Background: The nucleolus always shows delayed development and malfunction in somatic cell nuclear transfer (NT) embryos.
Results: NT embryos showed low rDNA activity and developmental competence when donor cells with low rDNA activity were used.
Conclusion: rDNA reprogramming efficiency in NT embryos was determined by the rDNA activity in donor cells from which they derived.
Significance: Developmental potential of NT embryos with rDNA activity in the donor cells was correlated.
Keywords: Embryo, Fibroblast, Mouse, Nucleolus, Ribosomal RNA (rRNA), Nucleolar Organizer Regions, rDNA Activation, rDNA Methylation, Somatic Cell Nuclear Transfer
Abstract
The well known and most important function of nucleoli is ribosome biogenesis. However, the nucleolus showed delayed development and malfunction in somatic cell nuclear transfer (NT) embryos. Previous studies indicated that nearly half rRNA genes (rDNA) in somatic cells were inactive and not transcribed. We compared the rDNA methylation level, active nucleolar organizer region (NORs) numbers, nucleolar proteins (upstream binding factor (UBF), nucleophosmin (B23)) distribution, and nucleolar-related gene expression in three different donor cells and NT embryos. The results showed embryonic stem cells (ESCs) had the most active NORs and lowest rDNA methylation level (7.66 and 6.76%), whereas mouse embryonic fibroblasts (MEFs) were the opposite (4.70 and 22.57%). After the donor cells were injected into enucleated MII oocytes, cumulus cells and MEFs nuclei lost B23 and UBF signals in 20 min, whereas in ESC-NT embryos, B23 and UBF signals could still be detected at 60 min post-NT. The embryos derived from ESCs, cumulus cells, and MEFs showed the same trend in active NORs numbers (7.19 versus 6.68 versus 5.77, p < 0.05) and rDNA methylation levels (6.36 versus 9.67% versus 15.52%) at the 4-cell stage as that in donor cells. However, the MEF-NT embryos displayed low rRNA synthesis/processing potential at morula stage and had an obvious decrease in blastocyst developmental rate. The results presented clear evidences that the rDNA reprogramming efficiency in NT embryos was determined by the rDNA activity in donor cells from which they derived.
Introduction
Eukaryotic ribosomal genes (rDNA) are arranged in clusters of tandem repeats known as nucleolar organizer regions (NORs),2 which locate on the short arm of acrocentric chromosomes (1). Active NORs are associated with ribosomal RNA (rRNA) transcription machinery including upstream binding factor (UBF) and RNA polymerase I (RPI). These regions often stain positive for silver and are, therefore, called Ag-NORs. rDNA repeat units in inactive NORs are highly methylated and are not associated with the RNA polymerase I machinery (2). Nucleoli form at the end of mitosis around the rDNA and result in a subnuclear compartment, which are constituted of rRNA transcription and processing machineries and responsible for generating ribosome subunits. In the full functional nucleolus, the rDNA transcription initiation complex (including RPI and UBF) is localized to tentative fibrillar centers, where proteins of the early (FBL) and late (nucleolin and nucleophosmin/B23) rRNA-processing machineries are distributed to presumptive fibrillar centers/dense fibrillar component and the full nucleolar compartment, respectively (3). rDNA is first transcribed to 47 S rRNAs by RPI, then the initial transcripts are subsequently cleaved to form the mature 28 S, 18 S, and 5.8 S rRNAs. After post-transcriptional modified through interaction with small nucleolar ribonucleoproteins and additional protein-processing factors, these rRNAs are finally assembled with ribosomal proteins and transported to the cytoplasm (4).
As an overall pattern, functional nucleoli are established in the oocyte upon activation of oocyte growth, which provided enough ribosome content for RNA and protein synthesis at a high level to support early development (5). By the end of oocyte growth, rRNA synthesis ceases. The nucleoli disassemble and change into non-functional nucleolar remnants (6, 7). It is intriguing that mouse oocytes destroy a large amount of rRNA, ribosomal protein, and mRNAs during maturation (8). So the maternal ribosome storage is evidently not sufficient to support embryonic development beyond the zygotic genome activation. Then in late 2-cell stage, the zygotic genome activation becomes apparent (9), and genes involved in ribosome biogenesis and assembly are included in the first gene activation burst (10). However, fibrillo-granular nucleoli with full rRNA synthesis, processing, and ribosome subunit assembly functions are only established at the 4-cell stage in mouse (11), which then provides a large amount of ribosomes for early development. Therefore, the reactivation of rDNA during zygotic genome activation and establishment of full functional nucleolus at the 4-cell stage are very important to preimplantation development of mouse embryos.
Actually, in mammalian cells only ∼50% rDNA is transcribed (1), with the remaining genes inactivated through a combination of different epigenetic mechanisms including methylation (12). With the new cytological technique, researchers indicated that rDNA is regulated allelically in a regional manner in human, with one parental copy of each NOR repressed in any individual cell, which is similar to X-chromosome inactivation in females (13). Whether the same mechanism exists in other species is unknown. When the somatic cell was fused into bovine enucleated oocyte, the nucleolus progressively transformed into nucleolar bodies and lost vacuoles structure (14). The NT embryo needs to rebuild full functional nucleolus, which is important to preimplantation development (15). However, the inactive rDNA copies in donor cells may become a barrier for full activation of rDNA in NT embryos. Recently, several studies reported that the nucleolus biogenesis in NT embryos was abnormal. In bovine granulosa cell nuclear transfer embryos, the reformation of fibrillo-granular nucleoli was initiated one cell cycle earlier than that in in vivo derived embryos. But the UBF localization to the nucleolar compartment was one cell cycle later, which indicated the NT embryos were lacking in development potentials (16). In pig, only half late 4-cell fibroblast NT embryos had transcriptionally active nucleoli, whereas in in vivo embryos the portion was 92% (17). Moreover, in mouse embryonic fibroblast or stem cell-cloned embryos, the activation of functional nucleoli was also one cell cycle-delayed (18).
Because the reprogramming competence of oocyte to somatic cell nuclear is limited, we wonder whether all those inactive rDNA/NORs could be fully activated at the 4-cell stage in mouse NT embryos when compared with normal ones. If not, will it impair ribosome synthesis and intracellular metabolism of early embryonic development? Besides, donor cells with a different differentiation status would yield different outcomes in somatic cell cloning experiments (19–21). We also want to know if the rDNA epigenetic status in donor cells will result in different rRNA synthesis and processing activities in NT embryos and, furthermore, affect preimplantation developmental competence.
In this study we chose mouse embryonic cells (ESCs), cumulus cells (CCs), and embryonic fibroblast cells (MEFs) as donor cells to reconstruct different NT embryos. Intracytoplasmic sperm injection (ICSI) embryos were used as control. The rDNA methylation level, active NORs numbers, and nucleolar-related gene expression were compared in donor cells and in corresponding NT embryos at different preimplantation development stages. The distribution of nucleolar protein (B23 and UBF) was also compared before and after NT.
