Background: TLP and p21 are involved in growth inhibition.
Results: TLP decreased cell growth and activated upstream promoter of p21 gene p53-dependently. TLP bound to p53. The promoter was associated with TLP and p53, and TLP enhanced recruitment of p53 to the promoter.
Conclusion: TLP is required for p53-dependent transcriptional activation of p21 upstream promoter.
Significance: A novel regulator for p53-p21 pathway was identified.
Keywords: p53, Transcription Coactivators, Transcription Factors, Transcription Regulation, Tumor Suppressor Gene, TLP, TRF2, p21
Abstract
TATA-binding protein-like protein (TLP) is involved in development, checkpoint, and apoptosis through potentiation of gene expression. TLP-overexpressing human cells, especially p53-containing cells, exhibited a decreased growth rate and increased proportion of G1 phase cells. TLP stimulated expression of several growth-related genes including p21 (p21Waf1/Cip1). TLP-mediated activation of the p21 upstream promoter in cells was shown by a promoter-luciferase reporter assay. The p53-binding sequence located in the p21 upstream promoter and p53 itself are required for TLP-mediated transcriptional activation. TLP and p53 bound to each other and synergistically enhanced activity of the upstream promoter. TLP specifically activated transcription from the endogenous upstream promoter, and p53 was required for this activation. Etoposide treatment also resulted in activation of the upstream promoter as well as nuclear accumulation of TLP and p53. Moreover, the upstream promoter was associated with endogenous p53 and TLP, and the p53 recruitment was enhanced by TLP. The results of the present study suggest that TLP mediates p53-governed transcriptional activation of the p21 upstream promoter.
Introduction
The cell cycle is regulated positively by multiple cyclin-dependent kinase·cyclin complexes and negatively by cyclin-dependent kinase inhibitors (CKIs)3 (1). CKI inhibits cell cycle progression to antagonize cell cycle engines, and its dysfunction and repressed gene expression result in immortalization of cells. Thus, CKI generally behaves as a tumor suppressor (2, 3). p21 (p21Waf1/Cip1) is one of the most representative and major CKIs for G1 arrest working from G1 to S phases (4, 5), and it binds to CDC4/6·cyclin D complexes needed for activation of a number of S phase-promoting regulatory proteins. p21 binds to proliferating cell nuclear antigen to inhibit DNA polymerase. It is also known that repressed p21 gene expression is required for propagation and maintenance of the undifferentiating property of induced pluripotent stem cells (6).
The p21 gene is enhanced by multiple transcription factors (7, 8), and its expression in a normal condition is regulated in accordance with cell cycle progression. When cells are exposed to genotoxic agents such as etoposide and UV light, p53 protein is activated via phosphorylation and binds to p53-responsive elements of the p21 gene (9). Then propagation of damaged cells is repressed at G1 phase by accumulated p21 protein. This arresting period is required for damaged cells to be repaired or to enter an apoptotic pathway. Therefore, p21 is regarded as a potent checkpoint factor like p53.
p53 is the most typical tumor suppressor that activates many genes via its transcriptional regulatory property as well as a stoichiometric fashion (10–12). p53-responsive genes include genes for apoptosis induction, DNA repair, and cell cycle repression such as p21. Normally, p53 is destabilized by association with MDM2 ubiquitin ligase, which brings p53 to the ubiquitin-proteasome pathway. When a genotoxin signal reaches a cell, a kinase cascade involving ATM/ATR and Chk1/Chk2 functions to phosphorylate p53, resulting in release of MDM2 from p53 (9), and the phosphorylated p53 comprises a homotetramer and binds to its target sequences of responsive genes (13). p53 forms a gene family including transactivating p63 (TAp63) and p73, all of which have the same consensus sequence (14). TAp63 has a role in differentiation as well as apoptosis induction and tumor suppression (15–17). We have found that TBP-like protein (TLP) enhances activity of the promoter of TAp63 gene, which induces apoptotic cell death, and we have presented a TLP-TAp63 pathway model to explain etoposide-triggered apoptosis (18).
TATA-binding protein (TBP) is one of the general transcription factors, and it has the ability to bind to TATA box elements of RNA polymerase II-driven genes (19–21). TBP comprises a gene family including TBP-related factor 1 (TRF1), TLP/TRF2, TRF3, and TRF4 (22–27). TLP exhibits 40% identity to the conserved region of TBP and binds to transcription factor IIA (TFIIA) more strongly than does TBP (28, 29). Originally, TLP was identified as a differentiation factor (30, 31). By using TLP knock-out chicken DT40 cells, we demonstrated that TLP inhibits G2-M transition (32). Although TLP has no obvious sequence-specific DNA binding activity, many lines of evidence indicate that TLP has a transcription-activating ability (33, 34). Through cell-based transient promoter assays, we have demonstrated that TLP activates several promoters that lack a TATA box, whereas TATA-containing promoters are repressed for an unknown reason (28, 34, 35). It has also been shown that TLP regulates cellular genes including cyclin G2 (32), TAp63 (18), wee1 (32, 36), proliferating cell nuclear antigen (32, 37), and NF1 (38), which are related to cell cycle regulation, apoptosis induction, tumor suppression, and DNA repair, implying that TLP works for cell integrity and growth control.
Chicken cells lacking the TLP gene exhibit high growth rates and apoptosis induction when they are exposed to genotoxic agents (32), suggesting that TLP serves as a checkpoint factor in chicken cells. However, it has not been determined whether TLP exhibits such functions in mammalian cells. In this study, we found that TLP exhibits a G1-arresting ability especially in p53-containing cells, and we identified p21 as one of the TLP-stimulated genes. Interestingly, we found that activation of the p21 upstream promoter by TLP absolutely depends on the p53-binding sequence and p53 itself. TLP could bind to p53. The chromosomal upstream promoter was also stimulated by etoposide, and p53 and TLP associated with that region and activated the p21 promoter synergistically. TLP enhanced the recruitment of p53 to the promoter. Consequently, we identified a novel regulatory factor of the human p21 gene. Interaction involving p53 and TLP is thought to be responsible for TLP-governed G1 arrest.
EXPERIMENTAL PROCEDURES
Cell Culture, Drug Treatment, and DNA Transfection
HeLa cells were maintained in Dulbecco's modified Eagle's medium with low glucose content (DMEM-low; Sigma-Aldrich) at 37 °C in the presence of 10% fetal calf serum and 5% CO2. Human HepG2 cells and HCT116 cells (wild-type and p53-lacking mutant cells) (39), and COS7 cells were similarly cultured in DMEM-high (Sigma-Aldrich). Etoposide dissolved in dimethyl sulfoxide (DMSO) was added to the medium for some experiments. Transfection of nucleic acids was performed by using Lipofectamine and Plus reagent (Invitrogen) according to the manufacturer's recommendations. If necessary, cells were irradiated by UV light 28 h after transfection.
Flow Cytometry
Trypsinized and magnesium/calcium-omitted PBS(PBS(−))-washed cells were fixed with ethanol, treated with RNase A, and stained with propidium iodide (5 μg/ml) as described previously (32). The proportion of cells was analyzed by FACSCalibur and Cell Quest Software (BD Biosciences).
Plasmids
Expression Plasmids Used in Mammalian Cells
FH-TLP, which is the same as pCIneo-FH-TLP described in a previous report (29), is a mouse TLP expression plasmid harboring FLAG and oligohistidine tags at the N terminus. NLS-TLP (same as pCIneo-FH-NLS-TLP) (32) is a derivative of pCIneo-FH-TLP that has a nuclear localization signal. Mouse and human TLPs have an identical amino acid sequence. A p53 expression plasmid, pcDNA-FLAG-p53, supplied by Addgene (Cambridge, MA) was modified to pcDNA-HA-p53 (referred to as HA-p53 in this study), which contains an HA tag at the N terminus.
