo mice causes susceptibility to otitis media. PLoS Genet. 2006;2:e149. doi: 10.1371/journal.pgen.0020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tetley TD. Inflammatory cells and chronic obstructive pulmonary disease. Curr Drug Targets Inflamm Allergy. 2005;4:607–618. doi: 10.2174/156801005774912824. [DOI] [PubMed] [Google Scholar]
- 15.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 16.Kleiman A, Tuckermann JP. Glucocorticoid receptor action in beneficial and side effects of steroid therapy: lessons from conditional knockout mice. Mol Cell Endocrinol. 2007;275:98–108. doi: 10.1016/j.mce.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Manson SC, Brown RE, Cerulli A, Vidaurre CF. The cumulative burden of oral corticosteroid side effects and the economic implications of steroid use. Respir Med. 2009;103:975–994. doi: 10.1016/j.rmed.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 19.Rothwarf DM, Karin M. The NF-kappa B activation pathway: a paradigm in information transfer from membrane to nucleus. Sci STKE. 1999;1999:RE1. doi: 10.1126/stke.1999.5.re1. [DOI] [PubMed] [Google Scholar]
- 20.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 21.Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M. Identification and characterization of an IkappaB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 23.Karin M. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene. 1999;18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 24.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem. 2003;278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 26.Hoberg JE, Popko AE, Ramsey CS, Mayo MW. IkappaB kinase alpha-mediated derepression of SMRT potentiates acetylation of RelA/p65 by p300. Mol Cell Biol. 2006;26:457–471. doi: 10.1128/MCB.26.2.457-471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 28.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimahara A, Yamakawa N, Nishikata I, Morishita K. Acetylation of lysine 564 adjacent to the C-terminal binding protein-binding motif in EVI1 is crucial for transcriptional activation of GATA2. J Biol Chem. 285:16967–16977. doi: 10.1074/jbc.M110.102046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuklinska D, Kilian M. Relative proportions of Haemophilus species in the throat of healthy children and adults. Eur J Clin Microbiol. 1984;3:249–252. doi: 10.1007/BF02014895. [DOI] [PubMed] [Google Scholar]
- 31.Murphy TF. Bacterial otitis media: pathogenetic considerations. Pediatr Infect Dis J. 2000;19:S9–15. doi: 10.1097/00006454-200005001-00003. discussion S15–16. [DOI] [PubMed] [Google Scholar]
- 32.Bluestone CD. Otitis media in children: to treat or not to treat? N Engl J Med. 1982;306:1399–1404. doi: 10.1056/NEJM198206103062305. [DOI] [PubMed] [Google Scholar]
- 33.Foxwell AR, Kyd JM, Cripps AW. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol Mol Biol Rev. 1998;62:294–308. doi: 10.1128/mmbr.62.2.294-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy TF, Sethi S. Bacterial infection in chronic obstructive pulmonary disease. Am Rev Respir Dis. 1992;146:1067–1083. doi: 10.1164/ajrccm/146.4.1067. [DOI] [PubMed] [Google Scholar]
- 35.Murphy TF. The role of bacteria in airway inflammation in exacerbations of chronic obstructive pulmonary disease. Curr Opin Infect Dis. 2006;19:225–230. doi: 10.1097/01.qco.0000224815.89363.15. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe T, Jono H, Han J, Lim DJ, Li JD. Synergistic activation of NF-kappaB by nontypeable Haemophilus influenzae and tumor necrosis factor alpha. Proc Natl Acad Sci U S A. 2004;101:3563–3568. doi: 10.1073/pnas.0400557101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 38.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 39.Hrabe de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, Reis A, Richter T, Alessandrini F, Jakob T, Fuchs E, Kolb H, Kremmer E, Schaeble K, Rollinski B, Roscher A, Peters C, Meitinger T, Strom T, Steckler T, Holsboer F, Klopstock T, Gekeler F, Schindewolf C, Jung T, Avraham K, Behrendt H, Ring J, Zimmer A, Schughart K, Pfeffer K, Wolf E, Balling R. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 40.Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, Washbourne R, Hough T, Greenaway S, Hewitt M, Liu X, McCormack S, Pickford K, Selley R, Wells C, Tymowska-Lalanne Z, Roby P, Glenister P, Thornton C, Thaung C, Stevenson JA, Arkell R, Mburu P, Hardisty R, Kiernan A, Erven A, Steel KP, Voegeling S, Guenet JL, Nickols C, Sadri R, Nasse M, Isaacs A, Davies K, Browne M, Fisher EM, Martin J, Rastan S, Brown SD, Hunter J. