Background: Nurr1 and FGFR1 are integrative nuclear factors participating in postmitotic dopaminergic neuron development.
Results: Both nuclear receptors show a functional interaction in co-immunoprecipitation, FRAP, ChIP, and luciferase gene reporter assay.
Conclusion: Cooperation of nuclear FGFR1 and Nurr1 offers a new mechanism in transcriptional regulation and integration.
Significance: This mechanism may channel diverse stimuli in developing and mature dopaminergic neurons, providing a potential therapeutic target.
Keywords: Fibroblast Growth Factor (FGF), Fibroblast Growth Factor Receptor (FGFR), Growth Factors, Nuclear Receptors, Nucleus, Nurr1, Orphan Receptor, Tyrosine Hydroxylase
Abstract
Experiments in mice deficient for Nurr1 or expressing the dominant-negative FGF receptor (FGFR) identified orphan nuclear receptor Nurr1 and FGFR1 as essential factors in development of mesencephalic dopaminergic (mDA) neurons. FGFR1 affects brain cell development by two distinct mechanisms. Activation of cell surface FGFR1 by secreted FGFs stimulates proliferation of neural progenitor cells, whereas direct integrative nuclear FGFR1 signaling (INFS) is associated with an exit from the cell cycle and neuronal differentiation. Both Nurr1 and INFS activate expression of neuronal genes, such as tyrosine hydroxylase (TH), which is the rate-limiting enzyme in dopamine synthesis. Here, we show that nuclear FGFR1 and Nurr1 are expressed in the nuclei of developing TH-positive cells in the embryonic ventral midbrain. Both nuclear receptors were effectively co-immunoprecipitated from the ventral midbrain of FGF-2-deficient embryonic mice, which previously showed an increase of mDA neurons and enhanced nuclear FGFR1 accumulation. Immunoprecipitation and co-localization experiments showed the presence of Nurr1 and FGFR1 in common nuclear protein complexes. Fluorescence recovery after photobleaching and chromatin immunoprecipitation experiments demonstrated the Nurr1-mediated shift of nuclear FGFR1-EGFP mobility toward a transcriptionally active population and that both Nurr1 and FGFR1 bind to a common region in the TH gene promoter. Furthermore, nuclear FGFR1 or its 23-kDa FGF-2 ligand (FGF-223) enhances Nurr1-dependent activation of the TH gene promoter. Transcriptional cooperation of FGFR1 with Nurr1 was confirmed on isolated Nurr1-binding elements. The proposed INFS/Nurr1 nuclear partnership provides a novel mechanism for TH gene regulation in mDA neurons and a potential therapeutic target in neurodevelopmental and neurodegenerative disorders.
Introduction
Ventral mesencephalic dopaminergic (mDA)5 neurons develop from self-renewing progenitors, which exit the cell cycle (mouse embryonic days (E) 11–15), leave the ventricular zone, and migrate toward the mantle zone close to the pial surface while progressively committing to the mDA fate (1). The differentiating mDA neurons start to build rostrally directed projections to their forebrain targets as soon as they begin to express tyrosine hydroxylase (TH) (2), the rate-limiting enzyme in dopamine synthesis. Two nuclear factors, Nurr1 and FGFR1, control terminal differentiation, maturation, and maintenance of mDA neurons (3, 4). Nurr1 (NR4A2) belongs to the subfamily of orphan nuclear receptors, which lack ligand binding capacity, typically present in other nuclear receptors (5). Nevertheless, Nurr1 transcriptional activity can be induced by hormones and growth factors (6, 7), by currently largely unknown mechanisms. In mDA neurons, Nurr1 acts as an essential transcription factor for expression of various genes, including TH, involved in dopamine synthesis and function (6, 8). Disruption of the Nurr1 gene in mice impairs development of TH-expressing mDA neurons and results in prenatal death (9).
FGFs mediate biological responses as extracellular proteins by activation of cell surface FGFRs (10). Additionally, newly synthesized FGFR1 translocates to the nucleus by β-importin, utilizing the nuclear localization signal (NLS) of its high molecular FGF-2 ligand (FGF-223) or other FGFR1-binding proteins (11). The nuclear translocation of FGFR1 is triggered by diverse developmental signals and regulates gene activities via integrative nuclear FGFR1 signaling (INFS) (12, 13). Mechanistically, nuclear FGFR1 releases CREB-binding protein (CBP), a common transcriptional co-activator, and RSK from an inactive complex (14), enabling their gene activating and chromatin remodeling functions. A direct interaction of intranuclear CBP with FGFR1 converts quickly diffusing nucleoplasmic FGFR1 into a more slowly moving chromatin-associated protein (15), which stimulates TH gene expression (16). Furthermore, disruption of FGFR1 signaling in developing postmitotic mDA neurons impairs their maturation and function (3), indicating a direct role of nuclear FGFR1 in postmitotic mDA neuronal development. Interestingly, FGF-2-deficient mice display an increase of mDA neurons expressing nuclear FGFR1 in the substantia nigra during terminal differentiation (60). The specific role of nuclear FGFR1 remains to be further elucidated in this system.
The present study shows that FGFR1 and Nurr1 (i) are both co-expressed in the nuclei of developing TH-expressing cells in the ventral midbrain of mouse embryos, (ii) associate in the same nuclear protein complexes, and (iii) cooperate in chromatin binding and transcriptional activation. These results integrate the INFS and Nurr1 into a common gene-activating mechanism important in dopaminergic cell development.
EXPERIMENTAL PROCEDURES
Plasmids
FLAG-tagged FGF-223 and FGF-218 isoforms were constructed by cloning FGF-223 and FGF-218 cDNA into the EcoRI/HindIII sites of p3XFLAG-CMV-14 (Sigma), resulting in clone pFGF-223-3XFLAG and pFGF-218-3XFLAG (18). Plasmid pcDNA3.1-FGFR1 expressing the IIIC form of FGFR1 was described previously (19). FGFR1 mutants FGFR1(TK−)-deleted tyrosine kinase domain and FGFR1(SP−/NLS) signal peptide replaced with the NLS from the SV40 large T antigen were described previously (16, 20). Nurr1 expressing pCAGGS-Nurr1-FLAG plasmid was generated from pCAGGS-empty provided by Dr. Hitoshi Niwa (RIKEN Center for Developmental Biology, Kobe, Japan) (21), which comprised the chicken β-actin promoter with CMV enhancer (CAG) promoter composed of CMV immediate early enhancer and chicken β-actin promoter. The generation of pCAGGS-3XFLAG plasmid was described previously (22). The full-length murine Nurr1 coding sequence was PCR-amplified from embryonic mouse brain cDNA; thereby an MfeI site followed by an Kozak sequence was introduced in front of the start codon. The stop codon was replaced by an XbaI site. The MfeI/XbaI Nurr1 coding sequence was inserted into the EcoRI/XbaI sites of pCAGGS-3XFLAG plasmid, resulting in pCAGGS-Nurr1–3XFLAG (R467) plasmid. The reporter plasmid TH-Luc containing −425/+25 bp fragment of bovine TH promoter was previously described (23). The reference reporter plasmid (pGL4.70 [hRluc] promoterless) was purchased from Promega. Plasmids NurRE3-Luc containing three Nur response elements and NBRE3-Luc containing three nerve growth factor (NGF)-binding response elements in minimal Pro-opiomelanocortin (POMC) gene promoter (−34/+63), were gifts from Dr. Jacques Drouin (Institut de Recherches Cliniques de Montréal) (24, 25).
Tissue Processing
The FGF-2-deficient mouse strain (FGF-2tm1Zllr) was maintained on C57BL/6 background (26). Wild-type (WT, FGF-2+/+) and knockout (KO, FGF-2−/−) littermates were obtained by cross-breeding of heterozygous FGF-2 mice and genotyped by PCR as described previously (27). Dissected embryonic brains (E14.5) were fixed in 4% paraformaldehyde in PBS overnight at 4 °C, cryoprotected overnight in 30% (w/w) sucrose, and embedded in Tissue Tec OTC compound (Sakura). Serial coronal cryosections of 20-μm thickness were sampled on slides. All of the experimental protocols followed German law on animal care and were approved by Bezirksregierung (Hannover, Germany; #08/1487).
Cell Culture and Transfections
The SK-N-BE(2) human neuroblastoma cell line was cultured in DMEM high glucose with 10% (v/v) FCS, 1 mm sodium pyruvate, 0.1 mg/ml penicillin/streptomycin. Transfection of cells for immunoprecipitation was performed at confluence of 60% in 75-cm2 culture flasks. 10 μg of plasmid-DNA encoding Nurr-1-FLAG and FGFR1, respectively, was delivered using Metafectene Pro reagent in antibiotic-free medium. After an overnight incubation with DNA-lipid complexes, the cells were differentiated for further 24 h after supplementation with 1 μm retinoic acid (28). Transfections in SK-N-BE(2) cells for luciferase assays were performed by Lipofectamine 2000 (Invitrogen) with cells of nearly 60% confluence in 24-well dishes. The cells were grown in DMEM/Ham's F-12 with 10% (v/v) fetal bovine serum. The cells were harvested 36 h after transfection. Each sample/well for transfection contained a total of 1 μg of DNA, including 0.3 μg of reporter plasmid, 0.3 μg of reference reporter plasmid (pGL4.70 [hRluc] without promoter), 0.3 μg of FGF-2 isoforms plasmid or FGFR1 mutants, 10–100 ng of pCAGGS-Nurr1 expression vector, and pCAGGS vector to make up the total amount.
The primary cells of ventral mesencephalon were dissected from E12.5 rat embryos and cultured as described previously (29). The cells were cultured for 2 days in proliferation medium and for either 1 or 4 days in vitro (DIV) in differentiation medium.
The SV40-immortalized rat ventral mesencephalic neuronal progenitor cells (SV40i-VM-NPCs) were described previously (30). The clone C2 was seeded in N2 medium (DMEM/Ham's F-12, 1% N2 supplement (100×), 0.25% BSA, 2 mm glutamine, 1 mm sodium pyruvate, 0.1 mg/ml penicillin/streptomycin) containing 3% FCS and afterward cultivated in serum-free N2 medium.
