Summary
Genetic diversification of Helicobacter pylori adhesin genes may allow adaptation of adherence properties to facilitate persistence despite host defenses. The sabA gene encodes an adhesin that binds sialyl-Lewis antigens on inflamed gastric tissue. We found variability in the copy number and locus of the sabA gene and the closely related sabB and omp27 genes due to gene conversion among 51 North American pediatric H. pylori strains. We determined that sabB to sabA gene conversion is predominantly the result of intra-genomic recombination and RecA, RecG, and AddA influence the rate at which it occurs. Although all clinical strains had at least one sabA gene copy, sabA and sabB were lost due to gene conversion at similar rates in vitro, suggesting host selection to maintain the sabA gene. sabA gene duplication resulted in increased SabA protein production and increased adherence to sialyl-Lewis antigens and mouse gastric tissue. In conclusion, gene conversion is a mechanism for H. pylori to regulate sabA expression level and adherence.
Keywords: Helicobacter pylori, sabA gene, adhesin, adherence, gene conversion, genetic diversity
Introduction
Helicobacter pylori, which infects the stomach of over half the world’s population, is generally acquired in childhood and can persist for the life of the infected host by evading host immune defenses and adapting to changing conditions in the stomach. H. pylori causes persistent gastritis (inflammation of the gastric mucosa) in all who are infected and this eventually leads to peptic ulcers and gastric cancer in a subset of infected individuals.
H. pylori exhibits a high level of genetic diversity, and genetic differences among H. pylori strains contribute to the differences in disease outcome among infected patients (Atherton et al., 1995; Blaser et al., 1995; Covacci et al., 1993; Gerhard et al., 1999; Nomura et al., 2002). Furthermore, genetic changes during the course of infection could allow H. pylori to adapt and persist in the face of host defenses, and thus would have an important role in the ability of H. pylori to maintain a life-long infection.
Genetic changes in H. pylori genes that encode outer membrane proteins (omp genes) are particularly likely to facilitate adaptation of H. pylori to the changing environment in the host. There are over 60 omp genes in the H. pylori genome and some are known to be adhesins (Ilver et al., 1998; Mahdavi et al., 2002; Odenbreit et al., 1999). The middle regions of the omp genes are variable, but the 5′ and 3′ ends are conserved. In some cases, the percent nucleotide identity at the 5′ and 3′ ends is high enough to allow for recombination between members of the gene family that result in gene conversions and switching of loci. This has been observed among the omp gene babA, which encodes the adhesin that binds Lewisb antigens on gastric tissue, and two closely related omp genes, babB and babC (Colbeck et al., 2006; Hennig et al., 2006; Pride and Blaser, 2002).
The omp gene sabA (omp17, hopP) encodes the adhesin that binds sialyl-Lewis antigens that are expressed on inflamed gastric tissue (Mahdavi et al., 2002). H. pylori infection induces gastritis and then is able to exploit receptors expressed by the inflamed tissue for close adherence. Adherence to the gastric epithelium can be beneficial to H. pylori because it helps the bacteria to avoid clearance and brings them closer to nutrients. However, adherence can also be detrimental in the presence of host immune responses that are most intense at the gastric epithelial surface. Therefore, H. pylori adherence properties may need to be dynamic for H. pylori to maintain a stable colonization. Genetic diversification of adhesin genes could allow H. pylori to adapt by constantly generating new variants.
The 5′ and 3′ ends of sabA share the highest nucleotide identity (80–100%) with two other omp genes, sabB (omp16, hopO) and omp27 (hopQ). Although the function of the sabB and omp27 genes is not known, both may also have a role in adherence (Loh et al., 2008; Yamaoka et al., 2002). Comparison of the gene content of fully sequenced H. pylori strains from different global regions found sabA, sabB, and omp27 copy number variability (Kawai et al., 2011), which appears could have resulted from recombination among these three genes. SabA and SabB expression can be switched on and off by slipped-strand mispairing in a 5′ dinucleotide repeat region, allowing translational phase variation (Goodwin et al., 2008). Genetic diversification resulting from recombination among the sabA, sabB, and omp27 genes could be another mechanism for H. pylori to regulate its adherence properties.
To better understand how H. pylori employs genetic variation to regulate production of the SabA adhesin, we investigated the copy number and locus of the sabA gene among North American pediatric H. pylori strains and found evidence of recombination with the sabB and omp27 genes. To explore the consequences of sabA gene duplication, we examined SabA protein production and binding to sialyl-Lewisx (sLex) as well as mouse gastric tissue in isogenic strains with gene configurations observed in our clinical strain collection. We also measured and compared the rates of gene conversion at the sabA, sabB, and omp27 loci in vitro and investigated the role of natural competence and recombination proteins as potential mediators of these gene conversion events.
Results
Variability in copy number and locus of sabA, sabB, and omp27 genes among clinical H. pylori strains
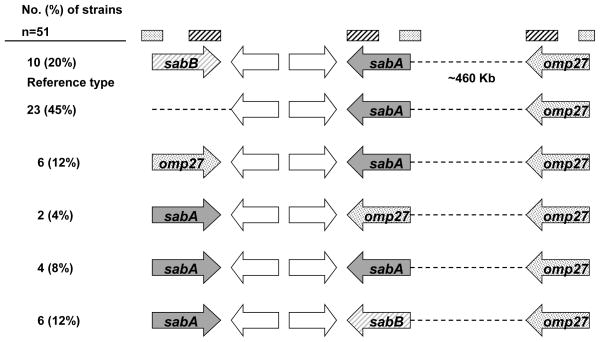
The gene content of the sabB (jhp0659), sabA (jhp0662), and omp27 (jhp1103) loci (as defined by the H. pylori reference strain J99) was characterized for 51 of the 53 clinical H. pylori strains collected in a study of North American pediatric strains (Talarico et al., 2009) (Figure 1). Two strains (both isolated from the same patient) failed to produce PCR product despite sufficient DNA quantity and quality. Of the 51 characterized strains, 10 (20%) had the sabB gene at the sabB locus, 23 (45%) did not have a gene at the sabB locus, and 18 (35%) had either the sabA or omp27 gene at the sabB locus. At the sabA locus, 43 (84%) of the 51 strains had sabA and 8 (16%) had either sabB or omp27. All 51 strains had the omp27 gene at the omp27 locus. The sabB gene was absent from the genome of 35 (69%) of the 51 strains.
Figure 1.
Configuration of the sabB (jhp0659), sabA (jhp0662), and omp27 (jhp1103) genes in the sequenced H. pylori reference strain J99 and H. pylori strain NSH57 and 51 clinical H. pylori strains. J99 and NSH57 share the same configuration denoted as reference type. Dotted and striped bars at the top represent the regions of homology (80 – 100% nucleotide identity) among these three omp genes that may direct recombination events.
Sequence comparison suggests recent intra-strain gene conversion events
For strains having two identified copies of the sabA or omp27 gene, the two copies were sequenced and compared to examine whether the second copy resulted from an intra-strain gene conversion event, indicated by a high level of nucleotide identity. Of the four strains with two identified sabA genes, two strains appeared to have had a recent intra-strain gene conversion event. One strain had two sabA genes that differed by one dinucleotide repeat and the other strain had two sabA genes that differed by 11 SNPs, all within the last 140 bp. The other two strains had two divergent sabA genes (126 and 304 SNPs, 4 and 26 indels, respectively), suggesting that either one copy resulted from recombination with a distantly related allele, perhaps from a super-infecting strain via natural transformation, or that gene conversion occurred relatively longer ago followed by sequence divergence. Of the eight strains that had two identified omp27 genes, six had one Type I allele and one Type II allele, two highly divergent allele types (Cao and Cover, 2002), indicating that one omp27 gene was not a duplication of the other. The other two strains both had two copies of an omp27 Type I allele that appeared to be the result of a recent intra-strain gene conversion event. One strain had two identical omp27 genes and the other strain had two omp27 genes that differed by four SNPs.