EXPERIMENTAL PROCEDURES
Animal
B6D2F1 (C57BL/6 × DBA/2) female/male mice were obtained at 8–10 weeks of age from Vital River (Beijing, China). Animals were conformed to the Guide for the Care and Use of Laboratory Animals. All animal experiments were performed under the Code of Practice Harbin Medicine University Ethics Committees.
Cell Culture and Treatment
Derivation and culture of mouse ESCs were according to a previous protocol (22). Cells were cultured in DMEM containing 15% FBS, 50 μg/ml penicillin/streptomycin (Invitrogen, 15140-148), 100 μm nonessential amino acids (Invitrogen, 11140-050), 100 μm β-mercaptoethanol (Sigma, M7522), and 1000 units/ml leukemia inhibitory factor (Chemicon, ESG1107). The medium were changed every day, and the cells were passaged every 2 days. The day before nuclear transfer, 3 μg/ml nocodazole (Sigma, M1404) was added to culture medium overnight to synchronize the cells to metaphase (23, 24). Then the cells were harvested and used as donor cells. All the ESCs used in this study were within 10 passages.
CCs were obtained during oocyte collection, then washed in HEPES-buffered CZB medium (HEPES-CZB) several times and cultured in DMEM containing 50 ng/ml FSH (Sigma, F2297) and 20 ng/ml EGF (Sigma, E4127). Partial cells were resuspended in HEPES-CZB containing 3% PVP (Polyvinylpyrrolidone, Sigma, PVP360) and used as donor cells (G0/G1) for NT directly.
MEFs were isolated from 13.5 post-coitum B6D2F1 mouse fetus as previous reported (25). Cells were cultured in DMEM containing 10% FBS under 5% CO2 in humidified air at 37 °C within three passages. Partial MEFs were treated with 10 μg/ml mitomycin C (Sigma, M4287) for 2.5 h then used as feed layers for ES cells. MEFs at the third passage were kept in DMEM containing 0.5% FBS for 72 h as serum starvation treatment and then used as NT donor cells (G0/G1).
Metaphase Chromosome Spreads Preparation, Karyotyping, and Silver-staining of Donor Cells
The ESCs, CCs, and MEFs were kept in culture medium containing 3 μg/ml nocodazole for 1, 12, and 12 h, respectively, to synchronize the cells to metaphase. Then the cells were made into metaphase chromosome spreads as previous reported (26). For karyotype observation, several slides of each group were stained with 4% Giemsa in PBS for 10 min. For silver-staining of active NORs, slides were first incubated in a humidity box at 37 °C for at least 30 min and then stained with fresh mixed silver-stain medium (2% agar medium with 1% formic acid, 50% AgNO3 medium (1:2)) in the dark at 40–50 °C for 8 min. The slides were washed, dried, mounted, and finally observed with a 100× oil-immersion objective.
Oocyte Recovery and Spermatozoa Preparation
Female B6D2F1 mice were superovulated by intraperitoneal injections of 5 IU pregnant mare serum gonadotropin (NSH, China) followed 48 h later by 5 IU of human chorionic gonadotropin (NSH, China). Oocytes were collected from the oviducts 14 h after human chorionic gonadotropin injection. After collection, cumulus cells were removed from oocytes with 300 μg/ml hyaluronidase (Sigma, H4272) in droplets of HEPES-CZB by gentle pipetting. Denuded oocytes with homogeneous ooplasm were selected and kept in new droplets of CZB medium containing 5.6 mm glucose (CZBG), covered with sterile mineral oil (Fisher, O121-20), then cultured at 37 °C in a 5% CO2 atmosphere until use. Spermatozoa were collected from the cauda epididymis of 8–12 weeks age of B6D2F1 males, then kept in CZB-HEPES medium and prepared for injection.
Generation of ICSI and NT Embryos
ICSI was carried out by a piezo-driven unit using the methods as described elsewhere (27, 28), except that our experiment was performed in HEPES-CZB containing 5 μg/ml cytochalasin B (Sigma, C6762) at room temperature (29). Only the sperm head was injected into oocyte. After 10–20 min of recovery, the ICSI-generated embryos were washed several times and cultured in K-modified simplex optimized medium at 37 °C in a 5% CO2 atmosphere. To eliminate any possible effects brought by NT methods to the following investigation, we adopted the same one-step micromanipulation technique to reconstruct ESCs nuclear transfer (ESNT), CCs nuclear transfer (CCNT), and MEFs nuclear transfer (MEFNT) embryos as described previously (30) with modifications. The outer diameters of injection pipettes used for ESCs, CCs, and MEFs were 13, 9, and 12 μm, respectively. Briefly, the donor cell membrane was broken with several Piezo pulses, then 5–10 cells were sucked into the injection pipette. The oocyte MII spindle was adjusted to 8–10 o'clock, then one donor cell was injected into the nearby plasma. The spindle was sucked into the injection pipette immediately and taken out of oocyte. One hour after NT, the reconstructed CCNT and MEFNT embryos were activated with 5 mm SrCl2 (Sigma, 439665) in Ca2+-free CZB containing 5 μg/ml CB for 6 h, whereas the ESNT embryos were just activated in SrCl2-added Ca2+-free CZB for 6 h. Then the embryos were washed in K-modified simplex optimized medium and cultured in the same conditions as ICSI embryos.
Immunofluorescent Detection of Nucleolar Proteins
Cells used for the immunofluorescent experiment were cultured in an 8-well glass slide (Thermo, 1256518). Embryos at 0, and 20-, 40-, and 60-min post donor cell/sperm injection and 4-, 6-, 8-, 24-, 48-, and 72-h post-activation/sperm injection (hpa) were collected. The cells and embryos were fixed in 4% paraformaldehyde for 40 min and permeabilized with 1% Triton X-100 for 30min. After incubation in Image-iTTM FX Signal Enhancer (Invitrogen, I36933) for 30 min, the cells and embryos were blocked with 1% bovine serum albumin (Sigma, A9418) in PBS for 1 h, then incubated with monoclonal anti-B23 antibody (Sigma, FC82291, 1:100) or monoclonal anti-UBF antibody (Sigma, WH00007343M1, 1:100) overnight at 4 °C followed by Alexa Fluor 488 donkey anti-mouse IgG (Invitrogen, A21202, 1:400) incubation for 1 h. After the nuclei were stained with 10 μg/ml Hoechst 33342, the cells were mounted on 8-well glass slides as per the manufacture's instruction, and the embryos were mounted on slides with DABCO (1,4-diazabicyclo(2.2.2)octane; Beyotime, P0126) and observed with laser-scanning confocal microscope (Zeiss, LSM700). For examination of UBF spots in 4-cell stage embryos at metaphase, the embryos were incubated in KOSM medium containing 3 μg/ml nocodazole for 6 h (started from 48 hpa) and then collected, and subjected to the same sample preparation procedures. Serial layer scanning was performed over the target metaphase plates.