Fluorescence Immunocytochemistry
For fluorescence immunocytochemistry, brain slices or cells fixed in 4% paraformaldehyde were blocked in 5% (v/v) normal goat serum (NGS), 1% (w/v) BSA, 0.3% (w/v) Triton X-100 in PBS for 1 h at room temperature, followed by primary antibody incubation overnight at 4 °C: mouse anti-TH (1:1000, Sigma, T1299), rabbit anti-Nurr1 (1:1000, Santa Cruz, sc-990), and rabbit anti-Lmx1a (1:6000, Millipore, AB10533) in 1% NGS, 1% BSA, 0.3% Triton X-100 in PBS. For mouse anti-FGFR1 (1:1000, Abcam, M19B2) and rabbit anti-FGFR1 (1:1000, Santa Cruz, sc-121) Soerensen's sodium phosphate buffer containing 10% (v/v) NGS and 0.3% (w/v) Triton X-100 was used. Secondary antibodies anti-mouse IgG1 Alexa 568 (1:200, Invitrogen, A11034), anti-mouse IgG Alexa 488 (1:2000, Molecular Probes), and anti-rabbit IgG Alexa 488 (1:500, Invitrogen, A11008) were applied for 1 h at room temperature. The nuclei were visualized by DAPI (Sigma-Aldrich, 1:1000 in PBS) staining. Images were taken with Olympus BX60 epifluorescence microscope equipped with ColorView 3 camera (Olympus) and CellP software (Olympus) or Leica TCS SP2 confocal microscope supplied with oil immersion objectives HCX PL APO BL (63×, numerical aperture 1.4).
Co-localization Analysis
Co-localization analysis was performed on single plane images of randomly selected cells (n = 17) using ImageJ software (National Institutes of Health) with intensity correlation analysis plug-in, as described previously (31, 32). Mander's overlap coefficient (R) represents the ratio of intersecting volume to total object volume (total pixels with intensity > 0). It ranges from one to zero, with one representing high co-localization. The number of objects in both channels was equal, with a red:green pixel ratio of 1.00 ± 0.3. Mander's coefficients (M1, FGFR1; and M2, Nurr1) represent the fraction of the pixels in which one signal overlaps the other. M1 and M2 range from one to zero with one for complete co-localization. M1 and M2 are not influenced by differences in absolute signal intensity in both channels, because they are normalized against total pixel intensity. The intensity correlation quotient (ICQ) represents the synchrony in which the pixel intensities of the two respective channels vary from the mean image intensities of both channels together. This is demonstrated by the product of the differences from the mean (PDM). Positive PDM (blue) represents the co-localized pixels, where both channels vary synchronically from the mean pixel intensity. Negative PDM represents the not co-localized pixels, where both channels vary asynchronously from the mean pixel intensity. The ICQ is based on the nonparametric sign test analysis of the PDM values and is equal to the ratio of the number of positive PDM (99,690.00 ± 35,588.46) values to the total number of pixels (202,647.00 ± 101,861.64). ICQ values are distributed between −0.5 and +0.5 where random staining gives an ICQ of ∼0, segregated asynchronous staining 0 > ICQ > −0.5 and dependent synchronous staining 0 < ICQ < +0.5 (31).
Immunoprecipitation and Western Blot Assay
The nuclear and cytoplasmic fractions were isolated as described previously (12, 14). Briefly, the cells were washed in ice-cold PBS and harvested in homogenization buffer (10 mm HEPES, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 2 mm DTT, 25 mm NaF, 1 mm NaVO3, PhosStop (Roche Applied Science), Complete protease inhibitor mixture (Roche Applied Science)). After incubation on ice for 15 min, the cells were lysed by the addition of 0.6% (v/v) Igepal CA-630 and vigorous vortexing. After centrifugation, the supernatant representing the cytoplasmic fraction was saved. The nuclear pellet was washed twice in homogenization buffer containing 0.6% (v/v) Igepal and dissolved by sonication in 1% (v/v) Triton X-100 buffer containing 50 mm HEPES, 150 mm NaCl, 5 mm EDTA, 1 mm DTT, and Complete protease inhibitor mixture. For immunoprecipitation, the nuclear extracts were adjusted to an overall protein concentration of 1–2 μg/μl in nuclear extraction buffer containing 150 mm NaCl and finally diluted 1:2 with radioimmune precipitation assay buffer (137 mm NaCl, 20 mm Tris-HCl, pH 7.5, 25 mm sodium glycerophosphate, 2 mm EDTA, 1 mm sodium orthovanadate, 1% (v/v) Triton X-100, 1% (w/v) sodium desoxycholate, Complete protease inhibitor mixture). Equal amounts of protein extracts (0.5–1 mg) were incubated with 2 μg of appropriate antibody overnight at 4 °C: rabbit anti-Nurr1 (Santa Cruz, sc-990), rabbit anti-FGFR1 (Flg C15, SantaCruz, sc-121), or rabbit IgG antibodies (DAKO, M737). The immunocomplexes were precipitated with protein A-fused Dynabeads (Invitrogen), washed, and analyzed with SDS-PAGE and Western blots. The following primary antibodies were used: rabbit anti-Nurr1 (1:1000, Santa Cruz, sc-990), rabbit anti-FGFR1 (Flg C15) (1:500, SantaCruz, sc-121), mouse anti-FGFR1 (1:400, mAb6), or mouse anti-FLAG (1:3000, Sigma, F1804).
In Vitro Protein-Protein Interaction
The in vitro protein interaction assay was performed in reticulocyte lysates using the TnT T7 quick coupled transcription/translation system (Promega) according to the manufacturer's instructions. Linearized plasmids containing Nurr1 in pGEM-T (NotI, T7 RNA polymerase) and IIIC form of FGFR1 in pcDNA3.1 (BamHI, T7 RNA polymerase) were used for expression. Recombinant FGF-2 was used as a positive control, and experiments were carried out under conditions previously described (33).
Fluorescence Recovery after Photobleaching (FRAP)
FRAP analysis in SK-N-BE(2) cells on μ-Dishes (Ibidi) was performed using an Olympus Fluoview FV1000 microscope equipped with an oil immersion objective (60×, 1.35 NA), a 6-fold zoom magnification, laserlines 405 and 491 nm, a dichroic mirror DM405/488/559/635, and an incubation chamber (37 °C and 5% CO2). For bleaching of EGFP-fused proteins, the laser output was set to 93% (405 nm). The size of the region for bleaching was the same for each cell covering the nucleus and the cytoplasm. Before bleaching, three images were acquired. After bleaching, images were taken every 0.5 s for 1.5 min, then every 1.5 s for 2 min, and finally every 3 s for 5 min. MacMaster Biophotonics Facility (MBF) ImageJ (1.43m) and Prism 4 (GraphPad Software, San Diego, CA) were used for data analysis. Normalized relative fluorescence intensities were determined using “FRAP profiler” (ImageJ), quantified by fitting one- and two-phase exponential association, and finally plotted using Prism 4. The two-phase exponential association was fitted using the equation,
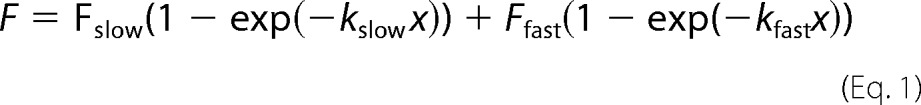 |
with Fslow representing the coefficient of a slow mobile population, whereas Ffast represents the coefficient of a fast mobile population. kslow and kfast are the population-specific values, which provide information about the recovery half-times (t½). The effects of Nurr-1 among populations of mobile and immobile FGFR1-EGFP, and recovery half-times (t½) were analyzed by using ANOVA, Bartlett, and t-tests. The parameters are shown as arithmetic means ± S.E.
TH Promoter Activation
Luciferase assays were performed with the dual luciferase (firefly and Renilla) reporter assay system (Promega Corp., Madison, WI). All of the reagents were prepared as described by the manufacturer. The 5× passive lysis buffer was supplied by the manufacturer and used for cell lysis. After washing twice, the NB-1 cells were removed directly from culture and transferred to 100 μl of 1× passive lysis buffer. After allowing lysis for 15–20 min, a 20-μl aliquot was used for luminescence measurements with a BioTek Plate Reader. The following steps were used for luminescence measurements: 100 μl of the firefly luciferase reagent (LARII) was added to the test sample; after equilibration for 10 s, measurement of luminescence was performed with a integration time of 2 s; followed by the addition of 100 μl of the Renilla luciferase reagent and firefly quenching (Stop & Glo) with the same equilibration time and measurement of luminescence. The data are represented as the means ± S.E. of the ratio of firefly to Renilla luciferase activity for two to four experiments, each performed in quadruplicate. The antibodies used were: anti-Nurr1 (Santa Cruz, sc-991) and anti-FGFR1 (C-term, Abcam, ab10646).
Chromatin Immunoprecipitation
The rats were sacrificed by CO2 asphyxiation followed by decapitation. The brains were quickly removed and dissected on ice into: brain cortex, cerebellum, olfactory bulbs, and ventral midbrain (substantia nigra region). The tissues were minced and incubated in cross-linking solution: cold phosphate-buffered saline with 1% (w/v) formaldehyde (Sigma) at room temperature for 15 min, rinsed twice with cold phosphate-buffered saline, and sonicated in phosphate-buffered saline with protease inhibitors. The lysates were centrifuged at 14,000 × g for 10 min at 4 °C. ChIP was performed in samples containing equal amount of genomic DNA using polyclonal antibodies: rabbit FGFR1 (ab10646, Abcam), Nurr1 (sc-5568, Santa Cruz), or control rabbit IgG provided by the MAGnify chromatin immunoprecipitation system. DNA was purified according manufacturer's instructions (Invitrogen). Sample PCR was then performed on the immunoprecipitated genomic DNA with primers for the response element containing regions of the TH genes. The primer sequences were as follows: rat TH NBS 1 forward, 5′-AGCTCATAAGAGCTTTCAGATTATC; rat TH NBS 1 reverse, 5′-CTGAGACAGGGTGGATCCCAG; rat TH NBS 2 forward, 5′-AGGTTATAGTTCTAACATGAG; and rat TH NBS 2 reverse, 5′-GCCTCCGTCCCATTAGATCTAATTG.
Quantitative PCR was used to determine relative amount of specific loci in IP, input, and IgG (preimmune) samples combined from two rats. Quantitative PCR was performed using iQ SYBR Green Supermix (Bio-Rad) on a Bio-Rad iCycler. Five μl of ChIP DNA and a 1:10 dilution of input DNA were used in duplicate reactions. The data are expressed as IP/input as follows.
The PCR assays were performed at least three times, and the results are combined and shown as the relative change means ± S.E.
