Background: GRASP proteins contain PDZ domains that mediate membrane tethering and are inhibited by mitotic phosphorylation.
Results: The crystal structure of a GRASP phosphomimic shows a propagation of conformational change from the phosphorylation site that shifts the internal PDZ ligand.
Conclusion: Mitotic phosphorylation alters the PDZ ligand to block membrane tethering.
Significance: The first structural mechanism of mitotic phosphoinhibition of membrane tethering is presented.
Keywords: Membrane Function, Membrane Fusion, Membrane Proteins, Phosphorylation, Structural Biology
Abstract
Mitotic phosphorylation of the conserved GRASP domain of GRASP65 disrupts its self-association, leading to a loss of Golgi membrane tethering, cisternal unlinking, and Golgi breakdown. Recently, the structural basis of the GRASP self-interaction was determined, yet the mechanism by which phosphorylation disrupts this activity is unknown. Here, we present the crystal structure of a GRASP phosphomimic containing an aspartic acid substitution for a serine residue (Ser-189) that in GRASP65 is phosphorylated by PLK1, causing a block in membrane tethering and Golgi ribbon formation. The structure revealed a conformational change in the GRASP internal ligand that prevented its insertion into the PDZ binding pocket, and gel filtration assays showed that this phosphomimic mutant exhibited a significant reduction in dimer formation. Interestingly, the structure also revealed an apparent propagation of conformational change from the site of phosphorylation to the shifted ligand, and alanine substitution of two residues (Glu-145 and Ser-146) at penultimate positions in this chain rescued dimer formation by the phosphomimic. These data reveal the structural basis of the phosphoinhibition of GRASP-mediated membrane tethering and provide a mechanism for its allosteric regulation.
Introduction
The mammalian Golgi apparatus consists of numerous stacked membranes, or cisternae, with analogous cisternae among the stacks interconnected to form a compartmentalized ribbon-like membrane network. The linkage of analogous cisternae is regulated, in part, by GRASP65 and GRASP55, which are localized to cis and medial Golgi cisternae, respectively (1, 2). The GRASP proteins share a highly conserved N-terminal GRASP domain capable of homodimerization to promote homotypic tethering (3, 4). The GRASP domain is composed of two circularly permuted PDZ domains, PDZ1 and PDZ2 (5). Tethering occurs when a GRASP molecule on one membrane presents the binding pocket of its PDZ1 to a surface-exposed internal ligand on PDZ2 of a second GRASP molecule on an adjacent membrane (5). Membrane anchoring of the GRASP domain appears to restrict its orientation to favor this trans interaction (6), and it is possible that GRASP molecules interdigitate as they bridge analogous cisternae to sustain the Golgi ribbon.
In preparation for cell division, the linkages that connect the Golgi stacks into a ribbon are disrupted, and this aspect of mitotically induced Golgi breakdown results from phosphoinhibition of the GRASP protein interaction (7). Phosphoinhibition appears to require two steps. GRASP65 and GRASP55 have sites outside the GRASP domain that are mitotically phosphorylated by CDK1 or ERK1 (8, 9). Evidence suggests that these sites recruit and activate PLK1 (10) such that PLK1 phosphorylates a site present in the GRASP domain (11). In the case of GRASP65, it is known that a phosphomimic mutation in the site phosphorylated by PLK1 (S189D) blocks GRASP dimer formation, tethering activity, and Golgi ribbon formation (11). Ser-189 is located in close proximity to the GRASP internal ligand, suggesting that its phosphorylation prevents the ligand from being able to bind the PDZ1 pocket. Nevertheless, the GRASP domain structure reveals that Ser-189 is separated from the ligand at residues 196–199 by a β-strand and a distance of about 24 Å, raising the fundamental question of how phosphorylation at this site regulates the activity of the internal PDZ ligand.
