Abstract
Epilepsy is a neurological disorder affecting approximately 1% of the worldwide population. Mutations in voltage-gated sodium channels have been identified in several monogenic epilepsy syndromes. Over 800 mutations have been identified in the voltage-gated sodium channel genes SCN1A and SCN2A in human epilepsies, including Genetic Epilepsy with Febrile Seizures Plus (GEFS+) and Dravet Syndrome. In GEFS+ families, affected members with the same mutation often display variability in clinical severity of the disease. This suggests that additional genes modify the effect of the primary mutation, resulting in the variable clinical presentation. The Scn2aQ54 transgenic mouse model has an epilepsy phenotype that varies depending on the genetic strain background. Scn2aQ54 mice congenic on the C57BL/6J strain exhibit delayed seizure onset and improved survival compared to (C57BL/6J × SJL/J)F1.Q54 mice. Two modifier loci of Scn2aQ54 seizure susceptibility were mapped and designated Moe1 (modifier of epilepsy) on chromosome 11 and Moe2 on chromosome 19. To confirm Moe1 and refine its position, we generated interval specific congenic (ISC) lines carrying C57BL/6J-derived chromosome 11 alleles on the SJL/J strain and refined the map position to 89–104 Mb. We then used RNA-Seq for candidate analysis in the modifier region. C57BL/6J and SJL/J male and female brain RNAs were sequenced, revealing numerous significant transcriptome differences and coding SNPs. Additional consideration of gene function and expression suggested several strong candidate modifier genes, including two voltage-gated calcium channel subunits, Cacna1g and Cacnb1, and the PAR bZIP transcription factor, Hlf.
Keywords: genetic modifier, seizures, epilepsy, voltage-gated sodium channels, voltage-gated ion channels, mouse models
Introduction
Mutations in voltage-gated sodium channels are responsible for several types of human epilepsy (Meisler & Kearney 2005). Mutations in SCN1A were first identified in patients with Genetic Epilepsy with Febrile Seizures Plus (GEFS+) (Escayg et al. 2000). Subsequently, SCN1A mutations were identified in patients with Dravet Syndrome (DS), a severe infant-onset epileptic encephalopathy (Claes et al. 2001). To date, over 800 epilepsy mutations in SCN1A have been reported. A smaller number of mutations in SCN2A have been identified in patients with Benign Familial Neonatal-Infantile Seizures (BFNIS), GEFS+ and DS (Meisler et al. 2010). Affected family members with the same sodium channel mutation often display variability in clinical severity of the disease, a common feature of genetic epilepsy. This suggests that epilepsy phenotype is influenced by other factors, which may include genetic modifiers.
A common feature of mouse seizure models, including sodium channel mutants, is that seizure susceptibility and severity vary significantly depending on the genetic strain background. This indicates that genetic modifiers contribute to variable phenotype expression. These mouse models provide a useful system for identifying modifier genes that may also contribute to variable expressivity in human epilepsy patients. Genes that influence a mutant phenotype can be identified systematically by evaluating the seizure phenotype on different background strains. Several strain-dependent seizure susceptibility loci and genes have been identified (Chaix et al. 2007; Ferraro et al. 2001, 2004, 2007a, 2007b, 2010b, 2011; Frankel et al. 1995; Gershenfeld et al. 1999; Legare et al. 2000; Maihara et al. 2000; Winawer et al. 2011).
The transgenic mouse model Scn2aQ54 has an epilepsy phenotype due to a mutation in Scn2a that slows channel inactivation and results in persistent sodium current (Kearney et al. 2001). Severity of the epilepsy phenotype in Scn2aQ54 mice is highly dependent on the genetic background. The phenotype is less severe on the resistant C57BL/6J (B6) strain, while it is more severe on the SJL/J (SJL) strain. We previously mapped two loci, Moe1 (Modifier of Epilepsy) on chromosome (chr) 11 and Moe2 on chr 19, that influence severity of the Scn2aQ54 phenotype (Bergren et al. 2005). Fine mapping of Moe2 on chr 19 identified Kcnv2 as a strong candidate gene (Bergren et al. 2009). Transgenic transfer of the modified phenotype in vivo supported the identification of Kcnv2 as a modifier gene in mice. Discovery of two novel human KCNV2 mutations in pediatric epilepsy patients suggested that KCNV2 may also contribute to human epilepsy susceptibility (Jorge et al. 2011).
In the present study, we generated interval specific congenic (ISC) lines to confirm and fine map the Moe1 locus on chr 11. Our results indicated the Moe1 locus is complex with at least two modifiers in the interval that exhibit sex-specific effects. We then used an RNA-Seq approach for candidate gene analysis within the Moe1 sub-intervals. Moe1 RNA-Seq data revealed numerous transcriptome differences and coding SNPs between sex and strain. We further evaluated and prioritized candidate modifier genes that may contribute to strain-dependent differences in seizure susceptibility in Scn2aQ54 mice.
Materials and methods
Mice
Scn2aQ54 transgenic mice were generated as previously described (Kearney et al. 2001). The congenic line C57BL/6J.Scn2aQ54 (B6.Q54) was established as described and is maintained by continued backcrossing of B6.Q54 hemizygous males to B6 females (Bergren et al. 2005). Mice were group-housed with access to food and water ad libitum. All studies were approved by the Vanderbilt University Animal Care and Use Committees in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Generation of ISC Lines
Four ISC lines were generated carrying B6-derived chr 11 segments in Moe1 on an SJL background. B6 males were crossed with SJL females to create F1 progeny which were then continually backcrossed to SJL to generate congenic lines. Whole genome genotyping was performed at generations N2 and N5 using the mouse MMDAP 768 SNP panel on the Illumina GoldenGate Platform (Moran et al. 2006). Animals retaining B6 alleles in Moe1 with the lowest percentage of B6 alleles in the rest of the genome were selected for breeding to the next generation. Once established, ISC strains were maintained by continued backcrossing to SJL and genotyping for chr 11 markers at all generations. All ISC lines were backcrossed for ≥10 generations prior to experiments.
Genotyping
Mice were tail biopsied on postnatal day 14 and genotyped. DNA was prepared from tail biopsies by phenol:chloroform extraction and ethanol precipitation or using the Gentra Puregene Mouse Tail Kit according to the manufacturer’s instructions (Qiagen). The Scn2aQ54 transgene was genotyped as previously described (Bergren et al. 2005). ISC lines were genotyped for chr 11 microsatellite markers by PCR and analyzed on 2% agarose or 7% non-denaturing polyacrylamide gels.