Reporter Plasmids for Luciferase Assay
Basically, pGL4.10 vector (Promega) for the luciferase reporter assay was used for plasmid construction. Human p21 (p21Waf1/Cip1) genomic DNA in pKM2L-ph21 was obtained from RIKEN DNA Bank (Tsukuba, Japan). In this study, the transcription start site (TSS) at +1 of the human p21 gene was assigned in accordance with TSS of p21 variant-1 transcript (NCBI accession number NM_00389.2). The promoter region of the p21 gene from −2677 to +66 was amplified by PCR and cloned with a Mighty TA-Cloning kit (Takara). Then the insert was transferred to a luciferase reporter vector between KpnI and XhoI sites for the 5′-end and 3′-end, respectively, to construct p21luc1 (see Fig. 3A). Other p21 promoter-reporter constructs were derived from p21luc1 as schematically illustrated in the figures. p21luc3 has a KpnI linker between −1875 and −1874 (see Fig. 4A). In p21luc18, the p53-binding sequence was changed from 5′-(−2261)GAACATGTCCCAACATGTTG to 5′-GAACCGTGAAACCACTGTTG. A fragment of 818 bp upstream from a BstEII site (located just upstream of the translation start site) that contains multiple TSSs of the human INK4a promoter was cloned in a pGL3-Basic vector (Promega) to construct pINK4aluc. A luciferase reporter for human BAX was described previously (40).
FIGURE 3.
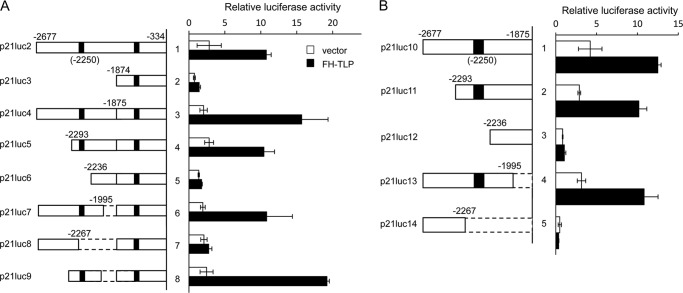
Activation of human p21 promoter by TLP. A, luciferase reporter assay for various human promoters. HeLa cells transfected with the indicated reporter plasmids together with FH-TLP or its vacant plasmids (vector) were examined for relative luciferase activity using the Dual-Luciferase Reporter Assay System. B, schematic illustration of human genomic DNA around the p21 gene from −2677 to +66 inserted in p21luc1 and its derivative plasmids. p21luc1 contains transcription start sites at −2230 for the upstream promoter and +1 for the downstream promoter. p21luc3 is a promoterless construct. Solid boxes represent reported p53-binding sites. C, COS7 cells were transfected with increasing amounts of FH-TLP, and luciferase activity derived from the p21 promoter in p21luc2 and BAX promoter was measured. Data are indicated as averages with error bars according to multiple experiments.
FIGURE 4.
Activation of p21 upstream promoter by TLP. p21luc2 containing the p21 upstream promoter and its truncation derivatives were analyzed by luciferase reporter assay with or without co-transfected FH-TLP. Results are displayed as relative activities. A, structures of inserts of p21 upstream promoter-derived DNA are schematically illustrated on the left side of the panel. Solid boxes and boxes with broken lines represent p53-binding sites and deleted sequences, respectively. Left panel, results of the luciferase assay. B, p21luc10 in which the downstream p53 target site was eliminated and its truncation derivatives were also analyzed. Data are indicated as averages with error bars according to multiple experiments.
Bacterial Expression Plasmids
Open reading frames of p53 and TLP were subcloned into a pET-3a vector (Novagen). Open reading frames of TLP, TBP, and p53 were subcloned into pGEX4T-1 (GE Healthcare) for production of GST fusion proteins.
Short Interfering RNA (siRNA)
siRNAs were prepared by using a Silencer siRNA Construction kit (Ambion) as described previously (41). Sequences of siRNA for human TLP contained 5′-UAACAGGGCCCAAUGUAAATT (sense) and 5′-UUUACAUUGGGCCCUAUUATT (antisense). A scrambled sequence of a part of human TFIIAαβ containing 5′-UGGCUGACGACUACUGCGCTT (sense) and 5′-GCGCAGUAGUCGUCAGCCATT (antisense) was used as a control siRNA.
Luciferase Assay
Cells were inoculated into a 24-well plate (3 × 104 cells/well). Twenty-four hours later, cells were transfected with 100 ng of a reporter plasmid and 200 ng of an effector plasmid and cultured for 24 h. If necessary, the total amount of transfected DNA was adjusted using pRL-TK (Promega). Cells were harvested and disrupted with Passive Lysis Buffer (Promega). Luciferase activity in lysates was determined using the Dual-Luciferase Reporter Assay System (Promega).
RT-PCR
Total cellular RNAs were prepared by using an RNeasy kit (Qiagen), and reverse transcription-PCR (RT-PCR) was performed as described previously (42). Briefly, cDNA synthesized from 500 ng of total RNA using Prime Script II (Takara) or avian myeloblastosis virus Reverse Transcriptase XL (Takara) was amplified by PCR using Paq5000 DNA polymerase (Stratagene) and appropriate primer sets. Amplified products were analyzed by 2% agarose gel electrophoresis.
Bacterially Expressed Recombinant Proteins
The pET series of expression plasmids and pGEX series of expression plasmids were transformed into Escherichia coli BL21 and DH5α, respectively. The recombinant proteins were induced by isopropyl 1-thio-β-d-galactoside. Cells for FH-tagged protein preparation were suspended in FH buffer (PBS(−), 0.5 m NaCl, 10% glycerol, 1% Triton X-100, 10 mm imidazole, and protease inhibitor (PI) mixture) (41). Cells for the GST-tagged proteins were suspended in buffer containing PBS(−), 0.5 m NaCl, 0.5 mm EDTA, 10% glycerol, 1% Triton X-100, 2 mm DTT, and PI mixture. Cells were disrupted by sonication and centrifuged at 9500 rpm for 20 min, and the supernatant fraction was collected as bacterial cell extracts.
GST Pulldown Assay
FH-tagged proteins in bacterial cell extracts were bound to nickel-nitrilotriacetic acid (Ni-NTA; Qiagen) and eluted with a buffer containing PBS(−), 0.5 m NaCl, 0.5 mm EDTA, 10% glycerol, 1% Triton X-100, 0.5 m imidazole, and PI mixture. GST-tagged proteins were bound to glutathione-Sepharose 4B beads (GE Healthcare). Purified FH-tagged proteins and protein-bound GST-tagged proteins were suspended in TNE buffer (50 mm Tris-HCl (pH 7.9), 150 mm EDTA, 1 mm EDTA, 10% glycerol, 0.1% Nonidet P-40, and PI mixture) and incubated at 4 °C for 3 h. The bound proteins were eluted with SDS sample buffer and detected by Western blotting as described before (43).
Immunoprecipitation of Intracellular Proteins
HeLa cells transfected with pcDNA-FLAG-p53 were suspended in TNE buffer, disrupted by sonication, and centrifuged at 13,000 rpm for 20 min. The supernatant fractions were collected as whole cell extracts. Protein concentration was determined using the BCA Protein Assay kit (Pierce). Three hundred micrograms of the extract was mixed with anti-FLAG M2 Affinity Gel (Sigma-Aldrich) for 3 h at 4 °C. IgG-Sepharose 6 Fast Flow (GE Healthcare) was used as a control antibody. Bound proteins were eluted with FLAG peptides, boiled for 5 min in SDS sample buffer, and analyzed by Western blotting.