RESULTS
FGFR1 and Nurr1 Localize to Nuclei of Developing DA Neurons
Nurr1 is a key transcription factor essential for signaling integration during terminal differentiation of mDA neurons (4). A similar function has been described for INFS activation and the nuclear translocation of FGFR1 during neuronal differentiation (11, 13). Both Nurr1 and nuclear FGFR1 stimulate TH gene expression, suggesting a potential partnership during mDA development.
To test this hypothesis, we first analyzed the expression patterns of both factors in the ventral midbrain of E14.5 wild-type and FGF-2-deficient mouse embryos, which display increased nuclear FGFR1 accumulation and supernumerary mDA neurons.6 Nurr1-positive cells were similarly distributed in wild-type (wt; Fig. 1, A and B) and FGF-2-deficient (ko; Fig. 1, E and F) embryos within the ventral midbrain at E14.5. Because Nurr1 expression is initiated in postmitotic mDA precursors (6), the Nurr1-positive cells were found in the subventricular zone (svz) and mantle zone (mz; Fig. 1A) but were absent in the ventricular zone (vz; Fig. 1A), the origin of proliferating, self-renewing neural stem cells. At E14.5, the majority of Nurr1-immunoreactive cells in the mantle zone began to express TH (Fig. 1, C, D, G, and H). Similarly, as in the whole brain (34–38), FGFR1 was abundantly expressed in the developing ventral midbrain including the TH-positive area in consecutive sections from wild-type (Fig. 1, I and L) and knockout (Fig. 1, M–P) embryos. By confocal microscopy, FGFR1 was detected in the nucleus of TH-positive mDA neurons of the mantle zone (Fig. 1, K, L, O, and P).
FIGURE 1.
Nurr1 and FGFR1 expression in the ventral midbrain of E14.5 embryos. A–H, Nurr1 is expressed in postmitotic dopaminergic precursors of subventricular (svz) and TH-expressing dopaminergic neurons of the mantle zone (mz) in wild-type (wt, A and B; scale bar, 200 μm) and FGF-2-deficient (ko, E and F: scale bar, 200 μm) animals, but not in the proliferative progenitors in the ventricular zone (vz). Higher magnification epifluorescence images of Nurr1/TH co-labeling (C, D, G, and H; scale bar, 10 μm) in the region outlined in F show Nurr1 (B) expression in the DAPI stained nucleus. I–P, FGFR1 was abundantly expressed in the VM of WT (I–L; scale bar, 200 μm) and knockout (ko, M–P; scale bar, 200 μm) embryos including the TH-positive mDA domain (J and N). Higher magnification confocal images of FGFR1/TH co-labeling in the regions outlined in J and N show FGFR1 expression in the nucleus (DAPI, blue) of the TH-positive cells located in the mantle zone in WT (K and L; scale bar, 5 μm) and knockout (ko, O and P; scale bar, 5 μm).
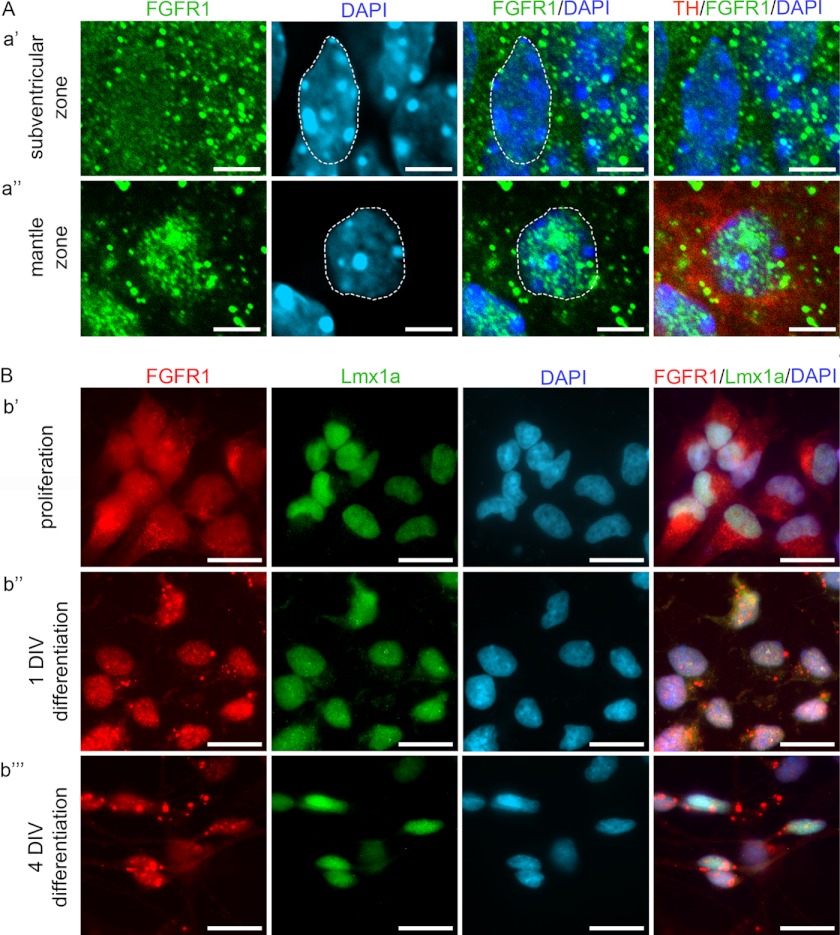
To further analyze the subcellular distribution of FGFR1 in vivo, we used midbrain sections of wild-type mouse embryos at E14.5 (Fig. 2A). Immunohistochemistry combined with confocal microscopy showed uniform expression of FGFR1 in the cytoplasm and nucleus of ventral mesencephalic precursor cells in the subventricular zone of the mDA area (Fig. 2A, panel a′). In the differentiated TH-positive neurons of the mantle zone, a predominantly nuclear localization of FGFR1 was observed (Fig. 2A, panel a″), which further emphasizes the role of nuclear function of FGFR1 in terminal differentiation and maturation of mDA neurons.
FIGURE 2.
Nuclear accumulation of FGFR1 during differentiation of VM cells. A, confocal microscope images showing subcellular localization of FGFR1 in cells of the mDA field in VM of E14.5 mouse embryos. In the subventricular zone (panel a′), where undifferentiated cells are located, FGFR1 (green) showed a nearly uniform distribution in the cytoplasm and nucleus (blue, dotted line). In the mantle zone (panel a″), FGFR1 (green) shows a prominent accumulation in the nuclear (blue, dotted line) proportion of the differentiated neurons expressing TH (red). Scale bar, 5 μm. B, epifluorescence microscope images showing the accumulation of FGFR1 during differentiation of primary neuronal cultures of rat ventral mesencephalic progenitor cells. During proliferation (panel b′) FGFR1 (red) was localized in the cytoplasm and nucleus (DAPI, blue) of Lmx1a-positive (green) progenitor cells. After differentiation for 1 DIV (panel b″), FGFR1 showed a mainly nuclear distribution in Lmx1a-positive cells. After 4 DIV in differentiation medium (b⁗), FGFR1 was accumulated in the nucleus and also present in the neurites of the neuron like-shaped cells expressing Lmx1a. Scale bar, 10 μm.
Nuclear Accumulation of FGFR1 in Differentiating Primary Ventral Midbrain Progenitor Cells
Consistent with these in vivo observations, in vitro experiments using a standard differentiation protocol for primary rat E12.5 ventral mesencephalic progenitor cells (29) showed a differential subcellular distribution of FGFR1 during the process of differentiation (Fig. 2B). Early, during the expansion of the plated cells in mitogen-containing (FGF-218) proliferation medium, FGFR1 was mainly found in the cytoplasm and to a lower extent in the nucleus of cells positive for Lmx1a (Fig. 2B, panel b′), an early marker for mDA progenitors (39). The replacement of proliferation medium with B27-supplemented differentiation medium resulted in an increased, mainly nuclear localization of FGFR1 already after 1 DIV (Fig. 2B, panel b″). After 4 DIV, FGFR1 remained enriched in the nucleus of Lmx1a-positive neurons (Fig. 2B, b⁗). These results demonstrate the importance of nuclear FGFR1 for mesencephalic progenitor differentiation.