Phenotyping
Chr 11 ISC females were crossed with hemizygous B6.Q54 males to generate (B6.Q54 × ISC.SJL/J).F1 offspring (abbreviated F1.Q54) carrying heterozygous or homozygous B6 alleles in Moe1. We used a similar phenotyping paradigm to that used for low-resolution mapping of the Moe1 locus (Bergren et al. 2005). Briefly, F1.Q54 offspring underwent 30 minute video-taped observations at three and six weeks of age. All observation sessions occurred between 1:00 and 4:00 PM. Based on prior extensive video-EEG monitoring, spontaneous behavioral seizures were assessed offline by a blinded observer using Observer XT software (Noldus) (Kearney et al. 2001). We counted the number of visible focal motor seizures with forelimb clonus and repetitive movements lasting 1–5 seconds (Racine seizure scale 3). Three and six week seizure counts were combined to obtain a frequency of seizures in 60 minutes for each animal.
Statistical Analysis
Average seizure frequencies were compared between groups by ANOVA with Fisher’s PLSD post-hoc tests (n ≥ 9 per group). Preliminary analysis indicated that males and females differed within the strains. Therefore, male and female data were analyzed separately.
RNA Isolation
Total RNA was isolated from whole brains of six week old B6 or SJL male and female mice with the TRIzol reagent according to the manufacturer’s instructions (Invitrogen). RNA integrity was assessed on an Agilent 2100 Bioanalyzer and all samples received RIN scores of ≥8.20. For each group, total RNAs from 4 mice were combined to generate a pooled sample.
Sample Preparation for RNA Seq
Starting with 3 μg of total RNA, samples underwent poly-A selection, chemical fragmentation, and first and second strand cDNA synthesis using the TruSeq RNA sample preparation kit (Illumina). Following second strand cDNA synthesis, samples were prepared for sequencing on the Illumina HiSeq 2000 platform in the Vanderbilt Genome Sciences Resource (http://gsr.vanderbilt.edu/) according to standard methods (Bentley et al. 2008). Two multiplexed lanes of 100 bp single-end sequencing were performed, resulting in 75 million mappable reads per lane.
RNA-Seq Data Analysis
Base calling and filtering of sequence reads were performed with the Illumina pipeline (Bentley et al. 2008). Subsequent analysis was performed using the Tuxedo Tools on the GALAXY open, web-based platform (Blankenberg et al. 2010; Giardine et al. 2005; Goecks et al. 2010). Tophat aligned RNA-Seq reads to the mm9 genome using Bowtie, allowing reads that span exon-exon boundaries to be aligned to the correct genomic location. Following alignment, novel splice variants were identified (Langmead et al. 2009a, 2009b; Trapnell et al. 2009). Cufflinks assembled transcripts, estimated abundance in fragments per kilobase of exon per million fragments mapped (FPKM) and tested for differential expression and regulation. Cuffcompare combined Cufflinks transcripts across sex and strains. Cuffdiff tested for statistically significant differences in total gene and transcript expression, splicing, transcription start sites (TSS) and promoter usage (Roberts et al. 2011a, 2011b; Trapnell et al. 2010). Parameter settings for each analysis tool are detailed in Supplemental Table 1.
Variant calls were performed by the Vanderbilt Computational Genomics Core using the Genome Analysis Toolkit (GATK) for SNP discovery. Quality filtering retained SNPs with a Phred score of >Q20. Additional manual filtering removed the following: SNPs that were different between male and female of the same strain, heterozygous or had missing sample data. SNPs that were homozygous alternate from the reference (C57BL/6J) genome were retained. VCF files were uploaded to Ensembl’s Variant Effect Predictor to reveal the functional consequences of identified variants (McLaren et al. 2010). Non-synonymous (NS) amino acid substitutions were uploaded to SIFT (http://sift.jcvi.org/) to predict the potential impact on protein function (Kumar et al. 2009).
Results
Interval Specific Congenics
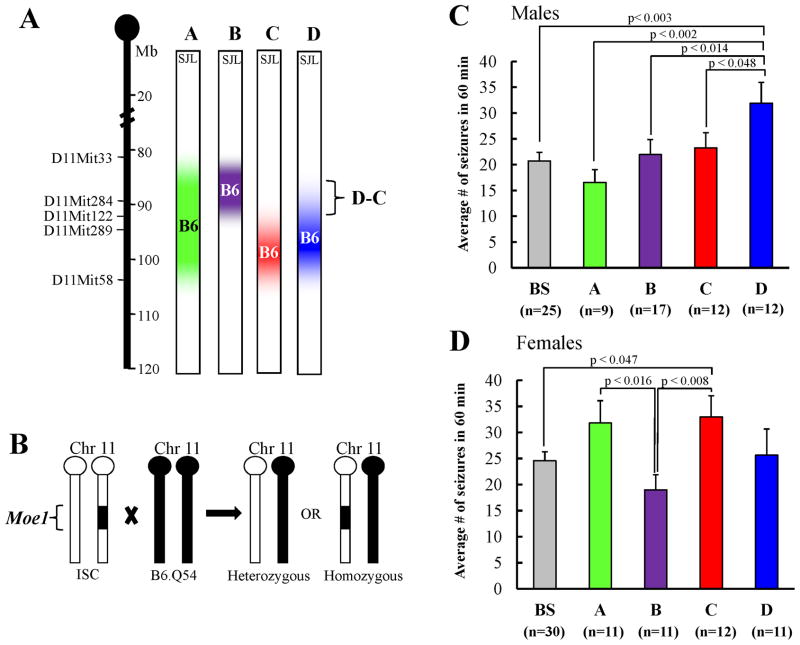
We previously mapped the Scn2aQ54 modifier Moe1 to mouse chr 11 with a peak at D11Mit289 (95 Mb) (Bergren et al. 2005). Inheritance of an SJL allele at this locus conferred increased seizure resistance, while homozygosity for B6 alleles conferred increased susceptibility. To confirm the initial mapping results and refine the Moe1 interval, we generated four ISC lines with varying B6-derived chr 11 segments covering the Moe1 1 LOD support interval on an SJL background. The resulting ISC lines were: ISC-A (D11Mit33-D11Mit58, 81–104 Mb); ISC-B (D11Mit33-D11Mit122, 81–93 Mb); ISC-C (D11Mit122-D11Mit58, 93–104 Mb); ISC-D (D11Mit284-D11Mit58, 89–104 Mb) (Fig. 1A).
Figure 1. Fine mapping of Moe1 on chr11 with interval specific congenic lines.