Gene conversion occurs at a similar rate at the sabA and sabB loci and at a lower rate at the omp27 locus in vitro
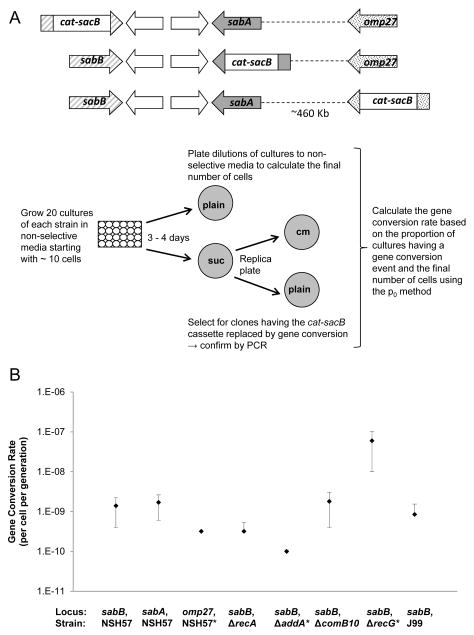
In the clinical strains, gene conversion involved duplication of the sabA or omp27 gene and loss of the sabB gene. This could be due to homologous recombination being favored at the sabB locus or to host selective pressures to preserve the sabA and omp27 genes. To investigate whether the pattern of gene conversion in the clinical strains is due to homologous recombination being favored at the sabB locus, the rate of gene conversion at the sabA, sabB, and omp27 loci in the absence of selection were compared by fluctuation analysis. For these studies the unique regions of sabA, sabB or omp27 were replaced by a cassette allowing both positive (cat; chloramphenicol resistance) and negative (sacB, sucrose sensitivity) selection. We grew parallel cultures in non-selective media starting from a very small number of cells for ~24 generations and then identified clones having spontaneous gene conversion (Figure 2A). sabA duplication at the sabB locus and sabB duplication at the sabA locus occurred at a similar rate, indicating that the sabA gene can be readily lost due to gene conversion in the absence of selective pressures. We did not observe omp27 duplication in any experiment and gene conversion at the omp27 locus was not observed in any of 29 cultures examined (Table 3, Figure 2B). Our in vitro gene conversion experiments were performed in strain NSH57, a derivative of the sequenced human clinical isolate G27 (Baldwin et al., 2007; Baltrus et al., 2009). To explore the generality of our findings, we repeated the gene conversion experiment in a second clinical isolate, J99 (Alm et al., 1999). The rate of sabA gene conversion at the sabB locus was similar between the J99 and NSH57 strains (Table 3, Figure 2B).
Figure 2.
Gene conversion rates and 95% confidence intervals as determined by fluctuation analysis. A) Experimental methods used to conduct fluctuation analysis. H. pylori strains were engineered with a cat-sacB cassette in place of the unique middle region of either the sabB, sabA, or omp27 gene. The 5′ and 3′ homologous ends were left to allow recombination to occur. Twenty cultures of the engineered strain were grown from ~10 cells in non-selective media for ~24 generations. At the end of the incubation period, dilutions of the cultures were plated onto non-selective (plain) plates to determine the final number of cells in the culture. Clones having a spontaneous gene conversion will be sucrose-resistant and chloramphenicol-sensitive due to replacement of the cat-sacB cassette by an omp gene. Clones having a gene conversion were identified by selection on media containing sucrose (suc) followed by replica plating onto non-selective media and media containing chloramphenicol (cm). PCR was used to confirm replacement of the cat-sacB cassette with a duplicated copy of an omp gene. Gene conversion rates were calculated using the p0 method (Foster, 2006). B) Gene conversion rates at the sabB locus (duplication of sabA, loss of sabB), the sabA locus (duplication of sabB, loss of sabA), and the omp27 locus in H. pylori strain NSH57 and at the sabB locus in a recA deletion mutant, an addA deletion mutant, a comB10 deletion mutant, a recG deletion mutant, and H. pylori strain J99. Gene conversion was not observed at the omp27 locus and the lower limit of detection for the assay was 3.2 × 10−10 gene conversions per cell per generation. Asterisk indicates that the 95% confidence interval does not overlap with the 95% confidence interval for the gene conversion rate at the sabB locus in the NSH57 background.
Table 3.
Gene conversion rates and 95% confidence limits as determined by fluctuation analysis using the p0 method
| Locus | Strain background | Experiment | Proportion of cultures having gene conversion | Final no. cells | Gene Conversion Rate (per cell per generation) | 95% confidence limits | |
|---|---|---|---|---|---|---|---|
| sabB | wild-type, NSH57 | 1 | 5 of 16 | 1.0 × 108 | 3.8 × 10−9 | 8.9 × 10−10 | 7.8 × 10−9 |
| sabB | wild-type, NSH57 | 2 | 4 of 17 | 3.6 × 108 | 7.5 × 10−10 | 9.5 × 10−11 | 1.6 × 10−9 |
| sabB | wild-type, NSH57 | combined | 9 of 33 | 2.3 × 108 | 1.4 × 10−9 | 5.6 × 10−10 | 2.4 × 10−9 |
| sabA | wild-type, NSH57 | 1 | 7 of 18 | 2.3 × 108 | 2.1 × 10−9 | 7.8 × 10−10 | 4.1 × 10−9 |
| sabA | wild-type, NSH57 | 2 | 4 of 19 | 1.9 × 108 | 1.2 × 10−9 | 1.5 × 10−10 | 2.6 × 10−9 |
| sabA | wild-type, NSH57 | combined | 11 of 37 | 2.1 × 108 | 1.7 × 10−9 | 7.7 × 10−10 | 2.8 × 10−9 |
| omp27 | wild-type, NSH57 | 1 | 0 of 10 | 1.0 × 108 | < 1.1 × 10−9 | ||
| omp27 | wild-type, NSH57 | 2 | 0 of 19 | 1.1 × 108 | < 4.9 × 10−10 | ||
| omp27 | wild-type, NSH57 | combined | 0 of 29 | 1.1 × 108 | < 3.2 × 10−10* | ||
| sabB | ΔcomB10, NSH57 | 1 | 4 of 19 | 6.3 × 107 | 3.8 × 10−9 | 4.4 × 10−10 | 8.0 × 10−9 |
| sabB | ΔcomB10, NSH57 | 2 | 3 of 20 | 1.5 × 108 | 1.1 × 10−9 | 4.3 × 10−11 | 2.4 × 10−9 |
| sabB | ΔcomB10, NSH57 | combined | 7 of 39 | 1.1 × 108 | 1.8 × 10−9 | 5.5 × 10−10 | 3.2 × 10−9 |
| sabB | ΔrecA, NSH57 | 1 | 0 of 19 | 8.0 × 107 | < 6.8 × 10−10 | ||
| sabB | ΔrecA, NSH57 | 2 | 2 of 20 | 1.3 × 108 | 8.1 × 10−10 | 2.4 × 10−10 | 2.0 × 10−9 |
| sabB | ΔrecA, NSH57 | 3 | 0 of 19 | 1.1 × 108 | < 4.9 × 10−10 | ||
| sabB | ΔrecA, NSH57 | combined | 2 of 58 | 1.1 × 108 | 3.2 × 10−10 | 1.1 × 10−10 | 7.7 × 10−10 |
| sabB | ΔrecG, NSH57 | 1 | 2 of 10 | 1.9 × 106 | 1.2 × 10−7 | 2.5 × 10−8 | 3.1 × 10−7 |
| sabB | ΔrecG, NSH57 | 2 | 5 of 20 | 5.6 × 106 | 5.1 × 10−8 | 1.1 × 10−8 | 1.0 × 10−7 |
| sabB | ΔrecG, NSH57 | combined | 7 of 30 | 4.4 × 106 | 6.0 × 10−8* | 1.9 × 10−8 | 1.1 × 10−7 |
| sabB | ΔaddA, NSH57 | 1 | 0 of 10 | 3.9 × 108 | < 2.7 × 10−10 | ||
| sabB | ΔaddA, NSH57 | 2 | 1 of 13 | 4.6 × 108 | 1.7 × 10−10 | 1.4 × 10−10 | 5.5 × 10−10 |
| sabB | ΔaddA, NSH57 | combined | 1 of 23 | 4.3 × 108 | 1.0 × 10−10* | 9.1 × 10−11 | 3.2 × 10−10 |
| sabB | wild-type, J99 | 1 | 3 of 18 | 1.5 × 108 | 1.2 × 10−9 | 3.7 × 10−11 | 2.8 × 10−9 |
| sabB | wild-type, J99 | 2 | 2 of 19 | 1.9 × 108 | 5.9 × 10−10 | 1.7 × 10−10 | 1.5 × 10−9 |
| sabB | wild-type, J99 | combined | 5 of 37 | 1.7 × 108 | 8.5 × 10−10 | 1.5 × 10−10 | 1.7 × 10−9 |
95% confidence interval does not overlap with the 95% confidence interval for the gene conversion rate at the sabB locus in the NSH57 background.
