Abstract
Viruses evade immune detection partly through immune-associated mutations. Analyses of HIV sequences derived from infected persons have identified numerous examples of HLA-associated mutations within or adjacent T cell epitopes, but the potential impact of most mutations on epitope production and presentation remains unclear. The multistep breakdown of proteins into epitopes includes trimming of N-extended peptides into epitopes by aminopeptidases before loading onto MHC-I molecules. Defining sequence signatures that modulate epitope production would lead to a better understanding of factors driving viral evolution and immune escape at the population level. Here we identified cytosolic aminopeptidases cleavage preferences in primary cells, its impact on HIV antigen degradation into epitopes in primary human cell extracts by mass spectrometry, and on epitope presentation to CTL. We observed a hierarchy of preferred amino acid cleavage by cytosolic aminopeptidases. We demonstrated that flanking mutations producing more or less cleavable motifs can increase or decrease epitope production and presentation by up to 14-fold. We found that the efficiency of epitope production correlates with cleavability of flanking residues. These in vitro findings were supported by in vivo population-level analyses of clinically-derived viral sequences from 1134 antiretroviral-naïve HIV-infected persons: HLA-associated mutations immune pressures drove the selection of residues that are less cleavable by aminopeptidases predominantly at N-flanking sites, leading to reduced epitope production and immune recognition. These results underscore an important and widespread role of antigen processing mutations in HIV immune escape and identify molecular mechanisms underlying impaired epitope presentation.
Introduction
Viruses such as HIV or HCV evolve rapidly within hosts and populations in order to evade immune recognition and to spread. They have developed multiple strategies to limit the presentation of viral epitopes from infected cells to CD8 T cells (1): mutations within epitopes that prevent their binding to HLA molecules or to the TCR (2–4), mutations triggering epitope degradation (5, 6), and flanking mutations which prevent the production of the epitopes of adequate length (7–9). Despite the enormous genetic diversity of HIV, population-level analyses of clinically-derived viral sequences indicate that mutational escape pathways are broadly reproducible in context of the HLA class I alleles expressed by the host (10–13). Indeed, a large number of HLA-associated polymorphisms have been identified across HIV, including many flanking epitope boundaries (14–17).
Epitopes displayed by MHC-I complexes are derived from intracellular degradation of proteins. Proteins or incomplete translation products are degraded by the proteasome in fragments of various lengths further degraded by endopeptidases and aminopeptidases (18, 19). N-extended versions of epitopes can be shortened by endopeptidases such as Nardilysin (20), insulin-degrading enzyme (21) or by cytosolic or ER-resident aminopeptidases. Leucine aminopeptidase (LAP) (22, 23), puromycin-sensitive aminopeptidase (24), ERAP1 (25, 26) and ERAP2 (27) play variable and complementary roles in producing or destroying epitopes. ERAP1 knocked-out mice infected with LCMV present different CD8 T cell responses compared to WT animals, highlighting the important role of aminopeptidases in epitope production (28, 29).
Cleavable residues have been identified for aminopeptidases in immortalized cells (30–32) but the impact of these flanking residues on immune escape was not assessed. Artificial or natural mutations flanking epitopes on the N-terminal side and altering epitope presentation have been identified for Hepatitis B (33), Influenza (34) and murine retroviruses (35) although the role of aminopeptidases in the production of these epitopes was not assessed. We identified a HLA-restricted mutation flanking an HIV epitope that prevents the trimming of N-extended epitope by ERAP1 and impairs recognition of infected cells by epitope-specific CTL (7). We reasoned that defining well-cleavable and non-cleavable motifs would allow us to identify antigen processing mutations in highly variable viruses at the population level.
After establishing the amino acid cleavage preferences of human PBMC, CD4 T cells and monocytes we demonstrate a strong correlation between the cleavability of N-flanking residues, the production of the adjacent epitope, and the endogenous processing and presentation of the epitope to CD8 T cells. These in vitro findings were supported by in vivo population-level analyses of HLA-associated polymorphisms from viral clinical isolates identified through analysis of HIV sequences from 1,134 patients with known HLA class I types. We show that at N-flanking sites of known CTL epitopes, HLA-restricted immune pressures in vivo drive the selection of residues that are less cleavable by aminopeptidases, thus reducing epitope production and presentation. These results provide the first demonstration of sequence signatures defined at the population level that lead to impaired epitope presentation and impaired immune recognition, supporting a critical role of antigen processing mutations in immune escape in HIV infection.
Material and methods
Peptides and reagents
Highly purified peptides (>98% pure) were purchased from Massachusetts General Hospital peptide core facility or from Biosynthesis, TX. All chemicals were purchased from Sigma and fluorogenic substrates from Enzo Life Sciences.
Study participants
Buffy coats from anonymous volunteers were purchased from Massachusetts General Hospital (MGH), Boston, MA. The use of buffy coats was approved under Protocol #2005P001218 by the Partners Human Research Committee, Boston, MA.
Cell sorting and cytosol preparation
CD4 T cells and monocytes were enriched by magnetic immunodepletion from freshly isolated PBMC as previously described (36). The purity of cell subsets was checked by FACS analysis and reached >96% for CD4 T lymphocytes and >85% for monocytes (unpublished observations). PBMC cytosol was purified detergent-free or by 0.0125% digitonin permeabilization (36, 37). Protein concentration was measured and checked by Western blot against various proteasome subunits (Enzo Life Sciences), actin (Abcam) and absence of ER markers. Cytosolic extracts were also checked for the absence of contamination by lysosomes through the measurement of cathepsin activities with a pan-cathepsin fluorogenic substrate (data not shown and (36, 37)).
Aminopeptidase activity measurement
Aminopeptidase activities in 3 ug cell extracts were measured in 96-well plates with 12.5uM X-amc fluorogenic substrate. Fluorescence emission was measured in the presence or absence of extracts at 37°C every 5 minutes during 1h on a Victor-3 plate fluorescence reader (Perkin Elmer, Boston). Fluorescence emission after peptide cleavage is proportional to the activity. The specificity of the reaction was checked by preincubation of extracts with inhibitors of proteasomes (MG132 10uM, lactacystin 10uM), aminopeptidases (Bestatin 12uM, puromycin 10uM), metallopeptidase (1–10 phenantroline 1uM), tripeptidylpeptidase II (butabindide 330nM), cysteine proteases (E64 50uM), cysteine/serine/asparatate protease (leupeptin 10uM). At each time point fluorescence values in the absence of extracts were subtracted to that measured in the presence of extracts. Substrate cleavage kinetics was reported as the maximum slope (i.e initial velocity) of this graph.
In vitro epitope degradation and peptide antigenicity assay
8 nmol of peptides were degraded with 40ug PBMC cytosol as described previously (36, 37). Peptides present in the digestion mix at designated times were purified by TCA precipitation, identified by mass spectrometry. The mix was diluted to 400–1600 fmol in 80% water, 15% acetonitril, 5% trifluoroethanol, fractionated by RP-HPLC on a C18 column (Nano-LC Ekisgent) and electrosprayed on an Orbitrap Discovery mass spectrometer (Thermo). Each peptide present in the mix, the area of the peak it generated and the proportion of peptides at each time point were calculated with Proteome Discoverer (Thermo) (Harvard Medical School Mass spectrometry facility and in-house mass spectrometer). The intensity of a peak generated by a given peptide is proportional to the amount of peptide (supplementary figure 2A). Each degradation time point was run on the mass spectrometer at least twice. Repeated injections of degradation products show strong reproducibility (supplementary figure 2B).
Antigenicity of degradation products
Peptides present in the digestion mix were purified, diluted in RPMI without serum (R+) and pH was readjusted to 7.4. Cells labeled with 51Chromium were pulsed with 0.02 ug/ml digestion products without serum and used as targets in killing assays with epitope-specific CTL clones at a 4:1 ratio as in (37). Lysis % were calculated as ((Cpm from CTL-mediated Cr release - spontaneous Cr release)/(maximum (induced with Triton X100 2.5%)– spontaneous Cr release)x100). Lysis % of cells pulsed with degradation products were compared to those of HLA-matched B cells pulsed with undigested long peptides or optimal epitopes at concentrations ranging from 0 to 0.4ug/ml.
mRNA preparation and transfection for epitope presentation
HIV-1 p24 synthetic genes were purchased from GenScript, Inc. Each construct included a T7 promoter, a Kozak sequence, antigen open reading frame, stop codon, and a polyA signal and synthetic mRNA were transcribed from linearized plasmid templates, quantified using a ND-1000 spectrophotometer as in (38). The size and purity of mRNA preparations were confirmed with the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA). 3 million B cells were electroporated using the following conditions: Square wave, 900V, 0.75ms on a BioRad GenePulser Xcell, 100/240V, 50/60Hz (BioRad, Hercules, CA). Transfection efficiency was >80% (38). 20 hours later cells were used as targets in 51Cr release assay.