A. ISC lines were generated with varying B6-derived chr 11 segments (colors) on an SJL background (white). Microsatellite markers used for ISC generation are identified along the chromosome. From the original mapping, the Moe1 peak was at D11Mit289. B. ISC lines were crossed with B6.Q54, generating F1.Q54 offspring. F1.Q54 mice will be heterozygous or homozygous for B6 alleles in Moe1. C. Seizure frequency in ISC males. Average number of seizures in 60 minutes for each genotype group is shown. Significant differences between groups were determined by ANOVA with Fishers PLSD post-hoc tests. D. Seizure frequency in ISC females. Average number of seizures in 60 minutes for each group is shown. Significant differences between groups were determined by ANOVA with Fishers PLSD post-hoc tests. Data are represented as mean ± SEM. Significant P-values determined by post-hoc Fishers PLSD are indicated in the figure.
ISC Phenotyping
Chr 11 ISC females were crossed with B6.Q54 males to generate F1.Q54 offspring that were heterozygous or homozygous for B6 alleles in Moe1 (Fig. 1B). F1.Q54 genotypes were obtained at the expected Mendelian ratios. F1.Q54 offspring underwent 30 minute video-taped observations for visible spontaneous focal motor seizures at three and six weeks of age. Average seizure frequencies (# seizures/60 minutes) were compared between ISC groups carrying homozygous B6 alleles and controls carrying heterozygous alleles. Preliminary analysis indicated within strain sex differences suggesting the possibility of sex-specific modifier effects.
Male mice carrying homozygous B6 alleles in the ISC-D interval (89–104 Mb) exhibited more seizures than heterozygous control littermates, while males with homozygous B6 alleles in the ISC-C interval (93–104 Mb) showed no difference compared to heterozygous control littermates (Fig. 1C) (F(4,70)= 3.321, p< 0.015). This suggests that the small region of non-overlap between ISC-C and ISC-D (D-C; 89–93 Mb) is likely responsible for increased seizure frequency in males.
Female mice carrying homozygous B6 alleles in the ISC-C interval (93–104 Mb) exhibited more seizures than heterozygous control littermates (Fig. 1D) (F(4,70)= 2.601, p< 0.043). This suggests that B6 homozygosity in the distal Moe1 interval is permissive in females. Surprisingly, females with homozygous B6 alleles in the ISC-D interval (89–104 Mb) showed no difference in seizure frequency compared to heterozygous control littermates. This suggests that B6 homozygosity in the small region of non-overlap between ISC-C and ISC-D (D-C; 89–93 Mb) may mask the effect of the distal permissive modifier in females.
In both sexes, B6 homozygosity in the large ISC-A interval (81–104 Mb) that spans the entire Moe1 1 LOD support did not result in a significant difference in seizure frequency compared to heterozygous controls. This suggests that B6 homozygosity in the proximal region of the Moe1 interval may mask the effect of more distal permissive modifiers in both males and females. However, B6 homozygosity in the small ISC-B proximal interval (81–93 Mb) alone does not significantly reduce seizure frequency compared to heterozygous controls.
Candidate Gene Analysis by RNA-Seq
Due to the complexity of the Moe1 interval and the high gene density of chr 11, we performed sequence-based transcriptome analysis in order to identify candidate modifier genes in Moe1. Focused analysis of Moe1 revealed numerous significant differences in total gene and transcript expression, TSS, alternate promoter usage and splicing (Supplemental Table 2). Our ISC results indicated that B6 homozygosity in the D-C interval (89–93 Mb) increased seizure frequency in males. Within the D-C interval, RNA-Seq identified significant gene or transcript expression differences between B6 and SJL males in three genes. Based on our ISC experiments, B6 homozygosity in the ISC-C interval (93–104 Mb) increased seizure frequency in females. In the ISC-C region, RNA-Seq analysis revealed significant gene or transcript expression differences between B6 and SJL females in 32 genes. A complete list of significant differences in the Moe1 region by RNA-Seq is provided in Supplemental Table 2.
To further evaluate candidate genes, we performed SNP analysis to identify genes with coding sequence polymorphisms between B6 and SJL. In region D-C (89–93 Mb), which contains 26 known and predicted genes, we identified 25 SNPs, including two NS SNPs in one gene. In interval C (93–104 Mb), which contains 534 known and predicted genes, we identified 630 SNPs, including 29 NS SNPs in 22 genes. The complete list of NS coding polymorphisms in Moe1 and the predicted impact on protein function are provided in Supplemental Table 3.
Following RNA-Seq analysis, we prioritized candidate genes based on the ISC mapping results, consideration of gene function, location and timing of expression and prior evidence of involvement in neuronal hyperexcitability (Table 1). Top candidate modifier genes were identified as Hlf in males and Cacna1g and Cacnb1 in females.
Table 1.
Top candidate modifier genes in Moe1.
| Interval | Priority† | Gene Symbol | Gene Name |
|---|---|---|---|
| B | * | Cltc | Clathrin |
| * | Dynll2 | Dynein Light Chain 2 | |
| D-C | *** | Hlf | Hepatic Leukemia Factor |
| C | *** | Cacna1g | Voltage-Dep. Ca2+ Channel, T type, α1G |
| *** | Cacnb1 | Voltage-Dep. Ca2+ Channel, β1 | |
| ** | Slc25a39 | Solute Carrier Family 25, Member 39 | |
| * | Pnpo | Pyridoxine 5′-Phosphate Oxidase | |
| * | Hap1 | Huntingtin-Associated Protein 1 |
Priority rating is based on biological function and prior association with neuronal hyperexcitability phenotypes.
highest priority;
moderate priority;
low priority.