To explore the potential structural effects of GRASP phosphorylation on the internal ligand, we solved the crystal structure of the GRASP domain phosphomimic (S189D). Significantly, the internal ligand of the phosphomimic displayed a conformational change and impaired self-association. The conformational change appeared to result not from propagation along the β-strand but rather through shifts in an α-helix running parallel to the β-strand, resulting in the rotation of two residues (Glu-145 and Ser-146) adjacent to the internal ligand. Mutation of these residues to alanines suppressed the ability of the phosphomimic change to regulate GRASP self-interaction, thereby providing experimental support for this allosteric mechanism of regulating an internal ligand binding to its PDZ-binding domain.
MATERIALS AND METHODS
Constructs
To generate His-tagged GRASP55, GRASP55 was inserted into pRSETB (Invitrogen) using the EcoRI site. Point mutations were introduced using the QuikChange protocol (Stratagene). Two sequential rounds of mutagenesis were used for double point mutations. Adding single point mutations with each round of mutagenesis made additional point mutants. For His-TEV-GRASP55(1–208), the tobacco etch virus recognition sequence ENLYFQG was inserted upstream of the start codon in His-GRASP55, and a stop codon was introduced after residue 208.
Protein Expression, Purification, Crystallization, and Structure Determination
Purified GRASP55 residues 1–208 containing the S189D mutation were purified as described previously for the wild type (5). The purified protein was crystallized by vapor diffusion using the sitting drop method. The sample was mixed with an equal volume of mother liquor containing 0.2 m ammonium acetate, 0.1 m Tris (pH 9.0), 0.1 m imidazole, 30% PEG 4000, 10% PEG 20000, and 10% glycerol. Crystals appeared in ∼2–3 days and reached full size in 1–2 weeks. The crystals were frozen in liquid nitrogen before mounting in a nitrogen stream at 100 K. Data were collected at the University of Pittsburgh School of Medicine Crystallography Facility using a Saturn 944 CCD detector (Rigaku) and processed with HKL2000 (12). The structure of the S189D phosphomimic was determined by molecular replacement using PHASER (13) with the wild-type GRASP55(1–208) structure as the input model (5). Model building was performed using Coot (14), and refinement was performed using REFMAC as implemented with CCP4i (14–16). Crystallographic and refinement statistics are given in Table 1.
TABLE 1.
Crystallographic data and refinement statistics
Values in parentheses correspond to the outer resolution shell. r.m.s.d., root mean square deviation.
| GRASP55(1–208) S189D | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Unit cell dimensions (Å) | a = 64.9, b = 82.5, c = 84.0 |
| Resolution (Å) | 58.9–1.90 |
| Outer shell (Å) | 1.95–1.90 |
| No. of reflections | |
| Unique | 33.386 |
| Total | 189,778 |
| Mean I/σ(I) | 22.2 (2.73) |
| Completeness (%) | 97.0 (95.4) |
| Rsym (%)a | 8.7 (64.6) |
| Refinement | |
| R factor/Rfreeb,c | 20.9/27.1 |
| Non-hydrogen atoms | |
| Total | 3487 |
| Solvent | 238 |
| r.m.s.d. from ideal geometry | |
| Bond lengths (Å) | 0.021 |
| Bond angles | 1.93° |
| Average isotropic B values (Å2) | 37.7 |
| Ramachandran plot (%) | |
| Most favorable region | 96.0 |
| Additionally allowed region | 4.0 |
| Generously allowed region | 0.0 |
| Disallowed region | 0.0 |
a Rsym = (|(ΣI − 〈I〉)|)/(ΣI), where 〈I〉 is the average intensity of multiple measurements.
b R factor = Σhkl‖Fobs(hkl)| − |Fcalc(hkl)‖/Σhkl|Fobs(hkl)|.
c Rfree is the cross-validation of the R factor for 5% of reflections against which the model was not refined.
Dimerization Assay
Purified GRASP55 protein was fractionated by gel filtration on an FPLC system equipped with a Superdex G-75 column to yield monomeric and dimeric peaks. The monomeric peak was isolated, concentrated to 10 mg/ml, and incubated for a minimum of 24 h in running buffer (12.5 mm HEPES (pH 7.4), 60 mm KCl, and 7 mm β-mercaptoethanol) before loading 50 μg of sample at 1 mg/ml onto the column for a second fractionation. Dimer formation was quantified for each construct by printing the UV chromatogram output on paper, cutting out the area encompassing the dimer peak, and weighing the piece on a scale.
