Background: The function of SbdP, a cytoplasmic rhodanese from Aquifex aeolicus, is unknown.
Results: SbdP is involved in sulfur energy metabolism via its interaction with key redox enzymes.
Conclusion: SbdP supplies long sulfur chains to enzyme-active sites.
Significance: Rhodaneses are part of the substrate traffic in sulfur energy metabolism.
Keywords: Energy Metabolism, Membrane Enzymes, Protein Complexes, Protein-Protein Interactions, Sulfur, Aquifex aeolicus, Rhodanese/Sulfurtransferase, Hyperthermophiles, Sulfur Oxygenase Reductase, Sulfur Reductase
Abstract
How microorganisms obtain energy is a challenging topic, and there have been numerous studies on the mechanisms involved. Here, we focus on the energy substrate traffic in the hyperthermophilic bacterium Aquifex aeolicus. This bacterium can use insoluble sulfur as an energy substrate and has an intricate sulfur energy metabolism involving several sulfur-reducing and -oxidizing supercomplexes and enzymes. We demonstrate that the cytoplasmic rhodanese SbdP participates in this sulfur energy metabolism. Rhodaneses are a widespread family of proteins known to transfer sulfur atoms. We show that SbdP has also some unusual characteristics compared with other rhodaneses; it can load a long sulfur chain, and it can interact with more than one partner. Its partners (sulfur reductase and sulfur oxygenase reductase) are key enzymes of the sulfur energy metabolism of A. aeolicus and share the capacity to use long sulfur chains as substrate. We demonstrate a positive effect of SbdP, once loaded with sulfur chains, on sulfur reductase activity, most likely by optimizing substrate uptake. Taken together, these results lead us to propose a physiological role for SbdP as a carrier and sulfur chain donor to these key enzymes, therefore enabling channeling of sulfur substrate in the cell as well as greater efficiency of the sulfur energy metabolism of A. aeolicus.
Introduction
Sulfur is a ubiquitous element in the earth's crust, well known as an essential component of life. It has come even closer to the center of attention in the light of current views of the origin of life (1, 2). In nature, elemental sulfur (S0, a cyclic chain of sulfur) and sulfur compounds are found in particularly high concentrations in extreme environments, usually volcanic or geothermically active (deep-sea vents and hot springs) (3). In these environments, the reduction and oxidation of these compounds are vital processes for many microorganisms that use sulfur as a basis for their energy metabolism (4, 5). Despite its central importance, however, the metabolism of sulfur is still not fully understood, probably because of the high reactivity of its compounds.
Aquifex aeolicus, isolated from a hydrothermal system at Vulcano in Italy, is one of the most hyperthermophilic bacteria known, with an optimum growth temperature of 85 °C (6, 7). It also represents the earliest branching order in the bacterial domain (8). A. aeolicus is able to grow chemolithoautotrophically and obtains energy for growth in the presence of H2, O2, and CO2, but it also requires the presence of a sulfur compound such as S0, sodium sulfide (Na2S), or sodium thiosulfate (NaS2O3) (9). After several years of study of the enzymes involved in the respiratory pathways of A. aeolicus growing in presence of S0, we have a better overall picture of its complex energy metabolism. Under these growth conditions, we have demonstrated the existence of three different respiratory pathways as follows: two aerobic from H2 to O2 and from H2S to O2 and one anaerobic from H2 to S0. Two of them (H2/S0 and H2S/O2) have been shown to be organized into supramolecular structures (9–11) that contain enzymes known to be involved in sulfur energy metabolism as follows: a sulfur reductase (SR)4 in H2/S0 pathway and a sulfide quinone reductase (Sqr) in H2S/O2 pathway. A. aeolicus is the sole organism known to date that possesses simultaneously these two complementary energy pathways (both reduction and oxidation of sulfur compounds), and we have proposed an energy coupling between these two pathways (11). A cytoplasmic sulfur oxygenase reductase (SOR), which catalyzes, in the presence of oxygen, the simultaneous oxidation and reduction of elemental sulfur into oxygen sulfur species and H2S, is also present in A. aeolicus (12). In addition, the presence of cytoplasmic sulfur globules of as yet unknown function has been observed in whole cells of A. aeolicus grown on S0 (9). The simultaneous presence of all these sulfur enzymes, sulfur compounds and sulfur-containing structures, is puzzling, and the interconnection between all these sulfur enzymes and pathways is our current main focus.
The use of S0 in energy metabolism has also been described in the archaeon Acidianus ambivalens. A model of its intricate sulfur energy metabolism has been proposed involving several sulfur oxidation or reduction enzymes, but the interconnection between all of the produced and consumed sulfur compounds, even though suggested, has never been demonstrated (13, 14). Another bacterium, Wolinella succinogenes, can reduce polysulfide (Sn2−), a soluble form of S0, with a membrane polysulfide reductase (Psr) enzyme but seems to have a simpler sulfur energy metabolism in comparison with A. aeolicus and A. ambivalens (15, 16). In W. succinogenes sulfur metabolism, membrane extract studies have suggested that a soluble periplasmic rhodanese-like protein (Sud) could act as a sulfur donor to the terminal electron acceptor, the periplasmic oriented Psr, revealing a potential protein involved in energy sulfur traffic (17).
Rhodaneses (PF00581) belong to the sulfurtransferase family of enzymes that are found in organisms from all three domains of life. They catalyze in vitro the transfer of a sulfur atom from thiosulfate (sulfur donor) to cyanide (sulfur acceptor) (18, 19). Rhodanese-like proteins share a common characteristic “rhodanese-fold” and catalyze a similar reaction using other sulfur donors (like polysulfide for Sud) to transfer sulfur to nucleophilic sulfur acceptors. Rhodanese and/or rhodanese-like proteins are known to play a critical role in sulfur traffic by delivering sulfur in a “safe” chemical species to biosynthetic pathways, using their labile persulfide group. Despite numerous studies, the physiological role of rhodaneses as well as their physiological substrates remain unclear and are still widely debated. It has been proposed that they accomplish essential cell functions as they may be involved in the maintenance of redox homeostasis (20), in the elimination of toxic cyanide (21), and in the biosynthesis of several other cellular metabolites such as vitamins, enzymes, and cofactors that include a step of sulfur transfer (22–25).
A. aeolicus is a convenient model for studying intricate sulfur energy metabolism and the potential role of rhodaneses in it. The presence of a rhodanese (Aq-477) of unknown function in the cytoplasmic compartment of A. aeolicus raised our curiosity about its potential role in this metabolism. Aq-477 presents in vitro the classical thiosulfate cyanide sulfurtransferase activity (26). Previous biochemical characterizations of Aq-477 have shown that it can also use in vitro at 85 °C tetrathionate (S4O62−) and polysulfide instead of thiosulfate (S2O32−) as sulfur donor (26). The existence of an equilibrium between monomers, dimers, and tetramers of Aq-477 has also been observed, and this oligomerization appears important for thermostability and thermoactivity, which is not surprising for a protein present in a hyperthermophilic bacterium (27, 28). Even though Aq-477 has been biochemically characterized, its function remains unknown. Unfortunately, no genetic tools are available in this organism to observe on growth the effect of the absence of Aq-477. In this study, we have accordingly used alternative approaches to describe the potential function of Aq-477 in A. aeolicus, and we took advantage of our ability to purify and produce different proteins involved in its energy pathways.
Analysis of the subcellular localization of Aq-477 as well as the characterization of its partners allow us to propose a potential role for this protein in the sulfur energy metabolism of A. aeolicus. A clear effect of Aq-477, which we rename SbdP (for sulfur-binding-donating protein), on the SR activity supports this model. SbdP might represent a prototype of rhodanese that uses its sulfurtransferase activity to transfer substrates to corresponding enzymes such as SR and SOR. A proposed mechanism for SbdP as a sulfur shuttle in the sulfur energy pathway is further discussed.
EXPERIMENTAL PROCEDURES
Organisms and Growth Conditions
A. aeolicus VF5 was grown at 85 °C in 2-liter bottles containing 400 ml of modified SME medium (29) at pH 6.8 in the presence of flowers of sulfur (7.5 g·liter−1) (which will be referred to as “S0” in this paper) and under 1.4 bar of H2/CO2 (80:20) as described previously (9). Cells were harvested in the late exponential growth phase and stored at −80 °C.
Escherichia coli BL21-CodonPlus (DE3)-RIPL, transformed with the pET22aq477 or pET22SOR plasmids, was grown in LB medium at 37 °C supplemented with 0.5 mm ampicillin (12, 26).
Cell Fractionation
Periplasmic fraction was obtained as described previously (30). After periplasmic extraction, cell material (spheroplasts) was broken. Debris and unbroken cells were removed by centrifugation (10,000 × g, 15 min). Membrane and soluble proteins were separated by ultracentrifugation at 170,000 × g. The membrane fraction was then washed with 50 mm Tris-HCl, pH 7.6, 5% (v/v) glycerol, 0.5% (w/v) aminocaproic acid (buffer A) to avoid cytoplasmic contamination. To dissociate poorly membrane-associated proteins from anchored membrane proteins, the resulting membrane pellet was washed with the same buffer plus 1.5 m NaBr for 30 min at room temperature under gentle shaking and ultracentrifuged as above. The supernatant was kept to identify poorly associated membrane proteins and is named “NaBr supernatant” in this study. NaBr-washed membranes (resulting pellet) were resuspended in buffer A to give a protein concentration of 10 mg·ml−1 and solubilized with 2% deoxycholic acid sodium salt (w/v) for a final concentration (37 °C for 1 h under gentle shaking). After solubilization of the membrane proteins, the supernatant called “solubilized NaBr-washed membranes” was recovered after ultracentrifugation. The fraction called “solubilized membranes” was obtained by the same procedure, with the exception of NaBr washing.