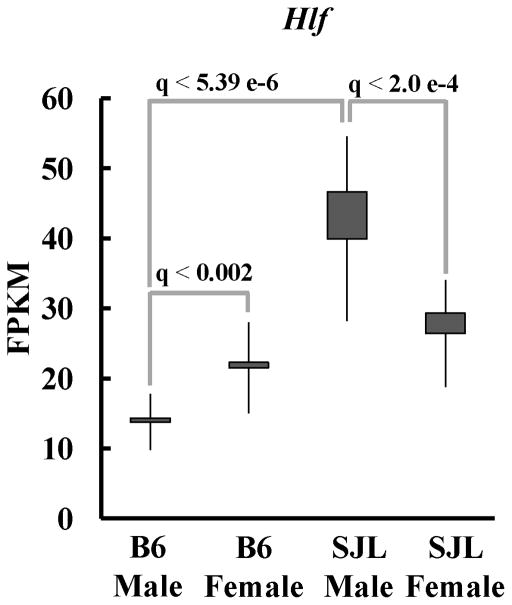
In the D-C interval (89–93 Mb) that contains a male-specific modifier, there were significant expression differences between B6 and SJL males in two genes, Hlf (hepatic leukemia factor) and Stxbp4 (syntaxin binding protein 4) and a processed pseudogene, GM11512. Stxbp4 functions in the insulin receptor signaling pathway and glucose transport (Min et al. 1999). Currently, there is no evidence linking Stxbp4 to neuronal hyperexcitability. Hlf is a member of the proline and acidic amino acid rich basic leucine zipper (PAR bZIP) transcription factor family which has previously been linked to epilepsy (Gachon et al. 2004). There was a significant between-strain Hlf transcript difference in males only, consistent with a male-specific effect (Fig. 2). Taken together, this suggests that Hlf is a strong candidate modifier gene in males (Table 1).
Figure 2. Expression of Hlf-002 transcript in B6 and SJL brain.
The RNA-Seq FPKM values and confidence interval for each group is shown. Boxes represent the range of values and whiskers represent the 95% confidence interval calculated by Cuffdiff. Q-values are false discovery rate-adjusted p-values. (Hlf-002=ENSMUST00000043658).
The ISC-C interval (93–104 Mb) which harbors a female-specific modifier contains numerous genes with expression differences between B6 and SJL females. Of the five genes with a total gene expression difference between females, three are involved in basic cellular processes and two have unknown function. RNA-Seq identified 32 genes with significant differences in transcript expression. A large fraction of these genes are involved in basic cellular processes, with no direct evidence connecting them to neuronal hyperexcitability. There was a significant strain difference in transcript expression of Slc25a39, which is a member of the mitochondrial solute carrier family 25 (SLC25) (Haitina et al. 2006). A family-based study of epilepsy with suggestive linkage to chr 17q12-24 demonstrated differential expression of SLC25A39 in affected family members (Siren et al. 2010). However, sequencing did not reveal a pathogenic variant in this gene. Based on its tenuous association with epilepsy, Slc25a39 was retained as candidate modifier gene with lower priority (Table 1).
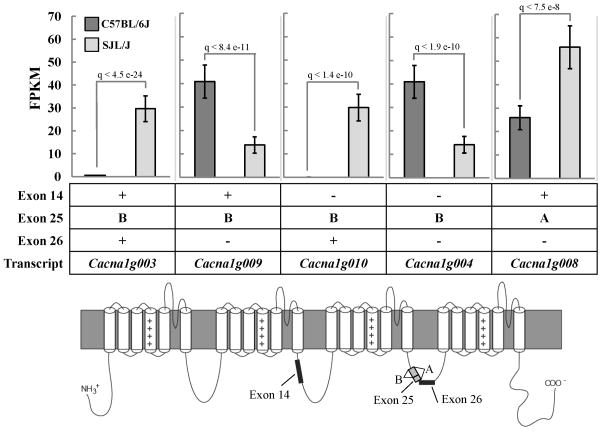
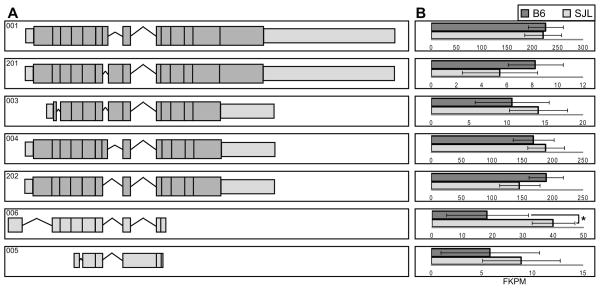
Cacna1g and Cacnb1 emerged as top candidate modifier genes in females based on their direct connection to epilepsy and the strong association between ion channels and epilepsy. Human mutations in both genes have been identified in epilepsy patients (Escayg et al. 1998; Ohmori et al. 2008; Singh et al. 2007). Differences in five major Cacna1g transcripts were identified between B6 and SJL females (Fig. 3). These transcripts have known differences in gating, kinetics and voltage-dependencies (Chemin et al. 2001; Emerick et al. 2006; Monteil et al. 2000). There were also several significant differences in promoter usage, TSS and splice variants between sex and strain. A Cacnb1 female-specific difference was identified between B6 and SJL in transcript Cacnb1-006, a processed transcript that is not translated (Fig. 4). There were also several significant differences in promoter usage, TSS and splice variants between sex and strain.
Figure 3. Differential expression of Cacna1g alternatively spliced transcripts in females.
Inclusion/exclusion of cassette exons 14 and 26 and alternate splicing of exon 25 are shown for the five transcripts that exhibited significant differences between B6 and SJL females. The location of alternatively spliced exons are shown on the Cacna1g channel protein (bottom). FPKM values and confidence intervals for B6 and SJL females are shown. Q-values are false discovery rate-adjusted p-values.
Figure 4. Alternative splicing and differential expression of Cacnbl.
A. Cacnb1 alternatively spliced transcripts are shown. Dark shaded boxes represent coding regions and light shaded boxes are non-coding regions. B. Respective FPKM values and confidence intervals for B6 and SJL female transcripts are shown (*q< 0.02). Q-values are false discovery rate-adjusted p-values.
Discussion
Genetic modifier loci influencing the Scn2aQ54 epilepsy phenotype were initially mapped to chrs 11 (Moe1) and 19 (Moe2) in a backcross between B6 and SJL strains (Bergren et al. 2005). We constructed ISC strains to confirm the presence of Moe1 on chr 11 and refine the critical interval. Our results revealed that the interval likely contains multiple modifiers that contribute to phenotype severity and exhibit sex-specific effects. Similar to our results, previous studies of epilepsy QTL in polygenic mouse models have demonstrated that the substructure of QTL can be complex with multiple genes contributing to the low-resolution mapping peak (Legare et al. 2000; Legare & Frankel 2000).
In females, B6 homozygosity in the ISC-C interval (93–104 Mb) conferred increased seizure frequency, suggesting the presence of a permissive modifier gene in this interval. In males, increased seizure frequency was associated with B6 homozygosity in the ISC-D (89–104 Mb) interval but not in the ISC-C interval (93–104 Mb). This suggests that a male-specific permissive modifier is located in the small non-overlapping interval between ISC-C and ISC-D (D-C; 89–93 Mb). Within the Moe1 interval we identified a significant number of transcripts that exhibited within-strain sex differences. These differences likely result from multiple different causes, which may include brain sexual dimorphism, epigenetics and endocrine influences.