Membrane Tethering Assay
Transfection was by jetPEI (VWR International) following the manufacturer's protocol on a coverslip on a 24-well plate. Cells were treated with brefeldin A (10 μg/ml) for 30 min prior to fixation in 3% paraformaldehyde. Analysis was restricted to cells expressing moderate levels. Quantification of clustering was performed on average value projections of confocal image stacks using the ImageJ radial profile plugin as described (4). Grayscale thresholds were set for each experiment, and the centroid of the fluorescence for each cell was determined using the “measure” function. The normalized fluorescence intensity in each concentric circle drawn from the determined centroid was obtained using the radial profile plug-in.
RESULTS
GRASP Phosphorylation Induces Conformational Change in Internal PDZ Ligand
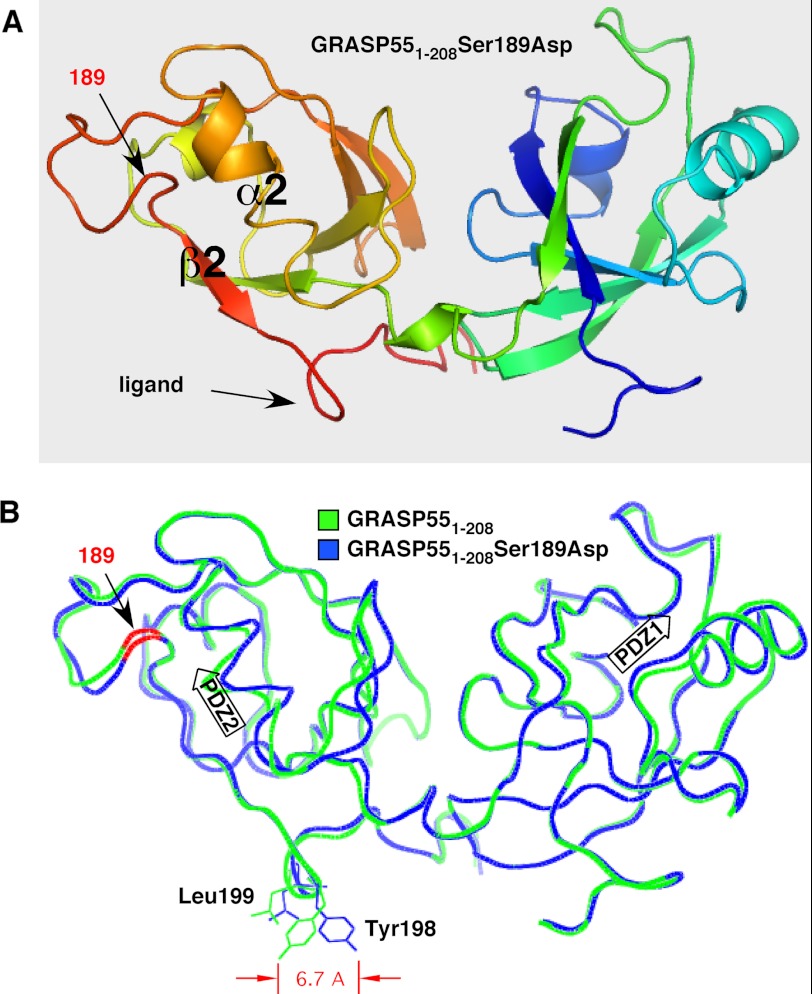
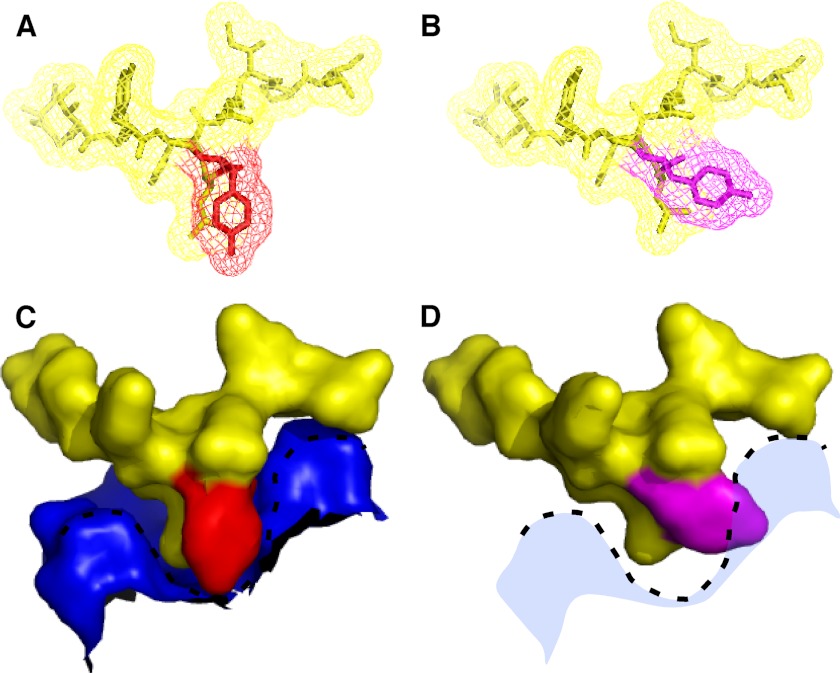
Mitotic phosphorylation blocks the self-interaction of GRASP proteins, and a site phosphorylated by PLK1 has been identified in the GRASP65 GRASP domain for which mutation to aspartic acid blocks tethering and alanine substitution prevents mitotic Golgi unlinking (11). Our recent structural analyses of the GRASP domain of GRASP55 revealed that the GRASP self-interaction is mediated through an interaction between a PDZ1 binding pocket and a prominent tyrosine-containing surface projection of PDZ2 that lies within the vicinity of the phosphorylation site (Ser-189). Thus, we wondered whether phosphorylation might induce a conformational change in the ligand causing phosphoinhibition. To test this idea, we solved the crystal structure of GRASP55(1–208) containing the S189D substitution to 1.9 Å resolution and aligned its backbone with the corresponding