qPCR Evaluation of Nucleolar-related Genes Expression
Embryos at 24, 48, 72, and 92 hpa were collected as at 2-cell, 4-cell, morula, and blastocyst stage, respectively. RNA extraction and qPCR evaluation were performed as described previously with modifications (31). Each embryo sample was added with 105 copies of XenoTM RNA from SYBR Green Cell-to-CTTM Control kit (Invitrogen, 4402959) during cell lysis as external control. The total RNA of cells and embryos was extracted by RNeasy Mini kit (Qiagen, 74104) according to the manufacturer's instructions. cDNA was synthesized from total RNA with a High Capacity cDNA Reverse Transcription kit (ABI, 4368814). qPCR reactions were performed based on 1.5 μl of cDNA sample, 10 μl of TransStartTM Top Green qPCR SuperMix (TransGen, AQ131), and gene-specific primers (supplemental Table 1) in a 20-μl reaction system on CFX96 Real-time System (Bio-Rad). H2afz, Hprt1, and ExnoTM were used as candidate references for embryos (31, 32). HP1γ, cyclophilin-A, and cyclophilin-B were used as candidate references for cells (13).
Detection of rDNA Methylation in Donor Cells and 4-Cell Embryos
The 5′-ETS sequence (81–507, GenBankTM BK000964.1) was used as the target sequence for detection of rDNA methylation levels (13). The experiment was conducted with the EZ DNA Methylation-DirectTM kit (ZYMO Research, Irvine, CA) according to the manufacturer's instruction. Around 10,000 cells for each cell group and 70–80 embryos for each 4-cell embryo group were used as samples. The samples were digested with proteinase K, then the unmethylated cytosines were converted to uracil by bisulfite. The target sequence was amplified by PCR and subjected to sequence analysis. The primers sequences used were TAGTTTATTTTTTTTATTGGTTTGG (forward) and TAACATAAACACTTAAACACCACAA (reverse).
Experiment Design and Statistical Analysis
Each experiment was repeated at least three times. More than 5 slides and 50 metaphase plates were checked for each group in karyotype and silver-stain experiment. More than five embryos in each group were selected for nucleolar protein observation by confocal microscope. More than 20 4-cell embryos blocked at metaphase in each group were observed for detection of UBF spots. The data were compared using the Least Significant Difference test after analysis of variance in SPSS 13.0. In qPCR experiments, 20 2-cell embryos, 15 4-cell embryos, 10 morulas, and 10 blastocysts of each group were used as one repeat. The raw data were first imported to qBasePLUS 2.2 software (Biogazelle) for reference gene selection and data calculation. Then the relative gene expression in ICSI embryos was set to 1. Finally, the relative amount of gene expression was compared using the Least Significant Difference test after analysis of variance in SPSS. A value of p < 0.05 was considered statistically significant.
RESULTS
rDNA Activity in ESCs, CCs, and MEFs
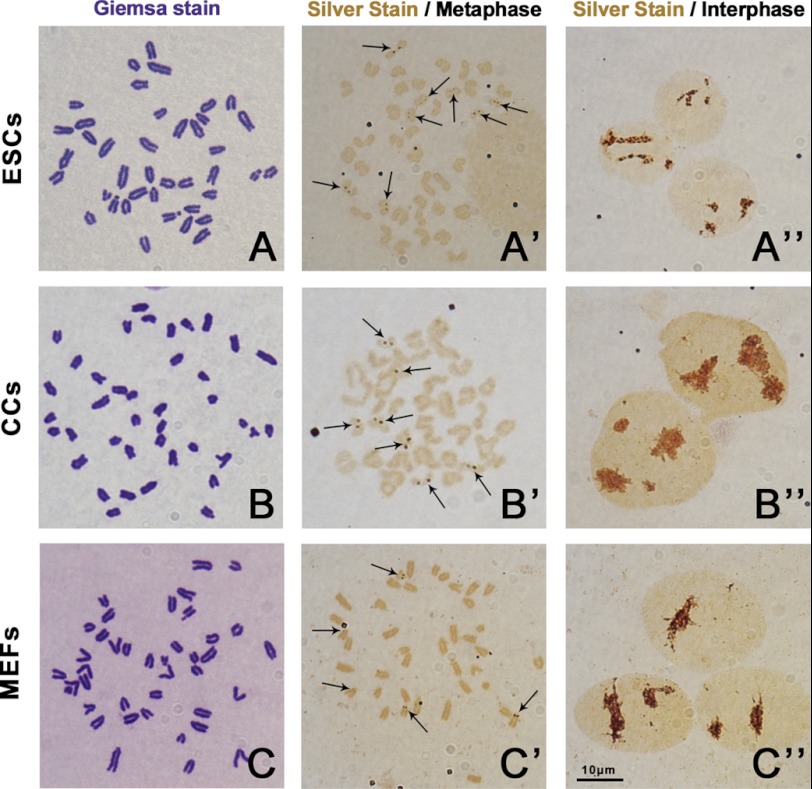
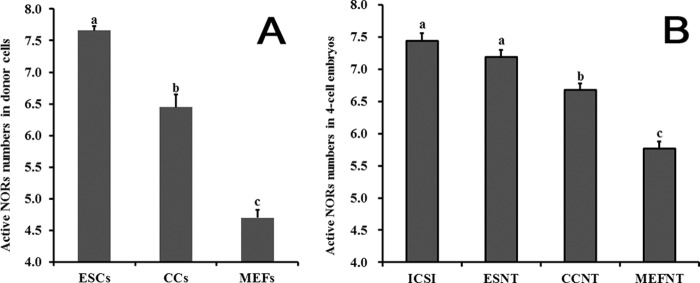
We first examined the karyotypes of ESCs, CCs, and MEFs. All the 3 cell lines showed normal karyotypes that contained 40 acrocentric chromosomes (Fig. 1, A–C). Then the metaphase slides were stained with AgNO3 to show the active NORs. At interphase, the silver-stain signals in three groups all showed as brown clusters with irregular shapes in the nucleus with no obvious differences (Fig. 1, A′′, B′′, and C′′). At metaphase, the silver-stain signals showed as brown spot pairs near centromeres of certain chromosomes (Fig. 1, A′, B′, and C′). The ESCs had significantly more chromosomes containing brown spot pairs than CCs and MEFs (7.66 versus 6.45 versus 4.70, p < 0.05) (Fig. 2A). After studying the pair numbers profile of these cell lines, we found that the majority of ESCs had eight pairs of spots. But for CCs, more than half of the cells had seven pairs of spots, and the rest were mainly below seven. However, MEFs had the least pair numbers, and most of the cells only had four or five pairs of spots (supplemental Fig. 1). Because the rDNA transcription machinery is still assembled during mitosis in active NORs and absent in inactive NORs (2), the spot pair numbers observed during metaphase plate could be directly considered as active NORs numbers. The result indicated that the three donor cells had different active NORs numbers during normal culture and passages.
FIGURE 1.