Protein Purification
Hydrogenase-Sulfur Reductase Complex Enrichment (“SR Complex”)
Solubilized membranes from 50 g of cells were obtained as described above but with the detergent n-dodecyl-β-d-maltoside (DDM) at 1% (w/v) used instead of sodium deoxycholate and without the NaBr washing step. Membranes were then applied on a DEAE-52 column (Whatman) equilibrated with buffer A with 0.05% DDM (buffer B). Proteins were eluted with a 50 mm step gradient of 0–500 mm NaCl in buffer B. The 100–200 mm NaCl fractions that contained sulfur reductase activity were dialyzed and loaded onto a hydroxyapatite column equilibrated with buffer B. Sulfur reductase activity was eluted with 500 mm potassium phosphate in the same buffer. After removal of phosphate and concentration, the SR complex was frozen with liquid nitrogen and stored at −80 °C. We obtained an enriched fraction containing SR clearly identified by mass spectrometry that we will call the SR complex for reasons of clarity.
Sqr Purification
Free Sqr was also purified from solubilized membranes of A. aeolicus as described previously (11).
SbdP Purification
Recombinant SbdP was obtained from E. coli as described previously (26). Previous studies showed that recombinant SbdP behaves similarly to SbdP from A. aeolicus (26).
SOR Purification
Recombinant SOR was obtained from E. coli as described previously (12).
Activity Measurements
Rhodanese (Thiosulfate Cyanide Sulfurtransferase) Activity
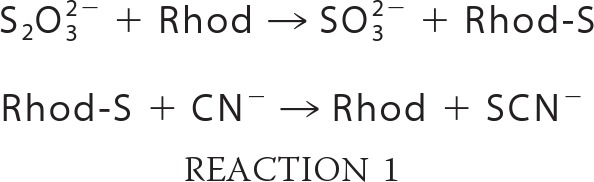
Rhodanese activity consists of the transfer of a persulfide group from thiosulfate to cyanide, leading to the formation of sulfite and thiocyanate (see Reactions 1). In vitro, this activity is followed by quantifying the product of the reaction, the thiocyanate (SCN), as described previously (26), in the absence of reducing agent.
 |
Sulfur Reductase Activity
Sulfur reductase activity was determined by quantifying the H2S formed by the reduction of S0 with H2, as described previously (9), but performed in 50 mm Hepes, pH 7.6. To study the influence of SbdP on SR activity, SbdP (1, 5, 10, 15, and 20 μg·ml−1, final concentrations) was added to the assay before the incubation at 85 °C. Measurements were made at different reaction times, with constant amounts of SR complex or solubilized NaBr-washed membranes (50 μg·ml−1 each, final concentration). Controls were done without SbdP, with SbdP alone and no SR, or with SbdP inactivated by cysteine alkylation treatment (0.3 mg·ml−1 SbdP was incubated with 0.5 mm iodoacetic acid overnight in 50 mm Tris-HCl, pH 7.6, 100 mm NaCl).
Analytical Procedures
Native Gel Electrophoresis
Proteins were loaded on 3–8% polyacrylamide gels (Mini-Protean II; Bio-Rad). After migration, proteins were either stained with PageBlue (Fermentas) or detected with specific antibodies, or the gels were used to determine hydrogenase or rhodanese activity.
Denaturing Gel Electrophoresis
Proteins were incubated at 95 °C for 5 min in the denaturing Laemmli buffer, and loaded on a 4% stacking, 5–20% running polyacrylamide gel (Mini-Protean II; Bio-Rad). After migration, gels were stained with PageBlue or used for Western blotting.
Western Blotting
After protein migration in gels, Western blot was performed using standard procedures (11). Anti-SbdP antibodies, anti-Sqr antibodies, and anti-SOR antibodies were used.
In Gel Enzymatic Activities
Hydrogenase activity on native gel was detected as described previously (31). Rhodanese activity was revealed at 85 °C by nitro blue tetrazolium chloride reduction by sulfite (a by-product of rhodanese, see Reaction 1) with phenazine ethosulfate as intermediate electron carrier. The reduction of nitro blue tetrazolium chloride allows the formation of a purple formazan precipitate visible on gel (32). The assay mixture contained 100 mm Tris-HCl, pH 9.0, 2 mm nitro blue tetrazolium chloride, 0.5 mm phenazine ethosulfate, 10 mm Na2S2O3, and 10 mm KCN.
Gel Shift Assay
SR complex (0.5 mg·ml−1) or Sqr (0.25 mg·ml−1) was incubated with SbdP (2.5 or 50 μg·ml−1, respectively) in 50 mm Tris-HCl, pH 7.6, for 30 min at 85 °C and loaded on a native gel.
Protein Concentrations
Protein Concentrations were determined with the modified Lowry method (33) and are expressed in milligrams·ml−1 for most samples. Molar concentrations cannot be expressed because stoichiometry of the active proteins/supercomplexes is yet unknown.
Cross-linking
In Vitro Cross-linking
After dialysis in 40 mm sodium phosphate, pH 7.4, to avoid cross-reaction with Tris, SbdP (2.5 μg·ml−1) and SOR (0.25 mg·ml−1) were incubated in 20 mm phosphate buffer, pH 5.7, in the presence of 15 mm 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) at 37 °C for 1 h under gentle shaking. The reaction was stopped by addition of Tris-HCl, pH 6.8, at a final concentration of 50 mm and then loaded on a native gel.
In Vivo Cross-linking
A. aeolicus cells grown on S0 were resuspended in 50 mm Hepes, pH 7.0, and were incubated with 1 mm dithiobis(succinimidyl propionate) for 30 min at 37 °C. The reaction was stopped with 100 mm Tris-HCl, pH 7.6. Cells were broken, and membranes were solubilized as described above. Membranes were loaded onto a 20–40% sucrose density gradient in 50 mm Tris-HCl, pH 7.6, and ultracentrifuged overnight at 185,000 × g (50.2 Ti rotor, Beckman-Coulter). The gradient was separated into 11 fractions. The fraction containing most of the rhodanese activity was dialyzed and loaded onto native gel. The band corresponding to SbdP was sliced and analyzed by ion trap mass spectrometry.
Surface Plasmon Resonance (SPR) Binding Experiments
A BIAcore T100 apparatus was used to investigate interaction between SbdP and SR complex, Sqr or SOR. SbdP and SR complexes were immobilized on a CM5 sensor chip (BIAcore) in 10 mm sodium acetate, pH 5.0, by amine coupling using a kit supplied by BIAcore. Running buffer, used when interactions were tested, contained 50 mm Hepes, pH 7.6, 50 mm NaCl, and 0.05% Tween 20 with a flow rate of 50 μl/min. Samples were diluted in the same buffer with 100 mm NaCl. Control experiments were done on sensor chips in the absence of immobilized proteins. Because of the difference between the index of refraction of the running buffer and the sample buffer, a burst phase could be observed at the beginning and the end of sample injection.
Mass Spectrometry (MS)
Protein Identification
Tryptic digestion of excised gel plugs, ion trap, and MALDI-TOF MS protein identification were done as described previously (10).
MS Analysis of Nondigested Proteins
To determine the binding of sulfur atoms on SbdP, the protein (2 mg·ml−1) was incubated in 100 mm Tris-HCl, pH 9.0, with thiosulfate (10 mm) or with flowers of sulfur (5% w/v) or without any sulfur source for 20 min at 85 °C and then subjected to mass analysis of nondigested proteins. MALDI-TOF analyses were performed on a Microflex II mass spectrometer (Bruker, Germany). A saturated solution of sinapinic acid in 50:50 acetonitrile/water, 0.1% trifluoroacetic acid was used as matrix. A 0.7 mg·ml−1 protein solution was used for the deposit according to the thin layer method; mixtures were dried at room temperature. Data were acquired in a positive linear mode, range was set from 2 to 20 kDa, and pulsed ion extraction was fixed to 200 ns. External mass calibration was done just before the acquisition of the sample using protein calibration standard I (Bruker Daltonics). Mass spectra were treated in Flex Analysis software by a Gaussian smoothing (width = 2 cycles = 2).
Microscopy
Spheroplasts Immunolabeling
For this experiment, spheroplast formation was optimized as described; fresh cells grown on S0 were resuspended in a buffer C containing 50 mm Tris-HCl, pH 7.5, 0.75 m sucrose, 1.5 mm EDTA. The cell suspension was frozen and thawed and centrifuged at 10,000 × g for 20 min to weaken the cells. The pellet was resuspended in buffer C supplemented with lysozyme from chicken egg (0.4 mg·ml−1) and then incubated at 37 °C for 24 h. The efficiency of cell wall degradation was monitored by microscopy. The spheroplast suspension was centrifuged at 1,500 × g for 20 min at 4 °C, and the pellet containing the spheroplasts was washed in buffer C. To be able to visualize the proteins of interest directly on the spheroplast preparation, purified immunoglobulins raised against Sqr, hydrogenase, or SbdP were labeled using the Alexa Fluor 488 protein labeling kit (Invitrogen). To determine the degree of labeling, the ratio of absorbance of Alexa Fluor 488 at 494 nm was compared with absorbance at 280 nm for proteins. We obtained a ratio between 5 and 7 mol of Alexa Fluor 488 dye per mol of antibody. The spheroplasts were then adsorbed on polylysine-treated slides, fixed with 2% paraformaldehyde, blocked with 5% BSA in PBS buffer, and immunolabeled with anti-hydrogenase- or anti-Sqr-conjugated antibodies for 1 h at room temperature in a moist chamber. Then the slides were washed twice in PBS buffer and mounted with VectaShield (Vector Laboratories). An Olympus confocal microscope (IX 81 with FluoView 1000) was used to visualize the immunolabeled spheroplast.