Western Blotting
Proteins were separated by 12.5% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to Immobilon-P PVDF membrane (Millipore), and detected by ECL (GE Healthcare) as described previously (43) by using specific antibodies and appropriate horseradish peroxidase-conjugated secondary antibodies such as anti (α)-rabbit IgG and α-mouse IgG. As primary antibodies, we used α-p53 antibody (Santa Cruz Biotechnology), α-GST antibody (Ambion), α-FLAG M5 antibody, α-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (Ambion), and antigen-purified α-TLP antibody as described previously (29).
Gel Shift Assay
FH-p53 was purified by essentially the same method as that described by Wu and Chiang (44). Proteins in extracts of FH-p53-overexpressing bacteria were precipitated in 30% ammonium sulfate and suspended in FH buffer. Proteins were bound to an Ni-NTA column and eluted by BC100 buffer (20 mm Tris-HCl (pH 7.9), 20% glycerol, 1 mm DTT, 0.2 mm EDTA, 0.1 m KCl, and PI mixture) supplemented with 0.4 m imidazole. Eluates were dialyzed against BC100 buffer. Other FH-tagged proteins were directly bound to the Ni-NTA column, eluted with 0.5 m imidazole-containing BC100 buffer, and dialyzed against BC100 buffer.
A gel shift assay was performed essentially in accordance with a method described by Hou et al. (45). A DNA fragment spanning from −2266 to −2237 of the p21 gene that contains a p53-binidng sequence was used as a probe (see Fig. 5). The DNA fragment was labeled with [γ-32P]ATP and T4 polynucleotide kinase and purified with Sephadex G-25 (GE Healthcare). The protein fraction was incubated in binding buffer (10 mm HEPES-KOH (pH 7.9), 4 mm NaCl, 0.2 mm EDTA, 5 mm DTT, 10% glycerol, 0.1 mg/ml bovine serum albumin, and 10 ng of poly(dA-dT)) at room temperature for 10 min. Then probe DNA (40,000 cpm) was incubated with the mixture at 37 °C for 30 min to allow binding of probe DNA to proteins. The protein·DNA complexes were separated by 4% native PAGE and detected by autoradiography. If necessary, cold probe DNA was added to the binding reaction as a competitor. Control competitor DNA with point mutations was prepared according to a method described previously (44).
FIGURE 5.
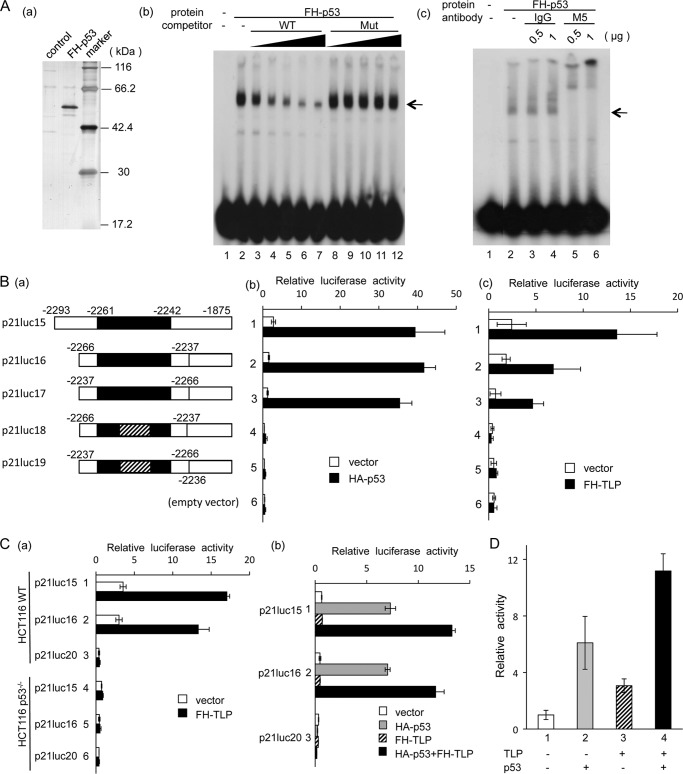
Role of p53 in TLP-activated transcription. A, gel shift assay of p53 for the p53-binding site located around −2250. Panel a, purified recombinant FH-p53 expressed in E. coli was separated by SDS-PAGE and stained with silver. control, protein sample prepared from normal E. coli by the same protocol. Panel b, purified FH-p53 was used for a gel shift assay (lane 2). Increasing amounts (4, 10, 25, 50, and 100 fmol) of cold probe DNA (WT; lanes 3–7) and its mutant DNA (Mut; lanes 8–12) were used as competitors. Sequences of the wild-type competitor (i.e. same as the probe) and mutant competitor were 5′-CTCAGGAACATGTCCCAACATGTTGAGCTC and 5′-GTCAGGAACCGTGAAACCACTGTTGAGCTC, respectively. The position of the specific shifted band is indicated by arrowheads. Panel c, M5 antibody against the FLAG tag and control IgG were included in the gel shift assay. B, luciferase assay for the p53-binding sequence around −2250 in the p21 upstream promoter. Panel a, structures of template DNAs and their nomenclature are schematically illustrated. Solid box, p53-binding site from −2261 to −2242. Reporter plasmids were derived from p21luc16. Box with oblique lines, region with point mutations in a core region of the p53-binding site. Luciferase activity for the reporter constructs was analyzed in HeLa cells co-transfected with HA-p53 expression plasmid (panel b) and FH-TLP-expression plasmid (panel c), and results are displayed as activities relative to the enzyme activity of lane 3 in vector-transfected cells. C, luciferase assay with HCT116 cells. Panel a, wild-type cells (lanes 1–3) and p53−/− cells (lanes 4–6) were transfected with p21luc15 (lanes 1 and 4), p21luc16 (lanes 2 and 5), and p21luc20 (lanes 3 and 6) and co-transfected with FH-TLP or vector DNA, and luciferase activity was measured. Panel b, p53−/− HCT116 cells transfected with the reporter plasmids indicated were co-transfected with vector DNA, HA-p53, FH-TLP, or HA-p53 + FH-TLP. D, synergy of TLP and p53 in the activation of the p21 upstream promoter. HeLa cells were transfected with 10 ng of HA-p53 or FH-TLP together with p21luc15. The total amount of effector plasmids was adjusted to 20 ng with vector DNA. Data are indicated as averages with error bars according to multiple experiments.
Chromatin Immunoprecipitation (ChIP)
A ChIP assay was performed as described previously with a few modifications (46). Chromatin of HCT116 cells was treated with formaldehyde, sheared by sonication, and incubated with a specific antibody. The sample was then immunoprecipitated with protein G-Sepharose (GE Healthcare), and the presence of a target DNA in the precipitates was analyzed by PCR.