Interaction of Nuclear FGFR1 and Nurr1
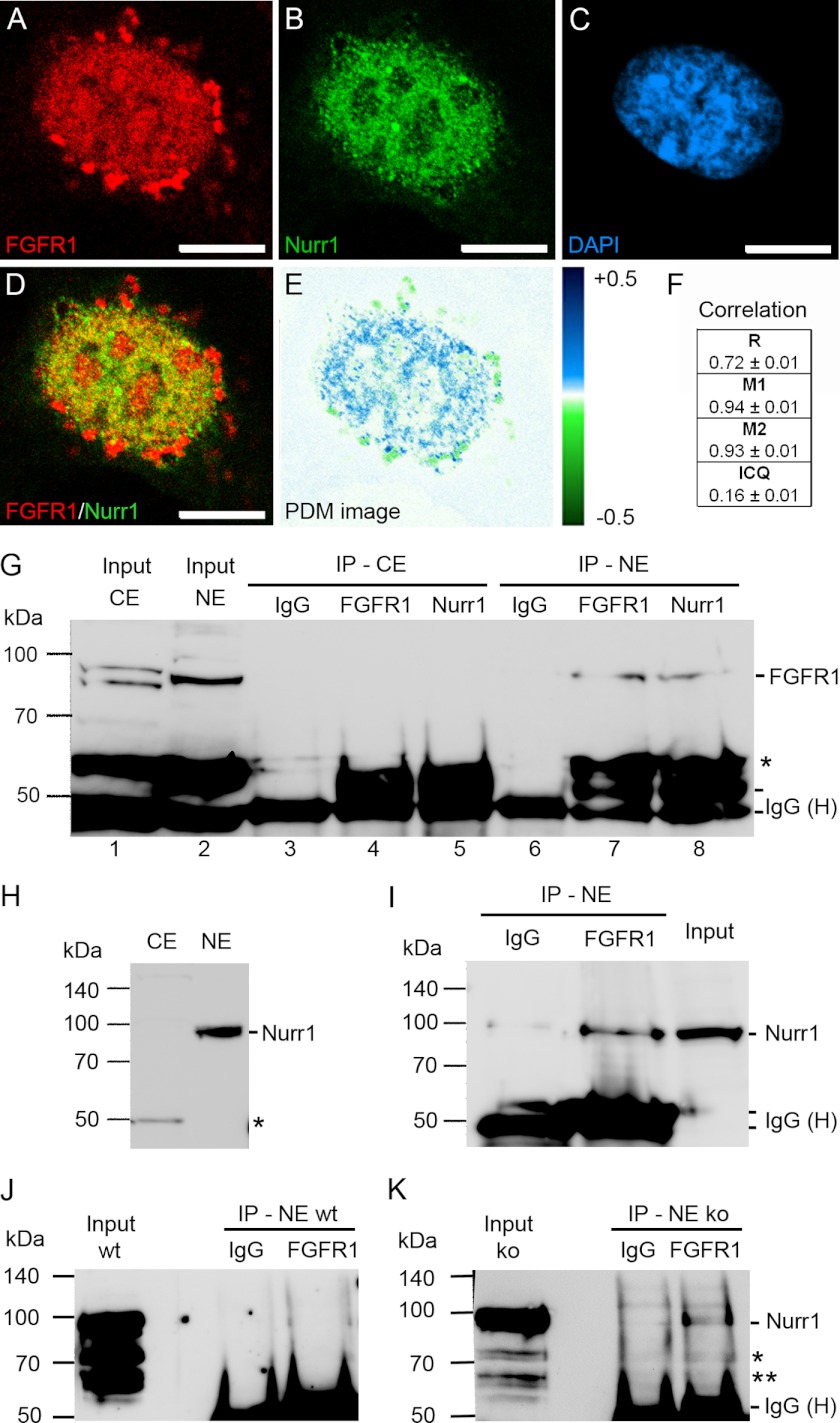
We used a SV40 immortalized ventral mesencephalic neuronal progenitor cell line (SV40i-VM-NPC) that expresses genes associated with mDA development and provides ample material for immunoprecipitation (30). The SV40i-VM-NPC cell line was previously established in our laboratory by stable transfection of the simian virus 40 (SV40) large T-antigen into primary neuronal ventral mesencephalic progenitor cells, derived from E12.5 rat embryos (30). In this cell line, Nurr1 and FGFR1 are endogenously expressed, and nuclear co-localization of both proteins was analyzed by immunocytochemistry and confocal microscopy (Fig. 3, A–D). Both FGFR1 and Nurr1 showed granular distribution within the nucleus. The overlay of the corresponding Nurr1 and FGFR1 confocal planes revealed co-localization of both proteins within the same nuclear speckle-like domains (Fig. 3D). We performed a detailed quantitative co-localization analysis using an intensity correlation algorithm (30, 31). FGFR1 and Nurr1 showed a mutual dependent localization (Fig. 3, E and F). The overlap of FGFR1 and Nurr1 staining is reflected in high Mander's overlap coefficient (r = 0.72 ± 0.01), indicating strong co-localization, high co-localization coefficients and a positive ICQ (Fig. 3F). This is demonstrated in a false color image (PDM image; Fig. 3E). Here, blue color represents dependent localization of both factors, whereas green shows segregated localization.
FIGURE 3.
Presence of FGFR1 and Nurr1 in the same nuclear protein complexes of ventral midbrain NPCs. A–F, co-localization analysis of SV40-VM-NPCs cultivated for 24 h in serum-free N2 medium. Confocal images showed granular distribution of FGFR1 (A, red) and Nurr1 (B, green) in the DAPI-stained nucleus (C, blue) of mDA progenitors. FGFR1 and Nurr1 showed co-localization in the nucleus as demonstrated in overlap (D) and PDM images (E, lut; blue, positive PDM and green negative PDM). F, co-localization analysis values with R, Mander's overlap coefficient (red:green pixel ratio = 1.00 ± 0.03); M1, Mander's co-localization coefficient for FGFR1; M2, Mander's co-localization coefficient for Nurr1; and ICQ. Scale bar, 5 μm. G–K, the IP with IgGs represented the negative control for co-precipitation. The input represented the loading control of 100 μg of pure denaturized nuclear protein extract free of denaturized IgG heavy chains (IgG (H)), which are present in IP lanes. Distinct nuclear (NE) ∼90-kDa and cytoplasmic (CE) ∼85- and ∼95-kDa bands of FGFR1 represented different glucosylation forms of the receptor and demonstrated the lack of cross-contamination between fractions (G, lane 1; *, truncated form of FGFR1). The precipitation with anti-Nurr1 resulted in co-precipitation of FGFR1 in the nuclear fraction (G, lane 8). Precipitation of nuclear FGFR1 with anti-FGFR1 is shown as a positive control (G, lane 7). The ∼90-kDa Nurr1 band may result because of post-transcriptional sumoylation of Nurr1 (59) and was present in the nuclear fraction but absent in the cytoplasmic fraction as determined by Western blot assay (H; *, antibody may recognize an additional splice isoform of Nurr1 (17)). Therefore precipitation was performed only in the nuclear fraction, which resulted in co-precipitation of Nurr1 with FGFR1 antibody (I). Precipitation of nuclear FGFR1 in lysates of VM from E14.5 FGF-2 knock out embryos resulted in co-precipitation of Nurr1 (K), whereas in IPs of VM lysates from E14.5 wild-type embryos, the signal was not clearly detectible (J; * and **, unmodified and/or splice forms of Nurr1). The amount of used material was limited by the availability of fresh tissue.
To verify that nuclear FGFR1 and Nurr1 may exist in a common protein complex, we examined whether both endogenous proteins can be co-immunoprecipitated with FGFR1 or Nurr1 antibodies. The nuclear presence of FGFR1 was verified by Western blot analysis of nuclear and cytoplasmic fractions using polyclonal anti-FGFR1 (Fig. 3G, lanes 1 and 2). In the nuclear fraction, one prominent signal of FGFR1 was detected. The cytoplasmic fraction showed two bands at ∼85 and ∼95 kDa (Fig. 3G, lane 1), shown to represent different glycosylation forms (40, 41). The full-length Nurr1 was present in the nuclear, but absent in the cytoplasmic fraction as determined by Western blot (Fig. 3H). As a negative control for co-immunoprecipitation, lysates were incubated with rabbit IgGs (Fig. 3, G, lanes 3 and 6, and I). As a positive control, precipitation of the endogenous ∼95-kDa FGFR1 protein with a polyclonal anti-FGFR1 antibody was confirmed (Fig. 3G, lane 7). Indeed, FGFR1 was co-precipitated by anti-Nurr1 in the nuclear fraction (Fig. 3G, lane 8). This result was confirmed in a reverse experiment, in which anti-FGFR1 antibody effectively precipitated Nurr1 (Fig. 3I). Thus, the immunoprecipitation experiments are consistent with the co-localization analysis and confirm that endogenous Nurr1 and FGFR1 belong to a common nuclear protein complex in ventral mesencephalic progenitors.
We further confirmed this interaction in vivo by co-immunoprecipitation of FGFR1 and Nurr1 in nuclear extracts of ventral mesencephalic tissue pooled from 8–11 E14.5 mouse embryos. No clear co-precipitation signal could be observed in extracts of wild-type embryos (Fig. 3J, NE wt). However, anti-FGFR1 antibody precipitated Nurr1 along with FGFR1 (Fig. 3K) from the nuclear protein lysates of the FGF-2-deficient embryos (Fig. 3K, NE ko), which display increased nuclear FGFR1 protein levels as shown previously.6 Thus, endogenous nuclear FGFR1 and Nurr1 interact in cultured immortalized ventral mesencephalic progenitors, as well as in the developing midbrain.
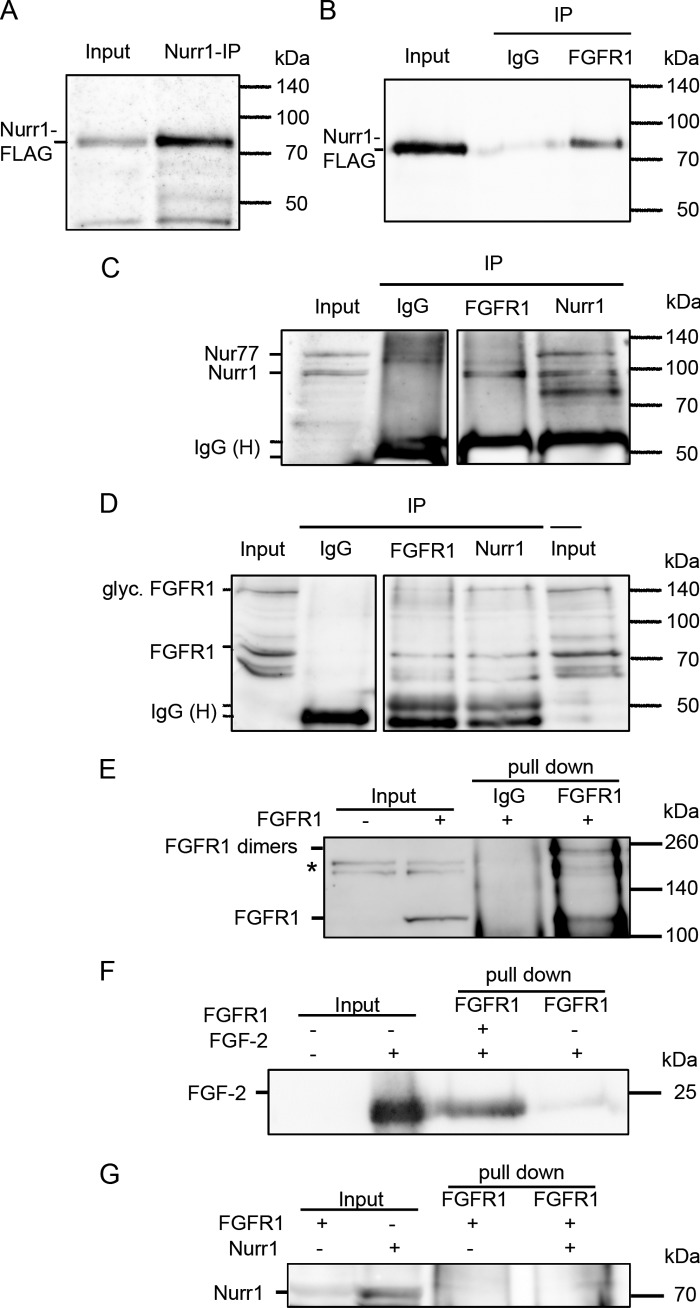
The interaction of FGFR1 and Nurr1 was further substantiated by co-immunoprecipitations of overexpressed proteins in the human neuroblastoma cell line SK-N-BE(2). These cells were used previously to study neuronal differentiation (28, 42–44), which occurs spontaneously or can be induced by treatment with retinoic acid. Therefore, neuroblastoma cells were transiently transfected with plasmids encoding FGFR1 and FLAG-tagged Nurr1 and subsequently treated with 1 μm retinoic acid, and nuclear extracts were harvested. The proteins were immunoprecipitated with anti-Nurr1, polyclonal anti-FGFR1 recognizing the FGFR1 C terminus, anti-FLAG for Nurr1, or control IgG. Consistent with co-precipitation of endogenous proteins (Fig. 3), co-precipitation of Nurr1 by anti-FGFR1 was detected with anti-Nurr1 as well as anti-FLAG (detecting Nurr1-FLAG) (Fig. 4, B and C). Furthermore, immunoprecipitation of Nurr1 with anti-Nurr1 antibody pulled down the ∼85-kDa form of FGFR1 (41) detected with monoclonal mcAb6 antibody, which recognized the N terminus of FGFR1 (Fig. 4D).