Karyotype and active NORs observation of mouse ESCs, CCs, and MEFs. The metaphase chromosome spreads were stained with 4% Giemsa for karyotyping or stained with silver stain medium to show active NORs. All the three cell lines had normal karyotypes (A, B, and C). The active NORs were stained by AgNO3 and are shown as brown spot pairs on certain chromosomes (arrows). ESCs had more spot pair numbers than CCs and MEFs at metaphase (A′, B′, C′). At interphase, the silver stain signal showed as big brown clusters in the nuclei in all the three cell lines (A′′, B′′, C′′). Bar = 10 μm.
FIGURE 2.
Active NORs in donor cells and in 4-cell embryos. Metaphase spreads of ESCS, CCs, and MEFs were stained with AgNO3, then the active NORs numbers were counted (A). ICSI, ESNT, CCNT, and MEFNT embryos at the 4-cell stage were blocked to metaphase by nocodazole treatment, then the UBF signal spots were labeled and counted (B). Columns with different letters indicate significant difference (p < 0.05). ESCs had significantly more active NORs than CCs and MEFs (7.66 versus 6.45 versus 4.70, p < 0.05), and MEFs had the least. ICSI and ESNT embryos had no significant difference in active NORs numbers at the 4-cell stage, but their active NORs numbers were significantly more than CCNT and MEFNT embryos (7.44 versus 7.19 versus 6.68 versus 5.77).
To find out whether rDNA methylation accounts for the difference among the cell lines in active NORs numbers, we detected the rDNA methylation levels of the cells at 5′-ETS of rDNA (13). The results showed ESCs had the lowest methylation level (6.67%), whereas MEFs had the highest (22.57%), and CCs were in the middle (13.59%) (Fig. 10), which indicated rDNA methylation did contribute to the inactivation of certain NORs.
FIGURE 10.
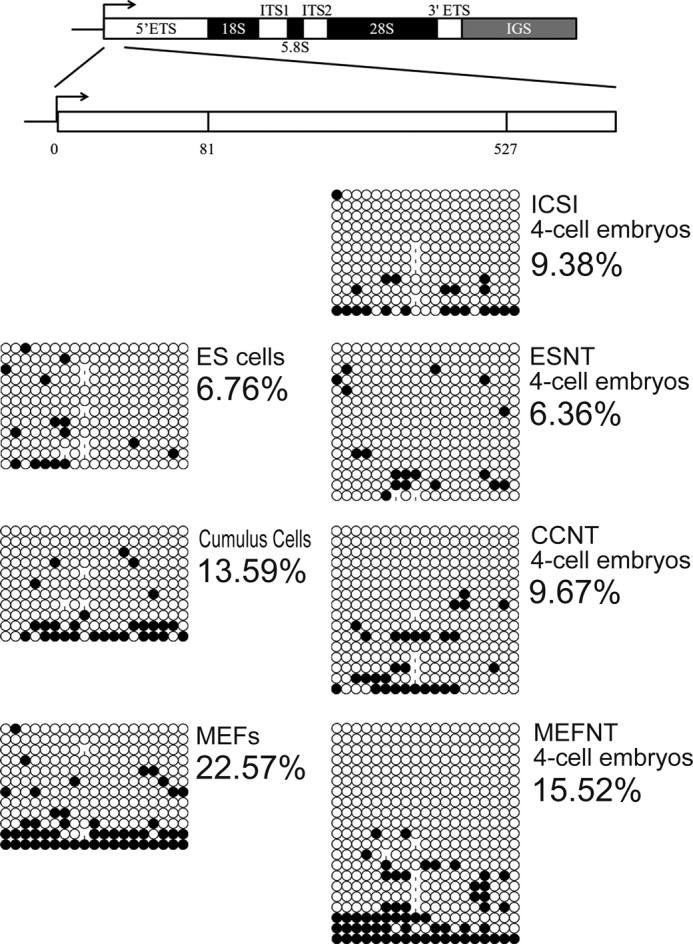
rDNA methylation detection of three donor cells and ICSI, ESNT, CCNT, MEFNT embryos at 4-cell stage. 5′-ETS (81–527) of the rDNA gene was chosen as the target sequence to evaluate the rDNA methylation levels in donor cells and 4-cell embryos. The methylated cytosines were counted and compared.
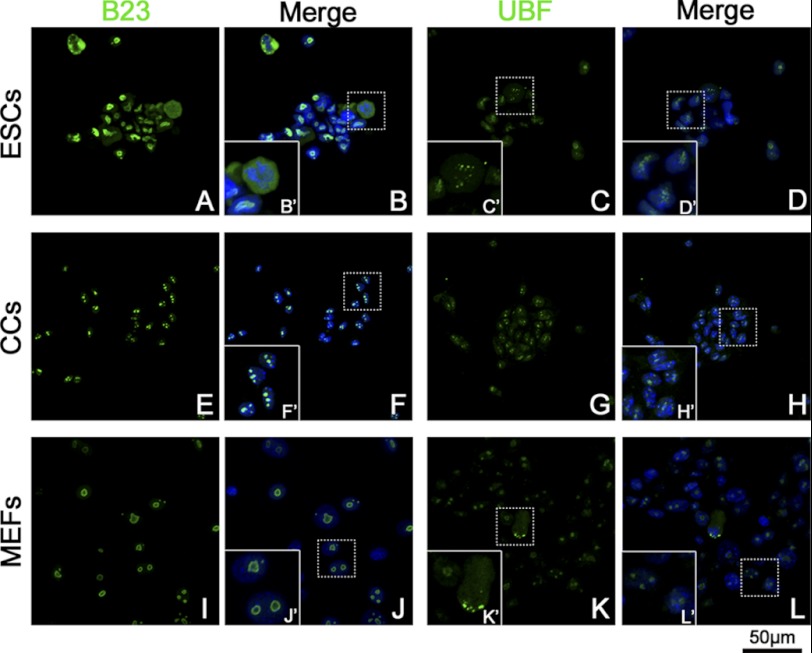
We then studied the distribution of nucleolar protein UBF and B23 in three types of cells that represent rDNA transcription and pre-rRNA-processing machineries, respectively. In CCs, B23 signals showed as three or four condensed clusters in nuclear (Fig. 3E). For MEFs, the B23 signal displayed as 1–3 bigger rings (Fig. 3I). The B23 signals in ESCs nuclei were much stronger than those in CCs or MEFs but irregular in shape (Fig. 3A). However, B23 signals in ESCs during metaphase distributed in the whole cytoplasm (Fig. 3B′) as mentioned previously (4). The UBF signal also showed as clusters within nuclear in three groups (Fig. 3, C, G, and K). During metaphase, the UBF signal aggregated into pairs of small spots on certain chromosomes (Fig. 3, C′ and K′), which presented a pattern similar to silver-staining, as UBF was one component of the transcription machinery.
FIGURE 3.