Cohort and identification of HLA-restricted mutations
We employed published, phylogenetically corrected methods (15) to identify HLA-associated viral polymorphisms in a cohort of 1134 chronically HIV-infected, treatment-naïve individuals from British Columbia, Canada for whom HLA class I types and nearly full genome bulk HIV-1 sequences were characterized (11). Analyses of all HLA class I alleles and their restricting epitopes were performed at 2-digit resolution. We identified HLA-associated polymorphisms occurring inside or within ±3 residues of all optimally defined CTL epitopes across the HIV-1 proteome except gp120 (39), in context of the restricting HLA only. A maximum likelihood phylogenetic tree is constructed from the HIV sequences in the dataset and a model of conditional adaptation is inferred for each observed amino acid at each codon (14, 15). In this model, the amino acid is assumed to evolve independently down the tree, until it reaches the terminal branch tips. On each tip, the selection pressure arising from HLA-associated immune pressures is directly modeled using a stochastic additive process where each HLA allele is treated as a binary variable. P-values are computed using the likelihood ratio test. Multiple comparisons are addressed using q-values, which estimate the expected proportion of false positives among results deemed significant at a given threshold (40). For example, at q≤0.2, we expect 20% false-positives among identified associations. Using this approach, we are able to identify two types of HLA-associated polymorphisms: (1) amino acids significantly depleted in the presence of the HLA allele in question (the “non adapted” form) and (2) amino acids significantly enriched in the presence of the HLA allele in question (“adapted” or “escape” form) (14, 15). At a q-value of 0.2, we identified a total of 102 HLA-associated polymorphisms occurring at positions −3 through 3 of known CTL epitopes for which a specific “adapted” form was identified for the restricting HLA, thus allowing us to calculate delta aminopeptidase scores between “non-adapted” and “adapted” forms. These escape pathways included 8, 8 and 10 HLA-associated polymorphisms occurring at codons −3, −2 and −1 of known CTL epitopes, respectively, and 21, 29 and 25 HLA-associated polymorphisms associations occurring at positions 1, 2 and 3 of known CTL epitopes, respectively. Out of 102 mutations, 14 mutations at position 2 were predicted to decrease HLA binding affinity by more than 10-fold (NetMHC3.2; http://www.cbs.dtu.dk/services/NetMHC/) (41); these were removed from analysis as they likely represented escape mutations whose main mechanism of action is abrogation of peptide-HLA binding. The published B57-associated A146P escape mutation occurring at position -1 of the B57-ISW9 (Gag) epitope (7)– primarily selected by HLA-B5703 whereas our cohort included mainly HLA-B5701 persons- was added to the list, bringing the final total of investigated HLA-associated escape pathways to 102.
Statistical analysis
Empirical data were analyzed using GraphPad Prism version 5.
Results
Cytosolic aminopeptidase substrate preferences in human primary cells
We first aimed to establish aminopeptidase substrate preferences in human PBMC, CD4 T cells and monocytes. Fluorogenic substrates composed of an amino acid (X, where X represents any amino acid) coupled to coumarin derivative (X-amc) were incubated with PBMC cytosol for 60 minutes and fluorescence emission was monitored every 5 minutes as done in (36) (figure 1A). As previously reported in immortalized cells Leucine-amc was very efficiently cleaved by cytosolic aminopeptidases whereas Tyrosine-amc was cleaved less efficiently (22, 30, 32). The specificity of substrate cleavage was checked by preincubation of extracts with inhibitors of aminopeptidases (bestatin, puromycin), metallopeptidases (1–10 phenantroline), proteasome (MG132 or lactacystin), TPPII (butabindide), cysteine proteases (E-64), or cysteine/serine proteases (leupeptin) (figure 1B). For each substrate the hydrolysis kinetics are measured as the maximum slope of fluorescence emission during a 1-hour experiment after subtraction of fluorescence in the absence of extracts. 100% represents the maximum slope of Leu-amc and Tyr-amc (928 and 316 respectively). It was reduced by >70% in the presence of inhibitors of aminopeptidases or metallopeptidases and much less (<22%) by inhibitors of proteasome, TPPII, serine/cysteine proteases.
Figure 1. Hierarchy of amino acid cleavage preference in human primary cells.
A. Differential cleavage of Leu-amc and Tyr-amc. Leu-amc (black triangles) or Phe-amc (black circles) were added to PBMC cytosol preincubated with (plain black line) or without bestatin (dotted grey lines) and incubated for 1h at 37°C during which fluorescence emission was monitored every 5 minutes. B. PBMC extracts were preincubated with various inhibitors (bestatin, 1–10 phenantroline, puromycin, MG132, butabindide, E64, leupeptin) before addition of Leu-amc or Tyr-amc. The maximum slope of fluorescence emission over 1h was calculated for each condition. 100% represents the maximum slope of fluorescence emission for each substrate in the absence of inhibitor (5591 for Leu-amc and 1804 for Tyr-amc). Average and SD of 3 experiments using cytosol from 3 different donors. C. Maximum slope of fluorescence emission by each substrate in a 1-hour incubation with PBMC cytosol. 100% corresponds to the slope of Leu-amc hydrolysis (fluorescence values after subtraction of those in the presence of inhibitor) (4214 average fluorescence emission). Average of 5 healthy donors. D. Amino acid cleavage efficiency by cytosol from PBMC, or CD4 T cells and monocytes sorted from PBMC. Average of 4 healthy donors. 100% represents the maximum slope of Leu-amc cleavage by each subset (3231 for PBMC, 964 for CD4 T cells and 5211 for monocytes).
We compared the hydrolysis of twenty X-amc substrates in PBMC cytosol from five different donors. The hydrolysis kinetics of aminopeptidase-specific activity is calculated after subtraction of fluorescence in the presence of bestatin (figure 1C). Leu-amc was he most efficiently cleaved substrate and its hydrolysis rate was used as 100% to rank other amino acids. Beside Leu-amc, nine X-amc were cleaved with variable efficiency: 3 amino acids (M, K, F) formed very efficiently cleavable bonds (slopes 77–85% of that of Leu-amc) whereas 6 amino acids (Y, A, R, C, W, I) formed bonds hydrolyzed at lower rates (11–53%). Ten amino acids (V, T, P, N, Q, D, E, G, H, S) formed bonds that were poorly or not cleaved by aminopeptidases (0–4.9%). The split between well- and poorly-cleavable residues is in accordance with published results in immortalized cells (31, 32). However the best aminopeptidase substrate in primary human cells was Leu-amc and not Lys-amc.
We had previously reported that monocytes have higher cytolosic aminopeptidase activities than CD4 T cells using Leu-amc substrate (36). Whether this difference remains for other amino acids is unknown. We compared the hydrolysis of 9 X-amc bonds representing highly to poorly cleaved residues in cytosolic extracts from PBMC, CD4 T cells or monocytes sorted from the same four donors (figure 1D). The ranking of the 9 amino acids was equivalent between PBMC, CD4 T cells and monocytes, with Leu-amc being the most efficiently cleaved. However, the cleavage rate for each substrate varied among subsets, with CD4 T cells having the lowest rate, and monocytes the highest. The largest difference in amino acid cleavage efficiency between monocytes and CD4 T cells (figure 1D inset) was for leucine (6.4-fold difference). Since Leu-amc is efficiently cleaved by LAP (22), we compared LAP expression in PBMC, CD4 T cells and monocytes by Western blot. The ratio of LAP over actin was significantly higher in monocytes than in donor-matched CD4 T cells (13 donors, p=0.0128; data not shown), suggesting that higher hydrolysis of Leu-amc in monocytes may involve higher LAP expression.
N-flanking residues alter the trimming of N-extended epitopes in vitro
We aimed to determine how good or poor aminopeptidase substrates affect the production of adjacent epitopes. We first identified HIV epitopes whose production requires aminopeptidase trimming (supplementary figure 1). HLA B57+ B cells were transfected with RNA encoding HIV p24 Gag as in (38) and used as targets in a killing assay with CTL specific for B57-ISW9 (ISPRTLNAW; as 15–23) or B57-KF11 (KAFSPEVIPMF; aa30–40), 2 epitopes targeted by HLA-B57 persons during chronic HIV infection (42). Preincubation of cells with a proteasome inhibitor or by aminopeptidase inhibitor reduced CTL-mediated lysis. However the combination of proteasome and aminopeptidase inhibitor most efficiently decreased CTL-mediated killing (2.4 and 5.2-fold for ISW9 and KF11 respectively). This indicates that the processing of B57-ISW9 and B57-KF11 is proteasome- and aminopeptidase-dependent and that these epitopes can be used to assess how various flanking motifs affect epitope processing and presentation.