RESULTS
TLP Negatively Affects G1-S Transition in Cell Cycle Progression of Human Cells
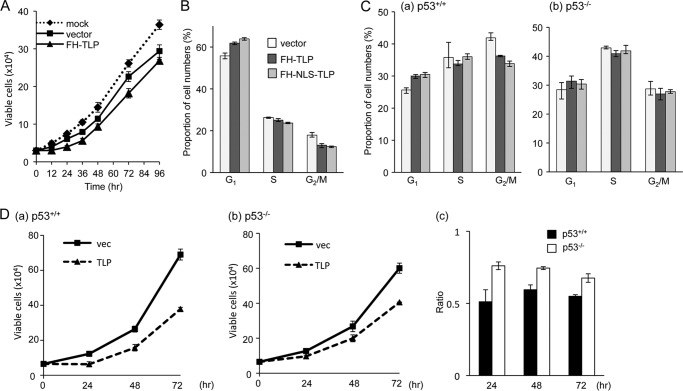
Although it is known that TLP inhibits cell cycle progression of chicken DT40 cells (32), it is not known how TLP behaves in mammalian cells. In this study, we addressed this issue. At first, we examined the effect of TLP on growth of HeLa cells. It is known that TLP is predominantly localized in the cytoplasm but is concentrated in the nucleus when cells are treated with genotoxins (32, 47). To amplify the effect of TLP, TLP expression plasmid-transfected cells were irradiated with UV light. Cells carrying TLP expression plasmids showed a slightly but significantly lower growth rate than that of vacant vector-transfected cells (Fig. 1A). The growth profile of these cells was analyzed by flow cytometry. TLP-overexpressing cells were found to contain a smaller proportion of G2/M phase cells, whereas the proportion of G1 cells was increased by about 10% (Fig. 1B). Much clearer results were obtained when cells were transfected with plasmid containing a nuclear localization signal-carrying TLP (FH-NLS-TLP).
FIGURE 1.
Growth inhibition of human cells by TLP. A, growth profile of TLP-overexpressing cells. HeLa cells (3 × 104) transfected with FH-TLP or vector DNA (200 ng) were irradiated with UV light (100 J/m2). The cells were replated in a 6-well plate, and their numbers were counted at the indicated times. Dotted line, mock-transfected cells. B, flow cytometry of TLP-overexpressing cells. HeLa cells transfected with DNA (vector, FH-TLP, or nuclear localization signal-containing FH-NLS-TLP) and irradiated with UV light (25 J/m2) were analyzed for cell cycle phases by flow cytometry. C, flow cytometry of TLP-overexpressing wild-type (p53+/+) (panel a) and mutant (p53−/−) (panel b) HCT116 cells. In this case, UV irradiation was omitted. D, growth profile of TLP-overexpressing HCT116 cells. Wild-type (panel a) and p53−/− cells (panel b) (8 × 104) transfected with FH-TLP or vector (vec) DNA (200 ng) were replated in a 6-well plate, and cells numbers were counted at the indicated times. Panel c, ratios of vector-transfected cells to TLP-transfected cells are displayed. Data are indicated as averages with error bars according to multiple experiments.
We performed similar experiments by using UV light-untreated HCT116 cells and obtained essentially the same results (Fig. 1C, panel a). However, when we used mutant (p53−/−) HCT116 cells, there was little G1 phase cell accumulation induced by TLP overexpression (Fig. 1C, panel b). We then analyzed the cell growth profile again using these HCT116 cells. Proliferation of p53+/+ HCT116 cells was considerably inhibited by TLP overexpression like HeLa cells, whereas there was little growth inhibition of p53−/− cells by TLP (Fig. 1D). Consequently, the results indicated that TLP is involved in inhibition of the G1-S transition of mammalian cells. These results also suggested that p53 is involved in this phenomenon.
TLP-activated Growth-inhibitory Genes of HeLa Cells
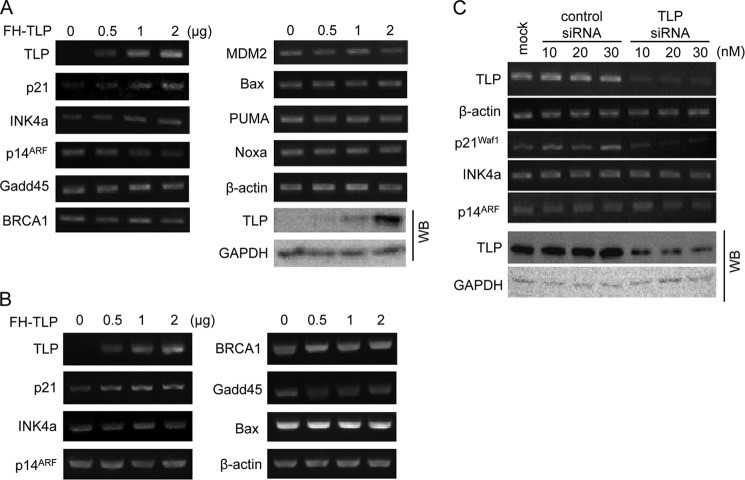
According to the above results, we searched for TLP-regulated genes. HeLa cells transfected with the TLP expression plasmid were analyzed by RT-PCR for mRNAs of several growth-related genes including G1-arresting genes such as p21 (p21Waf1/Cip1), INK4a (p16INK4a), and ARF (p14ARF). We further examined genes related to DNA repair (i.e. Gadd45 and BRCA1), p53 destabilization (i.e. MDM2), and apoptosis (i.e. Bax, PUMA, and Noxa). Results presented in Fig. 2A demonstrate that TLP overexpression stimulated p21 and INK4a among the genes examined. Other genes did not show any significant changes. A similar experiment was performed using HepG2 cells. Again, p21 was stimulated by TLP overexpression (Fig. 2B), whereas INK4a gene expression was not stimulated. We performed a gene knockdown experiment for TLP in HeLa cells. It was clearly shown that p21 gene expression was considerably reduced by TLP siRNA, whereas INK4a and ARF were not affected (Fig. 2C). The p21 gene has been shown to be repressed by TLP siRNA in Hep3B cells (18). Taken together, the results indicated that endogenous p21 gene expression is stimulated by TLP.
FIGURE 2.
Effect of TLP on endogenous gene expression of human cells. A, HeLa cells transfected with increasing doses of FH-TLP were harvested, and the amount of mRNA was determined by semiquantitative RT-PCR for the indicated genes. β-Actin and GAPDH were used as control genes for RT-PCR and Western blotting (WB), respectively. B, HepG2 cells transfected with FH-TLP were analyzed for mRNA as described above. C, mRNAs in HeLa cells transfected with increasing doses of TLP siRNA or control siRNA were analyzed for the genes indicated. The effect of siRNA was checked by Western blotting. mock, mock-infected cells.
p21 Promoter Is Potentiated by TLP
To investigate how TLP affects transcription, we performed promoter-reporter assays using luciferase reporter plasmids. Overexpressed TLP had no or little activating effect on BAX and INK4a promoters (Fig. 3A). We also constructed a p21 promoter-reporter plasmid (p21luc1). The insert of this plasmid contains two major functional promoters, referred to as downstream promoter and upstream promoter, that use the +1 TSS and −2258 TSS, respectively. The upstream and downstream promoters produce p21 alt-a and p21 variant-1 transcripts, respectively (Figs. 3B and 7A) (35). The upstream and downstream promoters are TATA-less and TATA-containing promoters, respectively. The p21 promoter(s) in p21luc1 was not activated by TLP but rather inhibited as observed previously for several TATA-containing promoters (28, 34). Because p21luc1 harbors two tandem-orienting promoters, the effect of TLP on the downstream promoter alone was not elucidated from this construct. On the other hand, p21luc2, which has the upstream promoter alone and weaker promoter function than that of p21luc1, was considerably activated by TLP. The same results were obtained when we used COS7 cells (Fig. 3C). The p21 upstream promoter was also stimulated greatly by TLP when it had been inserted into pGL3-Basic vector (data not shown). We found that the p21 upstream promoter is activated by TLP.
FIGURE 7.