Distribution of B23 and UBF in ESCs, CCs, and MEFs. B23 and UBF were labeled by monoclonal anti-B23 antibody/monoclonal anti-UBF antibody, respectively, and fluorescent conjunct second antibody (green). The nuclei/chromosomes were stained with Hoechst 33342 (blue). Typical areas (dotted line rectangle) were magnified to show the details (solid line rectangle). B23 showed as three or four condensed clusters within the nucleus in CCs (F′). In MEFs, B23 signals showed as 1–3 big rings (J′). ESCs showed much a stronger B23 signal with an irregular shape (A). At metaphase, the B23 signal distributed to the whole cytoplasm (B′). The UBF signal showed as several clusters in the nuclei of all the three groups of cells (C, G, and K). During mitosis, the UBF signal showed as pairs of small spots in metaphase plate (C′) or spindle polar (K′).
We also detected the nucleolar-related genes expression (47 S rRNA, 18 S rRNA, UBF, FBL, B23, RPI) by qPCR. HP1γ and, cyclophilin-A were stable enough to be applied as reference genes according to geNormPLUS analysis. The results demonstrated that ESCs had significant higher UBF, FBL, and B23 expression levels but lower 18 S rRNA levels. On the contrary, MEFs had high 47 S rRNA and 18 S rRNA expression levels but were low in UBF, FBL, and B23 levels. CCs had the highest RPI expression level, but its 47 S rRNA expression level was the lowest (Fig. 4). These data also indicated that ESCs owned higher rRNA synthesis, pre-rRNA processing, and ribosome assembly activities than CCs and MEFs, which was in accordance with its high proliferation and self-renew features.
FIGURE 4.
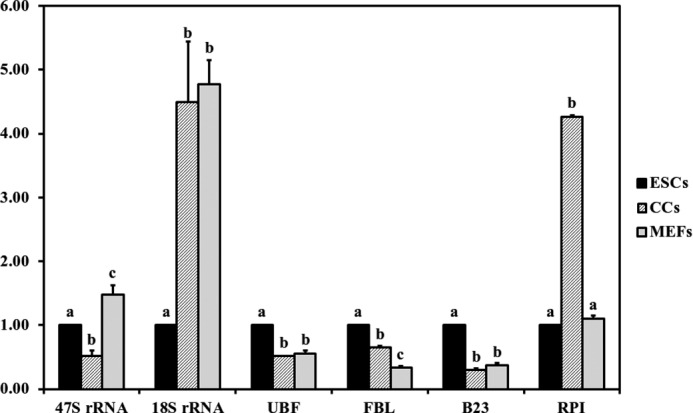
Nucleolar-related gene expression in ESCs, CCs, and MEFs. The expression level of ESCs was set as 1. Different letters indicate significant differences at p < 0.05.
B23 and UBF Distribution in Reconstructed Embryos 1 h after NT
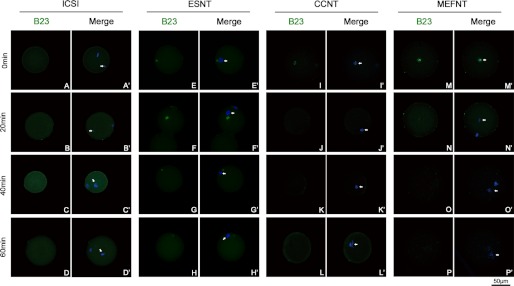
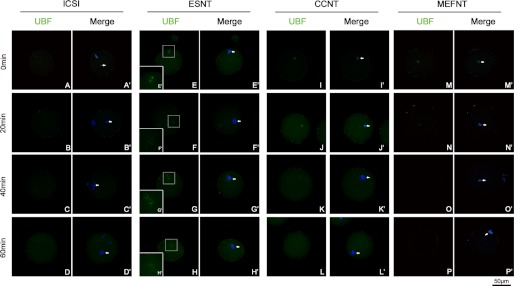
Because several differences were observed among the donor cells, we then checked the B23 and UBF distribution after NT to study whether they have different responses after being injected into ooplasm. Our pre-experiments showed the B23 and UBF signals of cumulus cell nucleoli disappeared within 1 h by 1 μg/ml actinomycin D (an RPI inhibitor) treatment (data not shown). We then fixed the embryos at 0, 20, 40, and 60 min post-NT/sperm injection. The results indicated that B23 and UBF signals co-localized within donor cell nuclear, except for B23 in the ESNT group (Fig. 5, E, F, G, and H). Immediately after injection, the B23 and UBF signals in reconstructed embryos displayed the same distributions as seen in cells. The B23 and UBF signals vanished 20 min after NT in CCNT and MEFNT embryos (Figs. 5, I, J, M, and N, and 6 I, J, M, and N), and indicated the rRNA transcription and processing machineries were immediately disassembled in ooplasm. In ESNT embryos, both signals could still be detected at 60 min post-NT (Figs. 5H and 6H), although the signals were getting weaker over time. As a comparison, no B23 or UBF signal could be detected at the injected sperm head or maternal chromosomes in ICSI embryos (Figs. 5, A–D, and 6, A–D).
FIGURE 5.
Changing of B23 signal in NT and ICSI embryos during a 1-h post-NT/sperm injection. B23s were labeled by monoclonal anti-UBF antibody and fluorescent conjunct second antibody (green). The donor cell chromatin was stained with Hoechst 33342 (blue). Arrowheads indicate the donor cell chromatin/sperm head. The B23 signals in CCNT and MEFNT were showed as several obvious clusters (I and M). But after 20 min, the signals got much weaker (J and N). No B23 signal could be detected after 40 min (K, L, O, and P). The B23 in ESNT group lasted for 60 min, although the signal got weaker over time, but the signal was not companied with chromatins (E–H). No B23 signal could be detected at sperm head or maternal chromatins (A–D).
FIGURE 6.
Changing of UBF signal in NT and ICSI embryos during a 1-h post-NT/sperm injection. UBFs were labeled by monoclonal anti-UBF antibody and fluorescent conjunct second antibody (green). The donor cell chromatin was stained with Hoechst 33342 (blue). Arrowheads indicate the donor cell chromatin/sperm head. The UBF signals in CCNT and MEFNT were showed as several obvious clusters (I and M). After 20 min, their signals got much weaker (J and N). Almost no B23 signal could be detected at and after 40 min (K, L, O, and P). The UBF in ESNT group showed as small bright spots in pairs and lasted for 60 min (E′–H′). No UBF signal could be detected at sperm head or maternal chromatins (A–D).