sabA gene conversion predominantly results from intra-genomic recombination and occurs at a rate influenced by RecA, AddA, and RecG
Since H. pylori is naturally competent for the uptake of exogenous DNA, sabA gene conversion may result from natural transformation with DNA from neighboring H. pylori cells. To assess the role of natural competence in sabA gene conversions, the rate of sabA gene conversion at the sabB locus of a comB10 mutant, which cannot take up exogenous DNA for natural transformation, was compared to that of wild-type. The comB10 mutant had a sabA gene conversion rate similar to that of wild-type (Table 3, Figure 2B) suggesting that sabA gene conversion is largely the result of intra-genomic recombination in the in vitro assay. To investigate the mediators of homologous recombination in sabA gene conversions, the rate of sabB to sabA gene conversion in a recA mutant, an addA mutant, and a recG mutant was compared to that of wild-type (Table 3, Figure 2B). RecA, which facilitates homology-dependent strand exchange, and AddA, a nuclease-helicase involved in double-strand break repair, have been shown to promote babA to babB gene conversion (Amundsen et al., 2008) and RecG has previously been implicated in limiting both inter-genomic and intra-genomic recombination (Kang et al., 2004). The recA mutant had a sabA gene conversion rate ~4-fold lower than that of wild-type, although sabA gene conversion was still observed indicating these recombination events can also be RecA-independent. The addA mutant had a sabA gene conversion rate ~14-fold lower and significantly different than that of wild-type, suggesting that these gene conversion events may be initiated by a double-strand break. The recG mutant had a sabA gene conversion rate ~40-fold higher and significantly different than that of wild-type, further supporting the role of RecG in limiting recombination.
Duplication of the sabA gene results in increased production of the SabA adhesion
sabA gene duplication that results in the second copy being expressed from the sabB promoter could result in SabA protein production that is less than or greater than two-fold because of nucleotide sequence differences between the sabB and sabA promoters including differences in the length of a poly-thymine tract. We thus investigated whether sabA expression levels varied depending on whether the sabB promoter was preserved during the recombination event that generated the second copy of sabA at the sabB locus. sabA expression level was measured by qRT-PCR for an NSH57 variant clone (ST4) and two J99 variant clones (ST83 and ST100) that had two sabA genes with the second copy at the sabB locus with the sabB promoter, and a J99 variant clone (ST101) that had two sabA genes with the second copy at the sabB locus with the sabA promoter. The NSH57 and J99 variant clones were selected from a small subset of chloramphenicol-resistant and sucrose-sensitive clones that arose during the gene conversion experiments for which frozen stocks were made prior to confirming a gene conversion event. Two sabA copies resulted in approximately two-fold expression relative to the parental strain regardless of whether the second copy was expressed from the sabA or sabB promoter in both the NSH57 and J99 strain backgrounds (ST4:2.3(2.1–2.6), ST83:1.6(1.1–2.5), ST100:1.6(1.0–2.8), ST101:2.0(1.2–3.1); fold-change(range)).
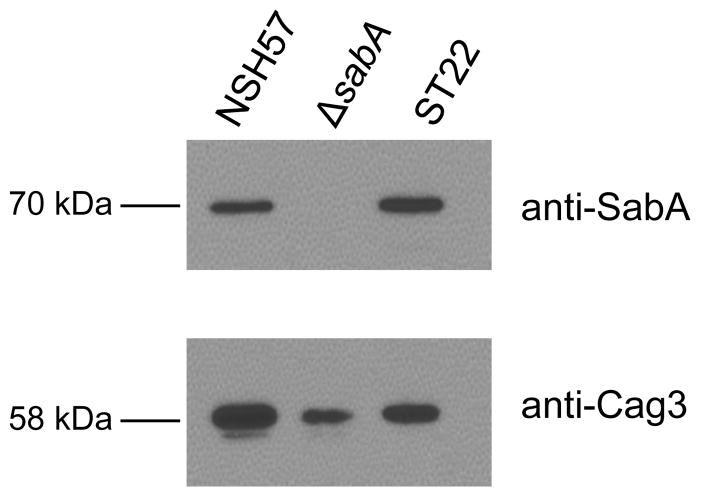
Given that gene conversions resulting in a second copy of the sabA gene resulted in two-fold higher sabA message, regardless of the promoter sequence driving sabA at the sabB locus, we wondered whether sabA gene duplication could be a mechanism for H. pylori to vary the amount of SabA adhesin produced. We were only able to test this in the NSH57 strain background because in the J99 variants one or both sabA genes in these strains were out of frame due to dinucleotide repeat changes within the 5′ end of the coding sequence that altered the reading frame. Immunoblotting was used to compare SabA protein production between NSH57 and an NSH57 variant clone (ST22) that arose during the gene conversion rate experiments. ST22 has two in-frame copies of the sabA gene both controlled by the sabA promoter, with the second sabA gene copy at the sabB locus. As expected, ST22 produced twice as much SabA protein as wild-type NSH57 (adjusted relative SabA density: 2.07±0.15, mean of three immunoblots ± standard deviation, Figure 3).
Figure 3.
Immunoblot analysis of SabA protein production of NSH57 having one sabA gene copy, a sabA deletion mutant, and ST22, a variant clone of NSH57 having two sabA gene copies with the second copy at the sabB locus. The relative amount of SabA protein produced was adjusted to the total protein loaded by stripping the blot and re-probing for Cag3 protein.