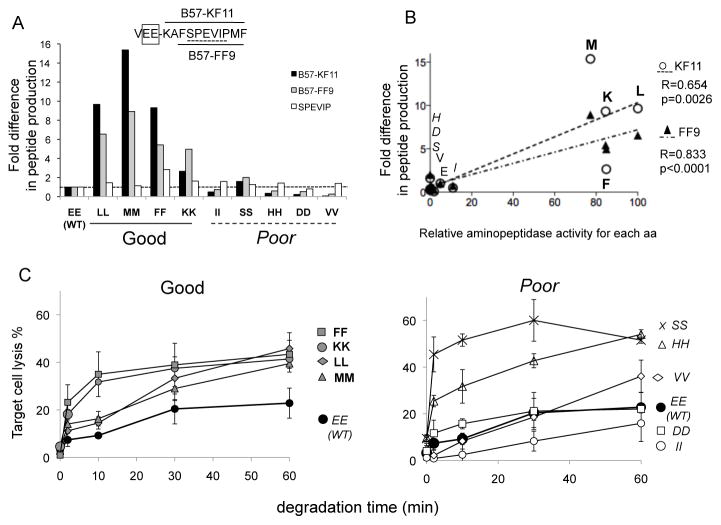
We compared the cytosolic processing of KF11 from N-extended peptides containing the WT N-extension (VEE) or modified N-extension including good (VLL, VMM, VFF, VKK) or poor aminopeptidase substrates (VII, VSS, VHH, VVV). The degradation products generated in 1 hour were identified by mass spectrometry as we previously described (5, 36, 37). The peak area corresponding to a sequence of given mass and charge is proportional to the amount of corresponding peptide (supplementary figure 2). We compared KF11 peak area generated from each mutated peptide over that of KF11 produced from the WT sequence (figure 2A). All four highly cleavable flanking motifs led to increased KF11 production (2.7- to 9.7-fold). In contrast the introduction of motifs as poorly cleavable as the WT sequence reduced or hardly affected the production of KF11. We also measured the production of FF9, another B57-restricted epitope within KF11 located two residues downstream of the KF11 start site (43). The production of FF9 was also strongly increased for peptides containing good aminopeptidase substrates. These results show that changes one to four residues upstream of a peptide can affect its production. To rule out any differences in degradation products recovery, mass spectrometer injection or peptide detection we quantified shorter peptides common to all sequences (i.e. included in KF11). The production of SPEVIP from mutated sequences was 0.8 to 1.4-fold that of WT sequence, showing that the higher production of KF11 and FF9 from peptides including good aminopeptidase motifs was not due to a technical discrepancy. The efficiency of cleavage of an amino acid established with fluorogenic substrates (figure 1C) correlated with the production of flanking epitope KF11 (R=0.64; p=0.0026) and downstream epitope FF9 (R=0.83; p<0.0001) (figure 2B), suggesting that changing aminopeptidase substrates impacts epitopes immediately adjacent to, or two residues downstream of the residue being cleaved. We also assessed the production of KF11 and FF9 from peptides containing a single flanking mutation toward better (L, M, F) or poor (I, D, H, V, S, T) aminopeptidase substrates. Mutations toward a good substrate increased KF11 production albeit less efficiently than with a double mutation (up to 6.2-fold) and replacement of E by another poorly cleavable residue did not alter epitope production (0.9 to 1.5-fold difference) (supplementary figure 3A). The only exception was poorly cleavable residue S for which KF11 production increased by 5.6-fold.
Figure 2. Flanking residues drive the amount and antigenicity of HIV peptides produced in PBMC cytosol.
A. 3-extended KF11 peptides (KAFSPEVIPMF; aa 30–40 in p24) with double mutations flanking KF11 (WT VEE or mutants LL, MM, FF, KK, II, SS, HH, DD, VV) were degraded for 1h in PBMC cytosol. Cleavable motifs are bolded and poor substrates are italicized. Peptides KF11 (black bars), FF9 (grey bars) and SPEVIP (white bars) were identified by mass spectrometry. The intensity of the peak of the 3 peptides was compared between WT and mutants. Representative of 3 experiments. B. Correlation between the fold increase production of KF11 (open circles) and FF9 (black triangles) and the relative PBMC aminopeptidase-specific hydrolytic activity of each amino acid. C. Antigenicity of trimmed KF11 peptides. Degradation products of each extended KF11 peptides preceded with WT or cleavable motifs (left panel: WT EE black circles, FF: grey squares, KK grey circles, LL grey diamonds, MM grey triangles), or poorly cleavable motifs (right panel: WT EE black circles, SS: x, HH: open triangles, VV open diamonds, DD open squares, II: open circles) were purified at different time points, pulsed onto HLA-B57+ B cells used as targets in cytolysis assay with KF11-specific CTL. Average of 3 experiments.
As an independent read-out of epitope production, we assessed the antigenicity of the degradation products generated over 1h as previously performed (5, 36, 37). Purified peptides were used to pulse an HLA-B57+ cell line used as targets in a 51Cr-based killing assay (figure 2C). At low peptide concentration extended KF11 peptide such as VEE-KF11 did not yield a KF11 CTL response (lysis <2%); however as the peptide N-extension was trimmed the antigenicity of the degradation products increased. Degradation products of VEE-KF11 became antigenic after a 10-minute degradation and antigenicity plateaued at 20.6–22.8% for products generated in 30 to 60 minutes. In contrast for peptides where KF11 was preceded by good aminopeptidase substrates (FF, KK, LL, MM; figure 2C left panel), antigenic products were generated faster and in higher quantity (maximum antigenicity around 40–50% compared to 22.8% for WT products). For peptides encompassing poor aminopeptidase substrates (figure 2C right panel), the antigenicity of degradation products from three peptides (II, DD, VV) was comparable to that of WT products (with VV generating more antigenic products at time 60 min) as expected from the in vitro production of KF11. However two peptides with poor aminopeptidase substrates (SS and HH) efficiently generated antigenic products (51.6 and 54% at 60 minutes, for HH and SS respectively). Out of 10 amino acids tested 8 behaved as hypothesized whereby poor/good aminopeptidase substrate flanking epitope led to low/high in vitro epitope production from N-extended epitopes, and low/high antigenicity of degradation products. Similarly for peptides carrying a single flanking mutation the antigenicity of the degradation products was increased for peptides including good aminopeptidase substrates and correlated with KF11 production measured by mass spectrometry (supplementary figure 3B). These results suggest that the 2 residues flanking KF11 drive the efficiency of epitope production.
The efficiency of endogenous epitope processing and presentation is driven by N-flanking motifs
We confirmed the relevance of good and bad aminopeptidase substrates during the endogenous processing and presentation of KF11 epitope by cells. HLA B57 cells were transfected with RNA vectors expressing p24 in which KF11 is flanked by various motifs (figure 3). In this experimental setting both cytosolic and ER-resident aminopeptidases can contribute to the production or degradation of KF11. We monitored the presentation of KF11 and of TW10, another B57-restricted epitope located in Gag p24 (figure 3A). We hypothesized that enhanced production of KF11 due to optimal flanking residues would lead to its improved presentation and an increase in the ratio of KF11/TW10 peptide presentation.
Figure 3. The efficiency of endogenous processing and presentation of an aminopeptidase-dependent HIV epitope to CD8 T cells is driven by epitope-flanking motifs.
A. Enhanced presentation of KF11 flanked by cleavable motifs. RNA encoding p24 with WT KF11 flanking motifs or mutated residues (LL, II) were transfected in HLA-B57+ B cells used as target in a cytolysis assay with KF11-specific CTL (black bars) or neighboring B57-restricted epitopes TW10 (TSTLQEQIGW; aa 108–117 in p24; grey bars). Cleavable motifs are bolded and poor substrates are italicized. Average of 3 transfection experiments. B. Ratio of KF11/TW10 epitope presentation by cells transfected with WT or mutated (LL, FF, FF, II, VV, DD, HH, SS) p24. For each transfection we calculated a ratio of an equivalent amount of exogenously pulsed KF11 or TW10 peptide needed to reach the same lysis percentage as those of transfected cells. Average of 3 transfection experiments. C. Correlation between the fold increase in KF11 presentation from various mutated p24 to CTL and the relative aminopeptidase activity against each residue. D. Correlation between the fold increase in KF11 presentation from various mutated p24 to CTL and the in vitro production of KF11 measured by mass spectrometry. Correlations with Spearman test.
