Abstract
The Fragile X-associated disorders (FXDs) and Friedeich ataxia (FRDA) are genetic conditions resulting from expansion of a trinucleotide repeat in a region of the affected gene that is transcribed but not translated. In the case of the FXDs, pathology results from expansion of CGG•CCG-repeat tract in the 5′ UTR of the FMR1 gene, while pathology in FRDA results from expansion of a GAA•TTC-repeat in intron 1 of the FXN gene. Expansion occurs during gametogenesis or early embryogenesis by a mechanism that is not well understood. Expansion then produces disease pathology in various ways that are not completely understood either. In the case of the FXDs, alleles with 55–200 repeats express higher than normal levels of a transcript that is thought to be toxic, while alleles with >200 repeats are silenced. In addition, alleles with >200 repeats are associated with a cytogenetic abnormality known as a fragile site, which is apparent as a constriction or gap in the chromatin that is seen when cells are grown in presence of inhibitors of thymidylate synthase. FRDA alleles show a deficit of the FXN transcript. This review will address the role of repeat-mediated chromatin changes in these aspects of FXD and FRDA disease pathology.
Keywords: Friedreich ataxia, Fragile X disorders, heterochromatin, repeat-mediated chromatin changes, gene silencing, chromosome fragility, epigenetic dysregulation
1. Introduction
Friedreich ataxia (FRDA; OMIM #229300) and the Fragile X-associated disorders (FXDs), Fragile X syndrome (OMIM #300624), Fragile X-associated tremor and ataxia syndrome (FXTAS; OMIM #300623) and Fragile X-associated primary ovarian insufficiency (FXPOI), are members of a group of human genetic disorders known as the Repeat Expansion Diseases (REDs). These diseases originate from an unstable short tandem repeat tract that is prone to expansion on intergenerational transfer. Pathology arises when the repeat exceeds a critical threshold. In most REDs, there is a direct relationship between repeat number and disease severity and in those disorders that are not congenital there is an inverse relationship between repeat number and the age of onset (see [1] for a more comprehensive review of these diseases).
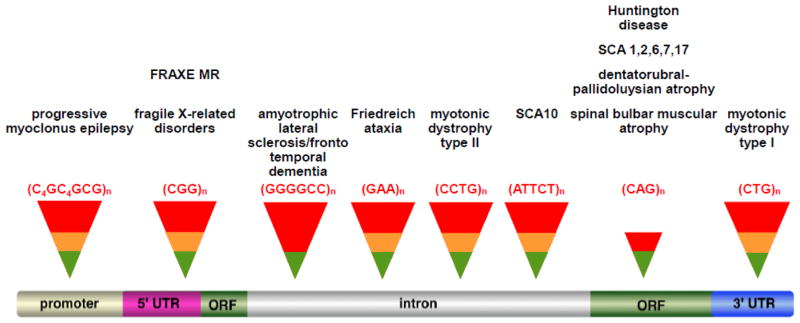
Most REDs arise from expansion of a CAG•CTG-tract that is located in an open reading frame (ORF) and encodes glutamine (Fig. 1). Pathology in these diseases arises primarily from toxicity of the resultant polyglutamine tract although contributions from other mechanisms are also possible (reviewed in [2]). In those disorders where the repeat is outside of the ORF, pathology can arise in a variety of ways some of which are not yet fully understood. The FXDs and FRDA belong to the latter group of diseases.
Fig. 1. Diagrammatic representation of the Repeat Expansion Diseases showing the location of the disease-causing repeat on a generic gene.
(see [1] for a more comprehensive description of these diseases). The diseases shown here arise from pathology that is known to be due either to the presence of the repeat in the open reading frame where it generates a polyglutamine tract in the protein that is toxic or in the non-coding portion of the gene where it can affect gene expression in a variety of ways. Three REDs are not shown, Spinocerebellar ataxia type 8 (SCA8), SCA12 and HD-like 2 (HDL2). The pathological effect in SCA8 is thought to be the result of a combination of having the repeats in the coding sequence of one transcript and in the non-coding region of a transcript synthesized in the antisense direction [136]. SCA12 results from a CAG•CTG-repeat in the 5′ region of the PP2R2B gene. However, there is no evidence that expansion results in polyglutamine production and the mechanism responsible for disease pathology is unknown [137]. In the case of HDL2, the disease is caused by a CTG•CAG expansion mutation in a variably spliced exon of junctophilin-3 in the CTG orientation [138]. This seems to exclude polyglutamine tract as the cause of disease pathology in this disorder as well. However, the source of the pathology remains unclear.
1.1. The Fragile X-associated Disorders (FXDs)
The FXDs arise from expansion of an unstable CGG•CCG-repeat tract in the 5′ untranslated region (UTR) of an X-linked gene called Fragile X mental retardation 1 (FMR1) [3–5]. The FMR1 gene encodes a protein, FMRP, important for learning and memory. One major role of FMRP is to offset the effect of mGluR5 receptor activation in the postsynaptic neuron via a negative effect on the translation of a subset of mRNAs [6].
The FXDs are named for a folate-sensitive fragile site (FS), a gap or constriction of the chromatin, coincident with the FMR1 gene [7, 8]. This site only becomes apparent when the repeat number exceeds 200. Alleles with this number of repeats are referred to as Full Mutation (FM) alleles. In addition to the eponymous FS, carriers of such alleles have FXS, the leading heritable cause of intellectual disability and the major known genetic cause of autism [3, 5]. Depression, anxiety and behavior problems are frequent co-morbid features. A recent study of 21,411 mother-newborn pairs from the general population suggests that the prevalence of FXS is ~1 in 6000 males and <1 in 12,000 females, with a very broad confidence interval [9]. Females with the FM are generally less severely affected because of the protective effect of the second X chromosome.
FMR1 alleles with 55–200 repeats are referred to as premutation (PM) alleles. Estimates of the frequency of PM alleles also varies widely ranging from ~1 in 500 to as many as 1 in 106 women [9]. PM carriers are at risk of two other disorders that are quite different from FXS. Males, in particular, are at risk of FXTAS, a late onset neurodegenerative disorder associated with gait and balance abnormalities as well as cognitive decline and dementia [10]. Female PM carriers are at risk of FXPOI, an ovarian dysfunction disorder that is thought to account for ~11% of familial cases of infertility and ~3.5% of idiopathic cases [11–13]. In addition to fertility problems, affected women often have early onset menstrual irregularities and an earlier than normal age at menopause. These women are also at risk of having a child with a FM allele that arises from the PM allele by repeat expansion either in the oocyte or very early embryo.
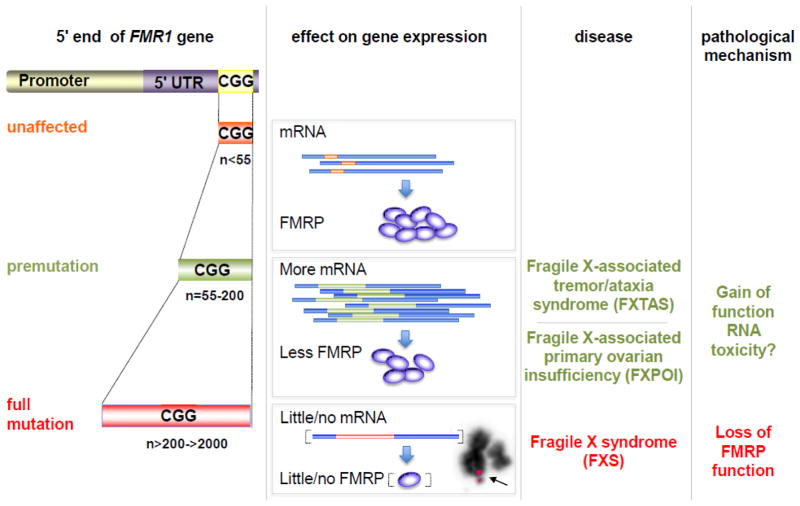
The very different pathologies seen in PM and FM carriers are thought to arise from differences in the effect of the repeats on FMR1 gene expression as illustrated in Fig. 2. In the PM range, FMR1 transcription is elevated and the transcript itself is thought to be deleterious, causing reduced viability of human cells [14], and neurodegeneration in both fly [15] and mouse models [16]. In contrast, in the FM range FMR1 transcripts, and thus FMRP levels, are drastically reduced.