FIGURE 4.
Nuclear FGFR1 and Nurr1 interaction after overexpression in human neuroblastoma cells. The human neuroblastoma cells were transfected with plasmids encoding for full-length FGFR1 protein, as well as Nurr1-protein fused to a 3×FLAG tag. 24 h after transfection, the cells were supplemented with 1 μm retinoic acid for a further 24 h. A–D, the nuclear extracts were immunoprecipitated with polyclonal anti-Nurr1, anti-FGFR1, and rabbit IgGs as negative control. Input represents 25 μg (A and C) and 100 μg (B and C), respectively, protein of the not precipitated nuclear extract. The additional band in C (compare Fig. 3K) may represent the closely related Nur77, which shows only a faint expression in VM (47). Precipitation with polyclonal Nurr1 and FGFR1 antibodies functioned properly as shown by detection of precipitated Nurr1 with the anti-FLAG antibody (A) and of precipitated FGFR1 with monoclonal anti-FGFR1 (mAb6) antibody (D), respectively. The FGFR1-IP resulted in co-precipitation of Nurr1 as recognized by anti-FLAG-tag antibody (B), as well as with anti-Nurr1 antibody (C, represents one blot). Correspondingly, the ∼85-kDa form of FGFR1 was able to co-precipitate with Nurr1, as detected by monoclonal anti-FGFR1 (mAb6) antibody (D, represents one blot). The negative controls, precipitated with rabbit IgGs, were missing the specific bands, confirming a specificity of the Nurr1 and FGFR1 immunoprecipitations. E, in vitro coupled transcription/translation of FGFR1 resulted in positive product at ∼120 kDa (input), which was missing in the control translation reaction without DNA template. The subsequent pull-down of FGFR1 with anti-FGFR1 antibody resulted in positive precipitates at ∼120 and ∼250 kDa, which would correspond to FGFR1 dimers. *, unspecific cross-reaction only observed in reticulocyte extracts. F, the positive interaction of FGF-2 was confirmed by subsequent pull-down along with FGFR1. G, the interaction of Nurr1 and FGFR1 seems to be indirect, because the subsequent pull-downs of Nurr1 and FGFR1 were negative.
To analyze whether Nurr1 and FGFR1 interact directly or within a complex, we performed cell-free in vitro transcription/translation of the binding partners in reticulocyte lysates combined with FGFR1 pull-down (Fig. 4E). As a positive control for a direct interaction with FGFR1, recombinant FGF-2 was employed. These two proteins displayed a direct interaction as expected (Fig. 4F). However, a direct interaction could not be detected for FGFR1 and Nurr1 (Fig. 4G). In conclusion, co-immunopreciptation experiments of endogenous and overexpressed proteins in different cellular systems reveal an indirect interaction between Nurr1 and FGFR1 within the nucleus.
Nuclear Mobility of FGFR1 Is Regulated by Nurr1
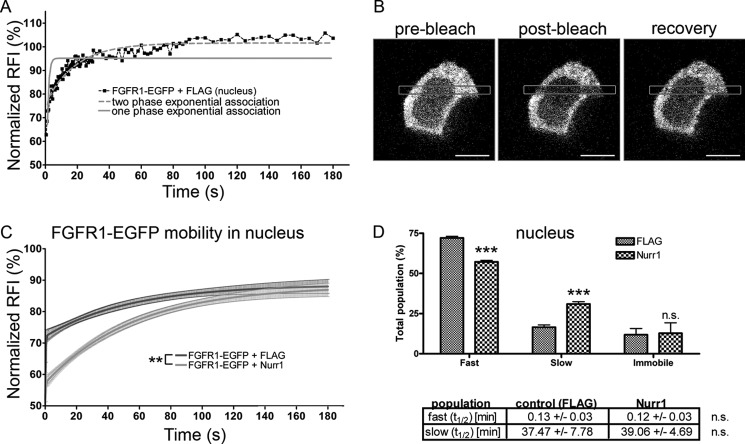
To analyze the FGFR1-Nurr1 interaction in live cells we used FRAP. Previous studies show that the fusion protein, FGFR1-EGFP, is expressed at similar levels and has a similar nuclear/cytoplasmic distribution as endogenous FGFR1 (15). Additionally, FGFR1-EGFP showed cAMP- or FGF-223 isoform-specific induced nuclear accumulation and gene activating functions as observed with the nonfused FGFR1. Previous FRAP measurements of transiently transfected FGFR1-EGFP demonstrated that the nuclear population of FGFR1 consists of the quickly diffusing (hyperkinetic) nucleoplasmic receptors, slowly diffusing (hypokinetic) chromatin-bound FGFR1, and an immobile nuclear matrix-bound fraction (15). To determine whether Nurr1 affects the mobility of FGFR1 in the nucleus, FRAP experiments were performed after co-transfection of FGFR1-EGFP with Nurr1-FLAG or a control plasmid expressing the FLAG tag. In both conditions, the cells displayed similar intensity of FGFR1-EGFP fluorescence. Co-transfection of Nurr1-FLAG significantly increased the slow fraction of FGFR1-EGFP in the nucleus compared with transfection of the control FLAG tag vector (Fig. 5, C and D). In previous studies, the slow FGFR1 population was shown to represent the chromatin-bound transcriptionally active population (15). The increase of hypokinetic FGFR1 was accompanied by a decrease in the fast FGFR1 population. However, the immobile population did not change, indicating that Nurr1 did not mobilize stored FGFR1 from the nuclear matrix but increased engagement of free diffusing FGFR1.
FIGURE 5.
FRAP of nuclear and cytoplasmic FGFR1-EGFP in neuroblastoma cells after co-transfection with Nurr1. A, one (gray line) and two exponential (gray dotted line) regression curves for data from one exemplary FRAP measurement in the nucleus of neuroblastoma cells transfected with FGFR1-EGFP. The regression analysis of recovery kinetics showed the best fit with a two exponential function. B, example of a single cell before and after photobleaching. Scale bar, 10 μm. C, after fitting two-exponential curves to FGFR1-EGFP, the recovery kinetics were significantly changed in the nucleus of neuroblastoma cells co-transfected with Nurr1–3×FLAG (n = 10). D, a significant shift of FGFR1-EGFP mobility was represented by a significant) decrease of the fast and a significant increase of the slow population in the nucleus of cells co-expressing Nurr1–3×FLAG compared with control cells co-expressing 3×FLAG. ***, p < 0.001. Recovery half-time (t½) values were not significantly altered (n.s., nonsignificant).
Cooperative TH Promoter Activation by Nurr1 and FGFR1
The expansion of the slow nuclear FGFR1 population by Nurr1 suggested that the Nurr1-FGFR1 complexes may co-engage in chromatin binding and gene transcription (Fig. 6). This was investigated further by measuring FGFR1 and Nurr1 interactions with the TH gene in vivo by ChIP.
FIGURE 6.
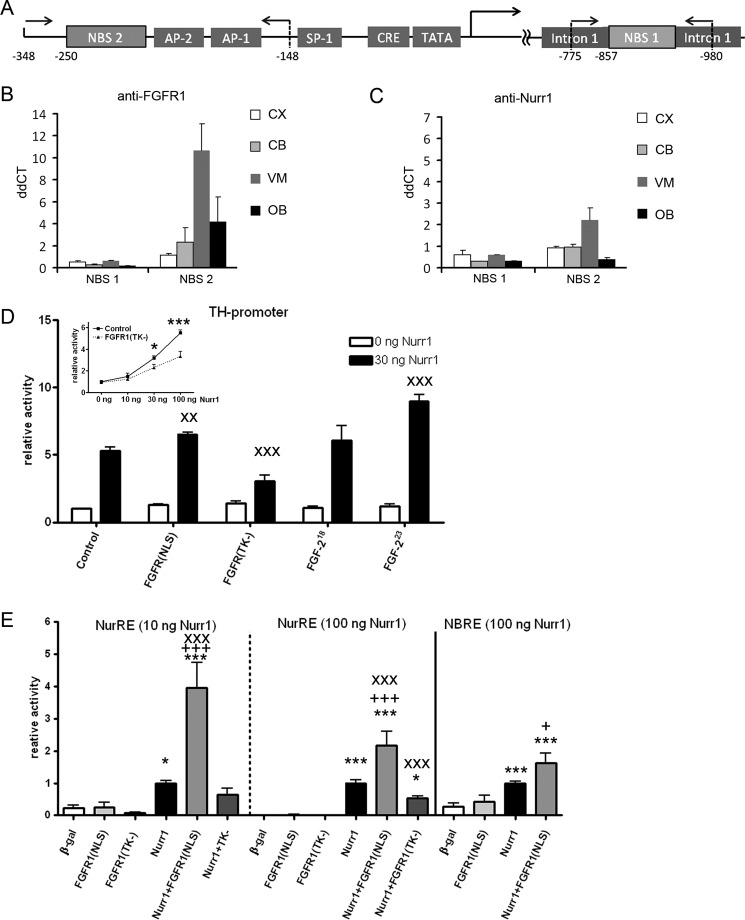
Nurr1 and FGFR1 bind to and cooperatively activate transcription from TH promoter. A, schematic of the TH gene promoter region. Binding sites for other factors are labeled accordingly. Arrows with numbers indicate positions of primers for ChIP. B and C, ChIP was performed with a panel of antibodies against FGFR1, Nurr1, and control IgG with subsequent quantitative PCR analyses of selected potential NBS on TH. IgG was used as a negative control. Graphs show ΔΔCt means ± S.E. of triplicate samples. CX, cortex; CB, cerebellum; VM, ventral midbrain (containing substantia nigra region); OB, olfactory bulb. D, co-transfection of human neuroblastoma cells with Nurr1–3×FLAG showed a dose-dependent increase of TH promoter-dependent luciferase reporter gene expression. The Nurr1-mediated TH promoter activity was significantly diminished by co-transfection of the dominant-negative form of FGFR1 lacking the tyrosine kinase activity [FGFR1(TK−)] starting at 30 ng of Nurr1-FLAG (inset). Co-transfection of FGFR1(NLS), FGFR1(TK−), as well as FGF-223 with Nurr1-FLAG, resulted in significant interaction altering Nurr1-dependent TH promoter activation. The noninteracting isoform FGF-218 did not influence TH promoter activity mediated by Nurr1. Two-way ANOVA on interaction with Nurr1: x, p < 0.05; xx, p < 0.01; xxx, p < 0.001. E, co-transfection of FGFR1(NLS) in neuroblastoma cells enhances Nurr1-NurRE and -NBRE-dependent luciferase transcription. The dose-dependent effects of NurRE activation by 10 and 100 ng of co-transfected Nurr1-FLAG are significantly potentiated by FGFR1(NLS), whereas the co-transfection of FGFR1(TK−) significantly inhibits 100 ng of Nurr1-NuRE activation. Co-transfection of FGFR1(NLS) also enhances Nurr1-dependent transcription from NBRE motive. One-way ANOVA significance to β-galactosidase (β-gal) is expressed as * and to Nurr1 as +. Two-way ANOVA displays Nurr1 interactions as x. Significance levels: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Nurr1 binds to DNA target sites either as a monomer to the canonical NBRE (45) or as homo-/heterodimer with other orphan nuclear receptors to the Nur response element (NurRE) (25). The TH gene promoter contains several (NBRE)-like binding sites with sequences similar to the canonical NBRE (46). One NBRE-like site upstream of the TATA-box (Fig. 6A) was shown to bind FGFR1 (61). Previous reports also suggest that first intron of the TH gene is critical for differential gene regulation among species (48) and can bind Nurr1 (49). In our study, we analyzed Nurr1 and FGFR1 binding to NBRE-like sequence in the TH gene intron-1 (NBS 1; Fig. 6A), as well as the TH promoter region, which contains a sequence homologous to one core element of NurRE (NBS 2; Fig. 6A).