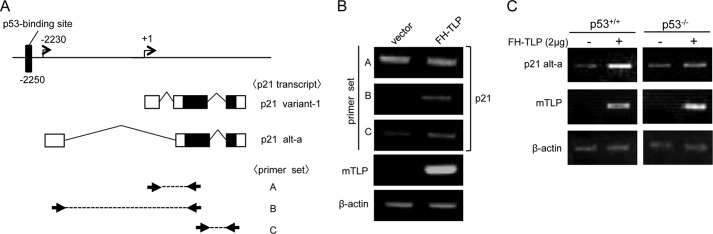
Role of endogenous p53 in TLP-activated transcription from chromosomal p21 upstream promoter. A, two kinds of p21 transcripts produced from the human p21 gene. Positions of exons of p21 variant-1 and p21 alt-a transcripts (48) and genomic DNA around the two p21 promoters are schematically illustrated. Open and solid boxes represent non-coding and coding exons, respectively. Primer sets A, B, and C indicated by thick arrows were used for RT-PCR to detect p21 variant-1, p21 alt-a, and both transcripts, respectively. B, cells were transfected with FH-TLP, and the amounts of p21 transcripts were determined by RT-PCR with each primer set. C, requirement of endogenous p53 for TLP-governed activation of p21 upstream promoter. Wild-type and p53−/− HCT116 cells (5 × 105) were transfected with FH-TLP (2 μg), and the amount of p21 alt-a transcripts was determined by using primer set B 24 h after transfection. mTLP, mouse TLP.
Effect of p53 Target Site and p53 Protein on TLP-activated Transcription from p21 Upstream Promoter
To delineate a TLP-responsive region in the p21 upstream promoter, we constructed truncated versions derived from p21luc2 and examined their TLP responsiveness. As shown in Figs. 3B and 4, the DNA sequence around the p21 upstream promoter in which the transcription start site is located at −2230 has two p53 target sites around −2250 and −1370. Among truncation versions, p21luc4, -5, -7, and -9 exhibited TLP-activated transcription (Fig. 4A). All TLP-activated promoter constructs were found to contain two p53 target sites. We prepared p21luc11 and further truncations (i.e. p21luc12, -13, and -14). Luciferase reporter assays revealed that promoter constructs containing both p53 target sites at −2250 and a transcription start site at −2230 were activated by TLP (Fig. 4B). Because the degree of activation of p21luc11 by TLP was comparable with that of p21luc2, the region required for TLP-activated transcription of p21luc2 DNA is most likely localized around the upstream p53 target site. To determine whether the upstream p53 target site participates in TLP-activated transcription, we conducted the following experiments.
First, we performed a gel shift assay to confirm that p53 actually binds to the upstream p53 target site. As seen in Fig. 5A, panel b, highly purified recombinant p53 forms a specific shifted band that is decreased by addition of cold probe DNA but not by corresponding control (mutant) DNA. This shifted band was not observed when a normal E. coli-derived protein preparation was used (data not shown). Moreover, the shifted band was hypershifted by specific antibody (Fig. 5A, panel c). Hence, the sequence around −2250 was confirmed to be a p53-binding site.
Second, we performed luciferase assays using reporter constructs that contain mutations within the p53-binding sequence. p21luc16 has a DNA cassette of 30 bp in which 20 bp of the p53-binding sequence (i.e. a core binding region and its flanking sequences) followed by the upstream promoter is included. The promoter DNA itself has no transcription-promoting ability (Fig. 5B, panels b and c, lane 6). However, the promoter responded strongly to both p53 and TLP when the promoter was followed by the p53 cassette in any direction (Fig. 5B, panels b and c, lanes 2 and 3). When the p53 cassette had a mutation within the p53-binding site, the reporter constructs did not respond to either p53 or TLP (Fig. 5B, panels b and c, lanes 4 and 5). Finally, we found that a TLP-responsive element cannot be separated from the p53-binding site.
Third, we performed luciferase reporter assays in HCT116 cells using the same reporter constructs as those mentioned above. Again, p21luc15 and p21luc16 responded to overexpressed TLP in wild-type cells but not in p53−/− cells (Fig. 5C, panel a). The TLP-activated transcription from p21luc15 and p21luc16 was rescued by co-transfection of the p53 expression plasmid (Fig. 5C, panel b, lanes 1 and 2). Taken together, the TLP-responsive cis element of the p21 upstream promoter was found to be the p53-binding sequence itself, and p53 protein is required for TLP-dependent transcriptional activation.
Mechanism of p53-dependent and TLP-activated Transcription
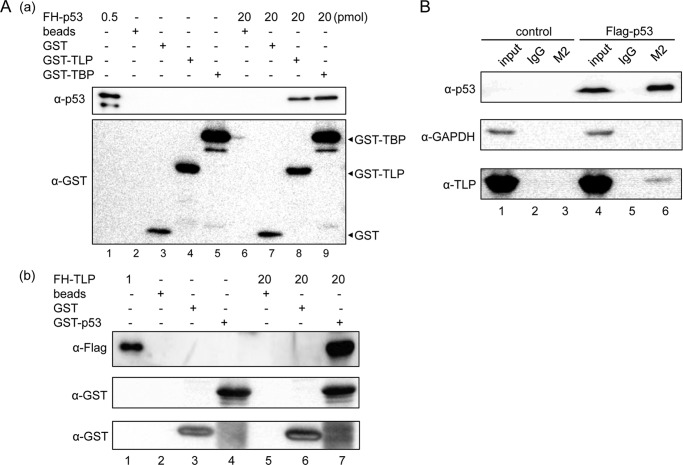
We investigated whether TLP potentiates p53-dependent transcriptional activation. p21luc15 showed 6- and 3-fold enhancement in HeLa cells by overexpression of p53 and TLP, respectively (Fig. 5D, lanes 2 and 3). However, co-transfection of both expression plasmids showed a much higher degree of activation (11-fold) (Fig. 5D, lane 4). This synergistic activation suggests physical interaction of these two proteins. We performed GST pulldown assays using bacterially expressed proteins. As seen in Fig. 6A, panel a, lane 9, GST-TBP was used as a positive control that binds to FH-p53 because p53 is one of the TBP-binding proteins (48). GST-TLP also binds to FH-p53, and GST-p53 could bind to FH-TLP (Fig. 6A), indicating that TLP binds to p53 in solution as efficiently as does TBP. Moreover, results of an immunoprecipitation assay demonstrated that FLAG-p53 and endogenous TLP in extracts of p53-overexpressing HeLa cells were co-immunoprecipitated (Fig. 6B, lane 6). These results indicate that p53 and TLP can interact in cells.
FIGURE 6.
Physical interaction of TLP and p53. A, GST pulldown assay for p53 and TLP. Panel a, FH-TLP, glutathione beads (beads), bead-bound GST, and bead-bound GST-tagged proteins (GTS-TLP or GST-TBP) were mixed as indicated, and bead-bound proteins were detected by Western blotting. Positions of GST tag-containing proteins are indicated by arrowheads. Panel b, GST pulldown assay was also performed as described above. In this experiment, FH-TLP and GST-p53-bound beads were used. B, immunoprecipitation for detection of intracellular p53-bound TLP. Extracts of HeLa cells overexpressing FLAG-p53 were immunoprecipitated with M2 beads, and endogenous TLP in precipitates was detected with the specific antibody. control, IgG.
Response of Upstream Promoter to TLP
A luciferase assay revealed that the upstream promoter for p21 alt-a transcript was activated by TLP in the presence of p53 as described above. We investigated which transcripts of p21 gene are affected by TLP. RT-PCR revealed that the upstream promoter-driven p21 alt-a was increased by FH-TLP expression (Fig. 7B), whereas FH-TLP had little effect on the downstream promoter-driven p21 variant. We further investigated the effects of endogenous p53 on the TLP-activated p21 upstream promoter. As seen in Fig. 7C, p21 alt-a transcripts were considerably increased by TLP in HCT116 p53+/+ cells, whereas such enhancement was limited in p53−/− cells. Consequently, we confirmed that the p21 upstream promoter in the chromosome is also activated by TLP in a p53-dependent manner.