B23 and UBF Distribution in Reconstructed Embryos during Preimplantation Development
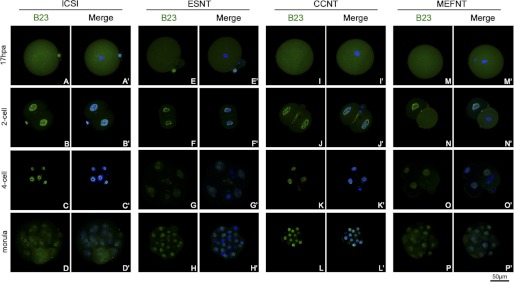
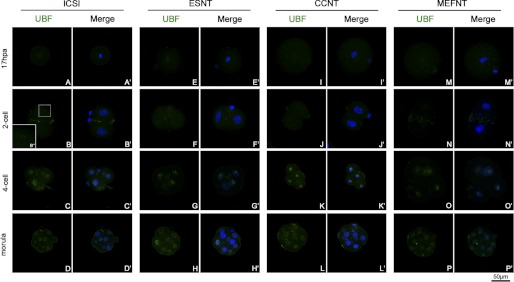
We also studied the distribution of B23 and UBF in ICSI, ESNT, CCNT, and MEFNT embryos at pronuclear, 2-cell, 4-cell, and morula stages. B23 recruited in pronuclei at 6 hpa, and the signal became stronger at 8 hpa because this protein was inherited and reused in early cleavage (33). There were no obvious differences among the four groups. Meanwhile, UBF could not be detected at pronuclear stage (supplemental Figs. 2 and 3). At metaphase of the first mitosis (17 hpa), B23 signals distributed to the whole cytoplasm (Fig. 7, A, E, I, and M). No UBF signals could be detected on metaphase chromosomes of each group (Fig. 8, A, E, I, and M). The B23 protein re-accumulated in nuclear at 2-cell stage (24 hpa). As blastomere started cleavage, B23 distributed to the whole plasma again (Fig. 7, B, F, J, and N). Meanwhile, no UBF signal could be detected in ESNT, CCNT, and MEFNT embryos at this stage (Fig. 8, F, J, and N). However, several ICSI embryos (3 in 5) showed weak UBF spots around the nucleoli (Fig. 8B′), which indicated they may start to synthesis rRNA earlier than NT counterparts. At 4-cell and morula stage, all groups showed ring-like B23 signals around nucleoli without any obvious differences (Fig. 7, C, D, G, H, K, L, O, and P). UBF signals showed as discontinuous small clusters around nucleoli at the 4-cell stage (Fig. 8, C, G, K, and O). At the morula stage, many blastomeres were under mitosis, and the UBF signals distributed in the whole cytoplasm with chromosomes or at the spindle poles (Fig. 8, D, H, L, and P). No obvious differences were observed among the four groups at the 4-cell or morula stages.
FIGURE 7.
Distribution of B23 in NT and ICSI embryos at difference preimplantation developmental stages. B23s were labeled by monoclonal anti-B23 antibody and fluorescent conjunct second antibody (green). The nuclei/chromosomes were stained with Hoechst 33342 (blue). The B23 distributed to the whole cytoplasm at metaphase of the first mitosis (A, E, I, and M) then recruited in the nuclei at the 2-cell stage (B, F, J, and N). When the blastomere started to cleavage, B23 distributed to the cytoplasm again (N). All the four groups of embryos showed ring-like B23 signals around the nucleoli at 4-cell and morula stages (C, D, G, H, K, L, O, and P).
FIGURE 8.
Distribution of UBF in NT and ICSI embryos at difference preimplantation developmental stages. UBFs were labeled by monoclonal anti-UBF antibody and fluorescent conjunct second antibody (green). The nuclei/chromosomes were stained with Hoechst 33342 (blue). No UBF signal could be detected at metaphase of the first mitosis (A, E, I, and M). At the 2-cell stage, no UBF signal could be detected in ESNT, CCNT, and MEFNT embryos (B, F, J, and N). But several ICSI embryos showed weak UBF spots around the nucleoli (B′′). At the 4-cell stage, the UBF signals showed as discontinuous small clusters around nucleoli (C, G, K, and O). At the morula stage, many blastomeres were under mitosis, and the UBF signals distributed in the whole cytoplasm with chromosomes or at the spindle polar (D, H, L, and P).
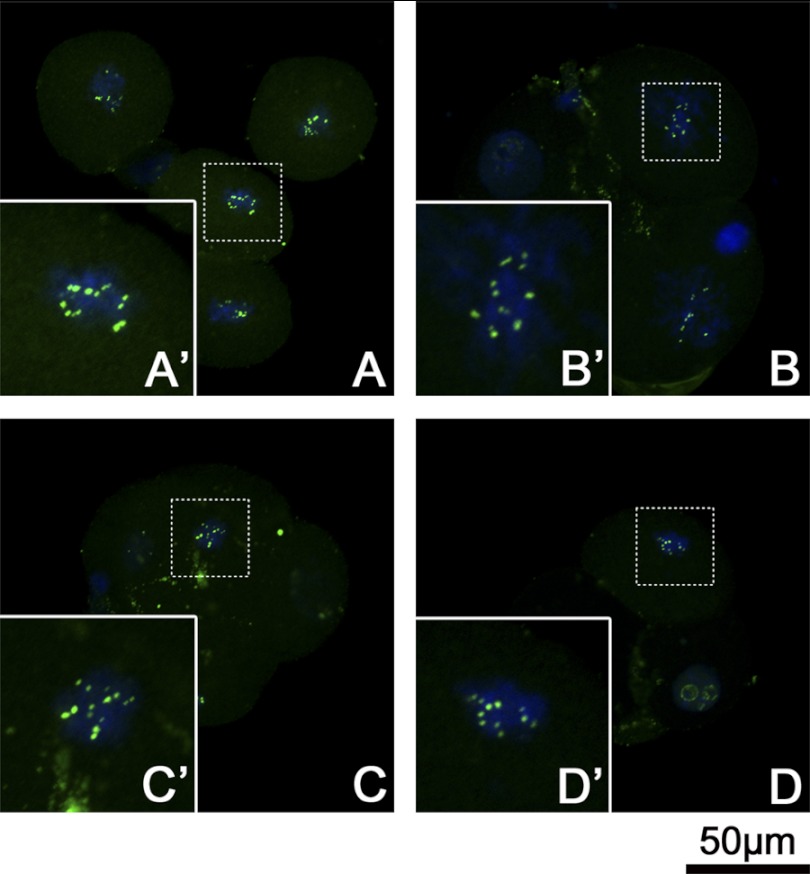
Active NORs Numbers and rDNA Methylation Level in 4-Cell Stage Embryos
Although silver stain is an effective method to distinguish the transcription active NORs from inactive ones, for 4-cell embryos it was difficult to prepare metaphase plates and conduct silver-stain procedures. Because UBF remain associated with active NORs throughout mitosis (5, 34), we then blocked the 4-cell embryos to metaphase by nocodazole and detected the UBF signals as an alternative way to count the active NORs. As shown in Fig. 9, in every blastomere at metaphase, the UBF signals strictly adhered to certain chromosomes and were revealed as bright signal spots in pairs. After comparison of the pair numbers of UBF spots in each group, we found ICSI and ESNT embryos owned significant more pair numbers than CCNT and MEFNT embryos (7.44 and 7.19 versus 6.68 versus 5.77, p < 0.05). However, there was no significant difference between ICSI and ESNT groups (Fig. 2B).