The endogenous processing and presentation of WT p24 led to efficient killing by TW10-specific CTL (58.5%) and KF11-specific CTL (24.2%), a 2.4-fold difference in CTL lysis. p24 containing II-KKF11, another poorly cleavable motif, led to similar lysis rates by each clone (23.8% and 45% by KF11 and TW10 CTL respectively, a 1.9-fold difference in lysis %). In contrast expression of p24 with LL-KF11, an efficiently cleavable motif, led to equivalent lysis by KF11 and TW10 clones (41.7% and 37.7%, a 0.9-fold difference in lysis), suggesting enhanced presentation of KF11 when preceded by a cleavable motif. In order to take into account possible differences in peptide avidity for HLA-B57 and TCR and variations in transfection efficiency among variants, we measured an equivalent amount of peptide presented by transfected cells by comparing the level of lysis of transfected cells to that of cells pulsed with increasing amounts of KF11 or TW10 as in (37). We then calculated a ratio of equivalent KF11/TW10 peptides presented by cells transfected by different p24 variants (figure 3B). The molar ratio of KF11/TW10 was 0.07 in cells transfected with WT p24. In cells expressing p24 containing KF11 preceded with efficiently cleavable motifs (LL and FF), the ratio of KF11/TW10 ratio reached 0.62 and 0.44 respectively, a 8.9- and 6.3-fold increase of KF11 presentation to CTL. In contrast cells expressing p24 in which KF11 is flanked by poorly cleavable motifs (II, DD, HH, SS) the KF11/TW10 ranged between 0.08 and 0.11 (1.1- to 1.6-fold increase compared to presentation of KF11 from WT), suggesting that replacement of poorly cleavable flanking motif by other poor substrates did not significantly change KF11 presentation. p24 containing VV-KF11 led to a 3.9-fold increase of KF11 presentation despite being identified as a poorly cleavable substrate. Accordingly in vitro epitope production led to production of antigenic KF11 peptide at a late time point (figure 2C, right panel). This may be indicative of slow KF11 production by peptidases other than aminopeptidases for which VV is an acceptable substrate.
We compared the fold increase in KF11 epitope presentation to aminopeptidase activity against each flanking residue (figure 3C), or to KF11 produced in vitro from VXX-KF11 peptides (figure 3D). We showed a strong correlation between endogenous processing and presentation of KF11 and the cleavability of the flanking residues (p=0.006, R=0.878) and the in vitro production of KF11 (p=0.0022, R=0.812). This demonstrates that the capacity of aminopeptidases to cleave epitope flanking residues plays a critical role in the amount of epitope produced and subsequent recognition by epitope-specific CTL. It also provides strong rationale for the use of in vitro epitope processing as a way to identify motifs altering epitope production in cells.
HLA-restricted flanking or intraepitopic mutations reduce HIV epitope processing and presentation
Since aminopeptidases play a critical role in trimming epitope precursors and have marked substrate preferences, HLA-restricted mutations occurring near the N-terminus of epitopes could significantly impact epitope presentation. We hypothesize that there could be two ways to reduce or eliminate epitope production in vivo. Firstly, selection of N-flanking mutations representing poor aminopeptidase substrates could lead to reduced or impaired trimming of epitopes. Conversely, selection of mutations within the epitope toward better aminopeptidase motifs could lead to “over-trimming” of the epitope. We analyzed the impact of 9 specific HLA-restricted flanking or intraepitopic mutations (positions −3 through 3) creating motifs predicted to be more or less cleavable by aminopeptidases on HIV epitope production (figures 4 and 5).
Figure 4. An HLA-restricted epitope-flanking mutation toward a poorly cleavable aminopeptidase motif impairs epitope production and presentation.
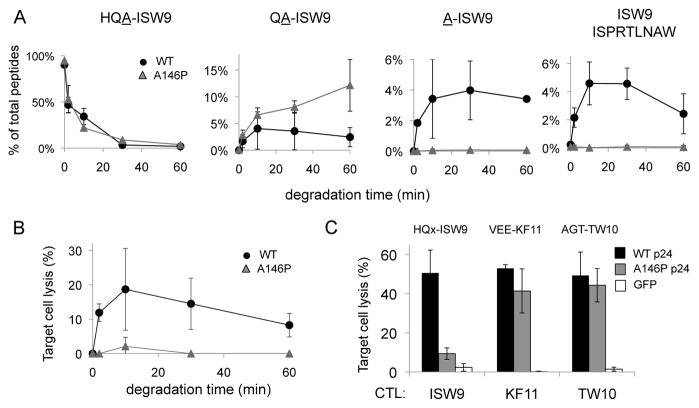
A. Peptide corresponding to HLA-B57-ISW9 with a 3-residue extension with WT (HQA-ISW9, black circles) or mutated HQP-ISW9 (grey triangles) were degraded in PBMC cytosol. The relative amount of 3-, 2-, 1-extended and epitope ISW9 (left to right) was identified by mass spectrometry over 60 minutes. The relative amount of each peptide in the total mix of degradation products is quantified at each time point. Average of 2 degradation experiments run twice on a mass spectrometer. 2 additional degradation experiments with less cytosol gave similar results albeit with slower degradation rates (not shown).
B. Degradation products from HQA-ISW9 (black circles) or HQP-ISW9 (grey triangles) of each time point were pulsed onto HLA-B57 cells used as targets in a chromium assay with ISW9-specific CTL. Average of 3 killing assays run in triplicates.
C. HLA-B57 cells were transfected with RNA encoding WT p24 (black bars), p24 with A146P mutation flanking ISW9 (grey bars) or GFP (white bars) and used as targets in a chromium-based killing assay with ISW9-, KF11, or TW10-specific CTL (3 epitopes located in p24 and restricted by HLA-B57). Epitope flanking sequences indicated above bars. Average of 3 transfections; killing assays run in triplicates.
Figure 5. HLA-restricted flanking mutations toward less cleavable motifs or intraepitopic mutations toward more cleavable aminopeptidase motifs reduce epitope production.
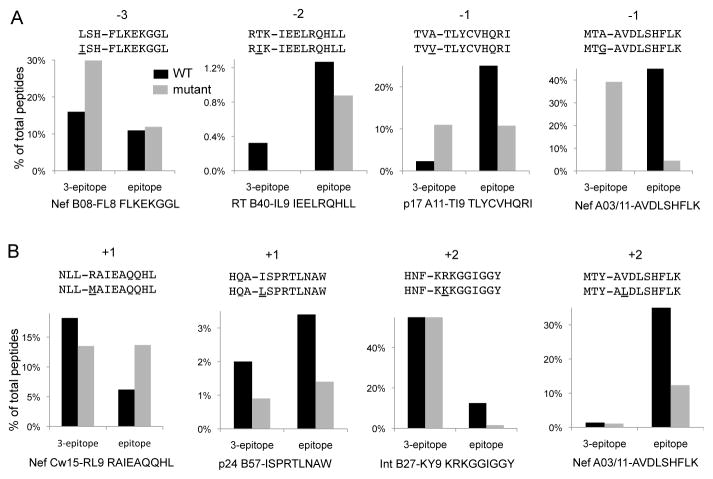
A. Peptides including an HIV epitope with a 3-residue N-terminal extension corresponding to WT sequences (black bars) or with a HLA-restricted mutation at positions −3, −2, −1 (grey bars) were degraded in PBMC cytosol for 60 minutes. All degradation peptides were identified by mass spectrometry and the relative amount of the 3-extended peptide and epitopes were calculated. Names, sequences and HLA-restriction of each epitope are indicated below each panel. N-extended sequences, mutations and their locations are indicated above each panel.
B. Same as A with HLA-restricted mutations located at positions 1 or 2 within epitopes. Average result of 3 mass spectrometry injections of one representative cytosolic degradation.
The HLA-B57-restricted Alanine 146 to Proline (A146P) escape mutation flanking Gag p24 epitope ISW9 whose processing is aminopeptidase-dependent is frequently observed in HLA-B57+ HIV-infected persons (7). We performed in vitro degradation of N-extended peptides (HQA-ISW9, HQP-ISW9 A146P) in PBMC cytosol and identified the degradation peptides produced over 1 hour by mass spectrometry (figure 4A and supplementary figure 4). The kinetics of degradation of HQA-ISW9 and HQP-ISW9 were similar (figure 4A left panel). The degradation of HQA-ISW9 led to the production of A-ISW9 and ISW9 (up to 4.6% of total peptides). In contrast the degradation of HQP-ISW9 led to the accumulation of QP-ISW9 (up to 11% of total amount of peptides) and no production of A-ISW9 or ISW9, likely due to the incapacity of cytosolic aminopeptidases to cleave the Q-P bound. We compared the antigenicity of the degradation products generated from the degradation of HQA-ISW9 or HQP-ISW9 (figure 4B). It reached up to 19% lysis at 10 minutes for WT where it was at 2% for HQP-ISW9, a 9-fold difference in equivalent ISW9 peptide production in accordance with the lack of production of ISW9 from HQP-ISW9. The production of ISW9 and antigenicity decreased later on, likely due to rapid cytosolic degradation of ISW9 (5).