Fig. 2. Illustration of the relationship between CGG•CCG-repeat number, the levels of FMR1 mRNA and FMRP, and disease pathology in the FXDs.
FX premutation alleles produce elevated levels of FMR1 mRNA [86]. However, FMRP levels can be lower than that seen in unaffected alleles due to difficulties in translating transcripts with long CGG-repeat tracts [139, 140]. Carriers of full mutation alleles make little or no protein. They also show a characteristic cytogenetic abnormality, a folate-sensitive fragile site (FS) indicated by the arrow, that is coincident with the repeat. In this case the FS was visualized by hybridization to a BAC (RP11-489K19) probe that spans the FMR1 locus. In the example shown, one sister chromatid has lost the telomeric end of one sister chromatid including any DNA homologous to the probe. A constriction or gap, typical of a fragile site, is seen on the other sister chromatid colocalizing with the FMR1 probe (pink signal).
1.2. FRDA
FRDA is a progressive neurodegenerative disorder with an early onset. It is the most common recessive ataxia with an incidence of 1 in 50,000. Loss of mobility is common during adolescence and there is a high early mortality due to hypertrophic cardiomyopathy. In addition to progressive gait and limb ataxia with associated limb muscle weakness, absent lower limb reflexes, extensor plantar responses, dysarthria, decreased vibratory sense and proprioception are commonly seen. Diabetes occurs in ~10% of FRDA patients (see [17] for full discussion of the clinical presentation). Most cases of FRDA result from expansion of a GAA•TTC-repeat located in the first intron of the Frataxin (FXN) gene [18]. The FXN gene product, frataxin, is active in the mitochondria where it is thought to play an important role in the biogenesis of iron-sulfur clusters. FRDA alleles express FXN at ~20–40% the levels seen in individuals with repeat numbers in the normal range.
It has long been known that the absence of FMRP in FXS results from heterochromatin-mediated gene silencing [4, 19] although the mechanism responsible is not completely understood. However, there is also evidence to suggest that altered chromatin is involved in other ways in these diseases. This review will discuss recent work done in our lab and elsewhere to better understand FXS gene silencing as well as the role of chromatin in repeat expansion, chromosome fragility and in the pathology of FXS, FXTAS, FXPOI and FRDA.
2. The expansion mechanism
The expansion mechanism is thought to be fundamentally different from the generalized microsatellite instability seen in certain cancers in that instability affects a single locus, expansions can be large, involving the addition of hundreds if not thousands of repeats within a single generation, and expansions far outnumber contractions. Indeed there is some evidence to suggest that expansions and contractions occur via different mechanisms [20–22]. The propensity to expand is thought to be related to the ability of all the disease-associated repeats to fold into hairpins, slipped-strand structures, triplexes and tetraplexes/quadruplexes [23].
Most work done to date on the expansion mechanism has involved CAG•CTG-repeats where a complicated and sometimes contradictory picture has emerged (see [24] for a recent review). Work in transgenic mouse models and human cells has implicated a number of chromatin remodeling/epigenetic factors in repeat expansion including DNMT1, the DNA methyltransferase responsible for maintenance methylation [25–27], histone deacetylases [28], CREB binding protein, CBP, a transcriptional coactivator with intrinsic histone acetylase activity [28] and CCCTC-binding factor, CTCF, a transcription factor which can also act to enforce chromatin boundaries [29]. The sequence context of the repeat has also been shown to be an important factor [30–32], as is cell type [30, 33–41]. However, a direct effect of chromatin structure on repeat expansion has not been demonstrated.
Other factors that have been implicated in repeat instability include the mismatch repair proteins MSH2/3 [42–46] and PMS2 [36]. OGG1 has also been implicated in somatic expansion in a mouse model of Huntingon Disease (HD) [142]. However, many of the details remain confusing, with evidence suggesting that the same repeat can expand by different mechanisms depending on the gender of the transmitting parent, the sequence context and the cell type.
In humans, methylated FM alleles are more stable than unmethylated ones [47, 48], a fact consistent with a role for chromatin structure in maintaining repeat stability or a role for transcription through the repeats. Our laboratory has been using a knock-in mouse model of the FX PM to try to understand the expansion mechanism [49]. Since mice and humans are syntenic in the region of the X chromosome in which the FMR1 gene is located, it may be that the expansion mechanism that operates in these animals is similar to that operating at the FMR1 locus in humans.
We have shown that mutations in ATM and ATR, key enzymes involved in the response to DNA damage and stalled replication forks, increase repeat expansion [21, 22]. Since ATM and ATR mutations have different effects on maternal and paternal expansions, our data suggest that there are at least two different expansion mechanisms that can operate at the Fmr1 locus in mice. We have also shown that oxidative damage increases the frequency of both paternally and maternally transmitted expansions [20]. This would be consistent with the observations from mouse models of other REDs that expansion is likely related to aberrant DNA repair. The underlying lesion responsible for repeat expansion may occur less frequently in heterochromatin either because the lesion occurs as a result of transcription or because heterochromatin is less vulnerable to things like oxidative damage. Alternatively, the repair process occurring in transcriptionally active regions may be different from that operating in transcriptionally silent regions. For example, transcription-coupled repair (TCR) is a repair process that is confined to the template strand of transcribed genes [50] that could potentially account for expansion in genes that are transcribed. However, evidence for a role for TCR in models of the CAG•CTG-diseases has been inconsistent [51, 52] and no data are available for CGG•CCG-repeats. Thus much work remains to be done in order to understand the mechanism responsible for repeat expansion in these disorders.
3. Chromosome fragility in FXS
The FS characteristic of FX FM alleles is apparent as a gap, constriction or break in the chromosome that colocalizes with the repeat [7, 8] as illustrated in Fig. 2. In some respects these sites are reminiscent of prematurely condensed chromatin. The human genome contains many other FSs. The common FSs (CFSs) are ubiquitous in the population, while the rare fragile sites are only seen in a subset of individuals. The FX fragile site (FRAXA) belongs to a group of at least 7 rare FSs that are induced by folate-stress or treatment with agents like fluorodeoxyuridine (FdU) that inhibit thymidylate synthase, an enzyme important for pyrimidine biosynthesis. These sites all consist of long CGG•CCG-repeats. Other sites like FRA10B, which is induced by BrdU [53], or FRA16B, which is induced by distamycin, consist of A+T-rich repeats [54]. Aphidicolin (APH) is responsible for the induction of many CFSs. These FSs span megabases of DNA with no specific sequences responsible for FS expression having been identified to date (see [55] for more detailed discussion).