Duplication of the sabA gene results in increased adherence to gastric tissue
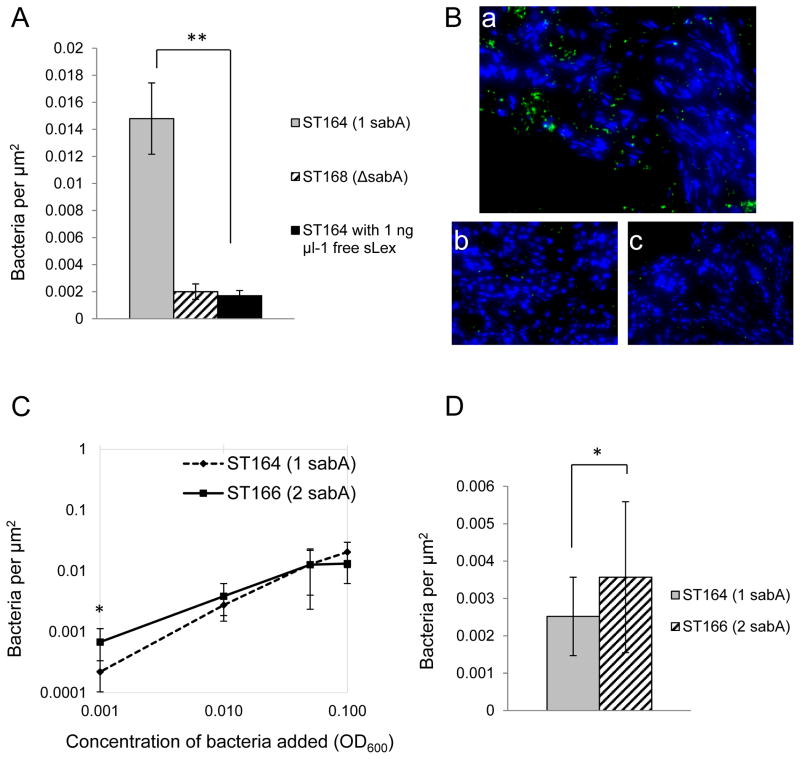
Varying the amount of SabA adhesin produced may allow H. pylori to adapt its adherence properties depending on the selective pressures in the host environment. To test whether a two-fold increase in SabA adhesin has an effect on H. pylori adherence to gastric tissue, an in vitro binding assay of H. pylori to mouse gastric tissue sections was performed. Previous evidence has suggested that H. pylori adherence to mouse gastric tissue is mediated by both the SabA adhesin and the BabA adhesin, which binds to host Lewisb antigens, although mice are known to lack an alpha-1,3/4 fucosyltransferase needed for biosynthesis of Lewisb (Magalhaes et al., 2009). Because BabA-mediated binding could potentially mask the adherence effects of a two-fold increase in SabA adhesin, we disrupted the babA gene of NSH57 and ST22 as well as a sabA deletion mutant to make ST164 (one sabA copy, babA mutant), ST166 (two sabA copies, babA mutant), and ST168 (sabA and babA mutant), respectively. ST168 showed an 86% decrease in adherence compared to ST164 demonstrating that the SabA adhesin contributes to adherence to mouse gastric tissue (ST164: 0.0148 ± 0.0026 adherent bacteria per μm2, mean of 8 regions of tissue ± standard deviation; ST168: 0.0020 ± 0.0006 adherent bacteria per μm2, mean of 5 regions of tissue ± standard deviation; Student’s t-test, p< 0.0001). This is further supported by an 89% decrease in adherence when ST164 is preincubated with sLex (ST164 with 1 ng μl-1 free sLex: 0.0017 ± 0.0004 adherent bacteria per μm2, mean of 5 regions of tissue ± standard deviation; Student’s t-test to compare to ST164, p<0.0001; Figure 4A–B). By incubating the tissue with different concentrations of H. pylori (OD600 0.1, 0.05, 0.01, and 0.001), we saw that adherence of ST164 and ST166 becomes saturated at OD600 0.1 and that a difference in the number of adherent bacteria can be seen at OD600 0.001 and OD600 0.01 (Figure 4C). When the tissue was incubated with H. pylori at OD600 0.001, the strain having two copies of sabA (ST166) had a 250% increase in adherent bacteria compared to the strain ST164 having one sabA copy (ST164: 0.0002 ± 0.0001 adherent bacteria per μm2, ST166: 0.0007 ± 0.0005 adherent bacteria per μm2; mean of two experiments ± standard deviation, Student’s t-test, p = 0.002). When the tissue was incubated with H. pylori at OD600 0.01, ST166 had a 39% increase in binding compared to ST164 (ST164: 0.0028 ± 0.0009 adherent bacteria per μm2, ST166: 0.0039 ± 0.0024 adherent bacteria per μm2; mean of two experiments ± standard deviation, Student’s t-test, p = 0.125). Because the magnitude of the increase in adherence at OD600 0.01 is small and the variation is large, we evaluated the difference in adherence at this bacterial concentration in further experiments to determine whether the difference was statistically significant. When we increased the sample size, the strain having two sabA copies (ST166) showed a 44% increase in adherence compared to the strain having one sabA copy (ST164) and this difference reached statistical significance (ST164: 0.0025 ± 0.0011 adherent bacteria per μm2, ST166: 0.0036 ± 0.0020 adherent bacteria per μm2; mean of 5 experiments ± standard deviation, Student’s t-test, p = 0.008; Figure 4D). These experiments show that, at both the lower concentrations of bacterial cells added, H. pylori having two-fold greater SabA protein production resulting from sabA gene conversion show an increase in adherence to mouse gastric tissue.
Figure 4.
Comparison of adherence to mouse gastric tissue of H. pylori strains ST164 (1 copy sabA, ΔbabA), ST166 (2 copy sabA, ΔbabA), and ST168 (ΔsabA, ΔbabA). A–B) Adherence of ST164 (a), ST168 (b), and ST164 with 1 ng μl-1 free sLex added immediately prior to incubation with the mouse gastric tissue (c). The concentration of H. pylori added to the tissue was OD600 0.1. The mean number adherent bacteria per μm2 was calculated from 8 different fields for ST164 and from 5 different fields for ST168 and ST164 with 1 ng μl-1 free sLex. Results are representative of three independent experiments. Magnification is 400x. C) Quantification of adherence of ST164 and ST166 at different concentrations (OD600 0.1, 0.05, 0.01, 0.001) of H. pylori incubated with the mouse gastric tissue. The mean number of adherent bacteria per μm2 was calculated by combining data from two independent experiments. D) Quantification of adherence of ST164 and ST166. The mouse gastric tissue was incubated with H. pylori at a concentration of OD600 0.01. The mean number of adherent bacteria per μm2 was calculated by combining data from five independent experiments. Statistical significance was evaluated using the Student’s t-test. p < 0.0001 (**) and p < 0.01 (*).