Immunogold Labeling Procedures
Harvested cells were fixed using 2% paraformaldehyde + 0.1% glutaraldehyde in 0.1 m phosphate buffer, pH 6.8, and washed. Cells were then pelleted in 2% agarose and cryoprotected with 2.3 m sucrose in 0.1 m phosphate buffer. Samples were frozen in liquid nitrogen and cryosubstituted during 72 h at −85 °C in methanol containing 1.5% uranyl acetate. Several washes were performed for 4 h until the temperature reached −40 °C. Then the samples were embedded in Lowicryl HM20. Polymerization was done during 48 h at −40 °C using UV light. Thin sections of 80 nm were cut with an ultramicrotome RMC MTX and collected onto nickel grids. They were incubated into 0.5 m ammonium chloride in TBS for 5 min. After washing in TBS, sections were incubated into TBS-saturating solutions containing 0.5% BSA for 10 min followed by a wash containing 0.1% BSA. Sections were then incubated for 3 h using primary antibody solution (anti-SbdP antibodies) diluted 1:100 in TBS containing 0.1% BSA. After washing in the same buffer, samples were labeled with 15 nm colloidal gold anti-rabbit IgG diluted 1:30 in the same buffer for 1 h as done previously. After several washes, fixation was performed with 2% glutaraldehyde for 5 min and washed in distilled water several times. Grids were examined using an electron microscope Zeiss EM 912, and digital images were acquired with CCD camera Gatan Bioscan model 792.
RESULTS
SbdP, a Soluble Rhodanese That Interacts with Membrane
SbdP has been previously characterized as a rhodanese protein with sulfurtransferase activity in vitro (26). It has been purified from the cytoplasmic compartment of A. aeolicus spheroplasts, but its exact subcellular localization had never been investigated. For this purpose and as a first approach, cells grown in the presence of S0 have been analyzed by immunogold labeling using specific SbdP antibodies in thin sections of intact cells (Fig. 1A). We confirmed a cytoplasmic localization of SbdP, and its presence near the membrane was less expected. However, it has been reported that a purely cytoplasmic protein is likely to be partially stained in a manner indistinguishable from a membrane-associated protein (34). To further confirm a potential localization of SbdP at the membrane, periplasmic extraction has been performed, and the resulting cells have been fractionated into membrane and cytoplasmic fractions. Western blot analysis with SbdP-specific antibodies was performed on these fractions after migration on denaturing gel. SbdP is mainly found in the cytoplasmic fraction with a main band corresponding to the monomeric form (Fig. 1B, 4th lane). Even in denaturing and reducing conditions, some tetrameric forms can be observed due to the high stability of the oligomeric state that we have previously shown to be thermostable (26). Once again, a non-negligible amount of SbdP is found in the solubilized membrane fraction (Fig. 1B, 1st lane) even though no evidence from the primary sequence of SbdP could explain this result. The absence of the tetrameric form in the membrane could either have a physiological signification, yet unknown, or could be due to experimental procedures (use of different buffers and SbdP concentrations). To better understand the potential interaction of SbdP with the membrane, the samples were treated with NaBr, a chaotropic reactant, that allowed the strength of the protein/membrane association to be estimated. Proteins interacting partially with the membrane are then usually separated from membrane-anchored proteins. Under these conditions, the majority of SbdP is released from the membrane (Fig. 1B, 2nd lane) and is found in the soluble washed fraction called the NaBr supernatant (Fig. 1B, 3rd lane). This result was confirmed by a quantitative measurement of the rhodanese activity determined in vitro in each fraction (Fig. 1C). This correlates perfectly with SbdP activity because no other active cytoplasmic rhodanese is present in this organism under these growth conditions (26).
FIGURE 1.
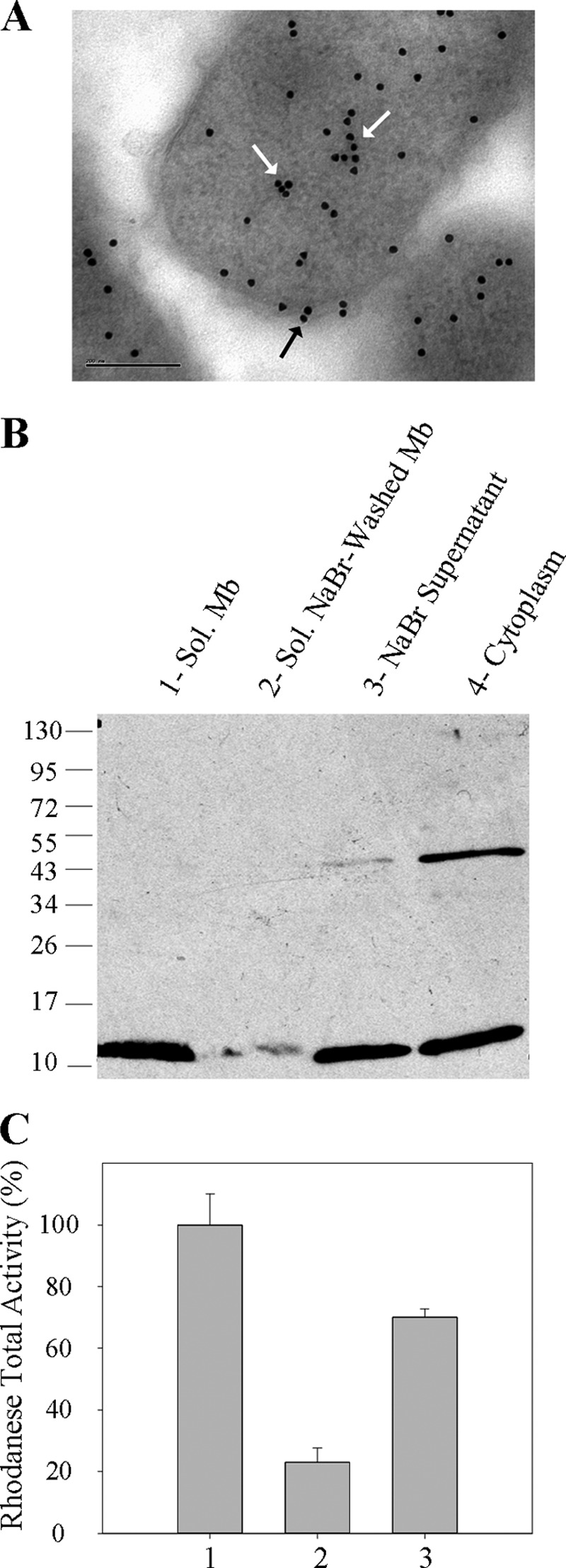
Localization of SbdP in A. aeolicus. A, immunogold labeling of SbdP on ultrathin sections of A. aeolicus. White arrows point to cytoplasmic SbdP, and the black arrow represents membrane-associated SbdP. The bar represents 200 nm. B, 30 μg of solubilized (sol) membranes (1st lane), solubilized NaBr-washed membranes (2nd lane), NaBr supernatant (3rd lane), or “cytoplasm” (4th lane) were loaded onto denaturing reducing gel, and SbdP (12.8 kDa) was detected using specific antibodies. The molecular mass markers are indicated in kDa on the left. C, rhodanese activity of solubilized membranes (column 1), solubilized NaBr-washed membranes (column 2), and NaBr supernatant (column 3). 100% represents the maximal rhodanese activity measured in the membrane fraction (200 μmol·min−1, specific activity = 4.8 μmol·min−1·mg−1) and corresponds to 10% of the total rhodanese activity. The results shown are averages of three independent experiments.
Overall, these results highlight the existence of a fraction of SbdP in contact with the membrane, which, to our knowledge, is the first experimental characterization of a membrane localization for a soluble rhodanese. This raises the question of the role of such interaction and the identity of the potential membrane protein partners of SbdP.
Identification of Potential Membrane Protein Partners of SbdP
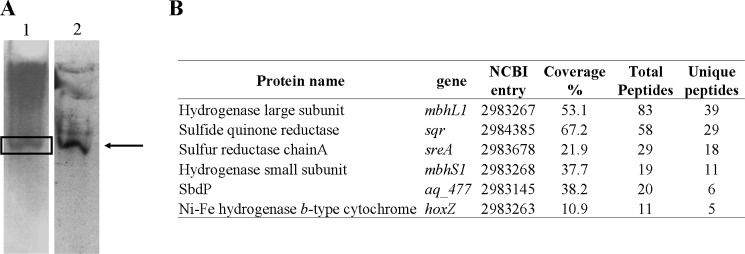
To determine the physiological membrane protein partners interacting with SbdP, we incubated cells with a cross-linking agent (dithiobis(succinimidyl propionate)) that has been shown to diffuse across the membrane. After treatment of the cells with dithiobis(succinimidyl propionate), the membrane fraction was isolated, solubilized, and separated on a sucrose gradient. Most of the membrane-bound SbdP proteins were found in the heavy fractions (high percentage of sucrose that contains heavy proteins, data not shown). To identify SbdP partners, the fraction containing SbdP activity was loaded on a native gel to allow in gel enzymatic activity detection. Hydrogenase activity could then be observed in the same band that was revealed by specific SbdP antibodies (Fig. 2A). To precisely define the composition of this potential complex, the protein band was cut out, digested with trypsin, and analyzed by ion trap MS. In this band, the presence of SbdP was confirmed, and several membrane proteins, either cross-linked to SbdP or just co-migrating with SbdP due to their intrinsic properties, were clearly identified (supplemental Fig. S1). By using this approach, we noticed that several identified proteins belong to a similar pathway as follows: SR (with its two subunits SreA and SreC), hydrogenase (which belongs to the SR supercomplex), and Sqr. These proteins are involved in sulfur energy metabolism (Fig. 2B) that makes them likely candidates as rhodanese partners. This first screen using intact cells for doing the cross-linking and trying to snapshot potential transient complexes allowed us to direct our research toward membrane proteins involved in sulfur energy metabolism as potential partners for the rhodanese SbdP.
FIGURE 2.