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- 41.Jeon KI, Xu X, Aizawa T, Lim JH, Jono H, Kwon DS, Abe J, Berk BC, Li JD, Yan C. Vinpocetine inhibits NF-kappaB-dependent inflammation via an IKK-dependent but PDE-independent mechanism. Proc Natl Acad Sci U S A. 2010;107:9795–9800. doi: 10.1073/pnas.0914414107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shuto T, Xu H, Wang B, Han J, Kai H, Gu XX, Murphy TF, Lim DJ, Li JD. Activation of NF-kappa B by nontypeable Hemophilus influenzae is mediated by toll-like receptor 2-TAK1-dependent NIK-IKK alpha /beta-I kappa B alpha and MKK3/6-p38 MAP kinase signaling pathways in epithelial cells. Proc Natl Acad Sci U S A. 2001;98:8774–8779. doi: 10.1073/pnas.151236098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim JH, Kim HJ, Komatsu K, Ha U, Huang Y, Jono H, Kweon SM, Lee J, Xu X, Zhang GS, Shen H, Kai H, Zhang W, Xu H, Li JD. Differential regulation of Streptococcus pneumoniae-induced human MUC5AC mucin expression through distinct MAPK pathways. Am J Transl Res. 2009;1:300–311. [PMC free article] [PubMed] [Google Scholar]
- 44.Komatsu K, Jono H, Lim JH, Imasato A, Xu H, Kai H, Yan C, Li JD. Glucocorticoids inhibit nontypeable Haemophilus influenzae-induced MUC5AC mucin expression via MAPK phosphatase-1-dependent inhibition of p38 MAPK. Biochem Biophys Res Commun. 2008;377:763–768. doi: 10.1016/j.bbrc.2008.10.091. [DOI] [PubMed] [Google Scholar]
- 45.Ishinaga H, Jono H, Lim JH, Kweon SM, Xu H, Ha UH, Koga T, Yan C, Feng XH, Chen LF, Li JD. TGF-beta induces p65 acetylation to enhance bacteria-induced NF-kappaB activation. EMBO J. 2007;26:1150–1162. doi: 10.1038/sj.emboj.7601546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu X, Ha CH, Wong C, Wang W, Hausser A, Pfizenmaier K, Olson EN, McKinsey TA, Jin ZG. Angiotensin II stimulates protein kinase D-dependent histone deacetylase 5 phosphorylation and nuclear export leading to vascular smooth muscle cell hypertrophy. Arterioscler Thromb Vasc Biol. 2007;27:2355–2362. doi: 10.1161/ATVBAHA.107.151704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu X, Jhun BS, Ha CH, Jin ZG. Molecular mechanisms of ghrelin-mediated endothelial nitric oxide synthase activation. Endocrinology. 2008;149:4183–4192. doi: 10.1210/en.2008-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raskatov JA, Meier JL, Puckett JW, Yang F, Ramakrishnan P, Dervan PB. Modulation of NF-kappaB-dependent gene transcription using programmable DNA minor groove binders. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1023–1028. doi: 10.1073/pnas.1118506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rana SN, Li X, Chaudry IH, Bland KI, Choudhry MA. Inhibition of IL-18 reduces myeloperoxidase activity and prevents edema in intestine following alcohol and burn injury. J Leukoc Biol. 2005;77:719–728. doi: 10.1189/jlb.0704396. [DOI] [PubMed] [Google Scholar]
- 50.Yu HP, Yang S, Hsieh YC, Choudhry MA, Bland KI, Chaudry IH. Maintenance of lung myeloperoxidase activity in proestrus females after trauma-hemorrhage: upregulation of heme oxygenase-1. Am J Physiol Lung Cell Mol Physiol. 2006;291:L400–L406. doi: 10.1152/ajplung.00537.2005. [DOI] [PubMed] [Google Scholar]
- 51.Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 53.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, Valk PJ, van der Poel-van de Luytgaarde S, Hack R, Slater R, Smit EM, Beverloo HB, Verhoef G, Verdonck LF, Ossenkoppele GJ, Sonneveld P, de Greef GE, Lowenberg B, Delwel R. High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood. 2003;101:837–845. doi: 10.1182/blood-2002-05-1459. [DOI] [PubMed] [Google Scholar]
- 54.Kurokawa M, Mitani K, Irie K, Matsuyama T, Takahashi T, Chiba S, Yazaki Y, Matsumoto K, Hirai H. The oncoprotein Evi-1 represses TGF-beta signalling by inhibiting Smad3. Nature. 1998;394:92–96. doi: 10.1038/27945. [DOI] [PubMed] [Google Scholar]
- 55.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 56.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 57.Kurt-Jones EA, Chan M, Zhou S, Wang J, Reed G, Bronson R, Arnold MM, Knipe DM, Finberg RW. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proc Natl Acad Sci U S A. 2004;101:1315–1320. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulevitch RJ, Tobias PS. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.