Function of Upstream Promoter in Etoposide-treated Cells
p21 transcripts in MCF-7 cells are known to be increased by etoposide treatment (49). We examined p21 alt-1 transcripts from the chromosomal p21 gene in etoposide-treated HCT116 cells by RT-PCR. The amount of p21 alt-a transcript was greatly increased in response to etoposide treatment (Fig. 8A, panel a). We examined intranuclear p53 and TLP of treated cells and found that both factors are concentrated in nuclei by etoposide as expected from previous work (32, 47, 50) (Fig. 8A, panel b). Because etoposide treatment can be regarded as a condition equivalent to that in which endogenous p53 and TLP are concentrated in nuclei for transcription activation, we further investigated the association of those two factors with the p21 upstream promoter by a ChIP assay using this etoposide-mediated gene activation condition. In wild-type HCT116 cells, in vivo binding of p53 and TLP to the p53-responsive element in the upstream promoter was increased in response to etoposide treatment (Fig. 8B). Then we performed a ChIP assay of p53 in the presence and absence of overexpressed TLP. Intriguingly, the amount of the p53-associated p21 upstream promoter was significantly increased by the exogenous TLP (Fig. 8C), implying that TLP positively works for recruitment of p53 to the upstream promoter. Exogenous FH-TLP also was confirmed to be associated with the same promoter region (Fig. 8C). These results are essentially consistent with those obtained by the luciferase reporter assay described above.
FIGURE 8.
Function of p21 upstream promoter in etoposide-treated HCT116 cells. A, effect of etoposide on the activity of the p21 upstream promoter and nuclear accumulation of TLP and p53. Cells were treated with 25 and 50 μm etoposide or vehicle (−) for 24 h. Panel a, RT-PCR for detection of p21 alt-a transcripts with a specific primer set. Panel b, Western blotting for detection of nuclear TLP and p21 protein. Nuclear extracts were prepared as described previously (46), and the amounts of three kinds of endogenous proteins were detected by Western blotting. TBP was analyzed as a constitutive nuclear protein. B, association of endogenous p53 (panel a) and TLP (panel b) with p21 upstream promoter. Wild-type HCT116 cells were treated with etoposide (E) or vehicle (V) and then analyzed by ChIP assay using specific antibodies for the p53-responsive element (RE) in the upstream promoter region (−2431 to −2196) and the far-upstream region (−7018 to −6833) as a negative control because this region had been shown not to bind to p53 (46). IgG and Ab, IgG- and specific antibody-precipitated materials, respectively; Inp, input. C, effect of overexpressed TLP on the association of p53 and TLP with the p21 upstream promoter. Etoposide-treated cells were transfected with FH-TLP (TLP) or vacant vector (vec) DNA, and association of p53 with the responsive element was analyzed by ChIP assay with a specific primer set as described above. IP, immunoprecipitation.
DISCUSSION
TLP Functions Generally as Growth Repressive Factor
In a previous study, we demonstrated that TLP works to repress the growth of chicken cells (32). Here, we further found that growth of mammalian cells is also repressed by TLP and that TLP functions to inhibit G1-S transition. Because this phenomenon was observed in multiple cell lines, growth inhibition is thought to be a common function of vertebrate TLP. Previous findings that TLP is a development factor (30, 31) are consistent with this view because cell differentiation is basically associated with growth arrest. For ordinary cells, the TLP-governed cell cycle stall may be required for checkpoint and/or apoptosis progression when cells are suffering from genotoxic stresses.
We identified p21 as one of the TLP-responsive genes. Cyclin G2, which works as a G1 phase cyclin, was also activated by TLP (data not shown). We have reported that TLP stimulates the TAp63 gene, which induces apoptotic cell death (18). Mammalian TLP also regulates tumor suppressor genes (i.e. TAp63 and FN1) (18, 38), and Drosophila TLP (TRF2) activates the proliferating cell nuclear antigen gene, which functions in DNA repair as well as replication (37). TLP can be regarded as a global growth-regulatory gene. Because the degrees of growth inhibition and G1 arrest by TLP were small or not clear in p53−/− cells compared with those in wild-type cells (Fig. 1, C and D), p53 is thought to be responsible for these phenomena. This situation is consistent with the fact that the growth-repressive p21 gene is activated by p53 and TLP as revealed in this study. Compared with wild-type HCT116 cells, the degree of growth inhibition of HeLa cells by TLP was significant but not great (i.e. 80–85% of vector-transfected cells in Fig. 1A). This difference may be due to limited amounts of functional intracellular p53 because p53 in HeLa cells is significantly repressed by papillomavirus proteins (45).
Through a search for TLP-stimulated endogenous genes, we identified p21 and INK4a. Because these genes are major CKIs for G1 arrest, a part of TLP-mediated growth inhibition seems to be responsible for activation of these genes. Although 14-3-3σ was also enhanced by TLP, the enhancement was weak (data not shown). This may be a reason why the proportion of G2-arrested cells was not increased by TLP overexpression (Fig. 1). Chicken DT40 cells are arrested at G2/M phase by TLP (32). We have found that some TLPs are concentrated in nuclei by genotoxin treatment (32, 47). In chicken DT40 cells, TLP accumulation in the nucleus induces G2 arrest but not G1 arrest (32). Differences in the proportions of TLP-sensitive regulatory factors in cells may be due to distinct responses to TLP.
p53-dependent and TLP-activated Transcription of p21 Upstream Promoter
The present study clearly demonstrated that the p21 gene is activated by TLP at the transcriptional level. Our study clarified that at least the upstream promoter is potentiated by TLP. No TLP-dependent activation was observed for the INK4a promoter even though TLP stimulates the chromosomal INK4a gene. The INK4a reporter plasmid may not contain a TLP-responsive region.
Purified p53 bound to the p53-binding sequence in the p21 upstream promoter (Fig. 5A), and mutation of the p53-binding sequence abolished TLP-directed transcriptional activation (Fig. 5B). TLP-activated transcription of the p21 promoter was not observed in p53−/− HCT116 cells but was rescued by ectopic p53 expression (Fig. 5C). Compared with wild-type HCT116 cells, TLP-mediated G1 arrest was not evident in p53−/− cells (Fig. 1, C and D). Consistent with the results of promoter-luciferase reporter assays shown in Figs. 4 and 5, TLP-governed stimulation of p21 alt-a transcripts was more evident in p53+/+ cells than in p53−/− cells in the case of the chromosomal p21 gene (Fig. 7C). Moreover, stimulation of the chromosomal p21 upstream promoter by etoposide occurred concordantly with nuclear accumulation and subsequent upstream promoter binding of endogenous p53 as well as TLP (Fig. 8). Thus, we concluded that p53 is an essential factor for TLP-activated transcription of the p21 upstream promoter. Among the p53 family genes, TAp63, whose gene product also binds to a p53 target site, is induced by TLP (18). Hence, TLP-induced TAp63 might activate the p21 gene in a TLP-overexpressing condition. However, this pathway is unlikely because the p21luc2 reporter was considerably activated in COS7 cells (Fig. 3C) in which the TAp63 gene is almost silent.4
It is well known that p53 activates genes for GADD45, MDM2, and BAX (40, 51). However, these genes were not stimulated by TLP in our experimental conditions, indicating that p53-mediated transcriptional activation is not always stimulated by TLP. Some unknown cis elements other than the p53-binding sequence may be required for TLP-dependent transcriptional activation. Alternatively, the core promoter may be involved in this specificity because TLP is known to bind to several general transcription factors including TFIIA (28, 29), TFIIB (28), and TBP (data not shown).