CGG•CCG-repeats exclude nucleosomes in vitro [56] as does FRA16B in the presence of distamycin [57]. Nucleosome exclusion in vivo potentially could account for the abnormal appearance of chromatin at these locations. However, it would be hard to explain the folate-sensitive nature of the FX fragile site on this basis. However, a common mechanism for chromosome fragility is suggested by the properties of the agents that induce fragility. Folate stress/FdU affects dCTP pools, a situation that may affect replication fork progression through G-rich regions. APH inhibits DNA polymerase α, δ, and ε [58]. Bromodeoxyruridine is a halogenated thymidine analog that can be incorporated into DNA while distamycin binds the minor groove of A+T-rich regions [59]. Thus agents that induce fragility all have the potential to affect the efficiency of DNA replication in some way. Heterochromatin is also a common feature of FSs [60]. Chromatin conformation can potentially affect DNA replication by altering the timing and efficiency of origins of replication [61]. It could also affect the efficiency of DNA repair [62].
Replication fork stalling has been reported for some of the CFSs [60, 63]. We have previously shown that the FX repeat forms hairpins and tetraplexes that block DNA synthesis in vitro very effectively [64]. These repeats are now known to block DNA replication in vivo as well [65]. It may be that reduced dCTP pools resulting from treatment with agents like FdU slows DNA synthesis on the CGG-rich template. This could create conditions that facilitate the formation of intrastrand structures that then further impede DNA synthesis. Since the normal FMR1 gene replicates late in S phase and the silenced allele replicates even later [66, 67] this could delay the completion of replication of this locus well into metaphase.
A stalled replication fork as the basis for chromosome fragility would be consistent with the observation that camptothecin (CPT), a topoisomerase I (Topo I) inhibitor, reduces fragility at many CFSs [68]. It also reduces the levels of proteins typically seen in the presence of single-stranded DNA (ssDNA). DNA polymerase inhibition by APH could lead to uncoupling of the polymerase-helicase-Topo I complex. This could result in the generation of long ssDNA regions that could form secondary structures that block replication [68]. S phase exit before replication can be restarted could result in the appearance of prematurely condensed chromatin at the FS.
However, recently it has been suggested that fragility at some CFS is simply a function of the late replication of a region in which there are few origins of replication (ORIs) [69]. It may be that some CFSs are located in regions that replicate so late and are so ORI poor, that no replication fork block is necessary in order to produce a FS. Other CFSs may replicate slightly earlier in the cell cycle and/or be in a more ORI-rich region, and thus require additional impediments to DNA replication in the form of a sequence or structural blocks to replication before they become apparent. A stalled replication fork may be even more important in the case of the much smaller, sequence specific FSs like FX.
Mutations in ATM and ATR affect chromosome fragility at both CFSs [70, 71] and FX [72]. However, there is some reason to think that the mechanisms responsible may differ. For example, both ATR and ATM are involved in preventing fragility at the CFSs. In contrast, we have shown that ATM is actually involved in the generation of the FX fragile site in the presence of FdU. However, ATM does seem to be involved in preventing a form of chromosome fragility that occurs spontaneously in tissue culture [72]. The role of ATM and ATR, along with the involvement of many DNA damage repair proteins in fragility at CFS [73–76], and the colocalization of γ-H2AX foci, a marker of DSBs with the FSs [72, 76], is consistent with a model in which the failure to complete DNA replication in a timely manner triggers DNA repair, with incomplete or error-prone repair leading to persistent gaps or breaks in the chromatin.
The formation of FSs can have important medical consequences. In particular, the FX fragile site may be responsible for the high incidence of Turner Syndrome (X chromosome monosomy) seen in female fetuses with the FM allele [77]: In all informative instances examined the lost chromosome corresponds to the one carrying the FM allele. Breakage in vivo would require the healing of the broken chromosome perhaps by sister chromatid fusion. Preferential migration of this fused chromosome to the spindle pole of one of the daughter cells during anaphase could account for the high frequency loss of the affected chromosome in these cases.
4. Gene silencing in FXS
Heterochromatin-mediated gene silencing has long been recognized as the cause of FXS [4, 19]. However, the silencing mechanism is not well understood. Work in our lab and elsewhere is beginning to shed light on this process. The FX allele is known to be active in embryonic stem cells and early embryos, with silencing occurring during differentiation [78]. In differentiated cells the 5′ end of FXS alleles is heavily methylated at the DNA level [4, 19]. FXS alleles are also hypoacetylated and enriched for dimethylated histone H3 lysine 9 (H3K9Me2) [79]. During differentiation of FX embryonic stem cells, H3K9 dimethylation on the FMR1 promoter is detected before DNA methylation [78]. Similarly, in rare FM carriers who do not show gene silencing, H3K9Me2 is present while DNA methylation is not [80]. These data illustrate that deposition of H3K9Me2 on the FMR1 gene is a relatively early event in the silencing process while DNA methylation occurs later or independently of this event.
More recently, we have shown that silenced alleles are enriched for marks of both facultative and constitutive heterochromatin [81]. The constitutive heterochromatin marks histone H3 trimethylated on lysine 9 (H3K9Me3) and H4 trimethylated on lysine 20 (H4K20Me3) are most highly concentrated on the repeat. In contrast, two marks of facultative heterochromatin H3K9Me2 and H3K27Me3 are evenly distributed across the locus merging with a zone of facultative heterochromatin that we have identified upstream of the FMR1 promoter in both normal and patient cells [81].
The concentration of the constitutive heterochromatin marks on the repeat suggests that the trigger for silencing may be intrinsic to the repeat. This trigger could be the repeats themselves either acting in DNA-mediated fashion or via an effect of the repeat-containing RNA as illustrated in Fig. 3. Treatment of FXS cells with the DNA methyltransferase inhibitor 5-azadeoxycytidine leads to gene reactivation [82, 83]. However, this compound is toxic. Histone deacetylase inhibitors like butyrate and trichostatin A (TSA), that target class I and class II histone deacetylases (HDACs), have only a modest effect on gene reactivation [82, 83]. However, we have shown that inhibition of the class III HDAC, the sirtuin SIRT1, results in comparable gene reactivation to that seen with azadC [84]. SIRT1 acts by deacetylating H3K9 and H4K16. Deacetylation of H3K9 occurs prior to DNA methylation, while deacetylation of H4K16 appears to be one of the last steps in the silencing process. Since a dominant negative mutation of hMOF, the enzyme responsible for acetylating H4K16, prevents SIRT1 inhibitors from reactivating the silenced allele [84], H4K16 deacetylation rather than DNA methylation per se, is vital for the silencing process. The ability of SIRT1 inhibitors to reactivate the FMR1 gene suggests that this class of drug may have therapeutic value in treating FXS, since it has the potential to be effective in post-mitotic cells like neurons, the cells in which the effect of aberrant FMR1 gene silencing is felt most acutely.
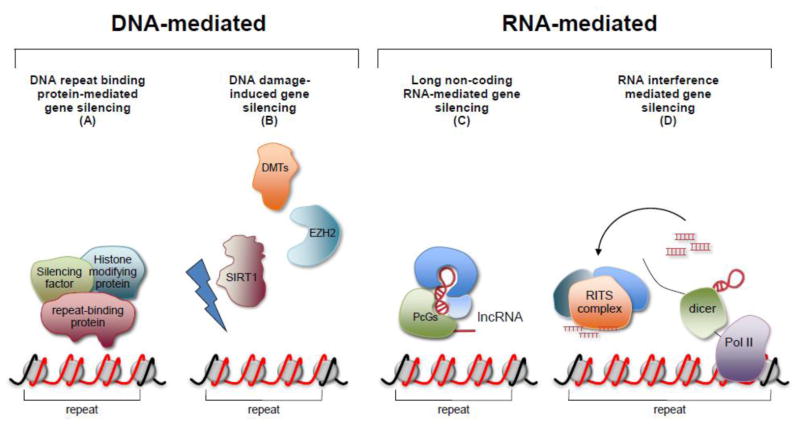
Fig. 3. Four models for repeat-induced gene silencing in FRDA and FXS.