RNA-Seq analysis identified several strong candidate modifiers that may contribute to severity of the seizure phenotype, including Cacna1g and Cacnb1 in females and Hlf in males. Although we identified significant transcript differences in these top candidate modifier genes, we did not identify SNPs within the coding regions. It is likely that non-coding SNPs contribute to the observed transcript differences. Future sequencing of the non-coding regions of Cacna1g, Cacnb1 and Hlf will determine the contribution of non-coding SNPs.
Cacna1g is a T-type Ca2+ channel α1 subunit which propagates low-voltage activated (LVA) Ca2+ currents (Ernst & Noebels 2009). T-type channels regulate neuronal firing by conducting Ca2+ during action potentials and can switch neurons between firing modes (Cain & Snutch 2010). T-type channels are expressed at high densities on thalamic neurons. These neurons innately oscillate during sleep, but can also inappropriately oscillate and cause a generalized seizure (McCormick & Contreras 2001; Perez-Reyes 2003). Rapid activation of T-type channels is responsible for the generation of low threshold spikes, which trigger epileptic “burst-firing” patterns. It is also responsible for abnormal spike-wave discharges (SWDs) in absence epilepsy that arise from synchronous firing of thalamocortical networks (Blumenfeld 2005; Contreras 2006; Ernst et al. 2009). Several spontaneous mouse models of absence epilepsy, including lethargic, tottering and stargazer, exhibit abnormalities in LVA Ca2+ currents, likely resulting in abnormal thalamocortical synchronization and absence epilepsy in the mutant animals (Zhang et al. 2002). Deletion of Cacna1g in mice suppressed GABAB receptor agonist-induced SWDs, indicating an essential role for this gene in seizure expression (Ernst et al. 2009; Kim et al. 2001). In humans, SNPs in CACNA1G have been associated with idiopathic generalized epilepsy (Singh et al. 2007).
Our RNA-Seq analysis identified expression, TSS, promoter usage and splicing differences in Cacna1g between sex and strain. Alternative splicing of T-type Ca2+ channels is known to contribute to their functional diversity. We identified significant strain differences between females in the expression of five major Cacna1g transcripts with known functional diversity. The inclusion of cassette exon 14 has been shown to shift steady-state inactivation in the hyperpolarized direction and accelerate activation and inactivation kinetics (Chemin et al. 2001; Emerick et al. 2006). Inclusion of cassette exon 26 contributes to more hyperpolarized steady state inactivation and slower recovery from inactivation (Chemin et al. 2001). Exon 25 has an internal donor site, resulting in a short exon 25 (B) or full length (A) (Chemin et al. 2001; Emerick et al. 2006; Mittman et al. 1999; Monteil et al. 2000). Transcripts with the 25A exon activate and inactivate at more hyperpolarized potentials, while 25B exon transcripts have slower activation and inactivation kinetics (Chemin et al. 2001). Based on this information, transcripts that exhibit significantly higher expression in SJL females have the most hyperpolarized steady-state inactivation potential and the fastest activation and inactivation kinetics (Fig. 3). We hypothesize that SJL females have a higher portion of Cacna1g channels that activate quickly, but inactivate rapidly and prematurely. This early inactivation state may lead to a low probability for neurons to reach synchronous oscillations, a common feature of epilepsy (Blumenfeld 2005; Contreras 2006; Ernst et al. 2009). Conversely, transcripts expressed at higher levels in B6 females compared to SJL are those with depolarized steady-state inactivation potentials and slower rates of activation and inactivation (Chemin et al. 2001) (Fig. 3). We hypothesize B6 females have an increased number of Cacna1g channels that are prone to hyperexcitability. This hyperexcitability emerges from an altered window current, resulting from inactivation shifts and rate changes, a known contributor to epilepsy (Vreugdenhil et al. 1998).
Cacnb1 encodes one of four homologous Ca2+ channel β subunits. β subunits modulate α1 channel trafficking, gating and kinetics (Buraei & Yang 2010; Dolphin 2003; Lie et al. 1999). Cacnb1 has four splice variants, which are highly expressed in the hippocampus and dentate gyrus (Buraei & Yang 2010; Dolphin 2003). β subunits have been previously linked to epilepsy in mice. Lethargic mice have a mutation in Cacnb4 that results in absence epilepsy, severe ataxia and loss of motor coordination (Burgess et al. 1997; Escayg et al. 1998). In humans, surgical resection samples from temporal lobe epilepsy patients exhibited an up-regulation of CACNB1 and CACNB2 (Lie et al. 1999). It was hypothesized that the increased expression may underlie enhanced Ca2+ currents commonly seen in rodent kindling and kainate models of temporal lobe epilepsy (Beck et al. 1998; Faas et al. 1996; Lie et al. 1999; Vreugdenhil & Wadman 1994). Several CACNB1 mutations have been identified in patients with a history of febrile seizures, juvenile myoclonic epilepsy, episodic ataxia, generalized epilepsy and praxis-induced seizures (Escayg et al. 1998; Ohmori et al. 2008).
RNA-Seq analysis determined that SJL females express significantly higher levels of transcript Cacnb1-006 than B6 females (Fig. 4). Although this transcript does not contain an open reading frame, it does undergo post-transcriptional processing, suggesting the possibility that it may act as a long non-coding RNA (lncRNA). Recent evidence demonstrates the importance of non-coding RNAs (ncRNA) in the brain (Berretta & Morillon 2009; Brosius 2005; Cao et al. 2006; Kapranov et al. 2007; Mehler & Mattick 2006). Contribution of ncRNAs has been demonstrated for neural differentiation and cell identity, hippocampal development, oligodendrocyte myelination and synaptic plasticity (Qureshi et al. 2010). Additionally, lncRNAs have been implicated in neurodevelopmental, neurodegenerative, neuroimmunological and psychiatric disorders and, relevant to our studies, epilepsy (Qureshi et al. 2010). Mice deficient in the lncRNA Evf2 had reduced numbers of GABAergic interneurons in the hippocampal formation and displayed reduced synaptic inhibition (Bond et al. 2009). This suggests the importance of a lncRNA in the formation of GABA circuitry and suggests a potential mechanism for hyperexcitability. The lncRNA BC1 has also been implicated in a seizure phenotype. BC1−/− homozygous knockout mice exhibited audiogenic seizures and it is hypothesized that BC1 lncRNA modulates hyperexcitability via the metabotropic glutamate receptor pathway (Zhong et al. 2009). It is possible that the Cacnb1-006 transcript may act as a lncRNA that directly or indirectly influences neuronal excitability.