Duplication of the sabA gene results in increased adherence to sialyl-Lewisx
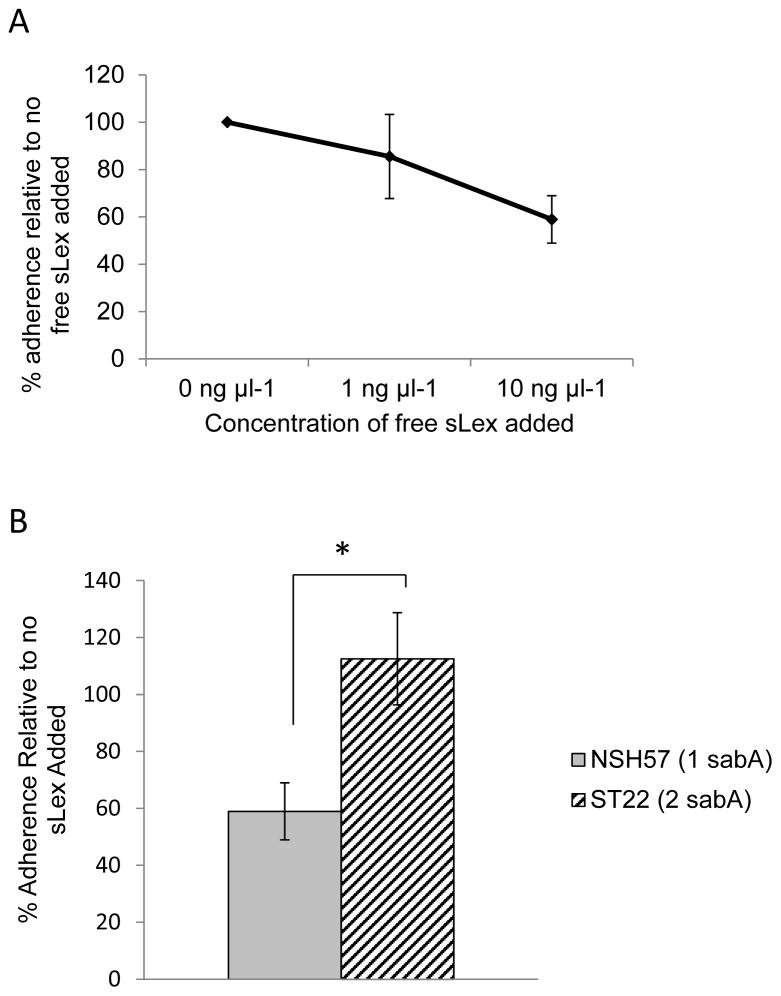
In the stomach Lewis antigens and sialyl-Lewis antigens are expressed on the epithelial cell surface proteins and lipids as well as secreted mucin molecules. In fact cleavage and release of cell associated mucin molecules may be an innate immune strategy to remove bacteria from the epithelium (Linden et al., 2009). To explore the effects of a two-fold increase in SabA adhesin on binding to surface associated sialyl-Lewis antigens in the presence of soluble sialyl-Lewis antigens, an ELISA-based adherence assay with immobilized sLex-coated wells was performed. In the absence of soluble sLex, NSH57 and ST22 both adhered to the immobilized sLex but no significant difference in adherence was detected (adherence ratios: NSH57 1.87±0.4, ST22 1.91±0.4; mean of 5 experiments ± standard deviation; Student’s t-test, p=0.87). Inhibition of H. pylori binding by addition of free sLex was then compared between NSH57 and ST22. Inhibition of NSH57 adherence increased with increasing concentrations of free sLex added (Figure 5A). Addition of 10 ng μl−1 free sLex resulted in a 41% ± 10% decrease (mean of four experiments ± standard deviation) in NSH57 adherence but no decrease in ST22 adherence (Figure 5B) indicating that a two-fold increase in SabA adhesin resulting from sabA gene duplication increases H. pylori adherence to immobilized sialyl-Lewisx.
Figure 5.
SabA adherence to sialyl-Lewisx (sLex) of wild-type H. pylori strain NSH57 having one sabA gene copy and ST22, a variant clone of NSH57 having two sabA gene copies with the second copy at the sabB locus. A) Inhibition of NSH57 adherence to immobilized sLex by addition of 1 ng μl−1 or 10 ng μl−1 free sLex to the H. pylori medium. B) Inhibition of NSH57 and ST22 adherence to immobilized sLex by addition of 10 ng μl−1 free sLex to the H. pylori medium. Percent adherence relative to adding no free sLex was calculated by first normalizing the sLex coated wells to the uncoated wells (sLex coated/uncoated) and then subtracting 1 to adjust the range of values so that a non-binding strain would have a value of 0. Mean ± standard deviation is calculated from three (NSH57 plus 1 ng μl−1 free sLex, ST22 plus 10 ng μl−1 free sLex) and four (NSH57 plus 10 ng μl−1 sLex) independent experiments. Statistical significance of difference in inhibition of adherence was evaluated using the Student’s t-test. P < 0.01 (*)
Discussion
We measured variability in the copy number and locus of the sabA gene among pediatric clinical H. pylori strains and among clones of a single strain resulting from recombination with the closely related sabB and omp27 genes. Changes in sabA copy number generate subpopulations of bacterial cells having differences in SabA adhesin production level that alters adherence to surface associated sialyl-Lewis antigens in the presence of soluble sialyl-Lewis antigens and to gastric tissue. While it has been shown that sabA expression can be turned on or off by translational phase variation (Mahdavi et al., 2002), sabA gene duplication by gene conversion represents an additional mechanism for regulating the amount of SabA protein produced on the bacterial surface. The ability of H. pylori to modulate its adherence properties is important since there are benefits and risks associated with adherence to the host epithelium. H. pylori may persistently infect the stomach by maintaining a balance between an adherent population and a population in the mucus layer.
We found that H. pylori having two-fold greater SabA protein production resulting from sabA gene conversion show an increase in adherence to mouse gastric tissue. To improve our ability to detect a difference in adherence, these experiments were performed in a babA mutant background because a previous study has shown that H. pylori adherence to mouse gastric tissue is mediated by both the SabA adhesin and the BabA adhesin despite the fact that mice are believed to lack the transferases necessary to synthesize Lewisb (Magalhaes et al., 2009). SabA protein production may be especially important in the absence of BabA-mediated adherence. BabA-mediated binding is absent in many human infections either due to the H. pylori strain lacking the babA gene or carrying a nonfunctional babA allele (Gerhard et al., 1999; Hennig et al., 2004) or the host not expressing the Lewisb antigen (Sakamoto et al., 1989). A study of H. pylori clinical isolates that uniformly expressed BabA found that SabA expression was associated with greater H. pylori density but only in patients having weak or no Lewisb expression (Sheu et al., 2006). Furthermore, expression of fucosylated antigens such as Lewisb were shown to reciprocally decrease while sialylated antigens increase during persistent H. pylori infection of Rhesus monkeys due to competition for carbohydrate chains by the sialyl and fucosyl transferases (Linden et al., 2008). Thus, SabA and BabA-mediated adherence may be important at different times during persistent H. pylori infection. These observations also suggest that variation of H. pylori adherence properties may allow this organism to adapt to changing selective pressures in the gastric environment including shifts in the available host receptors over time or upon transmission to a new host.
We also demonstrated that two-fold greater SabA protein production resulted in an increase in adherence to immobilized sialyl-Lewisx but only in the presence of free sialyl-Lewisx. This is biologically relevant because sialyl-Lewis antigens, in addition to being expressed by the gastric epithelium, are also present on mucin glycoproteins in the mucus layer of the stomach. Mucins are synthesized by the mucosal epithelial cells and can limit H. pylori colonization by steric hindrance as well as acting as a releasable decoy when shed into the mucus layer (Linden et al., 2009; McGuckin et al., 2007). Mucins containing sialyl-Lewis antigens in the mucus layer could limit H. pylori adherence to the host epithelium by competing with sialyl-Lewis antigens on the epithelial cells for binding to the SabA adhesin. Increasing SabA adhesin production by gene conversion may be a way for H. pylori to overcome this competitive binding and better adhere to host tissue.
We estimated the rate of sabA gene conversion at the sabB locus to be 1.4 × 10−9 gene conversions per cell per generation, similar to the mutation rates that have been reported for rifampicin resistance (2.98 × 10−8 per cell per division) and streptomycin resistance (4.34 × 10−10 per cell per division) in H. pylori (Baltrus et al., 2008). A previous study measured the frequency of babB gene conversion at the babA locus and estimated from that the rate of babA to babB gene conversions to be 3 × 10−6 gene conversions per cell division (Amundsen et al., 2008). It is not clear whether the difference in sabA and babB gene conversion rates is due to differences in experimental methods or whether recombination between babA and babB occurs at a much higher rate than recombination between sabA and sabB.