(A) The repeats in the chromosome may act as silencers by binding sequence-specific or structure-specific proteins that then recruit components of the silencing machinery. (B) DNA damage within the repeat may result in the recruitment of the deacetylase SIRT1, EZH2, a component of the repressive Polycomb group (PcG) complexes, and DNA methyltransferases. (C) The repeats in the chromosome may be targeted by PcGs directed to the locus by long non-coding RNA (lncRNA) [141] acting in cis or trans. (D) Silencing may occur via an RNA Interference based mechanism [134] with the long hairpins formed by RNA containing these repeats or duplexes formed by the sense and antisense transcript produced from both of these loci as the source of dsRNA. DMTs: DNA methyltransferases.; lncRNA: long non-coding RNA; PcGs: Polycomb Group Complexes; RITS: RNA-induced transcriptional silencing complex.
5. FMR1 hyperexpression in PM carriers
While repeat-mediated gene silencing is responsible for FM symptoms, not only are PM alleles not silenced, they actually make 2–6 times more FMR1 mRNA than normal alleles [85, 86]. The increase in transcription shows a linear association with repeat number. Since work in Drosophila suggests that the RNA pathology thought to be responsible for disease symptoms is a function both of the repeat number and the level of the RNA containing the repeats [15], the elevated level of mRNA seen likely contributes to the severity of the disorders seen in PM carriers.
In contrast to the hypoacetylation of FM alleles, PM alleles have 1.5–2 times the normal levels of acetylated H3 and H4 [87]. The basis of these chromatin changes is unknown. Should CGG•CCG-repeats turn out to exclude nucleosomes in vivo as they do in vitro [56], they could perhaps confer an initially more open chromatin structure on the FMR1 promoter. This could predispose PM alleles to increased transcription, perhaps by facilitating the increased usage of the more 5′ transcription start sites that occurs in PM carriers [88].
It is also possible that the observed chromatin abnormalities are the indirect result of the effect of CGG-repeat containing RNA on gene expression [89], perhaps by favoring the use of additional promoters or by affecting the expression of chromatin modifying proteins. The effect on gene expression could result from CGG-RNA's ability to act as a substrate for the RNA interference pathway [89] and thus to potentially affect the expression of genes containing the repeats. CGG-RNA is also thought to sequester proteins like SAM68, a splicing factor [90] and proteins like pur-α and hnRNP A2 [91], that have pleiotropic effects on gene expression.
Whatever the mechanism, histone acetylase transferase inhibitors like garcinol and anacardic acid have been shown to reduce FMR1 mRNA levels in patient cells [87]. Garcinol also reduces neurodegeneration and extends the life-span of flies expressing high levels of CGG-RNA, as does overexpression of different HDACs [87]. Whether or not this effect is mediated via a direct effect on the PM chromatin, it would suggest that the use of HAT inhibitors may help ameliorate disease symptoms in humans.
The paradox of hyperexpression of PM alleles and the hypoexpression of FM alleles remains unresolved. It does suggest that as the FM threshold represents some sort of turning point with respect to factors that affect gene expression. Since in the PM range, increasing repeat number is associated with the increased production of both sense and antisense transcripts, it is possible that somewhere close to the FM threshold the levels of one or both of these transcripts reaches a critical mass that allows the process of gene silencing to predominate.
6. FRDA as a chromatinopathy
At the time that the FXN gene was first identified in 1996, most work on gene silencing was focused on CpG-rich promoter regions that were hypermethylated. Since the FRDA repeat is located in an intron, contains no CpG-residues that could be methylated and FXN mRNA levels are still significant in patient cells, an alternative explanation for the FRDA transcription deficit was initially sought. This led to models in which the GAA•TTC-repeat, by virtue of its ability to produce secondary structures like triplexes, formed an impediment to transcription elongation [92–95]. Other models invoking altered splicing have also been proposed [96].
However, more recent work in our lab and elsewhere has shown that the region flanking the repeat in patient cells is hypermethylated [97] and associated with histone marks characteristic of transcriptionally silenced genes [98, 99]. We showed that both normal and patient alleles show hypermethylation of CpG residues upstream of the repeat. Methylation on normal alleles could be due to the spreading from Alu elements present in the vicinity including the Alu element from which the repeat has been suggested to have arisen. While hypermethylation is seen on normal alleles, patient alleles are more extensively methylated [97] and a relationship between the extent of methylation and disease severity has been demonstrated [100]. In addition, we showed that some residues that are rarely methylated on normal alleles are completely methylated in patient cells [97]. This suggests that some residues in the region upstream of the repeat are protected from methylation in normal cells perhaps because binding of proteins to that region blocks access of the CpG residues within the binding site to DNA methyltransferases. One of these regions binds a factor that is important for maximal promoter activity in reporter constructs [97]. Since DNA methylation does not extend into the promoter, it probably does not affect transcription initiation via an effect on promoter chromatin. However, our data suggests that DNA methylation may have an effect on transcription initiation via its ability to block binding of a factor important for optimal promoter activity. In addition, since intragenic methylation affects transcription elongation [101], methylation may contribute in additional ways to the FXN mRNA deficit in FRDA.
While there have been no reports of DNA methylation spreading into the promoter, there have been conflicting reports as to whether the repressive histone marks do. Some of the discrepancy may result from the use of different cell types, cells with different repeat numbers and the analysis of different DNA regions and chromatin marks. However, the elevated levels of H3K9Me2 in the brains of humans with FRDA [102] suggests that repeat-mediated changes may well extend into the promoter region in biologically relevant cells, thereby having the potential to affect transcription initiation in a way that could impact disease severity.
HDAC inhibitors have been shown to be effective at normalizing FXN expression in patient cells and in mouse models [103–105] and some of these compounds are now in clinical trials to test their efficacy in the treatment of FRDA. The effect of HDAC inhibitors would be consistent with the idea that the observed epigenetic changes seen on FRDA alleles are responsible for the reduced transcription. However, this work has been challenged by the observation that the compound BIX-01294 which inhibits dimethylation of H3K9, has no effect on the levels of transcript produced [106]. In this view, the chromatin modifications seen on patient alleles are not responsible for the transcriptional repression. They are either irrelevant or reflect a downstream consequence of the reduced transcription resulting from a block to transcription elongation formed by the repeats. However, it is possible to reconcile the HDAC inhibitor data and the BIX-01294 data with an epigenetic dysregulation model for FRDA, if it assumed that H3K9 dimethylation precedes or is independent of later events important for gene silencing, analogous to what we have observed in FXS [84].
Further support for an epigenetic model comes from work in our laboratory that showed that the level of the initiating form of RNA Polymerase II (Pol II) is reduced in patient cells in the vicinity of the major transcription start site [99]. Furthermore, trimethylation of H3K4, which is thought to occur cotranscriptionally on exon 1 in a manner dependent on the amount of initiating Pol II, is also lower in patient cells [99, 107]. In addition, H3K36Me3 [99, 106] and H3K79Me2 [107], marks of transcription elongation, are also reduced 5′ of the repeat in patient cells. Taken together the preponderance of evidence supports the idea that repeat expansion in FRDA, like repeat expansion in FXS, leads to the formation of heterochromatin that affects transcription initiation and elongation.
7. What is the molecular basis for these repeat-induced chromatin changes?
As with FXS and myotonic dystrophy type 1 (DM1), a CTG•CAG-repeat expansion disease that is also associated with heterochromatin formation [108], the heterochromatin marks in FRDA are highest in the vicinity of the repeat. This suggests that in all three disorders the trigger for heterochromatin formation is intrinsic to the repeat. The fact that all other identified long CGG•CCG-repeats in the human genome are also heterochromatinized [109–115], lends support to that idea. The tendency of these different repeats to become heterochromatinized could be related to their common ability to form unusual DNA and RNA structures, such as hairpins, that may affect a variety of biological processes [64, 94, 116–122].