We assessed the complete endogenous processing and presentation of HLA-B57-restricted epitopes by RNA transfection of WT or A146P p24 in HLA-B57+ B cells and assessed the presentation of 3 HLA-B57-restricted epitopes, ISW9, KF11 and TW10 (figure 4C). In this case both cytosolic and ER aminopeptidases may contribute to epitope processing. Cells transfected with WT or A146P p24 were equally recognized and killed by CTL specific for TW10 or KF11, showing that transfection and expression of WT or A146P p24 were similar. Mutation A146P flanking ISW9 reduced killing of transfected cells by ISW9-specific CTL by 5-fold (9.4 vs 50.6%), showing that a mutation creating a poorly cleavable motif (delta −50) flanking an epitope led to lack of production and presentation of the epitope and reduced killing by epitope-specific CTL.
We assessed the impact of 4 additional mutations upstream of HIV epitopes (figure 5A): HLA-B08-restricted L to I mutation at position −3 of B08-FL8 in Nef, HLA-B40-restricted T to I at position −2 of B40-IL9 in RT, HLA-A11-restricted A to V mutation at position −1 of A11-TI9 in p17, HLA-A03/11-restricted A to G at position −1 of A03/A11 AK9 in Nef (11, 12, 14). For each sequence we assessed the disappearance of WT and mutated 3-residue extended peptide and the appearance of the epitope in a one-hour cytosolic degradation. Mutation 3 residues upstream B08-FL8 in Nef did not affect epitope production and mutation at position −2 of B40-IL9 slightly reduced epitope production. Mutations directly flanking epitopes A11-TI9 and A03/A11 AK9 led to slower degradation of the N-extended peptides and reduced epitope production by 2.3-and 10.3-fold respectively.
Similarly we assessed the impact of 4 HLA-restricted intraepitopic mutations toward better aminopeptidase substrates (figure 5B): HLA-Cw15-restricted R to K mutation at position +1 of RL9 in Nef, HLA-B57-restricted I to L mutation in p24 B57-ISPRTLNAW in p24, HLA-B27-restricted R to K at position 2 in Integrase, HLA-A03-restricted V to L mutation at position 2 in A03-AK9 in Nef. Whereas the Cw15-restricted mutation increased epitope production by 2.2-fold, the B57-, B27- and A03 mutations at positions 1 or 2 reduced the production of the epitope by 2.4-, 7.8- and 3.0-fold respectively.
Altogether these results provide proof of concept that published escape mutations towards a poorly cleavable motif at epitope flanking sites, and towards more efficiently cleavable motifs within the epitope itself, reduce production of the relevant epitope. These results suggest that antigen processing mutations affecting peptide trimming occur frequently in vivo and that they can be predicted.
Aminopeptidase substrate preferences define HLA-restricted antigen processing mutations at the population level
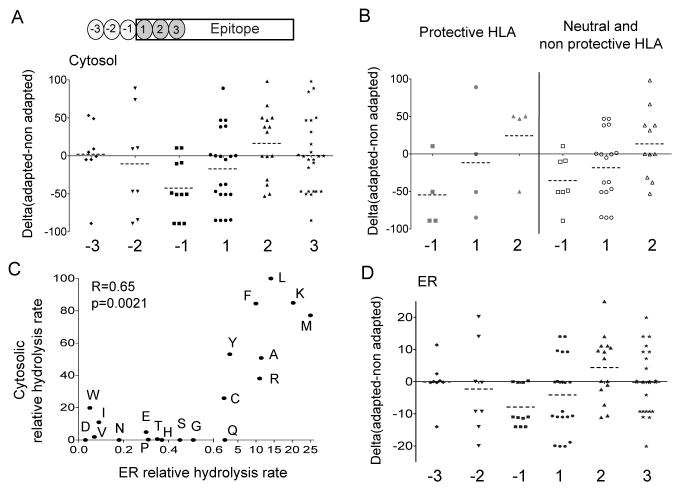
To further address whether alterations in aminopeptidase substrate preferences could represent a major mechanism underlying HLA-driven immune escape pathways in vivo, we analyzed linked HLA class I types and nearly full-genome HIV-1 sequences in a cohort of 1134 persons infected with HIV-1 clade B ((14, 44) and unpublished data). We aimed at identifying HLA-restricted mutations occurring in the 3 N-flanking residues (positions −3 through −1) and in the first 3 residues of the epitope (positions 1 through 3) for all optimally-described CTL epitopes in HIV-1 (except envelope gp120) (45). This analysis revealed a total of 102 distinct escape mutations significantly linked to an HLA allele and occurring at positions −3 through +3 of a published CTL epitope, among which 14 mutations at position affecting binding to MHC-I by more than 10-fold that were excluded from this analysis as impaired HLA binding may be the main escape mechanism at this site. For each escape mutation, we calculated the difference (delta; Δ) between the cleavage capacity of the epitope (or N-extended epitope precursor) containing the adapted (escape mutated) residue and that containing the non-adapted (usually consensus B) residue, based on the aminopeptidase rankings established in figure 1C. A negative delta is indicative of a mutation toward a worse aminopeptidase substrate whereas a positive delta indicates a mutation toward a better aminopeptidase substrate. At positions −3 through 3 of known CTL epitopes, a broad range of delta scores were observed (Figure 6A). Escape mutations occurring at position −1 of known CTL epitopes significantly deviated from a theoretical median delta score of 0 (Wilcoxon Signed-rank test p=0.018) and exhibited a strong trend towards poorer aminopeptidase substrates with 9 mutations out of 11 (81.8%) exhibiting negative deltas (mean −31.3; range −22.6 to −85). Within epitopes mutations did not significantly deviate from a median delta score of 0 although position 2 presented the highest proportion of escape mutations toward more cleavable residues (8 out of 15 (53%) mutations not affecting MHC-I binding). The comparison of mutations restricted by protective HLA alleles (HLA B57, B58, B27) vs neutral or non protective alleles also show similar, statistically significant tendencies towards less cleavable residues at position −1 (figure 6B). These results suggest that regardless of the HLA allele restricting the mutation, evolution toward residues poorly cleavable by aminopeptidases at N-flanking residues appears to be a hallmark of immune escape in HIV.
Figure 6. HIV evolution at the population level toward poorly cleavable flanking residues and more cleavable residues within epitopes.
A. HLA-associated escape pathways located at positions −3 (diamond), −2 (inverted triangles), −1 (squares) flanking epitopes or at position 1 (circles), 2 (triangles), or 3 (stars) were identified in all optimally defined CTL epitopes across the HIV-1 proteome (except gp120) in a cohort of 1134 chronically HIV-infected, antiretroviral-naïve persons. A total of 88 HLA-restricted escape pathways (after exclusion of 14 mutations affecting binding to MHC-I) were identified. For each given HLA-restricted escape pathway we calculated the difference (delta; Δ) between the aminopeptidase cleavage capacity of the adapted (escape mutated) residue and that of non-adapted (usually consensus B) residue, based on the ranking established in figure 1C. A negative delta is indicative of a mutation toward a worse aminopeptidase substrate whereas a positive value shows a mutation toward a better aminopeptidase substrate. Wilcoxon Signed-rank test for deviation from a theoretical median delta score of 0 at position −1 (p=0.018). B. Same as in A except mutations are compared at positions −1, 1 and 2 only, for HLA alleles associated with protection (HLA B27, B57, B58) (left; grey symbols) or neutral or associated with progression (right; open symbols). C. Comparison of amino acids hydrolysis rates by PBMC cytosolic aminopeptidase established in figure 1 and ER aminopeptidases established by Schatz et al, J. Immunology 2008. Spearman correlation. D. Similar to A with except that the difference (delta; Δ) between the aminopeptidase cleavage capacity of the adapted (escape mutated) residue and that of non-adapted residue is based on the ranking of ER aminopeptidases established by Schatz et al. Wilcoxon Signed-rank test for deviation from a theoretical median delta score of 0 at position −1 p=0.018.