While we observed that sabA and sabB are lost due to gene conversion at a similar rate in the absence of selection pressures in vitro, the sabB gene was lost due to an apparent gene conversion in 12 (24%) of the 51 clinical strains whereas all clinical strains had at least one sabA gene copy. This discrepancy suggests there are selective pressures during human infection to maintain the sabA gene. However, other studies have found that the sabA gene is not present in all strains. Twenty percent of 145 Taiwanese strains and 9% of 23 Japanese strains were sabA geno-negative (Shao et al., 2010; Sheu et al., 2006). This could be due to geographic differences among strains or different requirements for retention of sabA between adult and pediatric strains.
Interestingly, 45% of the clinical H. pylori strains did not have an omp gene at the sabB locus and thus would be less able to regulate sabA expression by gene conversion. This suggests that the genetic mechanisms for regulating the SabA adhesin, which binds sialyl-Lewis antigens produced during inflammation (Linden et al., 2008; Mahdavi et al., 2002), may be adapted to the pro-inflammatory nature of the strain. For example, having no omp gene at the sabB locus was correlated with absence of the inflammation-promoting cag pathogenicity island (PAI) (chi-square test, p<0.0001) in this North American pediatric population. In contrast, recent work showed 9/11 Asian strains lack sabB in spite of uniform presence of the cag PAI (Kawai et al., 2011). Although it might be possible for the sabA gene to duplicate by recombining at some other omp locus in the genome, this was not observed in these fully sequenced strains. Asian strains show near universal carriage of a cag PAI with enhanced cellular activity compared to cag PAI of Western strains (Hatakeyama and Higashi, 2005). Future studies are thus needed to examine the competitive advantage of strains having different available genetic mechanisms for regulating sabA expression, especially in the context of H. pylori strain differences in induction of inflammation.
Since the H. pylori genome encodes relatively few proteins that regulate transcription, genetic diversification is likely an important mechanism for H. pylori to modulate phenotypic properties in response to changing conditions in the stomach. This study improves our understanding of how H. pylori employs genetic diversification to adapt adherence properties and cause persistent active infection for decades despite a robust host response.
Experimental Procedures
Strain collection
The study sample consisted of 53 H. pylori strains isolated from 47 North American pediatric patients and was previously described (Talarico et al., 2009). Briefly, gastric biopsies from the antrum of the stomach were obtained during a diagnostic fiberoptic upper endoscopy performed at the discretion of the pediatric gastroenterologist because of the subjects’ persistent gastrointestinal symptoms. Culturing of biopsy samples was performed and single colonies were isolated and typed by RAPD-PCR fingerprinting. Informed consent was obtained from patients or their parents or guardians.
PCR and DNA Sequencing
The gene content of the sabB (jhp0659), sabA (jhp0662), and omp27 (jhp1103) loci of the 53 clinical H. pylori strains was examined by PCR using a combination of primer pairs in which one primer is specific for the gene flanking the locus and the other primer is inside and specific to the sabB, sabA, or omp27 gene (Table 1) and/or by PCR of the entire locus followed by DNA sequencing. The dcuA (jhp0660) and ansB (jhp0661) genes are inverted in some strains in comparison to the J99 reference genome. In these strains, the jhp0658F and the jhp0661F primers were used to amplify the sabB locus and the jhp0660R and jhp0663R primers were used to amplify the sabA locus. PCR with primers inside the sabB gene (sabBFor and sabBRev) was used to confirm absence of the sabB gene in the genome.
Table 1.
H. pylori strains used in this study
| Name | Genotype |
|---|---|
| NSH57 | wild-type H. pylori |
| ST1 | ΔsabB::cat-sacB in NSH57 |
| ST2 | ΔsabA::cat-sacB in NSH57 |
| ST3 | Δomp27::cat-sacB in NSH57 |
| ST27 | ΔcomB10::aphA3 in ST1 |
| ST40 | ΔrecG::aphA3 in ST1 |
| ST42 | ΔrecA::aphA3 in ST1 |
| SWH1 | ΔaddA::aphA3 in ST1 |
| ST4 | ΔsabB::sabA in NSH57 |
| ST22 | ΔsabB::sabA with sabA promoter in NSH57 |
| J99 | wild-type H. pylori |
| ST73 | ΔsabB::cat-sacB in J99 |
| ST83 | ΔsabB::sabA in J99 |
| ST100 | ΔsabB::sabA in J99 |
| ST101 | ΔsabB::sabA with sabA promoter in J99 |
| ST164 | ΔbabA::cat in NSH57 |
| ST166 | ΔbabA::cat in ST22 |
| ST168 | ΔbabA::cat, ΔsabA in NSH57 |
Prior to DNA sequencing, PCR products were purified using the Zymo Research DNA Clean and Concentrator™-5 Kit (Zymo Research). Sequencing was performed by the FHCRC Genomics Shared Resource and results were analyzed using FinchTV Version 1.4.0 (Geospiza) and BLAST (NCBI).
Construction of engineered strains and growth conditions
H. pylori knock-out mutants were generated as previously described (Humbert and Salama, 2008) using a vector-free allelic replacement strategy. Briefly, to generate knock-out mutants, 67–98% of the coding sequence of a gene was replaced by either a cat-sacB cassette, conferring chloramphenicol resistance and sucrose sensitivity, or the aphA3 cassette, conferring kanamycin resistance, by natural transformation with PCR product. The genotype of all mutants was confirmed by PCR and/or DNA sequencing. H. pylori strains (Table 1) were grown as previously described (Humbert and Salama, 2008).
Fluctuation analysis
To measure the gene conversion rate at the sabB, sabA, and omp27 loci, fluctuation analysis (Foster, 2006) was performed using H. pylori strains NSH57 (a derivative of G27) and J99 (Alm et al., 1999; Baldwin et al., 2007; Baltrus et al., 2009) engineered with a cat-sacB cassette in place of the unique middle region of either the sabB, sabA, or omp27 gene (Figure 2A). Between 277 and 368 bp of the 5′ and 3′ homologous ends were left to allow recombination to occur. Twenty cultures of the engineered strain were grown from ~10 cells in 350 μl Brucella broth containing 10% fetal bovine serum in a 24 well plate for 72 hours. A 10 μl sample was taken from four cultures and titred onto non-selective plates to determine the final CFU. The 20 cultures were each plated onto media containing 6% sucrose and sucrose-resistant colonies were then replica plated on non-selective media and media containing 15 μg ml−1 chloramphenicol. To confirm replacement of the cat-sacB cassette with a duplicated copy of an omp gene, chloramphenicol-sensitive and sucrose-resistant clones were first subject to PCR with primers in the flanking genes. For those having a PCR product size consistent with an omp gene in the locus, gene conversion was confirmed either by DNA sequencing or by PCR with one primer specific for the gene flanking the locus and the other primer specific for either sabA, sabB, or omp27, as appropriate. Gene conversion rates were calculated using the p0 method (Foster, 2006). Because a statistical test to compare the gene conversion rates is not available, differences in the gene conversion rates were assessed by comparing the 95% confidence intervals for overlap (Gould et al., 2007; Shaver and Sniegowski, 2003). For each strain, fluctuation analysis was performed at least twice. If the 95% confidence intervals were overlapping, the two rate estimates of the biological replicates were considered not significantly different and the data from the two experiments was combined. The rates of gene conversion at the different loci and in the different strain backgrounds were compared using the 95% confidence intervals for the gene conversion rate obtained from the combined data.