wild-type structure (Fig. 1, A and B). Although no gross conformational changes resulted from the phosphomimic mutation, a striking shift was observed at Tyr-198 in addition to a slight shift of Leu-199 in the opposite direction. These two residues principally compose the surface projection of the PDZ ligand. Consequently, the relatively narrow profile of the ligand in the wild-type structure became significantly enlarged in the phosphomimic. Modeling the ligand in its PDZ1 binding pocket clearly showed that this change caused steric constraints that would disfavor binding (Fig. 2, A–D). Thus, these observations suggest that the mechanism of phosphoinhibition may be attributed to a phosphorylation-induced alteration in the ligand configuration that prevents normal interaction of apposing GRASP molecules.
FIGURE 1.
Structure of GRASP domain phosphomimic S189D and comparison with non-phosphorylated GRASP domain. A, schematic structure of GRASP55(1–208) containing the S189D substitution showing the two PDZ domains PDZ1 (right) and PDZ2 (left), with key features of the latter highlighted. These are the α2-helix and β2-strand that define the pocket, the surface feature corresponding to the internal PDZ ligand, and the position of the aspartic acid that was substituted for serine. Coloring reflects N-terminal (blue) to C-terminal (red) progression. B, comparison of GRASP55(1–208) S189D (blue) and GRASP55(1–208) (green) shows high overall similarity with one another and a 6.7-Å displacement of Tyr-198. Together with Leu-199 (also shifted), Tyr-198 composes the surface protuberance of the internal PDZ ligand. The positions of the PDZ1 and PDZ2 binding pockets are indicated by arrows, and the location of residue 189 on the backbone is colored red.
FIGURE 2.