Peptides translocated into the ER can be further trimmed by ER aminopeptidases with defined substrate preferences (30–32). The comparison of the hydrolysis rate of amino acids in the PBMC cytosol established here and that of ER microsomes from various cell lines established by Schatz et al (32) showed strong correlation (p=0.0021), in accordance with some shared cleavable residues such as M, K, L and shared poorly cleavable substrates such as P, D, E (figure 6C). We analyzed the effect of HLA-restricted mutations on ER aminopeptidase hydrolytic rates. Similarly to trends observed for cytosolic peptidases, mutations at position −1 significantly evolved toward less cleavable residues by ER aminopeptidases (p=0.018) (figure 6D), suggesting that mutations could impair epitope processing in the cytosol and/or in the ER for peptides reaching this compartment. Our results support the notion that alterations in aminopeptidase substrate potential represents a key mechanism underlying in vivo HLA-restricted immune escape pathways identified at the population level.
Discussion
Identifying viral mutations that will impair epitope presentation to immune cells is critical to understand reasons for immune failure in chronic infections and to devise better therapeutic strategies. Although recent improvements in DNA sequencing and computation have facilitated the identification of immune-driven mutational pathways in rapidly evolving viruses such as HIV and HCV, the molecular mechanisms underlying these evolutionary pathways remain largely unknown. Understanding immune escape at the molecular level is not possible without first achieving clear understanding of basic mechanisms of epitope presentation. In this report we show that cytosolic aminopeptidases in the class I antigen processing pathway cleave substrates with varying efficiency, and that we can use this hierarchal pattern to define sequence signatures leading to impaired HIV epitope production. Importantly these signatures were consistent with in vivo immune-mediated escape pathways identified from patient-derived HIV sequences at the population level.
The degradation of proteins into epitopes presented by MHC-I relies on multiple peptidases located in several subcellular compartments. Aminopeptidases play a critical role in the post-proteasomal trimming of peptides into epitopes either in the cytosol or in the ER. Aminopeptidases located in the cytosol and in the ER share many cleavage preferences despite some ranking differences among compartments. As degradation peptides traffic from the cytosol to the ER and –for proteins in the exogenous processing pathway- from endo-lysosomes to ER or from endo-lysosomes to cytosol and ER before cross-presentation, mutations leading to altered peptide trimming by aminopeptidases may provide a common way to reduce epitope presentation regardless of the path followed by the antigen. Mutations with the strongest effect on aminopeptidase cleavage capacity appeared at position −1 in the cytosol and in the ER. HIV epitopes linked to these HLA-associated mutations may mainly rely on aminopeptidase trimming whereas epitopes associated with mutations at other positions may involve trimming by peptidases with different cleavage preferences. Aminopeptidase-dependent epitope B57-ISW9 is associated to mutations at position −1 (Alanine to Pro) preventing epitope production, and at position 1 (Isoleucine to Leucine) enhancing peptide degradation. Additionally aminopeptidases can cleave 1 or 2 residues at a time, thus not all mutations would have the same impact on aminopeptidase-mediated trimming. For other mutations impaired aminopeptidase trimming may not be the main force driving for viral evolution. Proteasomes and endopeptidases such as nardilysin also contribute to the processing of HIV epitopes and modifying their cleavage sites may also impair epitope processing (20, 46). Since different peptidases have different specificities with regards to lengths of the substrates and cleavable motifs, we expect that specific viral mutations will affect the activity of each class of enzymes that contribute to antigen processing and may explain some of the other HLA-restricted mutations. A comprehensive analysis of signatures of antigen processing mutations will require a better understanding of cleavage preferences of all peptidases. Proteasome-mediated destruction of HIV epitopes has been identified for a few HLA-associated mutations (6, 47). Peptides corresponding to HIV epitopes of optimal size are also subjected to cytosolic degradation prior to loading onto MHC-I. We have identified multiple intraepitopic HLA-restricted mutations that render HIV epitopes more sensitive to cytosolic degradation (5). These mutations create cleavage sites for cytosolic peptidases and were located at various positions throughout the epitope. Out of 25 mutated epitopes, 21 peptides (84%) saw their cytosolic stability reduced by 22–99% relative to WT, suggesting that multiple intraepitopic mutations lead to peptide degradation before loading onto MHC-I molecules. It is difficult –just from the location of a given HLA-restricted mutation- to determine if a mutation will affect epitope production, binding to MHC-I or to the TCR or be a compensatory mutation (3, 4). However our data provide a way to identify many mutations impairing epitope production and suggest that antigen processing mutations can occur at many positions within or outside or epitopes. It takes multiple trimming steps to produce an epitope from a protein, providing multiple opportunities for viral mutations outside epitopes that may interfere with its trimming and production. In addition a typical epitopic sequence contains 8 to 11 residues that could potentially be mutated to produce more cleavable targets and lead to epitope destruction in just one step. Even if HIV evolution is limited by structural and fitness constraints, antigen processing mutations provide multiple redundant options for HIV to escape immune recognition. Other variable viruses such as HCV also evolve antigen processing mutations to escape immune responses (3, 48, 49) although the contribution of flanking and intraepitopic mutations to immune failure remains to be clarified.
We showed that the higher aminopeptidase activity detected in monocytes was mainly due to higher expression and activity of leucine aminopeptidase or closely related aminopeptidases. This suggests the existence of cell type-specific expression of aminopeptidases as evidenced previously for a macrophage-specific serine protease (50). Differences in the pattern of expression and/or activities of aminopeptidases between different cell types may affect the kinetics and stoichiometry of epitope production within different cell types as we showed for CD4 T cells and monocytes (36) and should be assessed for additional cell types of interest to viral epitope presentation. Since both CD4 T cells and macrophages can be recognized by HIV-specific CTL (51), immune pressure may be exerted by CD8 T cells on viruses produced by both cell subsets. We propose that differences in peptidase activities among HIV-permissive cell types will be reflected in the mode of CTL-driven immune escape observed in HIV-1 sequences derived from patients. The combined effects of HLA-restricted mutations (impaired epitope processing or reduced binding to MHC) will tend to impair the ability of CTL to recognize infected cells. A comparison of early mutations occurring within the first year of infection with mutations occurring later during chronic infection in this large cohort of HIV-infected persons (using Gag, Pol and Nef sequences) show that this mechanism is operative during both acute and chronic phases of the infection (data not shown). Future studies of mutational escape in other HIV genes will be especially interesting in light of these results.
The identification of motifs leading to immune escape is critical to a better understanding of immune failure in chronic infection and to the identification of immune responses with strong antiviral potential. Immune responses driving antigen processing mutations may reduce viral fitness to variable extents (9, 52) although the fold-reduction in in vitro viral fitness required to substantially reduce viral replication in vivo remains to be determined. Additionally the degradation of mutated peptides may lead to the production of novel HIV epitopes and possibly to new CTL responses (53). Understanding and predicting how the virus and CD8 T cell responses evolve over time is critical to the development of novel vaccine strategies relying on the selection of various HIV sequences rather than HIV proteins of consensus sequences. It has been proposed to design immunogens that would elicit immune responses against both a consensus antigen and against the most commonly variants predicted to appear in vivo thereafter, pushing the virus to evolve toward unfit variants (54). Identifying sequence signatures leading to impaired epitope presentation -and distinguishing them from irrelevant mutations linked to fitness and antiviral drug resistance- will help to predict patterns of viral immune escape at the population level. In addition recent HIV vaccine design strategies explore the use of conserved areas of the virus as potential T cell-based immunogens (55–57). These fragments may need to be assembled –possibly with linkers- into a synthetic cassette encoding a protein that should remain immunogenic. The identification of flanking and intraepitopic motifs driving the efficiency of production and presentation of HIV epitopes to CD8 T cells provides a novel way to optimize epitope production from immunogens.
Supplementary Material
Acknowledgments
This project was funded by a development award P30 AI060354 from the Harvard Center for AIDS Research (Harvard CFAR) to SLG for initial studies, the Bill and Melinda Gates Foundation (Collaboration for AIDS Vaccine Discovery; to SLG and BDW) for the modulation of epitope production, by the NIAID (PO1 # AI074415-01A1 and AI074415-02S1 ARRA supplement; Special Emphasis Panel Vaccine Design and Acute HIV Infection; to SLG and BDW), and NIAID R01 AI084753 to SLG. ZLB is supported by a New Investigator Award from the Canadian Institutes for Health Research (CIHR). EM is supported by a Master’s Scholarship from the Canadian Association of HIV Research and Abbott Virology. CJB is supported by a Vanier Canada Graduate Scholarship from the CIHR.