Analysis of solubilized membranes from cross-linked intact cells. A, native gels loaded with heavy fraction of solubilized membranes from sucrose gradient. Lane 1, Hydrogenase activity; lane 2, SbdP detection via Western blot. The arrow indicates the position of SbdP and hydrogenase in the gel. B, protein identification of the band sliced from the native gel (marked with a box) using ion trap MS analysis. Only proteins identified with at least five unique peptides and involved in energy electron transfers are included in this table. Table headings are as follows: protein name, name in NCBI data base; NCBI entry, accession number; coverage %; protein sequence coverage by the matching peptides; total peptides, number of total peptides matching to protein sequence; unique peptides, number of different peptides matching the protein sequence.
SbdP and Sulfur Reductase, Two Proteins Involved in the Same Pathway?
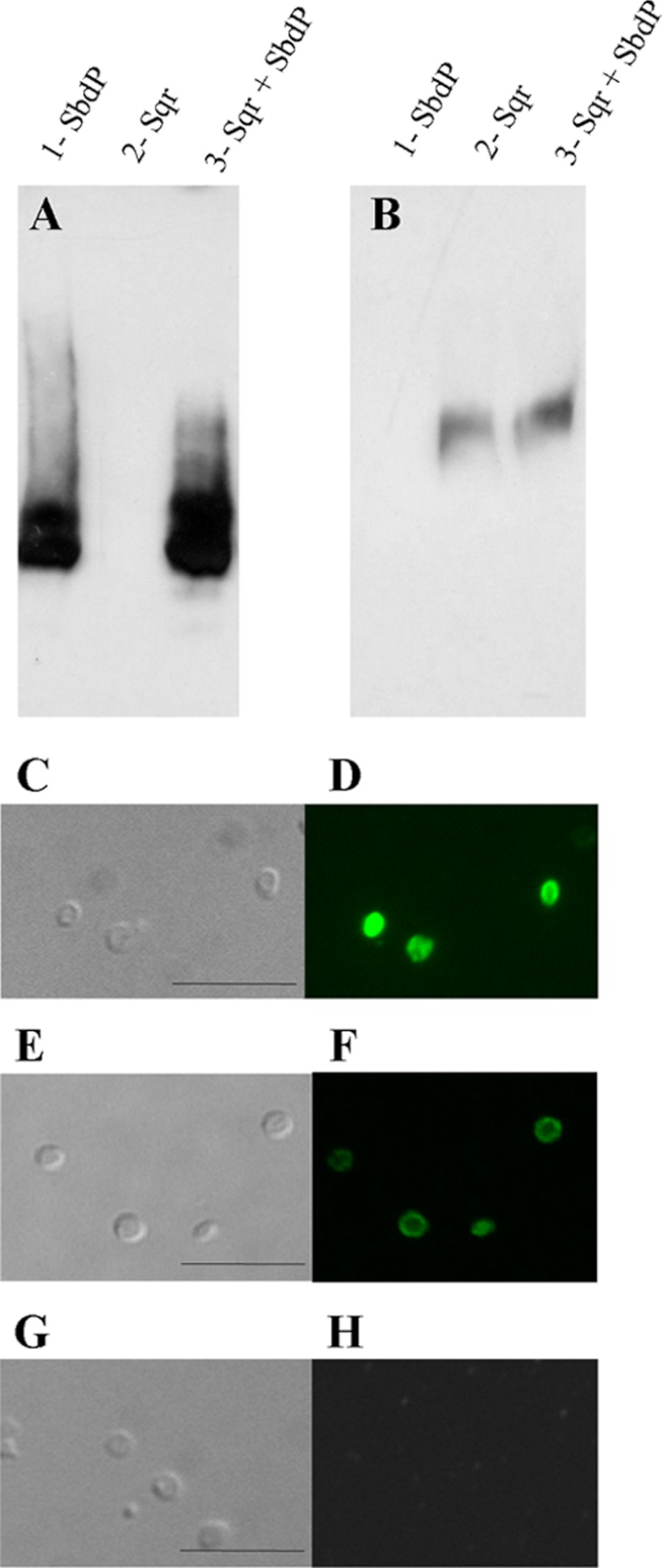
Its membrane localization and the identification of the SR in the in vivo cross-linked complex led us to test in vitro whether SbdP interacts with partially purified SR complex. Both proteins were preincubated at 85 °C for 30 min and subsequently loaded on a native gel (gel shift assay). The migration pattern of SbdP was then visualized with specific antibodies (Fig. 3A). Strikingly, a completely different migration pattern of SbdP was observed when SbdP was incubated with the SR complex. A gel shift of SbdP due to interaction with a protein present in the enriched fraction of SR was observed. The corresponding band had been cut out from the native gel, and MS allowed the identification of SbdP as well as of the subunit SreA of SR. As these two proteins co-migrate in native conditions, a direct interaction between the rhodanese SbdP and the sulfur reductase SR was strongly supported. To confirm this result, we decided to detect SbdP by following rhodanese activity on gel, a method previously described for other rhodaneses that we have adapted to our system (32). The presence of SbdP in the additional band is consistent with the previous result obtained by Western blot and MS (Fig. 3B). This also indicates that SbdP retains rhodanese activity when joined to SR.
FIGURE 3.

Interaction between SbdP and SR complex. After preincubation of 0.5 mg·ml−1 SR complex (lane 1), 2.5 μg·ml−1 SbdP (lane 2), or SbdP + SR complex (lane 3), proteins were loaded onto a native gel. SbdP was detected either by Western blot using specific antibodies (A) or by in gel rhodanese activity (B). The arrow represents the complex formed between SbdP and SR complex, which has been confirmed by proteins identification via MS. C, SPR experiment. SbdP was immobilized at 426 resonance units (RU) on a CM5 sensor chip, and increasing concentrations of the analyte (SR complex, 3.75, 7.5, 15, and 22.5 μg) were injected. As mentioned under “Experimental Procedures,” molar ratio between the two proteins could not be determined due to the existence of an equilibrium between multimeric forms for the different proteins.
A complementary approach, using SPR, was carried out to observe potential binding between these two partners in real time. We covalently immobilized SbdP on a chip, and we sequentially injected SR complex at different concentrations, in a continuous flow that passed over the chip and monitored binding events. The data show an unambiguous interaction between the two partners (Fig. 3C). Nevertheless, because of the presence of an equilibrium between multimeric forms of SbdP and the heterogeneity of SR samples, the sensorgrams obtained could not be fitted with the simplest 1–1 reaction model, preventing us from estimating kinetic constants.
The physical interaction between the SbdP and SR complex explains the finding of part of SbdP at the membrane. It also raises the question of the role of such an interaction.
SbdP, a Rhodanese Able to Bind Long Sulfur Chain
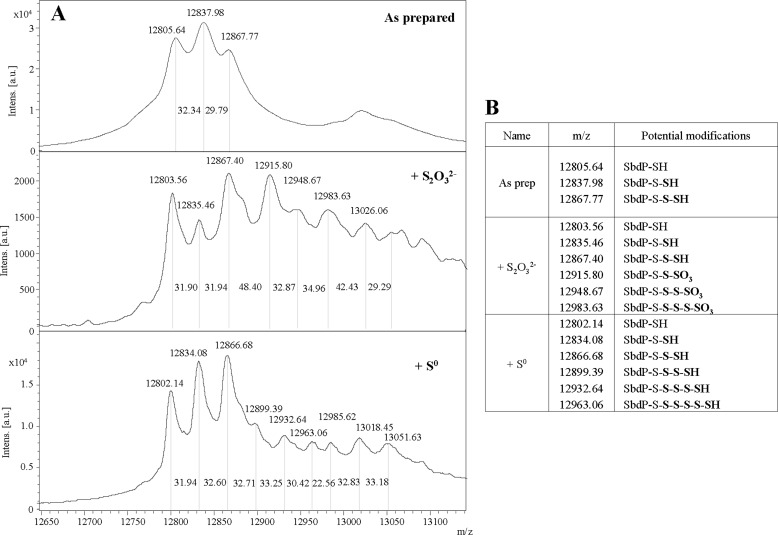
Sulfur reductase is known to reduce sulfur chains (such as S0 or polysulfide) to H2S. We hypothesized that soluble SbdP might bring the sulfur substrate to membrane SR. To fulfill such a function, SbdP should be able to load sulfur chains. In previous studies, SbdP has been shown to transfer sulfur atoms in vitro from S2O32− to the sulfur acceptor KCN. To better understand which SbdP form is involved in the presence of S2O32−, the molecular masses of both SbdP and SbdP incubated with this substrate were determined by MALDI-TOF MS. Additional sulfur atoms bound to SbdP catalytic cysteine was expected, as observed for other rhodaneses (35). As shown in Fig. 4, purified SbdP without any treatment (“as prepared”) already shows three distinct masses in approximately equivalent amounts. (i) An ion had been measured at m/z 12805.64 and could be assigned to unmodified SbdP (theoretical average m/z 12805). (ii) A population with an m/z of 12837.98 could correspond to one sulfur (mass of a sulfur atom, 32 Da) bound to SbdP (called SbdP-S-SH). (iii) The third population appeared at m/z 12867.77 and matched SbdP binding two sulfur atoms (SbdP-S-S-SH). After S2O32− incubation, all three initial species (unmodified SbdP-SH, SbdP-S-SH, and SbdP-S-S-SH) might react similarly with S2O32− because no drastic change in the proportions of these initial species could be observed. Additional masses were detected (Fig. 4A), which probably correspond to addition of oxygen and sulfur atoms (from the S2O32−) to the various initial SbdP species (Fig. 4B). As we were able to show that in vitro this rhodanese could catalyze the transfer of sulfur atom to KCN with S0 as sulfur donor (data not shown), an identical approach was performed using S0 as sulfur donor for SbdP. The MS results also show that this rhodanese can bind up to five sulfur atoms (m/z 12963.06) under our experimental conditions (Fig. 4). Additional peaks (with higher m/z) are clearly visible on the mass spectrum; however, it is difficult to know exactly which forms of SbdP they match.