Conformational change of GRASP internal PDZ ligand in phosphomimic. A, view of the wild-type internal ligand in stick and mesh representation, with Tyr-198 shown in red. B, same view for the phosphomimic of the internal ligand, with Tyr-198 shown in purple. C, surface representation of the PDZ1 pocket surface (blue) containing the internal ligand (yellow), with Tyr-198 shown in red. D, same view for the phosphomimic showing the clash of the internal ligand with the surface of the PDZ pocket.
Allosteric Regulation of Internal Ligand
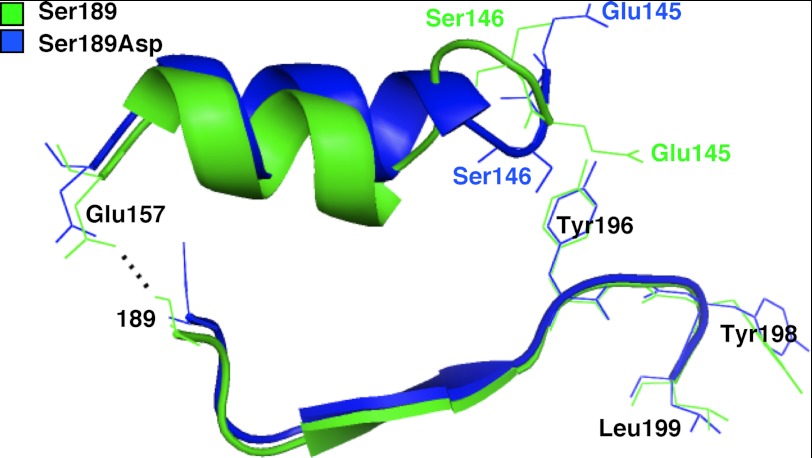
To determine how the conformation of the internal ligand might be affected by phosphorylation at an upstream serine residue, we analyzed the wild-type and phosphomimic structures for structural differences that may account for the conformational change. Surprisingly, we observed no significant differences along the β-strand that directly connects the phosphorylation site at position 189 to the surface projection of the internal ligand comprising residues 196–199 (Fig. 3). However, a polar contact formed in the wild-type structure between Ser-189 and Glu-157 at one end of the α-helix that lies parallel to the β-strand was broken by the S189D mutation in the phosphomimic (Fig. 3). This suggested that phosphorylation might induce perturbations along this α-helix, which projects toward the internal ligand. Examination of this region indeed revealed changes, as the backbones of the α-helices in the wild type versus the phosphomimic mutant became increasingly separated. This ultimately resulted in significant disruption of the loop containing Glu-145 and Ser-146, whose orientations were “flipped” in the phosphomimic (Fig. 3). Because these residues are located across from the internal ligand, this represented a potential mechanism by which phosphorylation at position 189 could allosterically regulate the conformation of the internal ligand by altering the interactions and/or repulsions of adjacent residues leading to the ligand.
FIGURE 3.
Chain of conformational change associated with ligand inhibition. Schematic views of the α2-helices and β2-strands in PDZ2 of GRASP55(1–208) S189D (blue) and GRASP55(1–208) (green) are aligned. The view shown highlights the bond disrupted by the S189D substitution (dotted black line), the shift in the α2-helix, and the reversals in the positions of Glu-145 and Ser-146 that correlate with the shift in position of the neighboring Tyr-198 in the internal ligand.