References
- 1.Loureiro J, Ploegh HL. Antigen presentation and the ubiquitin-proteasome system in host-pathogen interactions. Adv Immunol. 2006;92:225–305. doi: 10.1016/S0065-2776(06)92006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen TM, Altfeld M, Yu XG, O’Sullivan KM, Lichterfeld M, Le Gall S, John M, Mothe BR, Lee PK, Kalife ET, Cohen DE, Freedberg KA, Strick DA, Johnston MN, Sette A, Rosenberg ES, Mallal SA, Goulder PJ, Brander C, Walker BD. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erickson AL, Kimura Y, Igarashi S, Eichelberger J, Houghton M, Sidney J, McKinney D, Sette A, Hughes AL, Walker CM. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity. 2001;15:883–895. doi: 10.1016/s1074-7613(01)00245-x. [DOI] [PubMed] [Google Scholar]
- 4.Wolfl M, Rutebemberwa A, Mosbruger T, Mao Q, Li HM, Netski D, Ray SC, Pardoll D, Sidney J, Sette A, Allen T, Kuntzen T, Kavanagh DG, Kuball J, Greenberg PD, Cox AL. Hepatitis C virus immune escape via exploitation of a hole in the T cell repertoire. J Immunol. 2008;181:6435–6446. doi: 10.4049/jimmunol.181.9.6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazaro E, Kadie C, Stamegna P, Zhang SC, Gourdain P, Lai NY, Zhang M, Martinez SM, Heckerman D, Le Gall S. Variable HIV peptide stability in human cytosol is critical to epitope presentation and immune escape. J Clin Invest. 2011;121:2480–2492. doi: 10.1172/JCI44932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokomaku Y, Miura H, Tomiyama H, Kawana-Tachikawa A, Takiguchi M, Kojima A, Nagai Y, Iwamoto A, Matsuda Z, Ariyoshi K. Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection. J Virol. 2004;78:1324–1332. doi: 10.1128/JVI.78.3.1324-1332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draenert R, Le Gall S, Pfafferott KJ, Alasdair LJ, Chetty P, Brander C, Holmes EC, Chang SC, Feeney ME, Addo MM, Ruiz L, Ramduth D, Jeena P, Altfeld M, Prado JG, Kiepiela P, Martinez-Picado J, Walker BD, Goulder PJ. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J Exp Med. 2004;199:905–915. doi: 10.1084/jem.20031982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milicic A, Price DA, Zimbwa P, Booth BL, Brown HL, Easterbrook PJ, Olsen K, Robinson N, Gileadi U, Sewell AK, Cerundolo V, Phillips RE. CD8+ T cell epitope-flanking mutations disrupt proteasomal processing of HIV-1 Nef. J Immunol. 2005;175:4618–4626. doi: 10.4049/jimmunol.175.7.4618. [DOI] [PubMed] [Google Scholar]
- 9.Troyer RM, McNevin J, Liu Y, Zhang SC, Krizan RW, Abraha A, Tebit DM, Zhao H, Avila S, Lobritz MA, McElrath MJ, Le Gall S, Mullins JI, Arts EJ. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 2009;5:e1000365. doi: 10.1371/journal.ppat.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen TM, Altfeld M, Geer SC, Kalife ET, Moore C, O’Sullivan M, Desouza KI, Feeney ME, Eldridge RL, Maier EL, Kaufmann DE, Lahaie MP, Reyor L, Tanzi G, Johnston MN, Brander C, Draenert R, Rockstroh JK, Jessen H, Rosenberg ES, Mallal SA, Walker BD. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol. 2005;79:13239–13249. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, Kadie C, Bhattacharya T, Chui C, Szinger J, Mo T, Hogg RS, Montaner JS, Frahm N, Brander C, Walker BD, Harrigan PR. Evidence of Differential HLA Class I-Mediated Viral Evolution in Functional and Accessory/Regulatory Genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore CA, John M, James R, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 13.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, Kuse N, Oka S, Duda A, Prendergast A, Crawford H, Leslie A, Brumme Z, Brumme C, Allen T, Brander C, Kaslow R, Tang J, Hunter E, Allen S, Mulenga J, Branch S, Roach T, John M, Mallal S, Ogwu A, Shapiro R, Prado JG, Fidler S, Weber J, Pybus OG, Klenerman P, Ndung’u T, Phillips R, Heckerman D, Harrigan PR, Walker BD, Takiguchi M, Goulder P. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, Swenson LC, Tao I, Szeto S, Rosato P, Sela J, Kadie CM, Frahm N, Brander C, Haas DW, Riddler SA, Haubrich R, Walker BD, Harrigan PR, Heckerman D, Mallal S. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One. 2009;4:e6687. doi: 10.1371/journal.pone.0006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlson JM, Brumme ZL, Rousseau CM, Brumme CJ, Matthews P, Kadie C, Mullins JI, Walker BD, Harrigan PR, Goulder PJ, Heckerman D. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput Biol. 2008;4:e1000225. doi: 10.1371/journal.pcbi.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolland M, Carlson JM, Manocheewa S, Swain JV, Lanxon-Cookson E, Deng W, Rousseau CM, Raugi DN, Learn GH, Maust BS, Coovadia H, Ndung’u T, Goulder PJ, Walker BD, Brander C, Heckerman DE, Mullins JI. Amino-acid co-variation in HIV-1 Gag subtype C: HLA-mediated selection pressure and compensatory dynamics. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, Prendergast A, Matthews P, Payne R, Rolland M, Raugi DN, Maust BS, Learn GH, Nickle DC, Coovadia H, Ndung’u T, Frahm N, Brander C, Walker BD, Goulder PJ, Bhattacharya T, Heckerman DE, Korber BT, Mullins JI. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J Virol. 2008;82:6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Endert P. Post-proteasomal and proteasome-independent generation of MHC class I ligands. Cell Mol Life Sci. 2011 doi: 10.1007/s00018-011-0662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011 doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 20.Kessler JH, Khan S, Seifert U, Le Gall S, Chow KM, Paschen A, Bres-Vloemans SA, de Ru A, van Montfoort N, Franken KL, Benckhuijsen WE, Brooks JM, van Hall T, Ray K, Mulder A, Doxiadis II, van Swieten PF, Overkleeft HS, Prat A, Tomkinson B, Neefjes J, Kloetzel PM, Rodgers DW, Hersh LB, Drijfhout JW, van Veelen PA, Ossendorp F, Melief CJ. Antigen processing by nardilysin and thimet oligopeptidase generates cytotoxic T cell epitopes. Nat Immunol. 2011;12:45–53. doi: 10.1038/ni.1974. [DOI] [PubMed] [Google Scholar]
- 21.Parmentier N, Stroobant V, Colau D, de Diesbach P, Morel S, Chapiro J, van Endert P, Van den Eynde BJ. Production of an antigenic peptide by insulin-degrading enzyme. Nat Immunol. 2010;11:449–454. doi: 10.1038/ni.1862. [DOI] [PubMed] [Google Scholar]
- 22.Beninga J, Rock KL, Goldberg A. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J Biol Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 23.Towne CF, I, York A, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL. Leucine aminopeptidase is not essential for trimming peptides in the cytosol or generating epitopes for MHC class I antigen presentation. J Immunol. 2005;175:6605–6614. doi: 10.4049/jimmunol.175.10.6605. [DOI] [PubMed] [Google Scholar]
- 24.Towne CF, I, York A, Neijssen J, Karow ML, Murphy AJ, Valenzuela DM, Yancopoulos GD, Neefjes JJ, Rock KL. Puromycin-sensitive aminopeptidase limits MHC class I presentation in dendritic cells but does not affect CD8 T cell responses during viral infections. J Immunol. 2008;180:1704–1712. doi: 10.4049/jimmunol.180.3.1704. [DOI] [PubMed] [Google Scholar]
- 25.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 26.York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg A, Rock KL. The ER aminopeptidase ERAPI enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat Immunol. 2002;3:1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 27.Tanioka T, Hattori A, Masuda S, Nomura Y, Nakayama H, Mizutani S, Tsujimoto M. Human leukocyte-derived arginine aminopeptidase. The third member of the oxytocinase subfamily of aminopeptidases. J Biol Chem. 2003;278:32275–32283. doi: 10.1074/jbc.M305076200. [DOI] [PubMed] [Google Scholar]