FIGURE 4.
Loading of SbdP with sulfur atoms. A, analysis of covalent modification of SbdP by MS: SbdP as prepared (top) or incubated with either S2O32− (middle) or S0 (bottom). The mass (m/z) of each species is given above the corresponding peaks, and the distance (molecular mass) between peaks is also reported on the spectra. B, assignment of potential modifications of SbdP to some peaks observed on mass spectra. Atoms added on the catalytic cysteine of SbdP are shown in bold.
In summary, this experiment shows that SbdP can covalently bind several sulfur atoms on its unique catalytic cysteine. We confirmed it by using an alkylating agent that reacts specifically with cysteine residues. No sulfur fixation on SbdP could then be observed after S0 addition (supplemental Fig. S2). These overall data support our hypothesis of a potential role as sulfur donor to enzyme using long sulfur chains as substrate.
Positive Effect of SbdP to Key Enzyme of Sulfur Energy Metabolism
SPR was previously used to visualize the interaction in real time between the SbdP and SR complex (Fig. 3C). We postulate that if SbdP plays a critical role in transfering sulfur to SR, the presence of sulfur bound to SbdP may modify the kinetics or the level of interaction between these two proteins. To test it, we carried out an experiment that consisted of immobilizing the SR complex on the chip and injecting SbdP in the presence or not of a sulfur compound. As we could not use insoluble S0 with the BIAcore apparatus, S2O32− was chosen for the experiment. As observed in Fig. 5A, the interaction between SR complex and SbdP “loaded” with sulfur is drastically modified, with a faster complex formation and higher stability of the resulting complex. As SR cannot catalyze the reduction of S2O32− to H2S (9), we assume that the effect is mainly due to SbdP loaded with sulfur atoms that might stabilize the interaction of SbdP to SR.
FIGURE 5.
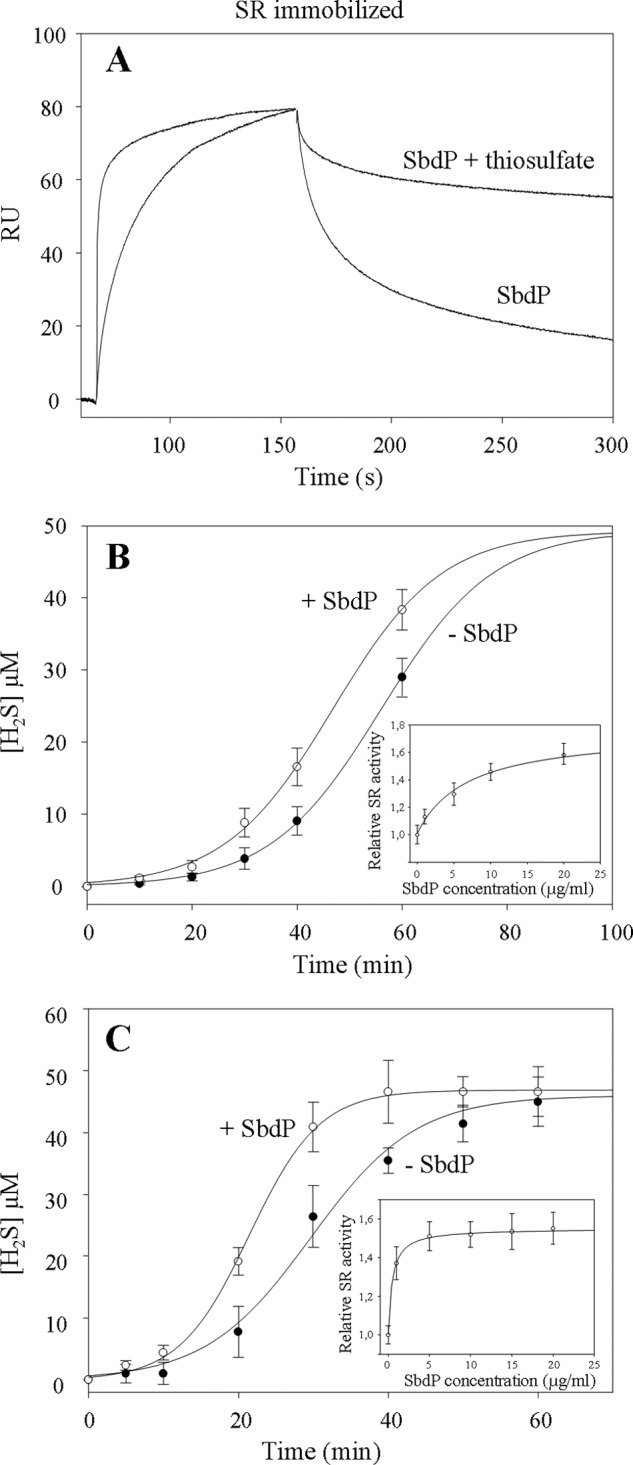
Investigation of SbdP-SR complex partnership. A, SPR experiment; SR complex was immobilized at 4710 RU on a CM5 sensor chip, and SbdP (0.6 μg) incubated or not with 10 mm thiosulfate (S2O32−) was injected. Thiosulfate alone was also injected as a control, without modification of the signal. B and C, sulfur reductase activity measurements of SR complex (50 μg·ml−1, final concentration) (B) or solubilized NaBr-washed membranes (50 μg·ml−1, final concentration) (C) performed with the insoluble S0 in presence (white circles) or absence (black circles) of exogenous SbdP (5 μg·ml−1, final concentration) as well as in presence of increasing concentrations of SbdP (1, 5, 10, 15, and 20 μg·ml−1, final concentration) (inset, B and C) after 40 min of incubation. Ratio between the different proteins, however, cannot be calculated because the oligomerization state of the active form of both proteins is yet unknown. A clear concentration dependence illustrates that we are most likely not in an excess of one over the other. Results presented correspond to an average of three independent experiments.
On the basis of this experiment, we proposed that SbdP could indeed act as a sulfur donor for SR once it is loaded with sulfur atoms. To test this potential functional role of SbdP on SR, we measured sulfur reductase activity in the presence of its usual substrate (S0) as well as in the presence of different amounts of SbdP. A difference observed in H2S production of SR due to SbdP might reflect a modification of SR-specific activity or a change of substrate accessibility to the SR-active site. As observed in Fig. 5B, whatever the presence of SbdP, the rate of H2S production increases steeply with time and reaches approximately the same plateau value. Interestingly, the curves were not superimposable; for some time points (30, 40, and 60 min), we clearly and reproducibly detected higher H2S production by SR in the presence of SbdP and in a concentration-dependent manner (as observed in Fig. 5B, inset). Comparison of the two curves shows that the slopes are quite similar (Fig. 5B). The observed difference seems due to a shorter lag phase in H2S production in the presence of SbdP. As we know that SbdP can bind sulfur compounds and that SbdP alone is not able to produce H2S in these conditions, we propose an active effect of SbdP on SR activity. In our experimental conditions, SbdP might increase the availability of the substrate toward the buried SR-active site. A similar trend was observed using membrane fractions and following SR activity in presence of exogenous SbdP (Fig. 5C). Slopes are, however, slightly different and are most likely due to additional proteins, yet unknown, present in the membrane extract that would optimize SbdP effect on SR. This experiment not only confirms the positive effect of SbdP on SR activity but also shows an amplified effect of SbdP compared with the purified SR complex, which is probably due to a better physiological environment for both proteins. Taken together, we observed a clear role of exogenous SbdP on SR activity. A control experiment was made using the alkylated form of SbdP that is inactive as a rhodanese in vitro and is not able to increase SR activity (supplemental Fig. S3), and this further demonstrates a role of SbdP in substrate presentation and is not a consequence of SbdP interaction to SR.
These in vitro experiments support the hypothesis of a role of SbdP as a sulfur donor to SR. In vivo, SbdP might play a role of a cytoplasmic shuttle that would not only convert the sulfur compound into a more “usable” substrate but also bring it to its membrane partner.
Link between SbdP and Another Membrane Protein Involved in Sulfur Metabolism?
Another protein, Sqr, involved in sulfur energy metabolism, has been identified in the in vivo cross-linking experiment (Fig. 2). It catalyzes the oxidation of H2S to S0 (11), and we hypothesized that SbdP could take care of the product of the reaction and per se would allow sulfur recycling. Interaction between Sqr and SbdP was therefore investigated using gel shift assay, as described previously. There was no significant difference in the migration pattern of SbdP or of Sqr when they were incubated alone or together (Fig. 6, A and B). The absence of interaction between the two proteins was further confirmed with SPR experiments (supplemental Fig. S4). In addition, A. aeolicus Sqr has been classified as an integral monotopic membrane protein, meaning that this protein is only exposed to one side of the membrane, for which the periplasmic orientation has been suggested (36). In parallel to our in vitro interaction studies, we investigated the orientation of Sqr; we consequently designed an optimized method adapted for this extremophile organism to create spheroplasts. As observed in Fig. 6, C, E, and G, the cells show a characteristic round shape after treatment due to the depletion of their external membrane and of their periplasmic compartment. We then incubated these cells with fluorescent antibodies directed against Sqr (Fig. 6D), hydrogenase (Fig. 6F), or SbdP (Fig. 6H). A fluorescent staining using confocal microscopy should be visualized only if the protein of interest is oriented toward the periplasm. Hydrogenase, known to be oriented toward the periplasm in A. aeolicus, was used as a positive control. In contrast to SbdP, fluorescence could be clearly detected, revealing the periplasmic orientation of Sqr (Fig. 6D). This experiment confirms that SbdP cannot bind Sqr in vivo and is consistent with the absence of interaction observed in vitro. This indicates that Sqr functions independently of SbdP.
FIGURE 6.