The repeats in the chromosome may act as silencers by binding sequence-specific or structure-specific proteins that then recruit components of the silencing machinery [123] as illustrated in Fig. 3A. Work in yeast has shown that replication pause sites are enriched for silencing factors [124]. Since work from our laboratory and elsewhere has shown that the FX, DM1 and FRDA repeats form blocks to DNA synthesis [64, 65, 125], it may be that this predisposes the repeats to silencing. Alternatively double strand breaks (DSBs) occurring in the repeat may lead to the recruitment of the deacetylase SIRT1, EZH2, a component of the repressive Polycomb group (PcG) complexes, and DNA methyltransferases as demonstrated for the G+C-rich promoter of the cadherin gene [126] (Fig. 3B). Our work demonstrating that the FMR1 gene co-localizes with γ-H2AX foci, a mark of DSBs, in patient cells [72], suggests that such a mechanism is possible. DSB formation could be related to the ability of these repeats to block DNA synthesis [64, 65, 125] and the resultant efforts to complete replication. Alternatively DSBs could arise from DNA damage or attempts to repair that damage. In this regard, it is worth noting that the hairpins formed by the DM1 repeat increases the sensitivity of the locus to oxidative damage [127].
It could also be that the repeats in the chromosome are targeted by PcGs directed to the locus by long non-coding RNA (lncRNA) as depicted in Fig. 3C. This lncRNA may act in cis, as in the case of the Kcnq1 gene cluster and the lncRNA, Kcnq1ot1 [128], or in trans, as in the case of the Hox gene clusters and the lncRNA, HOTAIR [129]. Long antisense transcripts have been described for all three disease loci [81, 130–132]. The lncRNA may act as a scaffold for the assembly of proteins necessary for heterochromatin formation as illustrated or as a guide for the recruitment of silencing complexes [133]. Since most PRC targets are G+C-rich, the CGG•CCG-repeats may be particularly prone to silencing by these complexes.
Finally, silencing may occur via an RNA interference-based mechanism as has been suggested for the centromeric repeats in fission yeast [134] (illustrated in Fig. 3D). The source of double-stranded RNA (dsRNA) for this pathway could be the long hairpins formed by RNA containing these repeats [89, 122, 135] that, in the case of the FX and DM1 repeats, have been shown to be substrates for Dicer [89, 135]. Alternatively, the source of dsRNA could be duplexes formed by the sense and antisense transcript produced from these loci [81, 130–132]. If a sense-antisense hybrid were involved, it would be necessary to invoke some special property of the region of the hybrid containing the repeat to account for the fact that the repressive histone marks appear to nucleate on the repeat. Perhaps the fact that the FRDA repeat is less G+C-rich than the FX and DM1 repeats, and thus less likely to form stable secondary structures, accounts for the fact that silencing is less complete on FRDA alleles than on FX or DM1 alleles.
8. Concluding remarks
Altered chromatin is a feature of many aspects of the FXDs and FRDA. Much work remains to understand the mechanism responsible for the repeat-mediated chromatin changes, to elucidate the role that these changes play in the repeat expansion that generates pathological alleles and how it relates to the chromosome fragility characteristic of FXS alleles. However, a large body of evidence clearly demonstrates that repeat-mediated changes in chromatin have deleterious consequences for gene expression that are responsible for or contribute to disease pathology. Since the coding sequences in both the FXDs and FRDA are unchanged in most affected individuals, identifying all of the events involved in the deposition of altered chromatin and understanding any common processes involved may facilitate the development of therapeutic approaches to treat these disorders.
Highlights.
The Fragile X disorders and Friedreich ataxia result from expansion of a tandem repeat tract.
In all of these diseases the repeat is transcribed but not translated.
Evidence suggests that repeat-mediated chromatin changes are responsible for disease pathology.
Acknowledgments
This work was made possible by funding from the Intramural Program of the NIH to KU.
Abbreviations
- FM
full mutation
- FRDA
Friedreich ataxia
- FS
fragile site
- FXDs
Fragile X associated disorders
- FXPOI
Fragile X-associated primary ovarian insufficiency
- FXTAS
Fragile X-associated tremor and ataxia syndrome
- FXS
Fragile X syndrome
- PM
premutation
- REDs
Repeat Expansion Diseases
- SCA
spinocerebellar ataxia
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum Mol Genet. 2010;19:R77–82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 4.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 5.Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG, Jr, Warren ST, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 6.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Turner G, Daniel A, Frost M. X-linked mental retardation, macro-orchidism, and the Xq27 fragile site. J Pediatr. 1980;96:837–841. doi: 10.1016/s0022-3476(80)80552-x. [DOI] [PubMed] [Google Scholar]
- 8.Proops R, Webb T. The ‘fragile’ X chromosome in the Martin-Bell-Renpenning syndrome and in males with other forms of familial mental retardation. J Med Genet. 1981;18:366–373. doi: 10.1136/jmg.18.5.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levesque S, Dombrowski C, Morel ML, Rehel R, Cote JS, Bussieres J, Morgan K, Rousseau F. Screening and instability of FMR1 alleles in a prospective sample of 24,449 mother-newborn pairs from the general population. Clin Genet. 2009;76:511–523. doi: 10.1111/j.1399-0004.2009.01237.x. [DOI] [PubMed] [Google Scholar]
- 10.Hagerman PJ, Hagerman RJ. Fragile X-associated tremor/ataxia syndrome (FXTAS) Ment Retard Dev Disabil Res Rev. 2004;10:25–30. doi: 10.1002/mrdd.20005. [DOI] [PubMed] [Google Scholar]
- 11.Allingham-Hawkins DJ, Babul-Hirji R, Chitayat D, Holden JJ, Yang KT, Lee C, Hudson R, Gorwill H, Nolin SL, Glicksman A, Jenkins EC, Brown WT, Howard-Peebles PN, Becchi C, Cummings E, Fallon L, Seitz S, Black SH, Vianna-Morgante AM, Costa SS, Otto PA, Mingroni-Netto RC, Murray A, Webb J, Vieri F, et al. Fragile X premutation is a significant risk factor for premature ovarian failure: the International Collaborative POF in Fragile X study--preliminary data. Am J Med Genet. 1999;83:322–325. [PMC free article] [PubMed] [Google Scholar]
- 12.Conway GS, Payne NN, Webb J, Murray A, Jacobs PA. Fragile X premutation screening in women with premature ovarian failure. Hum Reprod. 1998;13:1184–1187. doi: 10.1093/humrep/13.5.1184. [DOI] [PubMed] [Google Scholar]
- 13.Murray A, Webb J, Grimley S, Conway G, Jacobs P. Studies of FRAXA and FRAXE in women with premature ovarian failure. J Med Genet. 1998;35:637–640. doi: 10.1136/jmg.35.8.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handa V, Goldwater D, Stiles D, Cam M, Poy G, Kumari D, Usdin K. Long CGG-repeat tracts are toxic to human cells: implications for carriers of Fragile X premutation alleles. FEBS Lett. 2005;579:2702–2708. doi: 10.1016/j.febslet.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, Warren ST. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 16.Hashem V, Galloway JN, Mori M, Willemsen R, Oostra BA, Paylor R, Nelson DL. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum Mol Genet. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pandolfo M. Friedreich ataxia: the clinical picture. J Neurol. 2009;256(Suppl 1):3–8. doi: 10.1007/s00415-009-1002-3. [DOI] [PubMed] [Google Scholar]