- 28.York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci U S A. 2006;103:9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchard N, Kanaseki T, Escobar H, Delebecque F, Nagarajan NA, Reyes-Vargas E, Crockett DK, Raulet DH, Delgado JC, Shastri N. Endoplasmic reticulum aminopeptidase associated with antigen processing defines the composition and structure of MHC class I peptide repertoire in normal and virus-infected cells. J Immunol. 2010;184:3033–3042. doi: 10.4049/jimmunol.0903712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hearn A, I, York A, Bishop C, Rock KL. Characterizing the specificity and cooperation of aminopeptidases in the cytosol and endoplasmic reticulum during MHC class I antigen presentation. J Immunol. 2010;184:4725–4732. doi: 10.4049/jimmunol.0903125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hearn A, I, York A, Rock KL. The specificity of trimming of MHC class I-presented peptides in the endoplasmic reticulum. J Immunol. 2009;183:5526–5536. doi: 10.4049/jimmunol.0803663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schatz MM, Peters B, Akkad N, Ullrich N, Martinez AN, Carroll O, Bulik S, Rammensee HG, van Endert P, Holzhutter HG, Tenzer S, Schild H. Characterizing the N-terminal processing motif of MHC class I ligands. J Immunol. 2008;180:3210–3217. doi: 10.4049/jimmunol.180.5.3210. [DOI] [PubMed] [Google Scholar]
- 33.Del Val M, Schlicht HJ, Ruppert T, Reddehase MJ, Kosinowski UH. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 1991;66:1145–1153. doi: 10.1016/0092-8674(91)90037-y. [DOI] [PubMed] [Google Scholar]
- 34.Yellen-Shaw AJ, Wherry EJ, Dubois GC, Eisenlohr LC. Point mutation flanking a CTL epitope ablates in vitro and in vivo recognition of a full-length viral protein. J Immunol. 1997;158:3227–3234. [PubMed] [Google Scholar]
- 35.Ossendorp F, Eggers M, Neisig A, Ruppert T, Groettrup M, Sijts A, Mengede E, Kloetzel PM, Neefjes J, Koszinowski U, Melief C. A single residue exchange within a viral CTL epitope alters proteasome-mediated degradation resulting in lack of antigen presentation. Immunity. 1996;5:115–124. doi: 10.1016/s1074-7613(00)80488-4. [DOI] [PubMed] [Google Scholar]
- 36.Lazaro E, Godfrey SB, Stamegna P, Ogbechie T, Kerrigan C, Zhang M, Walker BD, Le Gall S. Differential HIV epitope processing in monocytes and CD4 T cells affects cytotoxic T lymphocyte recognition. J Infect Dis. 2009;200:236–243. doi: 10.1086/599837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. J Clin Invest. 2007;117:3563–3575. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kavanagh DG, Kaufmann DE, Sunderji S, Frahm N, Le Gall S, Boczkowski D, Rosenberg ES, Stone DR, Johnston MN, Wagner BS, Zaman MT, Brander C, Gilboa E, Walker BD, Bhardwaj N. Expansion of HIV-specific CD4+ and CD8+ T cells by dendritic cells transfected with mRNA encoding cytoplasm- or lysosome-targeted Nef. Blood. 2005 doi: 10.1182/blood-2005-04-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yusim K, Korber BTM, Brander C, Haynes BF, Koup RA, Moore JP, Walker BD, Watkins DI. HIV Molecular Immunology 2009. Los Alamos National Laboratory, Theoretical Biology and Biophysics; Los Alamos, New Mexico: 2009. [Google Scholar]
- 40.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundegaard C, Lund O, Nielsen M. Prediction of epitopes using neural network based methods. J Immunol Methods. 2011;374:26–34. doi: 10.1016/j.jim.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goulder PJ, Bunce M, Krausa P, McIntyre K, Crowley S, Morgan B, Edwards A, Giangrande P, Phillips RE, McMichael AJ. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res Hum Retrovir. 1996;12:1691–1698. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- 43.Frahm N, Adams S, Kiepiela P, Linde CH, Hewitt HS, Lichterfeld M, Sango K, Brown NV, Pae E, Wurcel AG, Altfeld M, Feeney ME, Allen TM, Roach T, St John MA, Daar ES, Rosenberg E, Korber B, Marincola F, Walker BD, Goulder PJ, Brander C. HLA-B63 presents HLA-B57/B58-restricted cytotoxic T-lymphocyte epitopes and is associated with low human immunodeficiency virus load. J Virol. 2005;79:10218–10225. doi: 10.1128/JVI.79.16.10218-10225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlson JM, Brumme ZL. HIV evolution in response to HLA-restricted CTL selection pressures: a population-based perspective. Microbes Infect. 2008;10:455–461. doi: 10.1016/j.micinf.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 45.Frahm N, Baker B, Brander C. Identification and Optimal Definition of HIV-Derived Cytotoxic T Lymphocyte (CTL) Epitopes for the Study of CTL Escape, Functional Avidity and Viral Evolution. In: Korber BT, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI, editors. HIV Molecular Immunology 2008. Los Alamos National Laboratory, Theoretical Biology and Biophysics; Los Alamos, New Mexico: 2008. p. 1-A. [Google Scholar]
- 46.Lopez D, Gil-Torregrosa BC, Bergman C, Del Val M. Sequential cleavage by metallopeptidases and proteasomes is involved in processing HIV-1 ENV epitope for endogenous MHC class I antigen presentation. J Immunol. 2000;164:5070–5077. doi: 10.4049/jimmunol.164.10.5070. [DOI] [PubMed] [Google Scholar]
- 47.Kimura Y, Gushima T, Rawale S, Kaumaya P, Walker CM. Escape mutations alter proteasome processing of major histocompatibility complex class I-restricted epitopes in persistent hepatitis C virus infection. J Virol. 2005;79:4870–4876. doi: 10.1128/JVI.79.8.4870-4876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seifert U, Liermann H, Racanelli V, Halenius A, Wiese M, Wedemeyer H, Ruppert T, Rispeter K, Henklein P, Sijts A, Hengel H, Kloetzel PM, Rehermann B. Hepatitis C virus mutation affects proteasomal epitope processing. J Clin Invest. 2004;114:250–259. doi: 10.1172/JCI20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timm J, Lauer GM, Kavanagh DG, Sheridan I, Kim AY, Lucas M, Pillay T, Ouchi K, Reyor LL, Zur Wiesch JS, Gandhi RT, Chung RT, Bhardwaj N, Klenerman P, Walker BD, Allen TM. CD8 epitope escape and reversion in acute HCV infection. J Exp Med. 2004;200:1593–1604. doi: 10.1084/jem.20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chateau MT, Robert-Hebmann V, Devaux C, Lazaro JB, Canard B, Coux O. Human monocytes possess a serine protease activity capable of degrading HIV-1 reverse transcriptase in vitro. Biochem Biophys Res Commun. 2001;285:863–872. doi: 10.1006/bbrc.2001.5252. [DOI] [PubMed] [Google Scholar]
- 51.Fujiwara M, Takiguchi M. HIV-1-specific CTLs effectively suppress replication of HIV-1 in HIV-1-infected macrophages. Blood. 2007 doi: 10.1182/blood-2006-07-037481. [DOI] [PubMed] [Google Scholar]
- 52.Schneidewind A, Brockman MA, Yang R, Adam RI, Li B, Le Gall S, Rinaldo CR, Craggs SL, Allgaier RL, Power KA, Kuntzen T, Tung CS, LaBute MX, Mueller SM, Harrer T, McMichael AJ, Goulder PJ, Aiken C, Brander C, Kelleher AD, Allen TM. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feeney ME, Tang Y, Pfafferott K, Roosevelt KA, Draenert R, Trocha A, Yu XG, Verrill C, Allen T, Moore C, Mallal S, Burchett S, McIntosh K, Pelton SI, St John MA, Hazra R, Klenerman P, Altfeld M, Walker BD, Goulder PJ. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J Immunol. 2005;174:7524–7530. doi: 10.4049/jimmunol.174.12.7524. [DOI] [PubMed] [Google Scholar]
- 54.Allen TM, Altfeld M. Crippling HIV one mutation at a time. J Exp Med. 2008;205:1003–1007. doi: 10.1084/jem.20080569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahirel V, Shekhar K, Pereyra F, Miura T, Artyomov M, Talsania S, Allen TM, Altfeld M, Carrington M, Irvine DJ, Walker BD, Chakraborty AK. Coordinate linkage of HIV evolution reveals regions of immunological vulnerability. Proc Natl Acad Sci U S A. 2011;108:11530–11535. doi: 10.1073/pnas.1105315108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nickle DC, Rolland M, Jensen MA, Pond SL, Deng W, Seligman M, Heckerman D, Mullins JI, Jojic N. Coping with viral diversity in HIV vaccine design. PLoS Comput Biol. 2007;3:e75. doi: 10.1371/journal.pcbi.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rolland M, Nickle DC, Mullins JI. HIV-1 group M conserved elements vaccine. PLoS Pathog. 2007;3:e157. doi: 10.1371/journal.ppat.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.