Absence of interaction between SbdP and Sqr. After preincubation of SbdP (lane 1), Sqr (lane 2), or Sqr + SbdP (lane 3), proteins were loaded onto a native gel. SbdP (A) or Sqr (B) was detected by Western blot using specific antibodies. C, E, and G, Nomarski contrast microscopy; D, F, and H, immunofluorescence (D, using anti-Sqr antibodies; F, using anti-hydrogenase antibodies; or H, using anti-SbdP antibodies) on spheroplasts. The bar represents 10 μm.
SOR, Another Partner of SbdP
We propose that SbdP plays a role of a shuttle transferring long sulfur chains to enzymes that require this substrate for fulfilling their enzymatic reactions. Another enzyme (SOR) uses S0 as substrate and has been proposed to be involved in the energy pathway in various Acidianus species (13, 14, 37). As SOR is present and active in A. aeolicus cytoplasm under our growth conditions, we first decided to test if SOR from A. aeolicus was also able to interact partially with the membrane to determine whether it could be part of the sulfur energy metabolism. As shown in Fig. 7A, a portion of SOR can be found in the membrane fraction and is released after treatment with a chaotropic agent.
FIGURE 7.
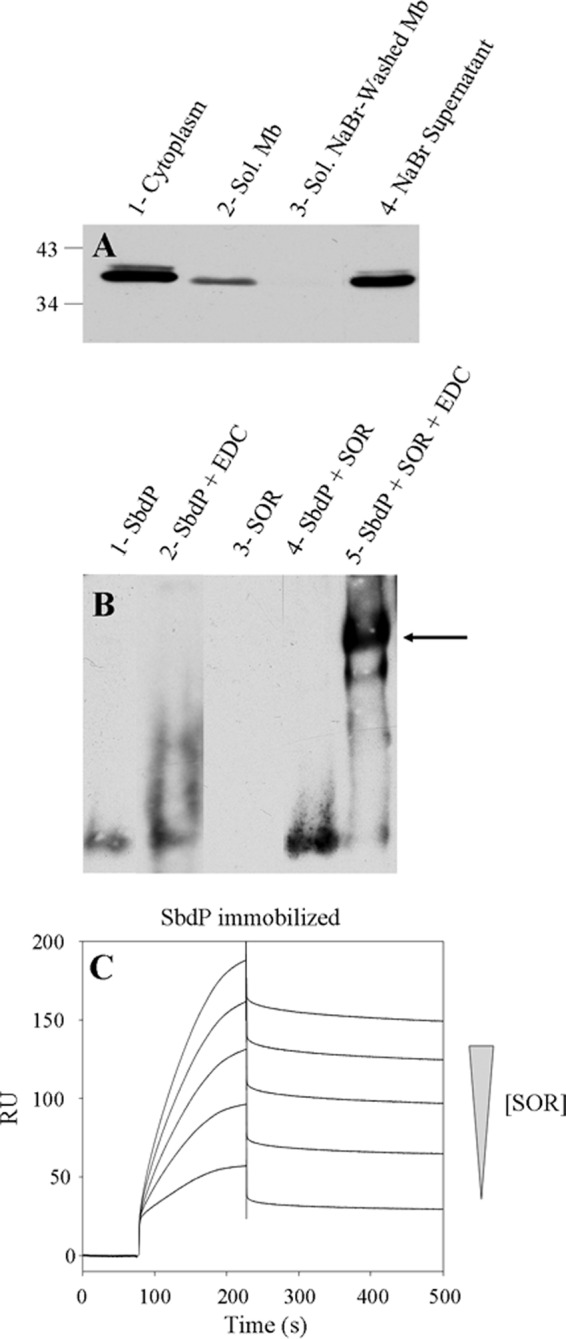
Interaction between SbdP and SOR. A, 30 μg of cytoplasm (lane 1), solubilized membranes (lane 2), solubilized NaBr-washed membranes (lane 3), and NaBr supernatant (lane 4) were loaded onto denaturing gel, and SOR was detected by Western blot with specific antibodies. The molecular mass markers are indicated in kDa on the left. B, cross-linking experiment; after preincubation of SbdP (lane 1), SbdP + EDC (lane 2), SOR (lane 3), SbdP + SOR (lane 4), SbdP + SOR + EDC (lane 5), proteins were loaded onto a native gel. SbdP was detected by Western blot using specific antibodies. The arrow represents the complex formed between SbdP and SOR complex. C, SPR experiment; SbdP was immobilized at 426 RU on a CM5 sensor chip, and increasing concentrations of the analyte (SOR, 0.1, 0.2, 0.3, 0.4, and 0.5 μg) were injected.
As SR and SOR use the same substrate (S0), we considered that SbdP might play a central role in sulfur energy metabolism and could supply sulfur substrate to the SOR as well. Unfortunately, gel shift assay does not give clear results (supplemental Fig. S5A). The formation of a very large complex (SOR being at least a 16-mer and SbdP oligomerizing), might prevent the incorporation into the gel under the conditions tested. We thus modified the physicochemical conditions (pH and temperature), and we used a cross-linking agent (EDC) to lock this complex and stabilize the interaction. As observed in Fig. 7B (5th lane), a band corresponding to the SbdP-SOR complex is clearly detected. The presence of SbdP and SOR in the cross-linked complex was not only observed using antibodies (supplemental Fig. S5, B and C) but also when rhodanese activity assay was performed in gel and showed a similar pattern (supplemental Fig. S5D). This result also shows that in these conditions, even in the presence of a cross-linking agent, SbdP in the complex is still active as a rhodanese. We decided to confirm the interaction between both purified proteins by SPR. Despite the fact that they present different oligomerization states making it difficult to obtain kinetic constants, the result obtained supports the conclusion of an interaction observed for the first time between SOR and a rhodanese (Fig. 7C). This interaction suggested a potential role of this rhodanese, as for SR, to bring SOR substrates and optimize their presentation to the active site of SOR.
DISCUSSION
In this study, we have described the use of in vitro approaches to determine the function of SbdP, a cytoplasmic rhodanese present in A. aeolicus. This enzyme has unique features as it can load, on its catalytic cysteine, long chains of sulfur atoms after incubation with S0. Another unusual feature among rhodaneses is that SbdP has two different partners, SR and SOR. Both enzymes use long sulfur chains as substrates, and both are key players in sulfur energy metabolism. The presence of SbdP would optimize substrate presentation to its partner enzymes. On the basis of these data, we propose that SbdP might play a central role in sulfur energy pathways as a shuttle in the transfer of sulfur substrate to catalytically active enzymes (SR or SOR). We present alternative approaches to describe rhodanese function. Indeed, characterizing rhodanese physiological function in an organism is a challenge due to the abundance of potential rhodanese/rhodanese-like proteins within the same genome. As an example, E. coli and Pseudomonas aeruginosa have, respectively, 9 and 10 annotated rhodaneses or rhodanese-like proteins in their genome (18, 35). This redundancy supports the notion that rhodaneses are involved in distinct cellular processes. However, this makes it difficult to attribute a defined in vivo function just by analyzing the phenotype of the corresponding mutant. Indeed, a different rhodanese can compensate for the absence of an other one (22, 35).
SbdP, a Rhodanese That Binds Long Sulfur Chains
Our attempt to determine the number of sulfurs bound to SbdP shows that, after incubation of SbdP in the presence of S0 and S2O32−, several species of the protein can be identified by MS. Some of them bind as many as five sulfur atoms. To our knowledge, this result has never been observed by MS for other rhodaneses, although binding of sulfur compounds has already been studied (35, 38). In most cases the presence of only one or two sulfur atoms bound to the catalytic cysteine has been observed. It is however noticeable that the rhodanese-like protein Sud from W. succinogenes has been suggested to be able to bind up to 10 sulfur atoms per monomer, based on biochemical studies (17). However, only two sulfur atoms bound to Sud were detected by MS (39) and five were observed after determination of the NMR structure (40). The authors explained this difference by suggesting that some sulfur atoms bind more tightly due to their protection by the active site, whereas a long chain of sulfur exposed might easily dissociate upon MS procedures (39). This explanation may also apply to SbdP, for which we did not see a homogeneous population binding five sulfur atoms. The docking of a pentasulfide-sulfur molecule on the modeled structure of SbdP (supplemental Fig. S6) shows that the sulfur chain is exposed, meaning that it could be used as a substrate for SR and SOR enzymes.
SbdP, a Shuttle for Carrying Reactive Sulfur Compounds to Enzymes of Sulfur Metabolism
The fact that SbdP has the potentiality to interact with at least two partners (SR and SOR) is another unusual feature for rhodaneses. The complex SR-SbdP is reminiscent of the Psr-Sud system of W. succinogenes, whereas the complex formed between SbdP and SOR has not yet been observed. SOR and SR have some common characteristics. One of them is the ability to use a long chain of sulfur atoms as substrates. In A. aeolicus, both enzymes are present simultaneously in the cell and have a similar location; SR is a three-subunit membrane enzyme oriented toward the cytoplasm, and SOR is a soluble cytoplasmic enzyme that we show to be partially located at the membrane. Moreover, both enzymes possess buried active sites. As an example, the structure of Psr, an SR homologous protein from Thermus thermophilus, has been resolved (41), and it has been shown that its active site is located in the subunit A buried at the end of a narrow funnel, as for other molybdoenzymes of this family. How the substrate can get access to this active site is still unclear (42). Likewise, the structure of A. ambivalens SOR shows that the SOR active site is also not very accessible and a chimney-like protrusion has been suggested to be the likely entry routes for a linear sulfur chain (43). We proposed that SbdP might act as a sulfur supplier by providing long sulfur chains to these buried active site enzymes. Once loaded with sulfur chain, SbdP could bind the catalytic subunits of its partners and the long chain could then be optimally presented to the cavity leading to the active site. The stabilization of the SbdP-SR complex observed in presence of the substrate supports this hypothesis (Fig. 5A). Overall, we proposed that SbdP might increase the efficiency of sulfur energy metabolism in this bacterium by optimizing the presentation of the substrate to its partner enzymes. We were able to strengthen this theory by demonstrating that SbdP presence accelerates H2S production of its partner enzyme SR (Fig. 5, B and C). This impressive effect due to an active role of SbdP might be explained by the following facts: (i) SbdP binds a chain of sulfur and by an appropriate interaction with SR, the substrate is correctly and efficiently presented to the active site of the SR; (ii) SbdP increases both the local concentration of substrate and the time of substrate presentation to the active site of the enzyme by forming a stable complex; and (iii) the substrate (sulfur chain) supplied by SbdP is more efficiently used by the enzyme than the initial cyclic sulfur S8 substrate, for example.