- 18.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 19.Hornstra IK, Nelson DL, Warren ST, Yang TP. High resolution methylation analysis of the FMR1 gene trinucleotide repeat region in fragile X syndrome. Hum Mol Genet. 1993;2:1659–1665. doi: 10.1093/hmg/2.10.1659. [DOI] [PubMed] [Google Scholar]
- 20.Entezam A, Lokanga AR, Le W, Hoffman G, Usdin K. Potassium bromate, a potent DNA oxidizing agent, exacerbates germline repeat expansion in a fragile X premutation mouse model. Hum Mutat. 2010;31:611–616. doi: 10.1002/humu.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Entezam A, Usdin K. ATR protects the genome against CGG.CCG-repeat expansion in Fragile X premutation mice. Nucleic Acids Res. 2008;36:1050–1056. doi: 10.1093/nar/gkm1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Entezam A, Usdin K. ATM and ATR protect the genome against two different types of tandem repeat instability in Fragile X premutation mice. Nucleic Acids Res. 2009;37:6371–6377. doi: 10.1093/nar/gkp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Usdin K. The biological effects of simple tandem repeats: lessons from the repeat expansion diseases. Genome Res. 2008;18:1011–1019. doi: 10.1101/gr.070409.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dion V, Lin Y, Hubert L, Jr, Waterland RA, Wilson JH. Dnmt1 deficiency promotes CAG repeat expansion in the mouse germline. Hum Mol Genet. 2008;17:1306–1317. doi: 10.1093/hmg/ddn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dion V, Lin Y, Price BA, Fyffe SL, Seluanov A, Gorbunova V, Wilson JH. Genome-wide demethylation promotes triplet repeat instability independently of homologous recombination. DNA Repair (Amst) 2008;7:313–320. doi: 10.1016/j.dnarep.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorbunova V, Seluanov A, Mittelman D, Wilson JH. Genome-wide demethylation destabilizes CTG.CAG trinucleotide repeats in mammalian cells. Hum Mol Genet. 2004;13:2979–2989. doi: 10.1093/hmg/ddh317. [DOI] [PubMed] [Google Scholar]
- 28.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 29.Libby RT, Hagerman KA, Pineda VV, Lau R, Cho DH, Baccam SL, Axford MM, Cleary JD, Moore JM, Sopher BL, Tapscott SJ, Filippova GN, Pearson CE, La Spada AR. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gourdon G, Radvanyi F, Lia AS, Duros C, Blanche M, Abitbol M, Junien C, Hofmann-Radvanyi H. Moderate intergenerational and somatic instability of a 55-CTG repeat in transgenic mice. Nat Genet. 1997;15:190–192. doi: 10.1038/ng0297-190. [DOI] [PubMed] [Google Scholar]
- 31.Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 32.Libby RT, Monckton DG, Fu YH, Martinez RA, McAbney JP, Lau R, Einum DD, Nichol K, Ware CB, Ptacek LJ, Pearson CE, La Spada AR. Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice. Hum Mol Genet. 2003;12:41–50. doi: 10.1093/hmg/ddg006. [DOI] [PubMed] [Google Scholar]
- 33.Chong SS, McCall AE, Cota J, Subramony SH, Orr HT, Hughes MR, Zoghbi HY. Gametic and somatic tissue-specific heterogeneity of the expanded SCA1 CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1995;10:344–350. doi: 10.1038/ng0795-344. [DOI] [PubMed] [Google Scholar]
- 34.Dragileva E, Hendricks A, Teed A, Gillis T, Lopez ET, Friedberg EC, Kucherlapati R, Edelmann W, Lunetta KL, MacDonald ME, Wheeler VC. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortune MT, Vassilopoulos C, Coolbaugh MI, Siciliano MJ, Monckton DG. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum Mol Genet. 2000;9:439–445. doi: 10.1093/hmg/9.3.439. [DOI] [PubMed] [Google Scholar]
- 36.Gomes-Pereira M, Fortune MT, Ingram L, McAbney JP, Monckton DG. Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum Mol Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 37.Ishiguro H, Yamada K, Sawada H, Nishii K, Ichino N, Sawada M, Kurosawa Y, Matsushita N, Kobayashi K, Goto J, Hashida H, Masuda N, Kanazawa I, Nagatsu T. Age-dependent and tissue-specific CAG repeat instability occurs in mouse knock-in for a mutant Huntington’s disease gene. J Neurosci Res. 2001;65:289–297. doi: 10.1002/jnr.1153. [DOI] [PubMed] [Google Scholar]
- 38.Kennedy L, Evans E, Chen CM, Craven L, Detloff PJ, Ennis M, Shelbourne PF. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 39.Lia AS, Seznec H, Hofmann-Radvanyi H, Radvanyi F, Duros C, Saquet C, Blanche M, Junien C, Gourdon G. Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum Mol Genet. 1998;7:1285–1291. doi: 10.1093/hmg/7.8.1285. [DOI] [PubMed] [Google Scholar]
- 40.Sato T, Oyake M, Nakamura K, Nakao K, Fukusima Y, Onodera O, Igarashi S, Takano H, Kikugawa K, Ishida Y, Shimohata T, Koide R, Ikeuchi T, Tanaka H, Futamura N, Matsumura R, Takayanagi T, Tanaka F, Sobue G, Komure O, Takahashi M, Sano A, Ichikawa Y, Goto J, Kanazawa I, et al. Transgenic mice harboring a full-length human mutant DRPLA gene exhibit age-dependent intergenerational and somatic instabilities of CAG repeats comparable with those in DRPLA patients. Hum Mol Genet. 1999;8:99–106. doi: 10.1093/hmg/8.1.99. [DOI] [PubMed] [Google Scholar]
- 41.Ueno S, Kondoh K, Kotani Y, Komure O, Kuno S, Kawai J, Hazama F, Sano A. Somatic mosaicism of CAG repeat in dentatorubral-pallidoluysian atrophy (DRPLA) Hum Mol Genet. 1995;4:663–666. doi: 10.1093/hmg/4.4.663. [DOI] [PubMed] [Google Scholar]
- 42.Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, Loring JF, Napierala M, Gottesfeld JM. Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foiry L, Dong L, Savouret C, Hubert L, te Riele H, Junien C, Gourdon G. Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum Genet. 2006;119:520–526. doi: 10.1007/s00439-006-0164-7. [DOI] [PubMed] [Google Scholar]
- 44.Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. Embo J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, te Riele H, Groenen PJ, Wieringa B. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 46.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 47.Wohrle D, Salat U, Glaser D, Mucke J, Meisel-Stosiek M, Schindler D, Vogel W, Steinbach P. Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J Med Genet. 1998;35:103–111. doi: 10.1136/jmg.35.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wohrle D, Salat U, Hameister H, Vogel W, Steinbach P. Demethylation, reactivation, and destabilization of human fragile X full-mutation alleles in mouse embryocarcinoma cells. Am J Hum Genet. 2001;69:504–515. doi: 10.1086/322739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Entezam A, Biacsi R, Orrison B, Saha T, Hoffman GE, Grabczyk E, Nussbaum RL, Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanawalt PC. Preferential DNA repair in expressed genes. Environ Health Perspect. 1987;76:9–14. doi: 10.1289/ehp.87769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kovtun IV, Johnson KO, McMurray CT. Cockayne syndrome B protein antagonizes OGG1 in modulating CAG repeat length in vivo. Aging (Albany NY) 2011;3:509–514. doi: 10.18632/aging.100324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hewett DR, Handt O, Hobson L, Mangelsdorf M, Eyre HJ, Baker E, Sutherland GR, Schuffenhauer S, Mao JI, Richards RI. FRA10B structure reveals common elements in repeat expansion and chromosomal fragile site genesis. Mol Cell. 1998;1:773–781. doi: 10.1016/s1097-2765(00)80077-5. [DOI] [PubMed] [Google Scholar]
- 54.Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, Eyre HJ, Lapsys N, Le Paslier D, Doggett NA, Sutherland GR, Richards RI. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997;88:367–374. doi: 10.1016/s0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]