Hlf is a member of the PAR bZIP transcription factor family. The loss of the PAR bZIP transcription factor family (Hlf, Dbp, Tef) results in mice with spontaneous, generalized tonic-clonic and absence seizures (Gachon et al. 2004). Microarray expression analysis performed to investigate contributors to the PAR bZIP knockout epilepsy phenotype, revealed a two-fold reduction in pyridoxal kinase (Pdxk), which encodes a coenzyme involved in the conversion of vitamin B6 to pyridoxal 5′-phosphate (PLP) (Gachon et al. 2004). PLP is a key coenzyme involved in amino acid and neurotransmitter metabolism (John 1995). Mice defective in PLP metabolism experience fatal seizures, found to be a consequence of significantly reduced GABA levels (Waymire et al. 1995). Mice maintained on a vitamin B6 deficient diet, resulting in reduced PLP levels, experienced increased audiogenic seizures (Coleman & Schlesinger 1965; Schlesinger R & Schreiber 1969; Schlesinger & Uphouse 1972). Human vitamin B6-dependent epilepsies can be successfully treated with PLP (Plecko & Stockler 2009). Our RNA-Seq analysis showed that B6 males have a significant reduction in Hlf expression compared to SJL males. Reduced expression of Hlf in B6 males may lead to reduced Pdxk and PLP levels and increased seizure susceptibility. Future studies testing the effect of vitamin B6 deficiency on B6.Q54 mice may help determine the contribution of this pathway to Scn2aQ54 epilepsy severity.
Based on our ISC and RNA-Seq data, along with further analysis of gene function and association with seizures, we nominate Cacna1g, Cacnb1 and Hlf as strong functional candidates for Moe1. Future testing of high priority candidate genes will be necessary to determine if they contribute to the epilepsy severity of Scn2aQ54 mice. Understanding the molecular basis of genetic modifiers in a mouse model can provide insight into human epilepsy. It has the potential to advance molecular diagnostic capabilities and identify novel therapeutic targets for the improved treatment of human patients.
Supplementary Material
Acknowledgments
We thank Avigile Baehr, Elizabeth Rutter and Rebecca Somershoe for technical assistance. We acknowledge RNA-Seq support from Travis Clark in The Vanderbilt Genome Sciences Resource and Christian Shaffer in The Vanderbilt Computational Genomics Core. This work was supported by National Institutes of Health grants R01-NS063097 (J.A.K.), T32-NS07491 (N.A.H.), F31-NS077700 (N.A.H.) and an Epilepsy Foundation Predoctoral Fellowship (N.A.H.).
References
- Beck H, Steffens R, Elger CE, Heinemann U. Voltage-dependent Ca2+ currents in epilepsy. Epilepsy Res. 1998;32:321–332. doi: 10.1016/s0920-1211(98)00062-x. [DOI] [PubMed] [Google Scholar]
- Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergren SK, Chen S, Galecki A, Kearney JA. Genetic modifiers affecting severity of epilepsy caused by mutation of sodium channel Scn2a. Mamm Genome. 2005;16:683–690. doi: 10.1007/s00335-005-0049-4. [DOI] [PubMed] [Google Scholar]
- Bergren SK, Rutter ED, Kearney JA. Fine mapping of an epilepsy modifier gene on mouse Chromosome 19. Mamm Genome. 2009;20:359–366. doi: 10.1007/s00335-009-9193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta J, Morillon A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 2009;10:973–982. doi: 10.1038/embor.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Von KG, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;19:21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenfeld H. Cellular and network mechanisms of spike-wave seizures. Epilepsia. 2005;46:21–33. doi: 10.1111/j.1528-1167.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. Waste not, want not--transcript excess in multicellular eukaryotes. Trends Genet. 2005;21:287–288. doi: 10.1016/j.tig.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Buraei Z, Yang J. The β subunit of voltage-gated Ca2+ channels. Physiol Rev. 2010;90:1461–1506. doi: 10.1152/physrev.00057.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DL, Jones JM, Meisler MH, Noebels JL. Mutation of the Ca2+ channel beta subunit gene Cchb4 is associated with ataxia and seizures in the lethargic (lh) mouse. Cell. 1997;88:385–392. doi: 10.1016/s0092-8674(00)81877-2. [DOI] [PubMed] [Google Scholar]
- Cain SM, Snutch TP. Contributions of T-type calcium channel isoforms to neuronal firing. Channels (Austin) 2010;4:475–482. doi: 10.4161/chan.4.6.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Chaix Y, Ferraro TN, Lapouble E, Martin B. Chemoconvulsant-induced seizure susceptibility: toward a common genetic basis. Epilepsia. 2007;48:48–52. doi: 10.1111/j.1528-1167.2007.01289.x. [DOI] [PubMed] [Google Scholar]
- Chemin J, Monteil A, Bourinet E, Nargeot J, Lory P. Alternatively spliced alpha(1G) (Ca(V)3.1) intracellular loops promote specific T-type Ca(2+) channel gating properties. Biophys J. 2001;80:1238–1250. doi: 10.1016/S0006-3495(01)76100-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes L, Del-Favero J, Ceulemans B, Lagae L, Van Broeckhoven C, De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am J Hum Genet. 2001;68:1327–32. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman DL, Schlesinger K. Effects of pyridoxine deficiency on audiogenic seizure susceptibility in inbred mice. Proc Soc Exp Biol Med. 1965;119:264–266. doi: 10.3181/00379727-119-30154. [DOI] [PubMed] [Google Scholar]
- Contreras D. The role of T-channels in the generation of thalamocortical rhythms. CNS Neurol Disord Drug Targets. 2006;5:571–585. doi: 10.2174/187152706779025526. [DOI] [PubMed] [Google Scholar]
- Dolphin AC. Beta subunits of voltage-gated calcium channels. J Bioenerg Biomembr. 2003;35:599–620. doi: 10.1023/b:jobb.0000008026.37790.5a. [DOI] [PubMed] [Google Scholar]