We propose that SbdP plays a similar function of substrate-donating protein for the SOR enzyme. However the impact of SbdP on the kinetics of SOR product formation is difficult to detect on account of the simultaneous reduction and oxidation of S0 to SO32−, S2O32−, and H2S. Moreover, some products of the reaction (SO32− and H2S) are unstable, and others (S2O32−) can be used by SbdP as a substrate.
Sulfur globules have been observed in A. aeolicus cytoplasm (9). These kind of sulfur structures are usually present either in the periplasmic compartment or in the extracellular environment (44). In any case, the chemical nature of sulfur in the globules is still a debated question, and it seems that three different forms could be found, depending on the approach used: cyclo-octasulfur (S8), polythionates ([(SnSO3)2]2−), and sulfur chains (45). These three forms can all be used as substrates by SbdP. In A. aeolicus, a role as sulfur storage has been suggested based on previous studies showing that in the absence of exogenous S0, these inclusions disappeared over time.5 The fact that A. aeolicus possesses two enzymes that use long sulfur chains in the cytoplasm allows us to speculate that sulfur globules might be one of their sources of substrate, with SbdP acting as a shuttle.
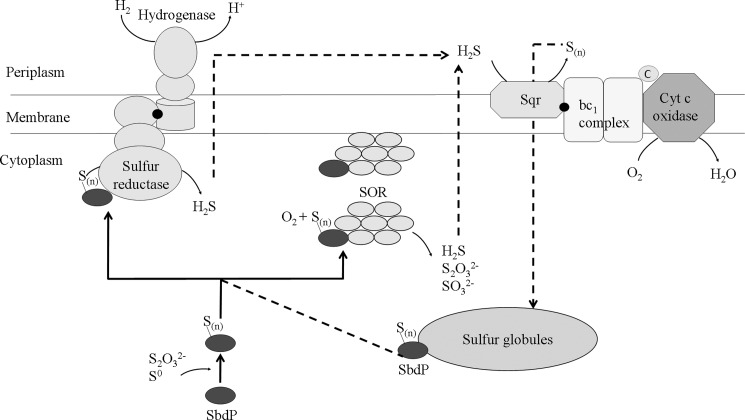
Sulfur Energy Metabolism of A. aeolicus, an Overview
As illustrated in Fig. 8, a broad picture of A. aeolicus sulfur energy metabolism is becoming visible. Pieces of this intricate puzzle of sulfur metabolism are now better defined by the association of complementary approaches (proteomics and enzymatic studies) with the main players, SR, Sqr, SOR, well characterized (as described in Fig. 8 legend) (9, 11, 12). A role of SOR in the sulfur energy pathway is strongly supported by its membrane location also observed in Acidianus tengchongensis (37), which would allow transfer of its products to membrane energy enzymes (13, 14). Here, we define the function of an additional protein involved in this complex sulfur energy metabolism of A. aeolicus. We propose a model in which SbdP would be at the central position of this sulfur energy network, as represented in Fig. 8, playing the role of a shuttle to carry a sulfur chain from different sources (such as the sulfur globules) to the cytoplasmic or membrane-located enzymes and allowing a better availability of the substrate to optimize the sulfur traffic. This substrate channeling toward its targets would also allow us to prevent any interference with other pathways, a reasonable possibility on account of the reactivity of sulfur compounds, in a similar way as metallochaperones for highly reactive metals (46). SbdP may represent the prototype of a new subfamily of rhodanese involved in sulfur traffic in cells. The periplasmic rhodanese-like protein Sud from W. succinogenes, which presents 32% of identity with SbdP, might as well belong to this family especially because biochemical data and metabolic context suggest that it might supply sulfur substrate to the Psr enzyme (17). In other organisms such as Acidithiobacillus ferrooxidans (47), A. ambivalens and some green sulfur bacteria (48), with similar sulfur metabolism, rhodanese or rhodanese-like proteins, might play an identical function.
FIGURE 8.
Central role of SbdP in the model of energy sulfur recycling in A. aeolicus. After growth with S0 as sulfur source, membrane-bound hydrogenase/SR (9) and Sqr/bc1 complex/cytochrome c) oxidase supercomplexes (11) are present and active, as well as cytoplasmic and membrane-associated SOR enzyme (Ref. 12 and this paper). H2S gas, produced by SR and SOR, might be used by Sqr as a substrate and converted into polysulfide or elemental sulfur that has been proposed to be incorporated into cytoplasmic sulfur globules (36). These sulfur globules (9), which probably serve as sulfur storage for the bacteria, might be in turn used by SbdP as an alternative sulfur source. Finally, sulfite and thiosulfate produced by SOR might be used by still unknown systems participating in sulfur energy metabolism as proposed previously (13, 14). By interacting with both SOR and SR, the rhodanese SbdP is at a key position in this scheme, delivering substrate as a long chain of sulfur atoms, a very suitable form of substrate for these enzymes and, finally, might increase the efficiency of the sulfur energy metabolism of A. aeolicus. Continuous lines represent the pathways demonstrated in this paper; dotted lines represent proposed sulfur traffic. Black points represent quinone molecules; Sn is for sulfur (the physiological substrate and the number of sulfur atoms in the molecule in vivo are not known); cyt c oxidase is for cytochrome c oxidase, and c is for monoheme cytochrome c555.
Finally, our findings not only allow a better understanding of the functional role of the rhodanese enzymes but also open the way to a deeper investigation of the sulfur compounds traffic and recycling between the actors of an intricate sulfur energy metabolism network.
Acknowledgments
We gratefully acknowledge the contribution of Marielle Bauzan (Fermentation Plant Unit, Institut de Microbiologie de la Méditerranée, Marseilles, France) for growing the bacteria; Alain Bernadac (Institut de Microbiologie de la Méditerranée, CNRS, Marseilles, France) and Jean-Paul Chauvin (Institut de Biologie du Développement de Marseille-Luminy, University Aix-Marseille II, Marseilles, France) for microscopy; Remy Puppo, Régine Lebrun, and Sabrina Lignon (Proteomic Analysis Center, Marseilles, France) for mass spectrometry analyses; Alexandre Ciaccafava for help with SbdP modeling; and Corinne Aubert for kindly supplying the pET22SOR plasmid. We also thank Laurence Prunetti, Valérie Belle, and Maria Luz Cardenas for helpful discussions and Athel Cornish-Bowden for reviewing the manuscript. The Proteomic Analysis Center of IFR88 is part of MaP (Marseille Protéomique, Infrastructure en Biologie Santé et Agronomie).

This article contains supplemental Figs. S1–S6 and additional references.
M.-T. Giudici-Orticoni, unpublished results.
- SR
- sulfur reductase
- Sqr
- sulfide quinone reductase
- SOR
- sulfur oxygenase reductase
- Psr
- polysulfide reductase
- SPR
- surface plasmon resonance
- RU
- resonance unit
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide.