FGFR1 and Nurr1 binding to TH gene in vivo was investigated by ChIP on tissues from the ventral midbrain (VM) region (containing TH-expressing substantia nigra), cerebellum, and cortex, which do not express TH, and in olfactory bulb containing TH-expressing cells. Little or no FGFR1 or Nurr1 binding was detected at the TH intron 1 region (NBS 1) in any of the tissues examined (Fig. 6, B and C). Both FGFR1 and Nurr1 showed binding to the NBS 2-containing TH promoter region (Fig. 6, B and C). The strongest binding occurred in the ventral midbrain tissue, which contains TH-expressing mDA neurons.
Given specific FGFR1/Nurr1 binding to the TH promoter in the ventral midbrain, we examined whether these proteins can affect TH promoter activity using a luciferase reporter transcriptional assay (Fig. 6D). We analyzed the effect of Nurr1 on TH promoter activity and whether the Nurr1-independent or -dependent promoter function is affected by nuclear FGFR1(SP−/NLS) or dominant-negative FGFR1(TK−). In addition, we analyzed the effects of the high molecular weight FGF-223 isoform, which binds and activates endogenous nuclear FGFR1 (15, 50).
Transfection of Nurr1 in neuroblastoma cells resulted in dose-dependent transcriptional activation of the luciferase gene initiated from the bovine TH promoter sequence (Fig. 6D). The Nurr1-dependent TH promoter activity was diminished by co-transfection of dominant-negative FGFR(TK−) mutant, which attained statistical significance at higher doses of transfected Nurr1-FLAG plasmid starting with 30 ng (Fig. 6D, inset). Co-transfection of FGFR1(SP-/NLS), as well as FGF-223, a potent activator of nuclear FGFR1 (15, 50), with Nurr1-FLAG resulted in enhancement of TH promoter-driven luciferase gene expression (Fig. 6D) shown by significant interaction between Nurr1 and FGFR1(SP-/NLS) or FGF-223 in two-way ANOVA. In contrast, co-transfection of Nurr1 with low molecular weight FGF-218, which neither interacts nor activates the nuclear FGFR1 (15, 50), had no effect on Nurr1-dependent or -independent TH promoter activity.
To determine whether FGFR1 and Nurr1 can act cooperatively at the isolated NurRE or NBRE motifs, we examined transactivation of a minimal promoter construct containing either NurRE or NBRE upstream of the TATA box fused to the luciferase reporter gene. In each experiment, the promoter activity was normalized to the Nurr1 condition, as the Nurr1-dependent initialization of transcription was investigated (Fig. 6E). Transfection with FGFR1(SP−/NLS) or its dominant-negative FGFR1(SP−/NLS/TK−) mutant lacking the tyrosine kinase domain did not affect the luciferase reporter gene expression compared with the control transfection with β-galactosidase. Transfection of 10 or 100 ng of Nurr1-FLAG plasmid significantly increased (p < 0.001) luciferase expression compared with control transfection with β-galactosidase in a dose-dependent fashion. Co-transfection of FGFR1(SP-/NLS) with Nurr1-FLAG significantly potentiated the NurRE driven expression of the luciferase gene compared with the single transfection of Nurr1 (two-way ANOVA; p < 0.001). The FGFR1(SP-/NLS) potentiating effect was more pronounced with the low (10 ng) dose of Nurr1 DNA (4-fold potentiation) than with the 100 ng of Nurr1 DNA (2-fold potentiation). The co-transfection of dominant-negative FGFR1(TK–) with 100 ng of Nurr1-FLAG significantly diminished the Nurr1 stimulation of the NuRE as shown by significant (p < 0.001) interaction of FGFR1(TK-) and Nurr1-FLAG in two-way ANOVA. However, the FGFR1(TK−) inhibition of the NurRE by a lower dose of Nurr1 was less effective. The transcriptional activity of Nurr1 on the isolated NBRE target motif was also significantly enhanced upon the addition of FGFR1(SP-/NLS). In conclusion, we have shown cooperative activity of Nurr1 and FGFR1 in activation of the TH gene promoter as well as at typical Nurr1-binding sites.
DISCUSSION
Nurr1 and FGFR1 are central nuclear integrators of diverse developmental signals (4, 11, 13). Both proteins have been implicated in postmitotic development, including the maturation and maintenance of mDA neurons (3, 51, 52). Although FGFR1 is ubiquitously expressed within the developing and the adult CNS (13, 36), Nurr1 expression is restricted to specific areas. During development, the mDA neurons are the only dopaminergic subtype expressing Nurr1 within the CNS (53). In the present study, we showed an overlapping expression pattern of both proteins in postmitotic precursors and maturating TH-expressing neurons of the mDA area at embryonic day 14.5.
The nuclear localization of FGFR1 in postmitotic ventral midbrain neurons observed in this study is consistent with previous observations in postnatal rats showing that in dopamine neurons of the substantia nigra, nuclear FGFR1 co-localizes with its transcriptional partner CBP (14). Furthermore, we observed the in vivo cytoplasmic localization of FGFR1 in the subventricular zone and in vitro in the expanding primary ventral mesencephalic progenitor cells. These observations are consistent with previous reports in which the cytoplasmic presence of FGFR1 is obvious in proliferating neuronal progenitors, and nuclear FGFR1 accumulation drives neuronal differentiation (13, 40). Thus, our present observations support the divergent role of FGFR1 in mDA development: canonical transmembrane FGFR1 signaling is relevant for mitogenically active ventral mesencephalic progenitors, whereas the nuclear form is associated with mDA differentiation. Further, we demonstrate here for the first time that both receptors, Nurr1 and FGFR1, co-localize to the nuclei of neuronal cells from ventral mesencephalon. The presence of FGFR1 and Nurr1 in the same nuclear protein complexes was demonstrated by co-immunoprecipitation from the nuclei of immortalized rat mDA progenitors and from the mouse ventral midbrain nuclei.
We have verified the Nurr1-FGFR1 nuclear interactions using transfected recombinant proteins in human neuroblastoma cells and showed that in live cells, the mobility of nuclear FGFR1-EGFP is reduced by Nurr1. Our earlier studies demonstrated that the slow, hypokinetic, nuclear FGFR1-EGFP population represents chromatin-bound FGFR1, which is expanded during FGFR1-dependent transcription activation and eliminated by transcription inhibitors (15). In contrast, the fast, hyperkinetic FGFR1 represents freely diffusing FGFR1 and its stochastic nonproductive collisions (15). In the present study, the Nurr1-induced conversion of the fast FGFR1 population to a slow hypokinetic pool suggests that the FGFR1 and Nurr1 are co-engaged in chromatin binding and gene regulation.
Both FGFR1 and Nurr1 are thought to control cell development and function through transcriptional activation and gene programming. An exemplary gene is TH, activated both by Nurr1 (6) and by nuclear FGFR1 in cooperation with CBP (14, 20). Nurr1 was shown to bind to the NBRE in the proximal TH promoter and activate transcription (46, 54). FGFR1 was shown to transactivate the adjacent CREB/CBP-binding CRE site (16). Our ChIP experiment showed an in vivo interaction of Nurr1 and FGFR1 further upstream on the TH promoter, which contains a potential Nurr1-binding site homologous to a core sequence related to the NurRE. Furthermore, luciferase reporter assays displayed a cooperative function of Nurr1 and nuclear FGFR1 on TH promoter-dependent transcriptional activation. Enhancement of Nurr1-mediated TH promoter activation is also observed after co-transfection of FGF-223, which, in contrast to FGF-218, activates the endogenous nuclear FGFR1 (20). The role of endogenous FGFR1 was further indicated by the dose-dependent inhibition of Nurr1 activity with a dominant-negative FGFR1(TK−), lacking the tyrosine kinase domain. The inhibitory function of FGFR1(TK−) mainly relies on competitive binding of the mutant to the CBP/RSK complex, preventing the release of CBP from this inactive complex. Although this mechanism seems to be independent of tyrosine kinase activity, it requires the presence of the tyrosine kinase domain to disrupt RSK/CBP binding (14). Furthermore, although the TH gene promoter contains a number of potential FGFR1-responsive elements in addition to NBRE- and NurRE-related sites, we were able to demonstrate the cooperative action of Nurr1 and nuclear FGFR1 on isolated canonical Nurr1-binding motifs. This finding indicates that nuclear FGFR1 enhances both Nurr1 monomer (NBRE)- and dimer (NurRE)-dependent transcription.