FIGURE 9.
UBF signals of 4-cell stage embryos at metaphase. ICSI embryos (A), ESNT embryos (B), CCNT embryos (C), and MEFNT embryos (D) at 48 h post-activation/sperm injection were blocked with 3 μg/ml nocodazole for 6 h to synchronize to the metaphase. UBF were labeled by monoclonal anti-UBF antibody and fluorescent conjunct second antibody (green). The chromosomes were stained with Hoechst 33342 (blue). Metaphase plate (dotted line rectangle) was magnified to show the details (solid line rectangle). The UBF signals showed as spot pairs on certain chromosomes. Four groups shared the same pattern but with different spot pair numbers.
rDNA methylation status of 4-cell embryos were also evaluated. The rDNA methylated level at 5′-ETS sequence of ICSI, ESNT, CCNT, and MEFNT embryos were 9.38, 6.36, 9.67, and 15.52 %, respectively (see Fig. 10). MEFNT embryos still had the highest methylation level, indicating those methylated rDNA copies in donor cells were not fully activated at 4-cell stage.
Preimplantation Development and Nucleolar-related Gene Expression of ICSI and NT Embryos
We then compared preimplantation development rates of the four groups of embryos. ICSI embryos had the highest development rate at every preimplantation developmental stage, reaching 97% in blastocyst rate. Among the NT groups, ESNT embryos had significantly higher morula and blastocyst rates (80.7 and 69.3%) than that of CCNT (72.2 and 62.0%) and MEFNT (60.2 and 35.6%) embryos (supplemental Table 2). MEFNT had the lowest development potential after the 2-cell stage and, moreover, showed an obvious drop of development rate from morula to blastocyst stage (Fig. 11).
FIGURE 11.
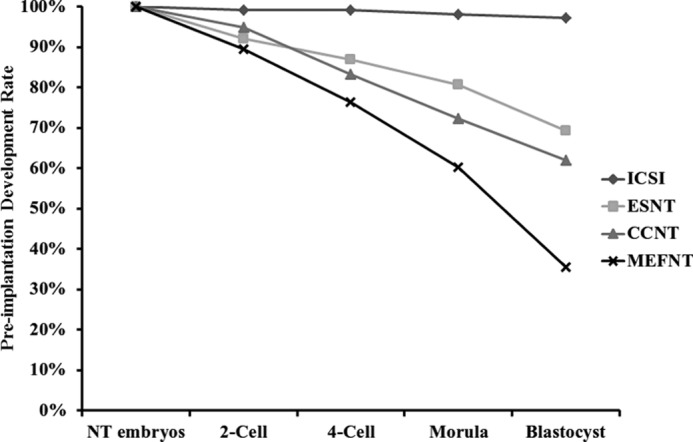
Preimplantation development of ICSI, ESNT, CCNT, and MEFNT embryos. Reconstructed embryo number was considered as 100% in each group. 4-Cell, morula, and blastocyst development rates were calculated based on 2-cell embryo numbers. ICSI embryos had the highest development rate at 4-cell, morula, and blastocyst stages. MEFNT embryos had significant lower development rates when compared with other groups at different stages and showed a dramatic drop from morula to blastocyst stage.
Finally, we compared the nucleolar-related gene expression of these embryos at different preimplantation developmental stages. Hrpt1 and ExnoTM were used as internal and external reference genes, respectively, according to geNormPLUS analysis. At the 2-cell stage, ICSI embryos had significantly higher UBF and RPI expression levels than NT groups, which was consistent with earlier immunofluorescent results that UBF could only be detected in ICSI embryos at the 2-cell stage. The ESNT group showed significant higher 47 S rRNA expression than CCNT embryos and significantly higher RPI levels than CCNT and MEFNT counterparts. There were no significant differences in 18 S rRNA, UBF, FBL, and B23 expression levels among the NT groups (supplemental Fig. 4). At the 4-cell stage, ICSI embryos had significant higher 47 S and FBL levels than the three NT groups. There were still no significant differences among ESNT, CCNT, and MEFNT groups at the expression levels of most genes except RPI (Fig. 12). When it came to the morula stage, we found MEFNT embryos had significant lower expression of FBL and 18 S rRNA compared with other groups. Moreover, its UBF and 47 S rRNA expression levels were significant lower than that of ICSI embryos (Fig. 13). At the blastocyst stage, the three NT groups had significant higher 47 S rRNA levels than that of the ICSI group. MEFNT blastocysts had significantly lower B23 levels than ICSI embryos, and the ESNT embryos had significantly lower RPI expression than the CCNT counterpart. No other differences existed between the four groups (supplemental Fig. 5).
FIGURE 12.
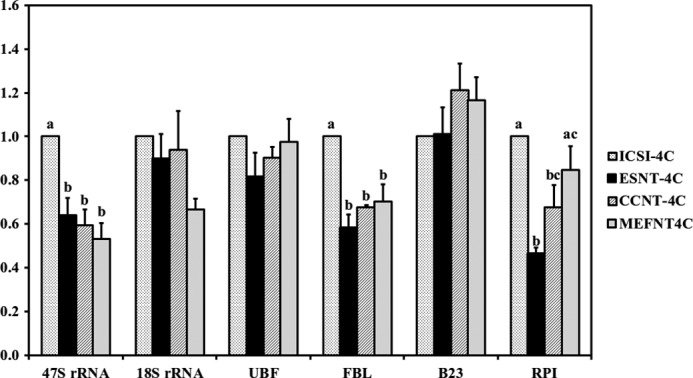
Nucleolar-related gene expression at 4-cell stage (48 hpa) of ICSI, ESNT, CCNT, and MEFNT embryos. The expression level of ICSI embryos was set as 1. Different letters indicate significant differences at p < 0.05.
FIGURE 13.
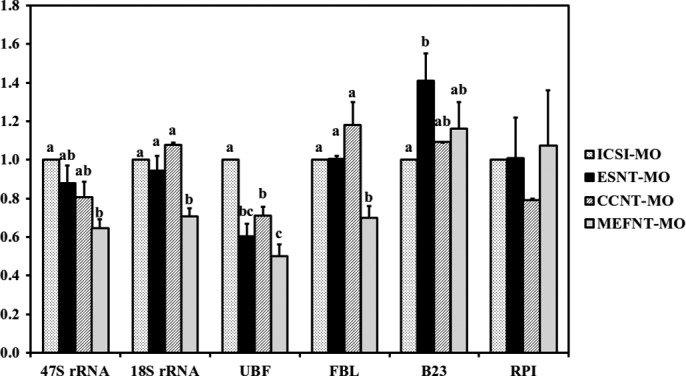
Nucleolar-related gene expression at morula stage (72 hpa) of ICSI, ESNT, CCNT, and MEFNT embryos. The expression level of ICSI embryos was set as 1. Different letters indicate significant differences at p < 0.05.