Reason Why p21 Promoter(s) in p21luc1 Was Not Enhanced by TLP Overexpression
Different from p21luc2, transcription from the p21 promoter in p21luc1 apparently was not activated by TLP (Fig. 3A). A critical factor of this phenomenon is probably the presence of a TATA box in the promoter. It is known that a TATA-containing promoter is repressed by overexpressed TLP in a promoter-reporter assay condition (28, 34, 35). Because TATA promoters in chromatin are not necessarily inhibited by TLP, a non-physiological chromatin structure may be related to this phenomenon. Because TLP strongly binds to TFIIA, excess amounts of TLP trap and inactivate TFIIA (28, 34), which may result in repression of TFIIA-dependent TFIID-driven TATA promoters in a reporter assay condition. The upstream and downstream p21 promoters are TATA-less and TATA promoters, respectively, and the same situation presumably occurred in the p21 downstream promoter. Moreover, transcription machineries at the downstream promoter probably interfere with elongation from upstream promoter-driven transcription in the p21luc1 plasmid. Ultimately, it is thought that the net amount of transcription in p21luc1 plasmid is depressed by TLP overexpression.
Mechanism of p53-dependent and TLP-activated Transcription
The present study suggests that p53 binding to the p21 upstream promoter is an essential condition for TLP-governed transcriptional activation because p53 binds to its functional target sequence (Fig. 5A) and associates with the upstream promoter region of the endogenous p21 gene (Fig. 8B). TLP increased in vivo association of p53 with its binding region (Fig. 8C), which suggests that TLP functions as a recruitment factor of p53. The binding capacity of p53 and TLP shown in Fig. 6 is consistent with this idea. However, we think that the binding affinity between p53 and TLP is not high in cells because the TLP signal in Fig. 6B, lane 6 is weak. Although TLP was also recruited to the p53-binding region together with p53 (Fig. 8B), it is speculated that TLP on the promoter does not bind tightly to the DNA but associates with p53 and DNA with some affinity. Actually, the gel shift assay could not show the stable binding of TLP to the p53-binding region (data not shown). Although TLP has been reported to bind to a DNA sequence directly in the NF1 promoter (38), the reported sequence was not found around the p53-binding sequence in the p21 upstream promoter. If stable DNA binding of TLP is indispensable for its function, competition between TLP and p53 for the p53-binding sequence must occur. However, we could not obtain such a result, and we could not separate a TLP-responsive element from a p53-binding sequence at −2250. Consequently, TLP may be able to work as a co-activator for p53. Drosophila TLP functions as a co-activator for the DNA replication-related element-binding factor (37). Evidence that TLP binds to several general transcription factors (see above) also supports the co-activator hypothesis.
General Aspects of p21 Gene Expression
We demonstrated that, among two promoters of the p21 gene, the upstream promoter is almost silent in a normal condition but is markedly activated by TLP, whereas the downstream promoter prominently works in a normal condition but is only slightly activated by TLP (Fig. 7B). Additionally, the upstream promoter is considerably stimulated by etoposide in accordance with nuclear accumulation and promoter association of p53 and TLP (Fig. 8), whereas there is little etoposide-mediated stimulation of the downstream promoter (data not shown). Hence, the downstream promoter is thought to be used for constitutive expression of the p21 gene. Multiple cis elements for constitutive transcription factors such as Sp1 and E-box elements in addition to a TATA box existing in the downstream promoter (7, 8) may be used mainly for downstream promoter-driven transcription. In contrast to the downstream promoter, the upstream promoter, whose function basically depends on p53 and TLP, is used in a condition in which cells are exposed to genotoxins. Because TLP is accumulated in nuclei by various kinds of genotoxins (32, 47), the p53/TLP-dependent upstream promoter is probably involved in the stress response. Moreover, we have demonstrated that etoposide induces the TLP gene itself (18), and the amount of etoposide-enhanced p21 alt-a transcripts is decreased by TLP siRNA (data not shown).
Acknowledgments
We thank Dr. Y. Suenaga, Dr. A. Nakagawara, and M. Ono for valuable discussions and technical assistance. We also thank Dr. B. Vogelstein for providing HCT116 cell lines.
H. Suzuki, R. Ito, K. Ikeda, and T. Tamura, unpublished observation.
- CKI
- cyclin-dependent kinase inhibitor
- TBP
- TATA-binding protein
- TLP
- TBP-like protein
- TRF
- TBP-related factor
- TSS
- transcription start site
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM- and Rad3-related
- TAp63
- transactivating p63
- TFII
- transcription factor II
- PI
- protease inhibitor
- FH
- FLAG and oligohistidine
- Ni-NTA
- nickel-nitrilotriacetic acid
- α
- anti
- NLS
- nuclear localization signal.
REFERENCES
- 1. Norbury C., Nurse P. (1992) Animal cell cycles and their control. Annu. Rev. Biochem. 61, 441–470 [DOI] [PubMed] [Google Scholar]
- 2. Ekholm S. V., Reed S. I. (2000) Regulation of G1 cyclin-dependent kinases in the mammalian cell cycle. Curr. Opin. Cell Biol. 12, 676–684 [DOI] [PubMed] [Google Scholar]
- 3. Bartek J., Lukas J. (2001) Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr. Opin. Cell Biol. 13, 738–747 [DOI] [PubMed] [Google Scholar]
- 4. Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]
- 5. Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. (1993) p21 is a universal inhibitor of cyclin kinases. Nature 366, 701–704 [DOI] [PubMed] [Google Scholar]
- 6. Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. (2009) Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gartel A. L., Tyner A. L. (1999) Transcriptional regulation of the p21WAF1/CIP1 gene. Exp. Cell Res. 246, 280–289 [DOI] [PubMed] [Google Scholar]
- 8. Jung Y. S., Qian Y., Chen X. (2010) Examination of the expanding pathways for the regulation of p21 expression and activity. Cell. Signal. 22, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McGowan C. H., Russell P. (2004) The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 16, 629–633 [DOI] [PubMed] [Google Scholar]
- 10. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 11. Vousden K. H. (2002) Activation of the p53 tumor suppressor protein. Biochim. Biophys. Acta 14, 47–59 [DOI] [PubMed] [Google Scholar]
- 12. Vousden K. H., Lu X. (2002) Live or let die: the cell's response to p53. Nat. Rev. Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 13. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 14. Stiewe T. (2007) The p53 family in differentiation and tumorigenesis. Nat. Rev. Cancer 7, 165–168 [DOI] [PubMed] [Google Scholar]
- 15. Flores E. R., Tsai K. Y., Crowley D., Sengupta S., Yang A., McKeon F., Jacks T. (2002) p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416, 560–564 [DOI] [PubMed] [Google Scholar]
- 16. Flores E. R., Sengupta S., Miller J. B., Newman J. J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T. (2005) Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7, 363–373 [DOI] [PubMed] [Google Scholar]
- 17. Müller M., Schleithoff E. S., Stremmel W., Melino G., Krammer P. H., Schilling T. (2006) One, two, three—p53, p63, p73 and chemosensitivity. Drug Resist. Updat. 9, 288–306 [DOI] [PubMed] [Google Scholar]