- Emerick MC, Stein R, Kunze R, McNulty MM, Regan MR, Hanck DA, Agnew WS. Profiling the array of Ca(v)3.1 variants from the human T-type calcium channel gene CACNA1G: alternative structures, developmental expression, and biophysical variations. Proteins. 2006;64:320–342. doi: 10.1002/prot.20877. [DOI] [PubMed] [Google Scholar]
- Ernst WL, Noebels JL. Expanded alternative splice isoform profiling of the mouse Cav3.1/alpha1G T-type calcium channel. BMC Mol Biol. 2009;10:53. doi: 10.1186/1471-2199-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst WL, Zhang Y, Yoo JW, Ernst SJ, Noebels JL. Genetic enhancement of thalamocortical network activity by elevating alpha 1g-mediated low-voltage-activated calcium current induces pure absence epilepsy. J Neurosci. 2009;29:1615–1625. doi: 10.1523/JNEUROSCI.2081-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, Jones JM, Kearney JA, Hitchcock PF, Meisler MH. Calcium channel beta 4 (CACNB4): human ortholog of the mouse epilepsy gene lethargic. Genomics. 1998;50:14–22. doi: 10.1006/geno.1998.5311. [DOI] [PubMed] [Google Scholar]
- Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C, Malafosse A. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- Faas GC, Vreugdenhil M, Wadman WJ. Calcium currents in pyramidal CA1 neurons in vitro after kindling epileptogenesis in the hippocampus of the rat. Neuroscience. 1996;75:57–67. doi: 10.1016/0306-4522(96)00254-0. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Dahl JP, Smith GG, Schwebel CL, MacDonald R, Lohoff FW, Berrettini WH, Buono RJ. Analysis of a quantitative trait locus for seizure susceptibility in mice using bacterial artificial chromosome-mediated gene transfer 4. Epilepsia. 2007a;48:1667–1677. doi: 10.1111/j.1528-1167.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Longman RL, Snyder RL, DeMuth D, Szpilzak I, Mulholland N, Eng E, Lohoff FW, Buono RJ, Berrettini WH. Quantitative genetic study of maximal electroshock seizure threshold in mice: evidence for a major seizure susceptibility locus on distal chromosome 1. Genomics. 2001;75:35–42. doi: 10.1006/geno.2001.6577. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Martin JF, Lohoff FW, Gieringer TA, Zamboni D, Schwebel CL, Press DM, Kratzer SO, Zhao H, Berrettini WH, Buono RJ. Fine mapping of a seizure susceptibility locus on mouse Chromosome 1: nomination of Kcnj10 as a causative gene. Mamm Genome. 2004;15:239–51. doi: 10.1007/s00335-003-2270-3. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Smith GG, Ballard D, Zhao H, Schwebel CL, Gupta A, Rappaport EF, Ruiz SE, Lohoff FW, Doyle GA, Berrettini WH, Buono RJ. Quantitative trait loci for electrical seizure threshold mapped in C57BLKS/J and C57BL/10SnJ mice. Genes Brain Behav. 2011;10:309–315. doi: 10.1111/j.1601-183X.2010.00668.x. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Smith GG, Schwebel CL, Doyle GA, Ruiz SE, Oleynick JU, Lohoff FW, Berrettini WH, Buono RJ. Confirmation of multiple seizure susceptibility QTLs on chromosome 15 in C57BL/6J and DBA/2J inbred mice. Physiol Genomics. 2010;42A:1–7. doi: 10.1152/physiolgenomics.00096.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro TN, Smith GG, Schwebel CL, Lohoff FW, Furlong P, Berrettini WH, Buono RJ. Quantitative trait locus for seizure susceptibility on mouse chromosome 5 confirmed with reciprocal congenic strains. Physiol Genomics. 2007b;31:458–462. doi: 10.1152/physiolgenomics.00123.2007. [DOI] [PubMed] [Google Scholar]
- Frankel WN, Johnson EW, Lutz CM. Congenic strains reveal effects of the epilepsy quantitative trait locus, El2, separate from other El loci. Mamm Genome. 1995;6:839–843. doi: 10.1007/BF00292432. [DOI] [PubMed] [Google Scholar]
- Gachon F, Fonjallaz P, Damiola F, Gos P, Kodama T, Zakany J, Duboule D, Petit B, Tafti M, Schibler U. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenfeld HK, Neumann PE, Li X, St Jean PL, Paul SM. Mapping quantitative trait loci for seizure response to a GABAA receptor inverse agonist in mice. J Neurosci. 1999;19:3731–3738. doi: 10.1523/JNEUROSCI.19-10-03731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitina T, Lindblom J, Renstrom T, Fredriksson R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics. 2006;88:779–790. doi: 10.1016/j.ygeno.2006.06.016. [DOI] [PubMed] [Google Scholar]
- John RA. Pyridoxal phosphate-dependent enzymes. Biochim Biophys Acta. 1995;1248:81–96. doi: 10.1016/0167-4838(95)00025-p. [DOI] [PubMed] [Google Scholar]
- Jorge BS, Campbell CM, Miller AR, Rutter ED, Gurnett CA, Vanoye CG, George AL, Jr, Kearney JA. Voltage-gated potassium channel KCNV2 (Kv8.2) contributes to epilepsy susceptibility. Proc Natl Acad Sci USA. 2011;108:5443–5448. doi: 10.1073/pnas.1017539108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kearney JA, Plummer NW, Smith MR, Kapur J, Cummins TR, Waxman SG, Goldin AL, Meisler MH. A gain-of-function mutation in the sodium channel gene Scn2a results in seizures and behavioral abnormalities. Neuroscience. 2001;102:307–317. doi: 10.1016/s0306-4522(00)00479-6. [DOI] [PubMed] [Google Scholar]
- Kim D, Song I, Keum S, Lee T, Jeong MJ, Kim SS, McEnery MW, Shin HS. Lack of the burst firing of thalamocortical relay neurons and resistance to absence seizures in mice lacking alpha(1G) T-type Ca(2+) channels. Neuron. 2001;31:35–45. doi: 10.1016/s0896-6273(01)00343-9. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Langmead B, Schatz MC, Lin J, Pop M, Salzberg SL. Searching for SNPs with cloud computing. Genome Biol. 2009a;10:134. doi: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009b;10:25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legare ME, Bartlett FS, Frankel WN. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 2000;10:42–48. [PMC free article] [PubMed] [Google Scholar]
- Legare ME, Frankel WN. Multiple seizure susceptibility genes on chromosome 7 in SWXL-4 congenic mouse strains. Genomics. 2000;70:62–65. doi: 10.1006/geno.2000.6368. [DOI] [PubMed] [Google Scholar]