REFERENCES
- 1. Philippot P., Van Zuilen M., Lepot K., Thomazo C., Farquhar J., Van Kranendonk M. J. (2007) Early Archaean microorganisms preferred elemental sulfur, not sulfate. Science 317, 1534–1537 [DOI] [PubMed] [Google Scholar]
- 2. Wächtershäuser G. (2000) Origin of life. Life as we don't know it. Science 289, 1307–1308 [DOI] [PubMed] [Google Scholar]
- 3. Montegrossi G., Tassi F., Vaselli O., Buccianti A., Garofalo K. (2001) Sulfur species in volcanic gases. Anal. Chem. 73, 3709–3715 [DOI] [PubMed] [Google Scholar]
- 4. Bonch-Osmolovskaya E. (September 2007) Microorganisms in high temperature sulfur environments. eLS. John Wiley and Sons Ltd., Chichester: els.net (DOI: 10.1002/9780470015902.a0000405.pub2) [DOI] [Google Scholar]
- 5. Amend J. P., Shock E. L. (2001) Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and bacteria. FEMS Microbiol. Rev. 25, 175–243 [DOI] [PubMed] [Google Scholar]
- 6. Huber R., Stetter K. O. (August 2002) eLS. John Wiley and Sons Ltd., Chichester: els.net (DOI: 10.1038/npg.els.0000442) [DOI] [Google Scholar]
- 7. Huber R., Wilharm T., Huber D., Trincone A., Burggraf S., Konig H., Rachel R., Rockinger I., Fricke H., Stetter K. O. (1992) Aquifex pyrophilus gen. nov., sp. Nov., represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. System Appl. Microbiol. 15, 340–351 [Google Scholar]
- 8. Olsen G. J., Woese C. R., Overbeek R. (1994) The winds of (evolutionary) change. Breathing new life into microbiology. J. Bacteriol. 176, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guiral M., Tron P., Aubert C., Gloter A., Iobbi-Nivol C., Giudici-Orticoni M. T. (2005) A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacterium Aquifex aeolicus. J. Biol. Chem. 280, 42004–42015 [DOI] [PubMed] [Google Scholar]
- 10. Guiral M., Prunetti L., Lignon S., Lebrun R., Moinier D., Giudici-Orticonit M. T. (2009) New insights into the respiratory chains of the chemolithoautotrophic and hyperthermophilic bacterium Aquifex aeolicus. J. Proteome Res. 8, 1717–1730 [DOI] [PubMed] [Google Scholar]
- 11. Prunetti L., Infossi P., Brugna M., Ebel C., Giudici-Orticoni M. T., Guiral M. (2010) New functional sulfide oxidase-oxygen reductase supercomplex in the membrane of the hyperthermophilic bacterium Aquifex aeolicus. J. Biol. Chem. 285, 41815–41826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pelletier N., Leroy G., Guiral M., Giudici-Orticoni M. T., Aubert C. (2008) First characterization of the active oligomer form of sulfur oxygenase reductase from the bacterium Aquifex aeolicus. Extremophiles 12, 205–215 [DOI] [PubMed] [Google Scholar]
- 13. Brito J. A., Sousa F. L., Stelter M., Bandeiras T. M., Vonrhein C., Teixeira M., Pereira M. M., Archer M. (2009) Structural and functional insights into sulfide:quinone oxidoreductase. Biochemistry 48, 5613–5622 [DOI] [PubMed] [Google Scholar]
- 14. Kletzin A., Urich T., Müller F., Bandeiras T. M., Gomes C. M. (2004) Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36, 77–91 [DOI] [PubMed] [Google Scholar]
- 15. Klimmek O. (2005) The biological cycle of sulfur. Met. Ions Biol. Syst. 43, 105–130 [DOI] [PubMed] [Google Scholar]
- 16. Hedderich R., Klimmek O., Kroger A., Dirmeier R., Keller M., Stetter K. O. (1999) Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol. Rev. 22, 353–381 [Google Scholar]
- 17. Klimmek O., Kreis V., Klein C., Simon J., Wittershagen A., Kröger A. (1998) The function of the periplasmic Sud protein in polysulfide respiration of Wolinella succinogenes. Eur. J. Biochem. 253, 263–269 [DOI] [PubMed] [Google Scholar]
- 18. Cipollone R., Ascenzi P., Visca P. (2007) Common themes and variations in the rhodanese superfamily. IUBMB Life 59, 51–59 [DOI] [PubMed] [Google Scholar]
- 19. Papenbrock J., Guretzki S., Henne M. (2011) Latest news about the sulfurtransferase protein family of higher plants. Amino Acids 41, 43–57 [DOI] [PubMed] [Google Scholar]
- 20. Remelli W., Cereda A., Papenbrock J., Forlani F., Pagani S. (2010) The rhodanese RhdA helps Azotobacter vinelandii in maintaining cellular redox balance. Biol. Chem. 391, 777–784 [DOI] [PubMed] [Google Scholar]
- 21. Cipollone R., Frangipani E., Tiburzi F., Imperi F., Ascenzi P., Visca P. (2007) Involvement of Pseudomonas aeruginosa rhodanese in protection from cyanide toxicity. Appl. Environ. Microbiol. 73, 390–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kessler D. (2006) Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol. Rev. 30, 825–840 [DOI] [PubMed] [Google Scholar]
- 23. Dahl J. U., Urban A., Bolte A., Sriyabhaya P., Donahue J. L., Nimtz M., Larson T. J., Leimkühler S. (2011) The identification of a novel protein involved in molybdenum cofactor biosynthesis in Escherichia coli. J. Biol. Chem. 286, 35801–35812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matthies A., Rajagopalan K. V., Mendel R. R., Leimkühler S. (2004) Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc. Natl. Acad. Sci. U.S.A. 101, 5946–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palenchar P. M., Buck C. J., Cheng H., Larson T. J., Mueller E. G. (2000) Evidence that ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis, may be a sulfurtransferase that proceeds through a persulfide intermediate. J. Biol. Chem. 275, 8283–8286 [DOI] [PubMed] [Google Scholar]
- 26. Giuliani M. C., Tron P., Leroy G., Aubert C., Tauc P., Giudici-Orticoni M. T. (2007) A new sulfurtransferase from the hyperthermophilic bacterium Aquifex aeolicus. Being single is not so simple when temperature gets high. FEBS J. 274, 4572–4587 [DOI] [PubMed] [Google Scholar]
- 27. Tanaka Y., Tsumoto K., Yasutake Y., Umetsu M., Yao M., Fukada H., Tanaka I., Kumagai I. (2004) How oligomerization contributes to the thermostability of an archaeon protein. Protein l-isoaspartyl-O-methyltransferase from Sulfolobus tokodaii. J. Biol. Chem. 279, 32957–32967 [DOI] [PubMed] [Google Scholar]
- 28. Vieille C., Zeikus G. J. (2001) Hyperthermophilic enzymes. Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baymann F., Tron P., Schoepp-Cothenet B., Aubert C., Bianco P., Stetter K. O., Nitschke W., Schütz M. (2001) Cytochromes c555 from the hyperthermophilic bacterium Aquifex aeolicus (VF5). 1. Characterization of two highly homologous, soluble and membranous, cytochromes c555. Biochemistry 40, 13681–13689 [DOI] [PubMed] [Google Scholar]
- 30. Giuliani M. C., Jourlin-Castelli C., Leroy G., Hachani A., Giudici-Orticoni M. T. (2010) Characterization of a new periplasmic single-domain rhodanese encoded by a sulfur-regulated gene in a hyperthermophilic bacterium Aquifex aeolicus. Biochimie 92, 388–397 [DOI] [PubMed] [Google Scholar]
- 31. Brugna-Guiral M., Tron P., Nitschke W., Stetter K. O., Burlat B., Guigliarelli B., Bruschi M., Giudici-Orticoni M. T. (2003) [NiFe] hydrogenases from the hyperthermophilic bacterium Aquifex aeolicus. Properties, function, and phylogenetics. Extremophiles 7, 145–157 [DOI] [PubMed] [Google Scholar]
- 32. Cannella C., Berni R., Ricci G. (1984) Determination of rhodanese activity by tetrazolium reduction. Anal. Biochem. 142, 159–162 [DOI] [PubMed] [Google Scholar]
- 33. Markwell M. A., Haas S. M., Bieber L. L., Tolbert N. E. (1978) A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87, 206–210 [DOI] [PubMed] [Google Scholar]
- 34. Bukau B., Reilly P., McCarty J., Walker G. C. (1993) Immunogold localization of the DnaK heat shock protein in Escherichia coli cells. J. Gen. Microbiol. 139, 95–99 [DOI] [PubMed] [Google Scholar]
- 35. Cheng H., Donahue J. L., Battle S. E., Ray W. K., Larson T. J. (2008) Biochemical and genetic characterization of PspE and GlpE, two single-domain sulfurtransferases of Escherichia coli. Open Microbiol. J 2, 18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marcia M., Ermler U., Peng G., Michel H. (2009) The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc. Natl. Acad. Sci. U.S.A. 106, 9625–9630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen Z. W., Jiang C. Y., She Q., Liu S. J., Zhou P. J. (2005) Key role of cysteine residues in catalysis and subcellular localization of sulfur oxygenase-reductase of Acidianus tengchongensis. Appl. Environ. Microbiol. 71, 621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Forlani F., Cereda A., Freuer A., Nimtz M., Leimkühler S., Pagani S. (2005) The cysteine-desulfurase IscS promotes the production of the rhodanese RhdA in the persulfurated form. FEBS Lett. 579, 6786–6790 [DOI] [PubMed] [Google Scholar]
- 39. Klimmek O., Stein T., Pisa R., Simon J., Kröger A. (1999) The single cysteine residue of the Sud protein is required for its function as a polysulfide-sulfurtransferase in Wolinella succinogenes. Eur. J. Biochem. 263, 79–84 [DOI] [PubMed] [Google Scholar]
- 40. Lin Y. J., Dancea F., Löhr F., Klimmek O., Pfeiffer-Marek S., Nilges M., Wienk H., Kröger A., Rüterjans H. (2004) Solution structure of the 30-kDa polysulfide-sulfurtransferase homodimer from Wolinella succinogenes. Biochemistry 43, 1418–1424 [DOI] [PubMed] [Google Scholar]
- 41. Jormakka M., Yokoyama K., Yano T., Tamakoshi M., Akimoto S., Shimamura T., Curmi P., Iwata S. (2008) Molecular mechanism of energy conservation in polysulfide respiration. Nat. Struct. Mol. Biol. 15, 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rothery R. A., Workun G. J., Weiner J. H. (2008) The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim. Biophys. Acta 1778, 1897–1929 [DOI] [PubMed] [Google Scholar]
- 43. Urich T., Gomes C. M., Kletzin A., Frazão C. (2006) X-ray structure of a self-compartmentalizing sulfur cycle metalloenzyme. Science 311, 996–1000 [DOI] [PubMed] [Google Scholar]
- 44. Dahl C., Prange A. (2006) in Inclusions in Prokaryotes (Shively J. M., ed) pp. 21–51, Springer, Berlin [Google Scholar]
- 45. George G. N., Gnida M., Bazylinski D. A., Prince R. C., Pickering I. J. (2008) X-ray absorption spectroscopy as a probe of microbial sulfur biochemistry. The nature of bacterial sulfur globules revisited. J. Bacteriol. 190, 6376–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robinson N. J., Winge D. R. (2010) Copper metallochaperones. Annu. Rev. Biochem. 79, 537–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Acosta M., Beard S., Ponce J., Vera M., Mobarec J. C., Jerez C. A. (2005) Identification of putative sulfurtransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome. Structural and functional characterization of the proteins. Omics 9, 13–29 [DOI] [PubMed] [Google Scholar]
- 48. Frigaard N. U., Bryant D. A. (2008) in Sulfur Metabolism in Phototrophic Organisms (Hell R., Dahl C., Knaff D. B., Leustek T., eds) pp. 337–355, Springer, Berlin [Google Scholar]