Immunoblotting
To detect SabA protein production, immunoblotting was performed as previously described (Pinto-Santini and Salama, 2009) except the primary antibody was SabA-specific antiserum (Sheu et al., 2006) used at a 1:10,000 dilution and the secondary antibody was goat anti-rabbit conjugated to horseradish peroxidase (Pierce) used at a 1:50,000 dilution. To adjust the relative amounts of SabA protein produced to the amount of protein loaded, the blots were stripped and re-probed with anti-Cag3 as the primary antibody. The relative densities of the SabA and Cag3 bands were determined using the ImageJ software (http://rsb.info.nih.gov/ij/) and SabA band density was normalized to Cag3 band density to compare samples. The mean adjusted relative SabA density was calculated from three technical replicates.
Reverse transcription quantitative PCR (RT-qPCR)
For RNA isolation, liquid cultures were grown to optical density at 600 nm (OD600) 0.4, then collected on 0.45 μm pore size membrane filters (Millipore) and frozen at −80°C prior to RNA extraction, which was performed as previously described (Merrell et al., 2003). Approximately 1 μg RNA was reverse transcribed in a standard reaction with Superscript II (Invitrogen) and 100 μM random octamer primers (Fisher). The resulting cDNA was then used for qPCR, which was performed in a standard reaction using SYBR green on an ABI prism 7900HT sequence detection system (Applied Biosystems). The primers used for qPCR are listed in Table 2. The quantity of sabA cDNA was normalized to the quantity of JHP0298 cDNA to compare samples. The fold-difference in sabA cDNA of the isogenic sabA gene variants were compared using the ΔΔCT Method (Livak and Schmittgen, 2001). For each strain, at least two biological replicates were performed and at least two technical replicates were performed on each biological replicate.
Table 2.
Oligonucleotides used in this study
| Primer | Sequence | Position |
|---|---|---|
| jhp0658F | tgggttgagatcatgcaagcat | nt 323 to 344 of J99 jhp0658 |
| jhp0660R | gatcatgcgtttttgatccctgg | nt 1249 to 1271 of J99 jhp0660 (dcuA) |
| jhp0661F | agggtgcttttacaactcgct | nt 925 to 945 of J99 jhp0661 (ansB) |
| jhp0663R | taggcaaacgcaaccgcttcaa | nt 746 to 767 of J99 jhp0663 |
| jhp1102F | gttaaacccgctctaaactcggtg | nt 42 to 65 ofJ99 jhp1102 |
| jhp1104R | gttatacgccacggcgatggaa | nt 552 to 573 of J99 jhp1104 (deoD) |
| sabB | ctgataaggcacagagattgcct | nt 526 to 548 of J99 jhp0659 (sabB) |
| sabA | aacaccgcgtattgcgttgggta | nt 749 to 771 of J99 jhp0662 (sabA) |
| omp27TypeI | tgccattctcatcggtgtagtg | nt 415 to 436 of J99 jhp1103 (omp27 allele Type I) |
| omp27TypeII | gttttaatggttacttccacc | nt 606 to 626 of Tx30a omp27 gene (allele Type II) |
| sabBFor | aagctcaaggcaatctctgtgc | nt 521 to 542 of HPG27_677 (sabB) |
| sabBRev | ccgcatagtcggttacaggaccag | nt 716 to 739 of HPG27_677 (sabB) |
| sabA_NSH57_RTFor | ctagaggcttgtttaccactgc | nt 389 to 410 of HPG27_680 (sabA) |
| sabA_J99_RTFor | aagtgagatagttccccgcat | nt 357 to 377 of J99 jhp0662 (sabA) |
| sabA_RTRev | acgccaacaacattgagctggt | nt 215 to 236 of HPG27_680 (sabA) |
| JHP0298For | gcgataaaacgccagatccaaagc | nt 845 to 868 of J99 jhp0298 |
| JHP0298Rev | ctgggtattgcggtgctaatggg | nt 1001 to 1023 of J99 jhp0298 |
In vitro Tissue Adherence Assay
An in vitro tissue adherence assay was performed as previously described (Magalhaes et al., 2009). Briefly, paraffin-embedded C57BL/6 mice gastric sections were deparaffinized and rehydrated followed by blocking for two hours at room temperature with blocking buffer (1% periodate-oxidized BSA in PBS containing 0.05% Tween 20). FITC-labeled H. pylori was diluted to an OD600 of either 0.1, 0.05, 0.01, or 0.001 in blocking buffer and 100 μl of the bacterial suspension was then incubated with the tissue section for two hours at room temperature. Inhibition of SabA-mediated bacterial adherence was tested by addition of 1 ng μl-1 sialyl-Lewisx (sLex, Dextra) to the bacterial suspension immediately prior to incubation with the tissue section. Slides were subject to three ten minute washes in PBS containing 0.05% Tween 20 with gentle shaking and stained with DAPI. Mean number of adhered bacteria per square micrometer of tissue was quantified by analyzing at least five different fields of each section under 400X magnification using the ImageJ software. For each strain and condition, at least two biological replicates were performed. Statistical significance of differences in number of adherent bacteria per square micrometer was tested using the Student’s t-test.
ELISA-based Adherence Assay
An ELISA assay to assess H. pylori binding to sLex was performed as previously described (Solnick et al., 2004). Briefly, Immulon 2HB 96 well plates (Thermo Scientific) were coated with immobilized sLex conjugated to human serum albumin and incubated with digoxigenin-labeled H. pylori. Adherent H. pylori were detected using anti-digoxigenin antibody conjugated to peroxidase and ABTS (Roche). Absorbance was measured using an EL808 microplate reader (Bio-Tek Instruments) at a wavelength of 405 nm and normalized to uncoated control wells. To examine the effect of competitive binding with free sLex, either 1 or 10 ng μl−1 sLex was added to the H. pylori media immediately prior to incubation in the sLex-coated 96 well plate. Percent adherence relative to adding no free sLex was calculated by first normalizing the sLex coated wells to the uncoated wells (sLex coated/uncoated) and then subtracting 1 to adjust the range of values so that a non-binding strain would have a value of 0. For each strain, at least two biological and technical replicates were performed.
Acknowledgments
We would like to thank Celso A. Reis and Ana Magalhães for sharing their mouse tissue adherence protocol and Jay Solnick and Lori Hansen for sharing their Lewisb adherence protocol. We would also like to thank Ilana Cohen for providing mouse gastric tissue sections and Marion Dorer for assistance in developing the protocol for the fluctuation analysis. This work was supported by grant AI054423 from the NIH (NRS) and the Anna D. Barker Fellowship in Basic Cancer Research from AACR (ST).