Dimerization and Membrane Tethering by GRASP Phosphomimic
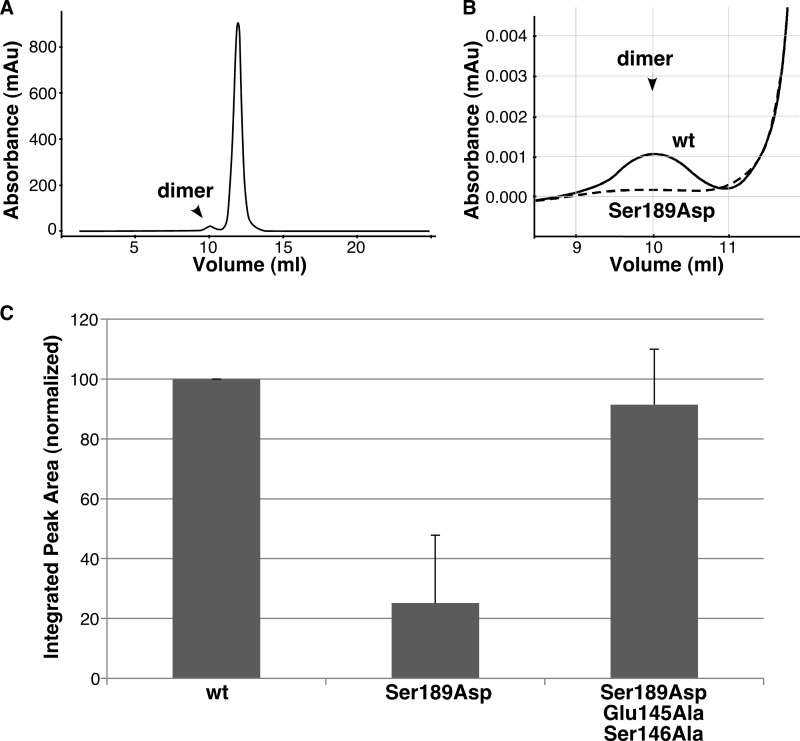
To test whether the two flipped residues (Glu-145 and Ser-146) were directly involved in the allosteric regulation of the internal ligand, we used two functional assays: GRASP dimer formation by gel filtration and GRASP-mediated membrane tethering of mitochondria. For the gel filtration assay, monomeric protein was incubated at 10 mg/ml and then refractionated. The GRASP domain of purified GRASP55 eluted largely as a single monomeric peak, with a small fraction forming a dimer (Fig. 4A). As expected, when the phosphomimic S189D was separated by gel filtration, its ability to form a dimer was significantly impaired (Fig. 4B), resulting in an ∼4-fold decrease relative to the wild type (Fig. 4C), consistent with an impaired ability of the displaced ligand to insert into the pocket. To determine what effect Glu-145 and Ser-146 had on dimer formation, we mutated Glu-145 and Ser-146 to alanines in the context of the phosphomimic (S189D/E145A/S146A) to nullify any aberrant interactions or repulsions caused by residue flipping and then tested whether we could restore dimer formation to wild-type levels. As shown (Fig. 4C), the alanine substitutions fully rescued the impaired dimer formation in the phosphomimic, suggesting that the conformation of the internal ligand is directly dependent on Glu-145 and Ser-146.
FIGURE 4.
Phosphomimic mutation disrupts dimerization by means of Glu-145 and Ser-146. A, Superdex G-75 elution profile of GRASP55(1–208), with the position of the minor dimer fraction indicated. mAu, milli-absorbance units. B, enlarged view of the dimer fraction for GRASP55(1–208) compared with that for GRASP55(1–208) S189D. C, quantified recovery of GRASP55(1–208), GRASP55(1–208) S189D, and GRASP55(1–208) S189D/E145A/S146A in the dimer fraction (mean ± S.D., n = 3). Note that the phosphomimic mutation blocked binding, but the block required the residues that sit between the phosphorylation site and the ligand and reverse position in the phosphomimic.