The nuclear role of FGFR1 in postmitotic development of mDA neurons is consistent with our recent observation of increased FGFR1 nuclear accumulation in ventral midbrain of FGF-2 deficient embryos, which was correlated with increased generation of TH-positive neurons (55).6 The binding of Nurr1 to nuclear FGFR1 in FGF-2-deficient mice observed in the present study, suggests that an excessive Nurr1-INFS co-signaling may lead to neuronal hyperplasia and an excessive TH-positive neuron production. In addition to the previously described differential abilities of the FGF-2 isoforms to bind to the FGFR1 and to induce its activation (15), they transactivate differently Nurr1-dependent gene transcription, as shown in the present study.
The Nurr1-FGFR1 interaction appears to be indirect, as indicated by in vitro transcription/translational assay, and may be mediated by other bridging proteins. One protein that binds FGFR1 and may engage Nurr1 is CBP, because cAMP is an effective inducer of mDA differentiation (14, 56–58). Smidt and Burbach (4) have hypothesized that Nurr1 may integrate multiple cellular events via presumed multifaceted interactions with several nuclear partners. The interaction with FGFR1, a central factor in the integrative cellular signaling (INFS), offers an alternative or additional mechanism for the broad developmental functions of Nurr1. In support of this mechanism, the INFS was recently shown to participate in retinoid and Nur mediated developmental gene programming of embryonic stem cells (61).
In conclusion, the newly discovered FGFR1-Nurr1 partnership in developing postmitotic and mature mDA provides a novel integrative mechanism for Nurr1-dependent transcriptional regulation and provides one plausible mechanism for growth factor-dependent induction of the orphan nuclear receptor, despite its missing ligand binding capacity. Further comprehensive elucidation of the signal integration and transcriptional regulation by the FGFR1-Nurr1 complex and their roles in neuronal differentiation may offer new therapies for abnormal dopaminergic neuronal development or degeneration.
Acknowledgments
We thank Kerstin Kuhlemann, Maike Wesemann, Silke Fischer, Natascha Heidrich, and Hella Brinkmann for excellent technical assistance.
This work was supported by New York State Stem Cell Science (NYSTEM) Grant N09G-271 (to M. K. S.) and by a Deutsche Forschungsgemeinschaft grant (to C. G.).
- mDA
- mesencephalic dopaminergic neuron
- CBP
- CREB-binding protein
- CREB
- cAMP-responsive element-binding protein
- FGFR
- FGF receptor
- FRAP
- fluorescence recovery after photobleaching
- INFS
- integrative nuclear FGFR1 signaling
- ko
- knock out
- FGF-2
- fibroblast growth factor deficient
- NBRE
- Nerve Growth Factor Inducible Factor-B (NGFI-B) response element
- NLS
- nuclear localization sequence
- NurRE
- Nur-responsive element
- SV40i-VM-NPC
- SV40-immortalized ventral mesencephalic neuronal progenitor cell
- TH
- tyrosine hydroxylase
- En
- embryonic day n
- DIV
- day(s) in vitro
- ICQ
- intensity correlation quotient
- PDM
- product of the differences from the mean
- EGFP
- enhanced GFP
- ANOVA
- analysis of variance
- IP
- immunoprecipitation
- NBS
- Nur-binding site
- VM
- ventral midbrain.
REFERENCES
- 1. Prakash N., Wurst W. (2006) Development of dopaminergic neurons in the mammalian brain. Cell Mol. Life Sci. 63, 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nakamura S., Ito Y., Shirasaki R., Murakami F. (2000) Local directional cues control growth polarity of dopaminergic axons along the rostrocaudal axis. J. Neurosci. 20, 4112–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klejbor I., Myers J. M., Hausknecht K., Corso T. D., Gambino A. S., Morys J., Maher P. A., Hard R., Richards J., Stachowiak E. K., Stachowiak M. K. (2006) Fibroblast growth factor receptor signaling affects development and function of dopamine neurons. Inhibition results in a schizophrenia-like syndrome in transgenic mice. J. Neurochem. 97, 1243–1258 [DOI] [PubMed] [Google Scholar]
- 4. Smidt M. P., Burbach J. P. (2009) Terminal differentiation ofmesodiencephalic dopaminergic neurons. The role of Nurr1 and Pitx3. Adv. Exp. Med. Biol. 651, 47–57 [PubMed] [Google Scholar]
- 5. Law S. W., Conneely O. M., DeMayo F. J., O'Malley B. W. (1992) Identification of a new brain-specific transcription factor, NURR1. Mol. Endocrinol. 6, 2129–2135 [DOI] [PubMed] [Google Scholar]
- 6. Wallén A., Perlmann T. (2003) Transcriptional control of dopamine neuron development. Ann. N.Y. Acad. Sci. 991, 48–60 [DOI] [PubMed] [Google Scholar]
- 7. Perlmann T., Wallén-Mackenzie A. (2004) Nurr1, an orphan nuclear receptor with essential functions in developing dopamine cells. Cell Tissue Res. 318, 45–52 [DOI] [PubMed] [Google Scholar]
- 8. Jacobs F. M., van der Linden A. J., Wang Y., von Oerthel L., Sul H. S., Burbach J. P., Smidt M. P. (2009) Identification of Dlk1, Ptpru and Klhl1 as novel Nurr1 target genes in meso-diencephalic dopamine neurons. Development 136, 2363–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zetterström R. H., Solomin L., Jansson L., Hoffer B. J., Olson L., Perlmann T. (1997) Dopamine neuron agenesis in Nurr1-deficient mice. Science 276, 248–250 [DOI] [PubMed] [Google Scholar]
- 10. Itoh N., Ornitz D. M. (2011) Fibroblast growth factors. From molecular evolution to roles in development, metabolism and disease. J. Biochem. 149, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stachowiak M. K., Maher P. A., Stachowiak E. K. (2007) Integrative nuclear signaling in cell development: A role for FGF receptor-1. DNA Cell Biol. 26, 811–826 [DOI] [PubMed] [Google Scholar]
- 12. Stachowiak M. K., Maher P. A., Joy A., Mordechai E., Stachowiak E. K. (1996) Nuclear accumulation of fibroblast growth factor receptors is regulated by multiple signals in adrenal medullary cells. Mol. Biol. Cell 7, 1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stachowiak M. K., Stachowiak E. K., Aletta J. M., Tzanakakis E. S. (2011) A common integrative nuclear signaling module for stem cell development, in Stem Cells: From Mechanisms to Technologies (Stachowiak M. K., Tzanakakis M. eds.), pp. 87–132, World Scientific Publishing Co. Inc., Hackensack, NJ [Google Scholar]
- 14. Fang X., Stachowiak E. K., Dunham-Ems S. M., Klejbor I., Stachowiak M. K. (2005) Control of CREB-binding protein signaling by nuclear fibroblast growth factor receptor-1. A novel mechanism of gene regulation. J. Biol. Chem. 280, 28451–28462 [DOI] [PubMed] [Google Scholar]
- 15. Dunham-Ems S. M., Lee Y. W., Stachowiak E. K., Pudavar H., Claus P., Prasad P. N., Stachowiak M. K. (2009) Fibroblast growth factor receptor-1 (FGFR1) nuclear dynamics reveal a novel mechanism in transcription control. Mol. Biol. Cell 20, 2401–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peng H., Myers J., Fang X., Stachowiak E. K., Maher P. A., Martins G. G., Popescu G., Berezney R., Stachowiak M. K. (2002) Integrative nuclear FGFR1 signaling (INFS) pathway mediates activation of the tyrosine hydroxylase gene by angiotensin II, depolarization and protein kinase C. J. Neurochem. 81, 506–524 [DOI] [PubMed] [Google Scholar]
- 17. Michelhaugh S. K., Vaitkevicius H., Wang J., Bouhamdan M., Krieg A. R., Walker J. L., Mendiratta V., Bannon M. J. (2005) Dopamine neurons express multiple isoforms of the nuclear receptor nurr1 with diminished transcriptional activity. J. Neurochem. 95, 1342–1350 [DOI] [PubMed] [Google Scholar]
- 18. Bruns A. F., van Bergeijk J., Lorbeer C., Nölle A., Jungnickel J., Grothe C., Claus P. (2009) Fibroblast growth factor-2 regulates the stability of nuclear bodies. Proc. Natl. Acad. Sci. U.S.A. 106, 12747–12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stachowiak E. K., Maher P. A., Tucholski J., Mordechai E., Joy A., Moffett J., Coons S., Stachowiak M. K. (1997) Nuclear accumulation of fibroblast growth factor receptors in human glial cells: Association with cell proliferation. Oncogene 14, 2201–2211 [DOI] [PubMed] [Google Scholar]
- 20. Peng H., Moffett J., Myers J., Fang X., Stachowiak E. K., Maher P., Kratz E., Hines J., Fluharty S. J., Mizukoshi E., Bloom D. C., Stachowiak M. K. (2001) Novel nuclear signaling pathway mediates activation of fibroblast growth factor-2 gene by type 1 and type 2 angiotensin II receptors. Mol. Biol. Cell 12, 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Niwa H., Yamamura K., Miyazaki J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 22. Ratzka A., Kalve I., Ozer M., Nobre A., Wesemann M., Jungnickel J., Koster-Patzlaff C., Baron O., Grothe C. (2012) The co-layer method as an efficient way to genetically modify mesencephalic progenitor cells transplanted into 6-OHDA rat model of Parkinson's disease. Cell Transplant 21, 749–762 [DOI] [PubMed] [Google Scholar]
- 23. Kim E. L., Esparza F. M., Stachowiak M. K. (1996) The roles of CRE, TRE, and TRE-adjacent S1 nuclease sensitive element in the regulation of tyrosine hydroxylase gene promoter activity by angiotensin II. J. Neurochem. 67, 26–36 [DOI] [PubMed] [Google Scholar]