- 55.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 56.Wang YH, Gellibolian R, Shimizu M, Wells RD, Griffith J. Long CCG triplet repeat blocks exclude nucleosomes: a possible mechanism for the nature of fragile sites in chromosomes. J Mol Biol. 1996;263:511–516. doi: 10.1006/jmbi.1996.0593. [DOI] [PubMed] [Google Scholar]
- 57.Hsu YY, Wang YH. Human fragile site FRA16B DNA excludes nucleosomes in the presence of distamycin. J Biol Chem. 2002;277:17315–17319. doi: 10.1074/jbc.M200901200. [DOI] [PubMed] [Google Scholar]
- 58.Cheng CH, Kuchta RD. DNA polymerase epsilon: aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry. 1993;32:8568–8574. doi: 10.1021/bi00084a025. [DOI] [PubMed] [Google Scholar]
- 59.Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- 60.Palakodeti A, Lucas I, Jiang Y, Young DJ, Fernald AA, Karrison T, Le Beau MM. Impaired replication dynamics at the FRA3B common fragile site. Hum Mol Genet. 2010;19:99–110. doi: 10.1093/hmg/ddp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Histone acetylation regulates the time of replication origin firing. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 62.Pandita TK, Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009;37:1363–1377. doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell. 2011;43:122–131. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 64.Usdin K, Woodford KJ. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat Struct Mol Biol. 2009;16:226–228. doi: 10.1038/nsmb.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hansen RS, Canfield TK, Lamb MM, Gartler SM, Laird CD. Association of fragile X syndrome with delayed replication of the FMR1 gene. Cell. 1993;73:1403–1409. doi: 10.1016/0092-8674(93)90365-w. [DOI] [PubMed] [Google Scholar]
- 67.Webb T. Delayed replication of Xq27 in individuals with the fragile X syndrome. Am J Med Genet. 1992;43:1057–1062. doi: 10.1002/ajmg.1320430633. [DOI] [PubMed] [Google Scholar]
- 68.Arlt MF, Glover TW. Inhibition of topoisomerase I prevents chromosome breakage at common fragile sites. DNA Repair (Amst) 2010;9:678–689. doi: 10.1016/j.dnarep.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 70.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 71.Ozeri-Galai E, Schwartz M, Rahat A, Kerem B. Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene. 2008;27:2109–2117. doi: 10.1038/sj.onc.1210849. [DOI] [PubMed] [Google Scholar]
- 72.Kumari D, Somma V, Nakamura AJ, Bonner WM, D’Ambrosio E, Usdin K. The role of DNA damage response pathways in chromosome fragility in Fragile X syndrome. Nucleic Acids Res. 2009;37:4385–4392. doi: 10.1093/nar/gkp391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Durkin SG, Arlt MF, Howlett NG, Glover TW. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene. 2006;25:4381–4388. doi: 10.1038/sj.onc.1209466. [DOI] [PubMed] [Google Scholar]
- 74.Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 75.Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB, Glover TW. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol Cell Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwartz M, Zlotorynski E, Goldberg M, Ozeri E, Rahat A, le Sage C, Chen BP, Chen DJ, Agami R, Kerem B. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005;19:2715–2726. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobkin C, Radu G, Ding XH, Brown WT, Nolin SL. Fragile X prenatal analyses show full mutation females at high risk for mosaic Turner syndrome: fragile X leads to chromosome loss. Am J Med Genet A. 2009;149A:2152–2157. doi: 10.1002/ajmg.a.33011. [DOI] [PubMed] [Google Scholar]
- 78.Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N, Ben-Yosef D. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 79.Coffee B, Zhang F, Warren ST, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 80.Tabolacci E, Moscato U, Zalfa F, Bagni C, Chiurazzi P, Neri G. Epigenetic analysis reveals a euchromatic configuration in the FMR1 unmethylated full mutations. Eur J Hum Genet. 2008;16:1487–1498. doi: 10.1038/ejhg.2008.130. [DOI] [PubMed] [Google Scholar]
- 81.Kumari D, Usdin K. The distribution of repressive histone modifications on silenced FMR1 alleles provides clues to the mechanism of gene silencing in fragile X syndrome. Hum Mol Genet. 2010;19:4634–4642. doi: 10.1093/hmg/ddq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pietrobono R, Pomponi MG, Tabolacci E, Oostra B, Chiurazzi P, Neri G. Quantitative analysis of DNA demethylation and transcriptional reactivation of the FMR1 gene in fragile X cells treated with 5-azadeoxycytidine. Nucleic Acids Res. 2002;30:3278–3285. doi: 10.1093/nar/gkf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiurazzi P, Pomponi MG, Willemsen R, Oostra BA, Neri G. In vitro reactivation of the FMR1 gene involved in fragile X syndrome. Hum Mol Genet. 1998;7:109–113. doi: 10.1093/hmg/7.1.109. [DOI] [PubMed] [Google Scholar]
- 84.Biacsi R, Kumari D, Usdin K. SIRT1 inhibition alleviates gene silencing in Fragile X mental retardation syndrome. PLoS Genet. 2008;4:e1000017. doi: 10.1371/journal.pgen.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tassone F, Beilina A, Carosi C, Albertosi S, Bagni C, Li L, Glover K, Bentley D, Hagerman PJ. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA. 2007;13:555–562. doi: 10.1261/rna.280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Todd PK, Oh SY, Krans A, Pandey UB, Di Prospero NA, Min KT, Taylor JP, Paulson HL. Histone deacetylases suppress CGG repeat-induced neurodegeneration via transcriptional silencing in models of fragile X tremor ataxia syndrome. PLoS Genet. 2010;6:e1001240. doi: 10.1371/journal.pgen.1001240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beilina A, Tassone F, Schwartz PH, Sahota P, Hagerman PJ. Redistribution of transcription start sites within the FMR1 promoter region with expansion of the downstream CGG-repeat element. Hum Mol Genet. 2004;13:543–549. doi: 10.1093/hmg/ddh053. [DOI] [PubMed] [Google Scholar]
- 89.Handa V, Saha T, Usdin K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 2003;31:6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD. Sticky DNA, a self-associated complex formed at long GAA*TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J Biol Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 93.Grabczyk E, Mancuso M, Sammarco MC. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35:5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grabczyk E, Usdin K. The GAA*TTC triplet repeat expanded in Friedreich’s ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grabczyk E, Usdin K. Alleviating transcript insufficiency caused by Friedreich’s ataxia triplet repeats. Nucleic Acids Res. 2000;28:4930–4937. doi: 10.1093/nar/28.24.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baralle M, Pastor T, Bussani E, Pagani F. Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing. Am J Hum Genet. 2008;83:77–88. doi: 10.1016/j.ajhg.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Greene E, Mahishi L, Entezam A, Kumari D, Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 99.Kumari D, Biacsi RE, Usdin K. Repeat expansion affects both transcription initiation and elongation in Friedreich ataxia cells. J Biol Chem. 2011;286:4209–4215. doi: 10.1074/jbc.M110.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Castaldo I, Pinelli M, Monticelli A, Acquaviva F, Giacchetti M, Filla A, Sacchetti S, Keller S, Avvedimento VE, Chiariotti L, Cocozza S. DNA methylation in intron 1 of the frataxin gene is related to GAA repeat length and age of onset in Friedreich ataxia patients. J Med Genet. 2008;45:808–812. doi: 10.1136/jmg.2008.058594. [DOI] [PubMed] [Google Scholar]