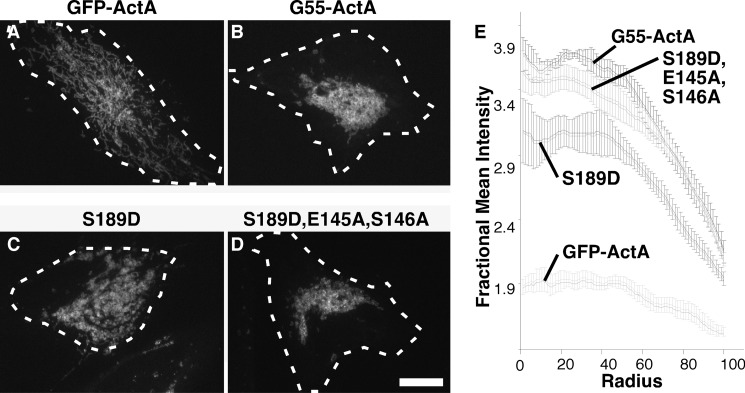
To assay membrane-tethering activity, constructs encoding full-length GRASP55 were modified to contain GFP and the C-terminal mitochondrial anchoring sequence from ActA (4). Whereas the control GFP-ActA construct yielded an extended lace-like mitochondrial network pattern (Fig. 5A), the mitochondrially anchored wild-type GRASP55-ActA construct converted this into a prominently juxtanuclear cluster (Fig. 5B). As confirmed by earlier work, GRASP-mediated clustering in this assay is due to its homotypic membrane-tethering activity (4). This tethering activity was diminished by introduction of the phosphomimic S189D substitution (Fig. 5C), and as in the dimerization assay, the inhibitory effect was nullified by introduction of the E145A and S146A substitutions, designed to disrupt the chain of conformational change from the phosphorylation site to the ligand (Fig. 5D). These effects were quantified using a radial profile plot, in which the fluorescence intensity starting from the fluorescence centroid is plotted as a function of radial distance (Fig. 5E). Peak height near the centroid indicates clustering of the mitochondrial membranes due to membrane tethering. Taken together, these two functional assays support the mechanism of phosphoinhibition revealed by the GRASP55 S189D structure.
FIGURE 5.
Phosphomimic mutation disrupts membrane tethering. A–D, HeLa cells transfected with the indicated GFP-tagged, mitochondrially targeted constructs were imaged. Thirty min prior to imaging, the cells were treated with brefeldin A to disperse Golgi membranes to ensure that clustering was independent of hypothetical interactions with a centrally positioned Golgi apparatus. Dashed white lines approximate the positions of the cell boundaries. Scale bar = 10 μm. E, the graphs record mitochondrial clustering induced by each construct as measured using the ImageJ radial profile plugin, in which the fluorescence intensity starting from the fluorescence centroid is plotted as a function of radial distance in the first 100 radials (mean ± S.E., n = 4, >15 cells/experiment).
DISCUSSION
The structural basis of GRASP domain membrane tethering has recently been described (5), but a structural understanding of its negative regulation by mitotic phosphorylation has not been obtained. Here, we have shown that phosphorylation alters the conformation of the internal ligand that binds to the pocket of the first PDZ domain and results in inhibition of the GRASP self-interaction. The conformational change is accompanied by disruption of residues along an α-helix connecting the ligand to the phosphorylation site, providing a structural mechanism underlying this allosteric regulation.
Although phosphoregulation of PDZ interactions is well documented, the available data indicate that the regulation involves either phosphorylation of a specific residue within the ligand or phosphorylation within the binding groove (17, 18) such that the introduced bulky phosphate presumably disrupts hydrogen bonding within the binding interface. This is a straightforward mechanism to explain a decreased PDZ interaction, although, in a few cases, an increase in binding is observed (19). Our study thus uncovers a unique allosteric phosphoinhibition of a PDZ-mediated interaction in which the introduction of a phosphate at a site spatially separated from the ligand initiates charge repulsion with an opposing glutamate residue that prevents a polar contact and physically displaces an associated α-helix. This chain of events eventually leads to disruption of upstream residues that are in close proximity to the ligand, which results in a conformational change that impedes the PDZ interaction.
The nature of the interaction between the surface projection of the GRASP internal ligand and the deep pocket of the PDZ1 interface suggests that precise orientation is critical for efficient binding of apposing GRASP molecules. Indeed, although the interaction involves multiple residues adjoining the surface projection, mutation of either the tyrosine or the leucine that together principally form the projection abolishes binding (11). Thus, even a minor conformational shift in the ligand would appear to be sufficient to regulate the interaction, and our results indicate that the ligand conformation is sensitive to perturbations in neighboring residues. Because PDZ domains exhibit relatively low binding affinities (20), this mechanism would provide a practical means of regulating the GRASP interaction during association and dissociation of GRASP binding partners. Moreover, regulation of the ligand conformation by phosphorylation at an alternative location avoids the need to access obscured binding sites buried within the deep depression of the PDZ1 pocket. It would be interesting to ascertain whether the topology of the ligand/groove-binding interface of a PDZ domain affects its mode of regulation, which perhaps could provide a structurally based method to analyze PDZ-mediated interactions.