- 24. Maira M., Martens C., Batsché E., Gauthier Y., Drouin J. (2003) Dimer-specific potentiation of NGFI-B (Nur77) transcriptional activity by the protein kinase A pathway and AF-1-dependent coactivator recruitment. Mol. Cell. Biol. 23, 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maira M., Martens C., Philips A., Drouin J. (1999) Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol. Cell. Biol. 19, 7549–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dono R., Texido G., Dussel R., Ehmke H., Zeller R. (1998) Impaired cerebral cortex development and blood pressure regulation in FGF-2-deficient mice. EMBO J. 17, 4213–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ratzka A., Baron O., Grothe C. (2011) FGF-2 deficiency does not influence FGF ligand and receptor expression during development of the nigrostriatal system. PLoS One 6, e23564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee J. H., Kim K. T. (2004) Induction of cyclin-dependent kinase 5 and its activator p35 through the extracellular-signal-regulated kinase and protein kinase A pathways during retinoic-acid mediated neuronal differentiation in human neuroblastoma SK-N-BE(2)C cells. J. Neurochem. 91, 634–647 [DOI] [PubMed] [Google Scholar]
- 29. Timmer M., Grosskreutz J., Schlesinger F., Krampfl K., Wesemann M., Just L., Bufler J., Grothe C. (2006) Dopaminergic properties and function after grafting of attached neural precursor cultures. Neurobiol. Dis. 21, 587–606 [DOI] [PubMed] [Google Scholar]
- 30. Nobre A., Kalve I., Cesnulevicius K., Ragancokova D., Ratzka A., Halfer N., Wesemann M., Krampfl K., Claus P., Grothe C. (2010) Characterization and differentiation potential of rat ventral mesencephalic neuronal progenitor cells immortalized with SV40 large T antigen. Cell Tissue Res. 340, 29–43 [DOI] [PubMed] [Google Scholar]
- 31. Li Q., Lau A., Morris T. J., Guo L., Fordyce C. B., Stanley E. F. (2004) A syntaxin 1, Gαo, and N-type calcium channel complex at a presynaptic nerve terminal. Analysis by quantitative immunocolocalization. J. Neurosci. 24, 4070–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Farías G. G., Vallés A. S., Colombres M., Godoy J. A., Toledo E. M., Lukas R. J., Barrantes F. J., Inestrosa N. C. (2007) Wnt-7a induces presynaptic colocalization of α 7-nicotinic acetylcholine receptors and adenomatous polyposis coli in hippocampal neurons. J. Neurosci. 27, 5313–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Claus P., Bruns A. F., Grothe C. (2004) Fibroblast growth factor-223 binds directly to the survival of motoneuron protein and is associated with small nuclear RNAs. Biochem. J. 384, 559–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heuer J. G., von Bartheld C. S., Kinoshita Y., Evers P. C., Bothwell M. (1990) Alternating phases of FGF receptor and NGF receptor expression in the developing chicken nervous system. Neuron 5, 283–296 [DOI] [PubMed] [Google Scholar]
- 35. Ota S., Tonou-Fujimori N., Tonou-Fujimori N., Nakayama Y., Ito Y., Kawamura A., Yamasu K. (2010) FGF receptor gene expression and its regulation by FGF signaling during early zebrafish development. Genesis 48, 707–716 [DOI] [PubMed] [Google Scholar]
- 36. Ozawa K., Uruno T., Miyakawa K., Seo M., Imamura T. (1996) Expression of the fibroblast growth factor family and their receptor family genes during mouse brain development. Mol. Brain Res. 41, 279–288 [DOI] [PubMed] [Google Scholar]
- 37. Wanaka A., Johnson E. M., Jr., Milbrandt J. (1990) Localization of FGF receptor mRNA in the adult rat central nervous system by in situ hybridization. Neuron 5, 267–281 [DOI] [PubMed] [Google Scholar]
- 38. Wanaka A., Milbrandt J., Johnson E. M., Jr. (1991) Expression of FGF receptor gene in rat development. Development 111, 455–468 [DOI] [PubMed] [Google Scholar]
- 39. Friling S., Andersson E., Thompson L. H., Jönsson M. E., Hebsgaard J. B., Nanou E., Alekseenko Z., Marklund U., Kjellander S., Volakakis N., Hovatta O., El Manira A., Björklund A., Perlmann T., Ericson J. (2009) Efficient production of mesencephalic dopamine neurons by Lmx1a expression in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 106, 7613–7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stachowiak M. K., Fang X., Myers J. M., Dunham S. M., Berezney R., Maher P. A., Stachowiak E. K. (2003) Integrative nuclear FGFR1 signaling (INFS) as a part of a universal “feed-forward-and-gate” signaling module that controls cell growth and differentiation. J. Cell. Biochem. 90, 662–691 [DOI] [PubMed] [Google Scholar]
- 41. Reid H. H., Wilks A. F., Bernard O. (1990) Two forms of the basic fibroblast growth factor receptor-like mRNA are expressed in the developing mouse brain. Proc. Natl. Acad. Sci. U.S.A. 87, 1596–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abemayor E., Sidell N. (1989) Human neuroblastoma cell lines as models for the in vitro study of neoplastic and neuronal cell differentiation. Environ. Health Perspect. 80, 3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ghigo D., Priotto C., Migliorino D., Geromin D., Franchino C., Todde R., Costamagna C., Pescarmona G., Bosia A. (1998) Retinoic acid-induced differentiation in a human neuroblastoma cell line is associated with an increase in nitric oxide synthesis. J. Cell. Physiol. 174, 99–106 [DOI] [PubMed] [Google Scholar]
- 44. Silvagno F., Guarnieri V., Capizzi A., Pescarmona G. P. (2002) Synergistic effect of retinoic acid and dehydroepiandrosterone on differentiation of human neuroblastoma cells. FEBS Lett. 532, 153–158 [DOI] [PubMed] [Google Scholar]
- 45. Murphy E. P., Dobson A. D., Keller C., Conneely O. M. (1996) Differential regulation of transcription by the NURR1/NUR77 subfamily of nuclear transcription factors. Gene Expr. 5, 169–179 [PMC free article] [PubMed] [Google Scholar]
- 46. Kim K. S., Kim C. H., Hwang D. Y., Seo H., Chung S., Hong S. J., Lim J. K., Anderson T., Isacson O. (2003) Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. J. Neurochem. 85, 622–634 [DOI] [PubMed] [Google Scholar]
- 47. Saucedo-Cardenas O., Conneely O. M. (1996) Comparative distribution of NURR1 and NUR77 nuclear receptors in the mouse central nervous system. J. Mol. Neurosci. 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 48. Romano G., Macaluso M., Lucchetti C., Iacovitti L. (2007) Transcription and epigenetic profile of the promoter, first exon and first intron of the human tyrosine hydroxylase gene. J. Cell. Physiol. 211, 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kelly B. B., Hedlund E., Kim C., Ishiguro H., Isacson O., Chikaraishi D. M., Kim K. S., Feng G. (2006) A tyrosine hydroxylase-yellow fluorescent protein knock-in reporter system labeling dopaminergic neurons reveals potential regulatory role for the first intron of the rodent tyrosine hydroxylase gene. Neuroscience 142, 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Claus P., Doring F., Gringel S., Muller-Ostermeyer F., Fuhlrott J., Kraft T., Grothe C. (2003) Differential intranuclear localization of fibroblast growth factor-2 isoforms and specific interaction with the survival of motoneuron protein. J. Biol. Chem. 278, 479–485 [DOI] [PubMed] [Google Scholar]
- 51. Calò L., Spillantini M., Nicoletti F., Allen N. D. (2005) Nurr1 co-localizes with EphB1 receptors in the developing ventral midbrain, and its expression is enhanced by the EphB1 ligand, ephrinB2. J. Neurochem. 92, 235–245 [DOI] [PubMed] [Google Scholar]
- 52. Kadkhodaei B., Ito T., Joodmardi E., Mattsson B., Rouillard C., Carta M., Muramatsu S., Sumi-Ichinose C., Nomura T., Metzger D., Chambon P., Lindqvist E., Larsson N. G., Olson L., Björklund A., Ichinose H., Perlmann T. (2009) Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J. Neurosci. 29, 15923–15932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bäckman C., Perlmann T., Wallén A., Hoffer B. J., Morales M. (1999) A selective group of dopaminergic neurons express Nurr1 in the adult mouse brain. Brain Res. 851, 125–132 [DOI] [PubMed] [Google Scholar]
- 54. Iwawaki T., Kohno K., Kobayashi K. (2000) Identification of a potential nurr1 response element that activates the tyrosine hydroxylase gene promoter in cultured cells. Biochem. Biophys. Res. Commun. 274, 590–595 [DOI] [PubMed] [Google Scholar]
- 55. Timmer M., Cesnulevicius K., Winkler C., Kolb J., Lipokatic-Takacs E., Jungnickel J., Grothe C. (2007) Fibroblast growth factor (FGF)-2 and FGF receptor 3 are required for the development of the substantia nigra, and FGF-2 plays a crucial role for the rescue of dopaminergic neurons after 6-hydroxydopamine lesion. J. Neurosci. 27, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stachowiak E. K., Fang X., Myers J., Dunham S., Stachowiak M. K. (2003) cAMP-induced differentiation of human neuronal progenitor cells is mediated by nuclear fibroblast growth factor receptor-1 (FGFR1). J. Neurochem. 84, 1296–1312 [DOI] [PubMed] [Google Scholar]
- 57. Tremblay R. G., Sikorska M., Sandhu J. K., Lanthier P., Ribecco-Lutkiewicz M., Bani-Yaghoub M. (2010) Differentiation of mouse Neuro 2A cells into dopamine neurons. J. Neurosci. Methods 186, 60–67 [DOI] [PubMed] [Google Scholar]
- 58. Malmersjö S., Liste I., Dyachok O., Tengholm A., Arenas E., Uhlén P. (2010) Ca2+ and cAMP signaling in human embryonic stem cell-derived dopamine neurons. Stem Cells Dev. 19, 1355–1364 [DOI] [PubMed] [Google Scholar]
- 59. Galleguillos D., Vecchiola A., Fuentealba J. A., Ojeda V., Alvarez K., Gómez A., Andrés M. E. (2004) PIASγ represses the transcriptional activation induced by the nuclear receptor Nurr1. J. Biol. Chem. 279, 2005–2011 [DOI] [PubMed] [Google Scholar]
- 60. Ratzka A., Baron O., Stachowiak M. K., Grothe C. (2012) Fibroblast growth factor 2 regulates dopaminergic neuron development in vivo. J. Neurochem. doi: 10.1111/j.1471-4159.2012.07768.x [DOI] [PubMed] [Google Scholar]
- 61. Lee Y. W., Terranova C., Birkaya B., Narla S., Kehoe D., Parikh A., Dong S., Ratzka A., Brinkmann H., Aletta J. M., Tzanakakis E. S., Stachowiak E. K., Claus P., Stachowiak M. K. (2012) A novel nuclear FGF Receptor-1 partnership with retinoid and Nur receptors during developmental gene programming of embryonic stem cells. J. Cell. Biochem. doi: 10.1002/jcb.24170 [DOI] [PubMed] [Google Scholar]