References
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Amundsen SK, Fero J, Hansen LM, Cromie GA, Solnick JV, Smith GR, Salama NR. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol Microbiol. 2008;69:994–1007. doi: 10.1111/j.1365-2958.2008.06336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton JC, Cao P, Peek RM, Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- Baldwin DN, Shepherd B, Kraemer P, Hall MK, Sycuro LK, Pinto-Santini DM, Salama NR. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun. 2007;75:1005–1016. doi: 10.1128/IAI.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA, Guillemin K, Phillips PC. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution. 2008;62:39–49. doi: 10.1111/j.1558-5646.2007.00271.x. [DOI] [PubMed] [Google Scholar]
- Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–2115. [PubMed] [Google Scholar]
- Cao P, Cover TL. Two different families of hopQ alleles in Helicobacter pylori. J Clin Microbiol. 2002;40:4504–4511. doi: 10.1128/JCM.40.12.4504-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbeck JC, Hansen LM, Fong JM, Solnick JV. Genotypic profile of the outer membrane proteins BabA and BabB in clinical isolates of Helicobacter pylori. Infect Immun. 2006;74:4375–4378. doi: 10.1128/IAI.00485-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci U S A. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard M, Lehn N, Neumayer N, Boren T, Rad R, Schepp W, Miehlke S, Classen M, Prinz C. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci U S A. 1999;96:12778–12783. doi: 10.1073/pnas.96.22.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin AC, Weinberger DM, Ford CB, Nelson JC, Snider JD, Hall JD, Paules CI, Peek RM, Jr, Forsyth MH. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology. 2008;154:2231–2240. doi: 10.1099/mic.0.2007/016055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould CV, Sniegowski PD, Shchepetov M, Metlay JP, Weiser JN. Identifying mutator phenotypes among fluoroquinolone-resistant strains of Streptococcus pneumoniae using fluctuation analysis. Antimicrob Agents Chemother. 2007;51:3225–3229. doi: 10.1128/AAC.00336-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama M, Higashi H. Helicobacter pylori CagA: a new paradigm for bacterial carcinogenesis. Cancer Sci. 2005;96:835–843. doi: 10.1111/j.1349-7006.2005.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig EE, Mernaugh R, Edl J, Cao P, Cover TL. Heterogeneity among Helicobacter pylori strains in expression of the outer membrane protein BabA. Infect Immun. 2004;72:3429–3435. doi: 10.1128/IAI.72.6.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig EE, Allen JM, Cover TL. Multiple chromosomal loci for the babA gene in Helicobacter pylori. Infect Immun. 2006;74:3046–3051. doi: 10.1128/IAI.74.5.3046-3051.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert O, Salama NR. The Helicobacter pylori HpyAXII restriction-modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components. Nucleic Acids Res. 2008;36:6893–6906. doi: 10.1093/nar/gkn718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- Kang J, Tavakoli D, Tschumi A, Aras RA, Blaser MJ. Effect of host species on RecG phenotypes in Helicobacter pylori and Escherichia coli. J Bacteriol. 2004;186:7704–7713. doi: 10.1128/JB.186.22.7704-7713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Furuta Y, Yahara K, Tsuru T, Oshima K, Handa N, Takahashi N, Yoshida M, Azuma T, Hattori M, Uchiyama I, Kobayashi I. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiol. 2011;11:104. doi: 10.1186/1471-2180-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden S, Mahdavi J, Semino-Mora C, Olsen C, Carlstedt I, Boren T, Dubois A. Role of ABO secretor status in mucosal innate immunity and H. pylori infection. PLoS Pathog. 2008;4:e2. doi: 10.1371/journal.ppat.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden SK, Sheng YH, Every AL, Miles KM, Skoog EC, Florin TH, Sutton P, McGuckin MA. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLoS Pathog. 2009;5:e1000617. doi: 10.1371/journal.ppat.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loh JT, Torres VJ, Algood HM, McClain MS, Cover TL. Helicobacter pylori HopQ outer membrane protein attenuates bacterial adherence to gastric epithelial cells. FEMS Microbiol Lett. 2008;289:53–58. doi: 10.1111/j.1574-6968.2008.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes A, Gomes J, Ismail MN, Haslam SM, Mendes N, Osorio H, David L, Le Pendu J, Haas R, Dell A, Boren T, Reis CA. Fut2-null mice display an altered glycosylation profile and impaired BabA-mediated Helicobacter pylori adhesion to gastric mucosa. Glycobiology. 2009;19:1525–1536. doi: 10.1093/glycob/cwp131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi J, Sonden B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA, Altraja S, Wadstrom T, Kersulyte D, Berg DE, Dubois A, Petersson C, Magnusson KE, Norberg T, Lindh F, Lundskog BB, Arnqvist A, Hammarstrom L, Boren T. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin MA, Every AL, Skene CD, Linden SK, Chionh YT, Swierczak A, McAuley J, Harbour S, Kaparakis M, Ferrero R, Sutton P. Muc1 mucin limits both Helicobacter pylori colonization of the murine gastric mucosa and associated gastritis. Gastroenterology. 2007;133:1210–1218. doi: 10.1053/j.gastro.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Merrell DS, Thompson LJ, Kim CC, Mitchell H, Tompkins LS, Lee A, Falkow S. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect Immun. 2003;71:6510–6525. doi: 10.1128/IAI.71.11.6510-6525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura AM, Perez-Perez GI, Lee J, Stemmermann G, Blaser MJ. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am J Epidemiol. 2002;155:1054–1059. doi: 10.1093/aje/155.11.1054. [DOI] [PubMed] [Google Scholar]
- Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol Microbiol. 1999;31:1537–1548. doi: 10.1046/j.1365-2958.1999.01300.x. [DOI] [PubMed] [Google Scholar]
- Pinto-Santini DM, Salama NR. Cag3 is a novel essential component of the Helicobacter pylori Cag type IV secretion system outer membrane subcomplex. J Bacteriol. 2009;191:7343–7352. doi: 10.1128/JB.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT, Blaser MJ. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J Mol Biol. 2002;316:629–642. doi: 10.1006/jmbi.2001.5311. [DOI] [PubMed] [Google Scholar]
- Sakamoto S, Watanabe T, Tokumaru T, Takagi H, Nakazato H, Lloyd KO. Expression of Lewisa, Lewisb, Lewisx, Lewisy, siayl-Lewisa, and sialyl-Lewisx blood group antigens in human gastric carcinoma and in normal gastric tissue. Cancer Res. 1989;49:745–752. [PubMed] [Google Scholar]
- Shao L, Takeda H, Fukui T, Mabe K, Han J, Kawata S, Ootani K, Fukao A. Genetic diversity of the Helicobacter pylori sialic acid-binding adhesin (sabA) gene. Biosci Trends. 2010;4:249–253. [PubMed] [Google Scholar]
- Shaver AC, Sniegowski PD. Spontaneously arising mutL mutators in evolving Escherichia coli populations are the result of changes in repeat length. J Bacteriol. 2003;185:6076–6082. doi: 10.1128/JB.185.20.6076-6082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu BS, Odenbreit S, Hung KH, Liu CP, Sheu SM, Yang HB, Wu JJ. Interaction between host gastric Sialyl-Lewis X and H. pylori SabA enhances H. pylori density in patients lacking gastric Lewis B antigen. Am J Gastroenterol. 2006;101:36–44. doi: 10.1111/j.1572-0241.2006.00358.x. [DOI] [PubMed] [Google Scholar]
- Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci U S A. 2004;101:2106–2111. doi: 10.1073/pnas.0308573100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarico S, Gold BD, Fero J, Thompson DT, Guarner J, Czinn S, Salama NR. Pediatric Helicobacter pylori isolates display distinct gene coding capacities and virulence gene marker profiles. J Clin Microbiol. 2009;47:1680–1688. doi: 10.1128/JCM.00273-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y, Kita M, Kodama T, Imamura S, Ohno T, Sawai N, Ishimaru A, Imanishi J, Graham DY. Helicobacter pylori infection in mice: Role of outer membrane proteins in colonization and inflammation. Gastroenterology. 2002;123:1992–2004. doi: 10.1053/gast.2002.37074. [DOI] [PubMed] [Google Scholar]