In closing, this study provides a mechanistic description of the heretofore poorly understood process of phosphoinhibition of the GRASP self-association. We hope that the data not only shed light on the regulation of Golgi membrane tethering, but also help to elucidate the mechanisms that regulate PDZ-mediated function.
Acknowledgment
We are grateful to Debrup Sengupta for contributing expression constructs.
Footnotes
The work was supported, in whole or in part, by National Institutes of Health Grant GM095549 (to A. D. L. and M. R. M.).
REFERENCES
- 1. Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. (2006) GM130- and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 8, 238–248 [DOI] [PubMed] [Google Scholar]
- 2. Feinstein T. N., Linstedt A. D. (2008) GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell 19, 2696–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y., Satoh A., Warren G. (2005) Mapping the functional domains of the Golgi stacking factor GRASP65. J. Biol. Chem. 280, 4921–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sengupta D., Truschel S., Bachert C., Linstedt A. D. (2009) Organelle tethering by a homotypic PDZ interaction underlies formation of the Golgi membrane network. J. Cell Biol. 186, 41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Truschel S. T., Sengupta D., Foote A., Heroux A., Macbeth M. R., Linstedt A. D. (2011) Structure of the membrane-tethering GRASP domain reveals a unique PDZ ligand interaction that mediates Golgi biogenesis. J. Biol. Chem. 286, 20125–20129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bachert C., Linstedt A. D. (2010) Dual anchoring of the GRASP membrane tether promotes trans pairing. J. Biol. Chem. 285, 16294–16301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feinstein T. N., Linstedt A. D. (2007) Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol. Biol. Cell 18, 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y., Seemann J., Pypaert M., Shorter J., Warren G. (2003) A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 22, 3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jesch S. A., Lewis T. S., Ahn N. G., Linstedt A. D. (2001) Mitotic phosphorylation of Golgi reassembly stacking protein 55 by mitogen-activated protein kinase ERK2. Mol. Biol. Cell 12, 1811–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Preisinger C., Körner R., Wind M., Lehmann W. D., Kopajtich R., Barr F. A. (2005) Plk1 docking to GRASP65 phosphorylated by Cdk1 suggests a mechanism for Golgi checkpoint signaling. EMBO J. 24, 753–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sengupta D., Linstedt A. D. (2010) Mitotic inhibition of GRASP65 organelle tethering involves Polo-like kinase 1 (PLK1) phosphorylation proximate to an internal PDZ ligand. J. Biol. Chem. 285, 39994–40003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 13. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 14. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 15. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 16. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 17. Chung H. J., Huang Y. H., Lau L. F., Huganir R. L. (2004) Regulation of the NMDA receptor complex and trafficking by activity-dependent phosphorylation of the NR2B subunit PDZ ligand. J. Neurosci. 24, 10248–10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Voltz J. W., Brush M., Sikes S., Steplock D., Weinman E. J., Shenolikar S. (2007) Phosphorylation of PDZ1 domain attenuates NHERF-1 binding to cellular targets. J. Biol. Chem. 282, 33879–33887 [DOI] [PubMed] [Google Scholar]
- 19. Adey N. B., Huang L., Ormonde P. A., Baumgard M. L., Pero R., Byreddy D. V., Tavtigian S. V., Bartel P. L. (2000) Threonine phosphorylation of the MMAC1/PTEN PDZ-binding domain both inhibits and stimulates PDZ binding. Cancer Res. 60, 35–37 [PubMed] [Google Scholar]
- 20. Hung A. Y., Sheng M. (2002) PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 277, 5699–5702 [DOI] [PubMed] [Google Scholar]