- 18. Suenaga Y., Ozaki T., Tanaka Y., Bu Y., Kamijo T., Tokuhisa T., Nakagawara A., Tamura T. A. (2009) TATA-binding protein (TBP)-like protein is engaged in etoposide-induced apoptosis through transcriptional activation of human TAp63 gene. J. Biol. Chem. 284, 35433–35440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Orphanides G., Lagrange T., Reinberg D. (1996) The general transcription factors of RNA polymerase II. Genes Dev. 10, 2657–2683 [DOI] [PubMed] [Google Scholar]
- 20. Davidson I. (2003) The genetics of TBP and TBP-related factors. Trends Biochem. Sci. 28, 391–398 [DOI] [PubMed] [Google Scholar]
- 21. Thomas M. C., Chiang C. M. (2006) The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 [DOI] [PubMed] [Google Scholar]
- 22. Ohbayashi T., Makino Y., Tamura T. A. (1999) Identification of a mouse TBP-like protein (TLP) distantly related to the Drosophila TBP-related factor. Nucleic Acids Res. 27, 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berk A. J. (2000) TBP-like factors come into focus. Cell 103, 5–8 [DOI] [PubMed] [Google Scholar]
- 24. Persengiev S. P., Zhu X., Dixit B. L., Maston G. A., Kittler E. L., Green M. R. (2003) TRF3, a TATA-box-binding protein-related factor, is vertebrate-specific and widely expressed. Proc. Natl. Acad. Sci. U.S.A. 100, 14887–14891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reina J. H., Hernandez N. (2007) On a roll for new TRF targets. Genes Dev. 21, 2855–2860 [DOI] [PubMed] [Google Scholar]
- 26. Deato M. D., Marr M. T., Sottero T., Inouye C., Hu P., Tjian R. (2008) MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol. Cell 32, 96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. D'Alessio J. A., Wright K. J., Tjian R. (2009) Shifting players and paradigms in cell-specific transcription. Mol. Cell 36, 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teichmann M., Wang Z., Martinez E., Tjernberg A., Zhang D., Vollmer F., Chait B. T., Roeder R. G. (1999) Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 96, 13720–13725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakadai T., Shimada M., Shima D., Handa H., Tamura T. (2004) Specific interaction with transcription factor IIA and localization of the mammalian TATA-binding protein-like protein (TLP/TRF2/TLF). J. Biol. Chem. 279, 7447–7455 [DOI] [PubMed] [Google Scholar]
- 30. Kaltenbach L., Horner M. A., Rothman J. H., Mango S. E. (2000) The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol. Cell 6, 705–713 [DOI] [PubMed] [Google Scholar]
- 31. Zhang D., Penttila T. L., Morris P. L., Teichmann M., Roeder R. G. (2001) Spermiogenesis deficiency in mice lacking the Trf2 gene. Science 292, 1153–1155 [DOI] [PubMed] [Google Scholar]
- 32. Shimada M., Nakadai T., Tamura T. (2003) TATA-binding protein-like protein (TLP/TRF2/TLF) negatively regulates cell cycle progression and is required for the stress-mediated G2 checkpoint. Mol. Cell. Biol. 23, 4107–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohbayashi T., Shimada M., Nakadai T., Tamura T. (2001) TBP-like protein (TLP/TLF/TRF2) artificially recruited to a promoter stimulates basal transcription in vivo. Biochem. Biophys. Res. Commun. 285, 616–622 [DOI] [PubMed] [Google Scholar]
- 34. Ohbayashi T., Shimada M., Nakadai T., Wada T., Handa H., Tamura T. (2003) Vertebrate TBP-like protein (TLP/TRF2/TLF) stimulates TATA-less terminal deoxynucleotidyl transferase promoters in a transient reporter assay, and TFIIA-binding capacity of TLP is required for this function. Nucleic Acids Res. 31, 2127–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore P. A., Ozer J., Salunek M., Jan G., Zerby D., Campbell S., Lieberman P. M. (1999) A human TATA binding protein-related protein with altered DNA binding specificity inhibits transcription from multiple promoters and activators. Mol. Cell. Biol. 19, 7610–7620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tanaka Y., Nanba Y. A., Park K. A., Mabuchi T., Suenaga Y., Shiraishi S., Shimada M., Nakadai T., Tamura T. (2007) Transcriptional repression of the mouse wee1 gene by TBP-related factor 2. Biochem. Biophys. Res. Commun. 352, 21–28 [DOI] [PubMed] [Google Scholar]
- 37. Hochheimer A., Zhou S., Zheng S., Holmes M. C., Tjian R. (2002) TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420, 439–445 [DOI] [PubMed] [Google Scholar]
- 38. Chong J. A., Moran M. M., Teichmann M., Kaczmarek J. S., Roeder R., Clapham D. E. (2005) TATA-binding protein (TBP)-like factor (TLF) is a functional regulator of transcription: reciprocal regulation of the neurofibromatosis type 1 and c-fos genes by TLF/TRF2 and TBP. Mol. Cell. Biol. 25, 2632–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan T. A., Hwang P. M., Hermeking H., Kinzler K. W., Vogelstein B. (2000) Cooperative effects of genes controlling the G2/M checkpoint. Genes Dev. 14, 1584–1588 [PMC free article] [PubMed] [Google Scholar]
- 40. Ozaki T., Okoshi R., Sang M., Kubo N., Nakagawara A. (2009) Acetylation status of E2F-1 has an important role in the regulation of E2F-1-mediated transactivation of tumor suppressor p73. Biochem. Biophys. Res. Commun. 386, 207–211 [DOI] [PubMed] [Google Scholar]
- 41. Shiraishi S., Zhou C., Aoki T., Sato N., Chiba T., Tanaka K., Yoshida S., Nabeshima Y., Nabeshima Y., Tamura T. (2007) TBP-interacting protein 120B (TIP120B)/cullin-associated and neddylation-dissociated 2 (CAND2) inhibits SCF-dependent ubiquitination of myogenin and accelerates myogenic differentiation. J. Biol. Chem. 282, 9017–9028 [DOI] [PubMed] [Google Scholar]
- 42. Jogo M., Shiraishi S., Tamura T. (2009) Identification of MAFbx as a myogenin-engaged F-box protein in SCF ubiquitin ligase. FEBS Lett. 583, 2715–2719 [DOI] [PubMed] [Google Scholar]
- 43. Shiraishi S., Tamamura N., Jogo M., Tanaka Y., Tamura T. (2009) Rapid proteasomal degradation of transcription factor IIB in accordance with F9 cell differentiation. Gene 436, 115–120 [DOI] [PubMed] [Google Scholar]
- 44. Wu S. Y., Chiang C. M. (2009) Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J. 28, 1246–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hou S. Y., Wu S. Y., Chiang C. M. (2002) Transcriptional activity among high and low risk human papillomavirus E2 proteins correlates with E2 DNA binding. J. Biol. Chem. 277, 45619–45629 [DOI] [PubMed] [Google Scholar]
- 46. Saramäki A., Banwell C. M., Campbell M. J., Carlberg C. (2006) Regulation of the human p21waf1/cip1 gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res. 34, 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Park K. A., Tanaka Y., Suenaga Y., Tamura T. A. (2006) TATA-binding protein-related factor 2 is localized in the cytoplasm of mammalian cells and much of it migrates to the nucleus in response to genotoxic agents. Mol. Cells 22, 203–209 [PubMed] [Google Scholar]
- 48. Truant R., Xiao H., Ingles C. J., Greenblatt J. (1993) Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J. Biol. Chem. 268, 2284–2287 [PubMed] [Google Scholar]
- 49. Radhakrishnan S. K., Gierut J., Gartel A. L. (2006) Multiple alternate p21 transcripts are regulated by p53 in human cells. Oncogene 25, 1812–1815 [DOI] [PubMed] [Google Scholar]
- 50. Inoue T., Wu L., Stuart J., Maki C. G. (2005) Control of p53 nuclear accumulation in stressed cells. FEBS Lett. 579, 4978–4984 [DOI] [PubMed] [Google Scholar]
- 51. Sherr C. J., McCormick F. (2002) The RB and p53 pathways in cancer. Cancer Cell 2, 103–112 [DOI] [PubMed] [Google Scholar]