- Lie AA, Blumcke I, Volsen SG, Wiestler OD, Elger CE, Beck H. Distribution of voltage-dependent calcium channel beta subunits in the hippocampus of patients with temporal lobe epilepsy. Neuroscience. 1999;93:449–456. doi: 10.1016/s0306-4522(99)00162-1. [DOI] [PubMed] [Google Scholar]
- Maihara T, Noda A, Yamazoe H, Voigt B, Kitada K, Serikawa T. Chromosomal mapping of genes for epilepsy in NER: a rat strain with tonic-clonic seizures. Epilepsia. 2000;41:941–949. doi: 10.1111/j.1528-1157.2000.tb00276.x. [DOI] [PubMed] [Google Scholar]
- McCormick DA, Contreras D. On the cellular and network bases of epileptic seizures. Annu Rev Physiol. 2001;63:815–846. doi: 10.1146/annurev.physiol.63.1.815. [DOI] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS. Non-coding RNAs in the nervous system. J Physiol. 2006;575:333–341. doi: 10.1113/jphysiol.2006.113191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler MH, Kearney JA. Sodium channel mutations in epilepsy and other neurological disorders. J Clin Invest. 2005;115:2010–2017. doi: 10.1172/JCI25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisler MH, O’Brien JE, Sharkey LM. Sodium channel gene family: epilepsy mutations, gene interactions and modifier effects. J Physiol. 2010;588:1841–1848. doi: 10.1113/jphysiol.2010.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Okada S, Kanzaki M, Elmendorf JS, Coker KJ, Ceresa BP, Syu LJ, Noda Y, Saltiel AR, Pessin JE. Synip: a novel insulin-regulated syntaxin 4-binding protein mediating GLUT4 translocation in adipocytes. Mol Cell. 1999;3:751–760. doi: 10.1016/s1097-2765(01)80007-1. [DOI] [PubMed] [Google Scholar]
- Mittman S, Guo J, Agnew WS. Structure and alternative splicing of the gene encoding alpha1G, a human brain T calcium channel alpha1 subunit. Neurosci Lett. 1999;274:143–146. doi: 10.1016/s0304-3940(99)00716-8. [DOI] [PubMed] [Google Scholar]
- Monteil A, Chemin J, Bourinet E, Mennessier G, Lory P, Nargeot J. Molecular and functional properties of the human alpha(1G) subunit that forms T-type calcium channels. J Biol Chem. 2000;275:6090–6100. doi: 10.1074/jbc.275.9.6090. [DOI] [PubMed] [Google Scholar]
- Moran JL, Bolton AD, Tran PV, Brown A, Dwyer ND, Manning DK, Bjork BC, Li C, Montgomery K, Siepka SM, Vitaterna MH, Takahashi JS, Wiltshire T, Kwiatkowski DJ, Kucherlapati R, Beier DR. Utilization of a whole genome SNP panel for efficient genetic mapping in the mouse. Genome Res. 2006;16:436–440. doi: 10.1101/gr.4563306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori I, Ouchida M, Miki T, Mimaki N, Kiyonaka S, Nishiki T, Tomizawa K, Mori Y, Matsui H. A CACNB4 mutation shows that altered Ca(v)2.1 function may be a genetic modifier of severe myoclonic epilepsy in infancy. Neurobiol Dis. 2008;32:349–354. doi: 10.1016/j.nbd.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Perez-Reyes E. Molecular physiology of low-voltage-activated t-type calcium channels. Physiol Rev. 2003;83:117–161. doi: 10.1152/physrev.00018.2002. [DOI] [PubMed] [Google Scholar]
- Plecko B, Stockler S. Vitamin B6 dependent seizures. Can J Neurol Sci. 2009;36:S73–S77. [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32:281–294. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011a;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011b;12:22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger RK, Schreiber RA. Interaction of drugs and pyridoxine deficiency on central nervous system excitability. Ann NY Acad Sci. 1969;166:281–287. doi: 10.1111/j.1749-6632.1969.tb54278.x. [DOI] [PubMed] [Google Scholar]
- Schlesinger K, Uphouse LL. Pyridoxine dependency and central nervous system excitability. Adv Biochem Psychopharmacol. 1972;4:105–140. [PubMed] [Google Scholar]
- Singh B, Monteil A, Bidaud I, Sugimoto Y, Suzuki T, Hamano S, Oguni H, Osawa M, Alonso ME, gado-Escueta AV, Inoue Y, Yasui-Furukori N, Kaneko S, Lory P, Yamakawa K. Mutational analysis of CACNA1G in idiopathic generalized epilepsy. Hum Mutat. 2007;28:524–525. doi: 10.1002/humu.9491. [DOI] [PubMed] [Google Scholar]
- Siren A, Polvi A, Chahine L, Labuda M, Bourgoin S, Anttonen AK, Kousi M, Hirvonen K, Simola KO, Andermann E, Laiho A, Soini J, Koivikko M, Laaksonen R, Pandolfo M, Lehesjoki AE. Suggestive evidence for a new locus for epilepsy with heterogeneous phenotypes on chromosome 17q. Epilepsy Res. 2010;88:65–75. doi: 10.1016/j.eplepsyres.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreugdenhil M, Faas GC, Wadman WJ. Sodium currents in isolated rat CA1 neurons after kindling epileptogenesis. Neuroscience. 1998;86:99–107. doi: 10.1016/s0306-4522(98)00041-4. [DOI] [PubMed] [Google Scholar]
- Vreugdenhil M, Wadman WJ. Kindling-induced long-lasting enhancement of calcium current in hippocampal CA1 area of the rat: relation to calcium-dependent inactivation. Neuroscience. 1994;59:105–114. doi: 10.1016/0306-4522(94)90102-3. [DOI] [PubMed] [Google Scholar]
- Waymire KG, Mahuren JD, Jaje JM, Guilarte TR, Coburn SP, MacGregor GR. Mice lacking tissue non-specific alkaline phosphatase die from seizures due to defective metabolism of vitamin B-6. Nat Genet. 1995;11:45–51. doi: 10.1038/ng0995-45. [DOI] [PubMed] [Google Scholar]
- Winawer MR, Gildersleeve SS, Phillips AG, Rabinowitz D, Palmer AA. Mapping a mouse limbic seizure susceptibility locus on chromosome 10. Epilepsia. 2011;52:2076–2083. doi: 10.1111/j.1528-1167.2011.03256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mori M, Burgess DL, Noebels JL. Mutations in high-voltage-activated calcium channel genes stimulate low-voltage-activated currents in mouse thalamic relay neurons. J Neurosci. 2002;22:6362–6371. doi: 10.1523/JNEUROSCI.22-15-06362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Chuang SC, Bianchi R, Zhao W, Lee H, Fenton AA, Wong RK, Tiedge H. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J Neurosci. 2009;29:9977–9986. doi: 10.1523/JNEUROSCI.3893-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.