- 101.Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 102.Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, Trabzuni D, Pook M. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 103.Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Coppola G, Geschwind DH, Gottesfeld JM, Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS One. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu C, Soragni E, Chou CJ, Herman D, Plasterer HL, Rusche JR, Gottesfeld JM. Chemical probes identify a role for histone deacetylase 3 in Friedreich’s ataxia gene silencing. Chem Biol. 2009;16:980–989. doi: 10.1016/j.chembiol.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rai M, Soragni E, Chou CJ, Barnes G, Jones S, Rusche JR, Gottesfeld JM, Pandolfo M. Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich’s ataxia patients and in a mouse model. PLoS One. 2010;5:e8825. doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Punga T, Buhler M. Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol Med. 2010;2:120–129. doi: 10.1002/emmm.201000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim E, Napierala M, Dent SY. Hyperexpansion of GAA repeats affects post-initiation steps of FXN transcription in Friedreich’s ataxia. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Filippova GN, Thienes CP, Penn BH, Cho DH, Hu YJ, Moore JM, Klesert TR, Lobanenkov VV, Tapscott SJ. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 109.Parrish JE, Oostra BA, Verkerk AJ, Richards CS, Reynolds J, Spikes AS, Shaffer LG, Nelson DL. Isolation of a GCC repeat showing expansion in FRAXF, a fragile site distal to FRAXA and FRAXE. Nat Genet. 1994;8:229–235. doi: 10.1038/ng1194-229. [DOI] [PubMed] [Google Scholar]
- 110.Sarafidou T, Kahl C, Martinez-Garay I, Mangelsdorf M, Gesk S, Baker E, Kokkinaki M, Talley P, Maltby EL, French L, Harder L, Hinzmann B, Nobile C, Richkind K, Finnis M, Deloukas P, Sutherland GR, Kutsche K, Moschonas NK, Siebert R, Gecz J. Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics. 2004;84:69–81. doi: 10.1016/j.ygeno.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 111.Winnepenninckx B, Debacker K, Ramsay J, Smeets D, Smits A, FitzPatrick DR, Kooy RF. CGG-repeat expansion in the DIP2B gene is associated with the fragile site FRA12A on chromosome 12q13.1. Am J Hum Genet. 2007;80:221–231. doi: 10.1086/510800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nancarrow JK, Kremer E, Holman K, Eyre H, Doggett NA, Le Paslier D, Callen DF, Sutherland GR, Richards RI. Implications of FRA16A structure for the mechanism of chromosomal fragile site genesis. Science. 1994;264:1938–1941. doi: 10.1126/science.8009225. [DOI] [PubMed] [Google Scholar]
- 113.Jones C, Penny L, Mattina T, Yu S, Baker E, Voullaire L, Langdon WY, Sutherland GR, Richards RI, Tunnacliffe A. Association of a chromosome deletion syndrome with a fragile site within the proto-oncogene CBL2. Nature. 1995;376:145–149. doi: 10.1038/376145a0. [DOI] [PubMed] [Google Scholar]
- 114.Ritchie RJ, Knight SJ, Hirst MC, Grewal PK, Bobrow M, Cross GS, Davies KE. The cloning of FRAXF: trinucleotide repeat expansion and methylation at a third fragile site in distal Xqter. Hum Mol Genet. 1994;3:2115–2121. doi: 10.1093/hmg/3.12.2115. [DOI] [PubMed] [Google Scholar]
- 115.Knight SJ, Voelckel MA, Hirst MC, Flannery AV, Moncla A, Davies KE. Triplet repeat expansion at the FRAXE locus and X-linked mild mental handicap. Am J Hum Genet. 1994;55:81–86. [PMC free article] [PubMed] [Google Scholar]
- 116.Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 117.Gacy AM, Goellner GM, Spiro C, Chen X, Gupta G, Bradbury EM, Dyer RB, Mikesell MJ, Yao JZ, Johnson AJ, Richter A, Melancon SB, McMurray CT. GAA instability in Friedreich’s Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 118.Mitas M, Yu A, Dill J, Kamp TJ, Chambers EJ, Haworth IS. Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res. 1995;23:1050–1059. doi: 10.1093/nar/23.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu A, Dill J, Mitas M. The purine-rich trinucleotide repeat sequences d(CAG)15 and d(GAC)15 form hairpins. Nucleic Acids Res. 1995;23:4055–4057. doi: 10.1093/nar/23.20.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu A, Barron MD, Romero RM, Christy M, Gold B, Dai J, Gray DM, Haworth IS, Mitas M. At physiological pH, d(CCG)15 forms a hairpin containing protonated cytosines and a distorted helix. Biochemistry. 1997;36:3687–3699. doi: 10.1021/bi9625410. [DOI] [PubMed] [Google Scholar]
- 121.Suen IS, Rhodes JN, Christy M, McEwen B, Gray DM, Mitas M. Structural properties of Friedreich’s ataxia d(GAA) repeats. Biochim Biophys Acta. 1999;1444:14–24. doi: 10.1016/s0167-4781(98)00267-x. [DOI] [PubMed] [Google Scholar]
- 122.Heidenfelder BL, Makhov AM, Topal MD. Hairpin formation in Friedreich’s ataxia triplet repeat expansion. J Biol Chem. 2003;278:2425–2431. doi: 10.1074/jbc.M210643200. [DOI] [PubMed] [Google Scholar]
- 123.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 124.Dubarry M, Loiodice I, Chen CLL, Thermes C, Taddei A. Tight protein-DNA interactions favor gene silencing. Gene Dev. 2011;25:1365–1370. doi: 10.1101/gad.611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich’s ataxia (GAA)n repeats in vivo. Mol Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jarem DA, Wilson NR, Delaney S. Structure-dependent DNA damage and repair in a trinucleotide repeat sequence. Biochemistry. 2009;48:6655–6663. doi: 10.1021/bi9007403. [DOI] [PubMed] [Google Scholar]
- 128.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Molecular Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 129.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 131.Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 132.De Biase I, Chutake YK, Rindler PM, Bidichandani SI. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One. 2009;4:e7914. doi: 10.1371/journal.pone.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 135.Krol J, Fiszer A, Mykowska A, Sobczak K, de Mezer M, Krzyzosiak WJ. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol Cell. 2007;25:575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 136.Ikeda Y, Daughters RS, Ranum LP. Bidirectional expression of the SCA8 expansion mutation: one mutation, two genes. Cerebellum. 2008;7:150–158. doi: 10.1007/s12311-008-0010-7. [DOI] [PubMed] [Google Scholar]
- 137.Holmes SE, O’Hearn E, Margolis RL. Why is SCA12 different from other SCAs? Cytogenet Genome Res. 2003;100:189–197. doi: 10.1159/000072854. [DOI] [PubMed] [Google Scholar]
- 138.Holmes SE, O’Hearn E, Rosenblatt A, Callahan C, Hwang HS, Ingersoll-Ashworth RG, Fleisher A, Stevanin G, Brice A, Potter NT, Ross CA, Margolis RL. A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat Genet. 2001;29:377–378. doi: 10.1038/ng760. [DOI] [PubMed] [Google Scholar]
- 139.Feng Y, Zhang F, Lokey LK, Chastain JL, Lakkis L, Eberhart D, Warren ST. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995;268:731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 140.Tassone F, De Rubeis S, Carosi C, La Fata G, Serpa G, Raske C, Willemsen R, Hagerman PJ, Bagni C. Differential usage of transcriptional start sites and polyadenylation sites in FMR1 premutation alleles. Nucleic Acids Res. 2011;39:6172–6185. doi: 10.1093/nar/gkr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 142.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–52. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]