DISCUSSION
The successful reprogramming of the donor cell genome to the state of totipotency requires both the continued silencing of the somatic-specific genes and the activation of essential embryonic genes, enabling further embryonic development (35–37). Moreover, the developing preimplantation embryo has a profound need for synthesis of proteins supporting both housekeeping and cell differentiation. So it is not surprising that the rRNA genes are among the earliest genes being activated. A proper activation of these genes is crucial for continued embryonic development (38, 39).
In this study, ESCs, CCs, and MEFs were chosen as donor cells as they were easy to achieve, widely used in NT experiments, and were supposed to have different differentiation status. rDNA clusters (NORs) commonly exist on 5 chromosomes (12, 15, 16, 18, and 19) in standard inbred mouse strains. These strains may have 3–5 pairs of chromosomes with silver-stained active NORs (40, 41). For outbred F1 strains, they may have half of the maternal/paternal NOR-bearing chromosomes (42). However, the reported NORs numbers for C57BL/6J and DBA/2 strains varied due to different methods and samples used (40, 41, 43, 44). Moreover, there was still no available data of the NOR numbers in B6D2F1 strain. Previous studies indicated that >90% of the NORs in ES cells were marked with both H3Ac and H3K4me, which means almost all the NORs had transcription activity (13, 45). In our system 68% of the ESCs had 8 active NORs at metaphase, and the active NORs numbers in CCs, MEFs, and all the four groups of embryos did not exceed 8. We speculated that the outbred B6D2F1 mouse used in our study had eight NOR-bearing chromosomes in total. The diploid genome of mouse contains about 400 rDNA copies whose expression depends on the demand of cell biosynthetic activity. The ratio of active and inactive rRNA genes is tissue-specific and is stably propagated through the cell cycle (46). As a kind of somatic cell, the relative more active NORs in CCs may reflect its high proliferation and secretion activity during oocyte growth and maturation. MEFs had the least active NORs, which is in accordance with its high rDNA-methylated status. Therefore, the three cell lines had different rDNA activities that were intimately linked with rDNA methylation level as previously reported (12). It then provided a good model for evaluating the influence of donor cell rDNA activity on NT embryos in the following experiments.
The nucleolar protein distributions in donor cell nucleoli before and after NT have never been demonstrated in mice before. Previous work on bovine NT embryos showed that after bovine skin fibroblast was fused into enucleated MII oocyte, the donor cell nucleoli degenerated, as indicated by B23 and FBL signals. The same phenomenon could be simulated by treating the donor cells with 5 μg/ml actinomycin D, a kind of rDNA transcription inhibitor. However, the two proteins could still be detected at 2 h post-fusion (14). In our study, UBF and B23 were used as indicators of rRNA transcription and processing machineries. They both vanished 20 min post-NT in CCNT and MEFNT embryos, which was much faster than that in bovine NT embryos. This result indicated that the donor cell nucleoli lost rRNA synthesis and processing activities quickly in ooplasm and may undergo degeneration. However, UBF and B23 signals in ESCs were more resistant to ooplasm as they were still detectable 60 min after NT. Because almost all the CCs and MEFs used in NT experiments were at G0/G1 phase but ESCs were at metaphase before NT, the difference may come from the cell cycle phase of donor cell at the time of entering ooplasm. During mitosis, the nucleolar disassembly starts with the loss of RPI subunits from the fibrillar centers, and no nascent rRNA transcripts are synthesized (34, 47). So the ESNT embryos had no rDNA transcription activity after NT, like the other two somatic cell NT counterparts. No sign of rRNA transcription activity was detected during pronucleus formation and the first mitosis (17 hpa), as previously reported (48). Because UBF remaining associated with chromosomes at active NORs is a typical pattern during mitosis (49), lacking a UBF signal during the first mitosis in NT embryos indicated that donor cell chromosomes had undergone deep remodeling. Until 2-cell stage, the ICSI embryos took the lead to transcribe rRNA, and then all the groups started to transcribe and process rRNAs. Thus, we speculated that all the embryos lost rRNA transcription and processing capacity shortly after NT. The different rDNA epigenetic status and rRNA synthesis activities in donor cells would barely have any impact on NT embryos before 2-cell stage.
ESNT embryo had more RPI and 47 S rRNA expressed at the 2-cell stage than that of the CCNT and MEFNT counterparts. More opened NORs in chromosomes may result in earlier or more rRNA transcription. Apart from this, no other obvious differences were observed among the NT groups at 2-cell and 4-cell stages in nucleolar proteins distribution and nucleolar-related gene expression. However, ESNT embryos had almost all the NORs activated and with no significant difference compared with ICSI embryos, which indicated their rDNA activity was near the normal embryos. The active NORs numbers in MEFNT embryos obviously increased when compared with MEFs but were still much less than being sufficiently activated as in ICSI or ESNT embryos. The rDNA methylation levels in three NT groups slightly decreased when compared with the donor cells from which they derived. The result indicated that partial methylated rDNA were unmethylated during NT reprogramming. Moreover, they repeated exactly the same trend as what was seen in the donor cells; that is, ESNT embryos had the lowest rDNA methylation level, MEFNT had the highest, and CCNT embryos were in the middle. In summary, the rDNA activity in 4-cell NT embryos directly reflected the rDNA activity in donor cells from which they derived.
Full functional nucleolus is established at the 4-cell stage in in vitro fertilization mouse embryos, accompanied with formation of fully fibrillo-granular nucleolus in ultra-structure (11), which is the most important step of nucleolus biogenesis during preimplantation development. However, researchers demonstrated that mouse MEFNT embryos had poor nucleolus development at the 4-cell stage, as the fibrillo-granular structure only appeared at the surface of the nucleolus (18). This phenomena could be partially explained by our results that MEFNT embryos had the least active NORs and the functional nucleoli formation was based on the active NORs (4). As a kind of somatic cell NT embryo, CCNT embryos had relatively more active NORs and almost the same nucleolar-related genes expression level when compared with ESNT embryos. This may be one of the reasons why CCs always yielded better cloning outcomes than other somatic cells and were wildly used in NT experiments of different animals (50). Because the oocytes were supposed to have the same reprogramming competence against different donor cells, we believed that the rDNA methylation level and active NOR numbers in 4-cell embryos were determined by the rDNA epigenetic status of donor cells.
When it came to the morula stage, the MEFNT embryos showed obvious weakness in 47 S rRNA, 18 S rRNA, UBF, and FBL expressions. We examined the rRNA transcripts level from 2-cell to blastocyst in ICSI embryos and found they reach the peak at morula stage (supplemental Fig. 6). The immunofluorescent results also revealed that many blastomeres were undergoing mitosis at this stage. These indicated the embryos may have the highest proliferation and metabolism level around morula stage. Because the rRNA transcription, pre-rRNA processing, and ribosome assembly are well linked and coordinated (51), we could speculate that the low rDNA activity in MEFNT embryos would result in insufficient rRNA transcription, processing, and ribosome assembly, which could not fulfill the high ribosome requirement around morula stage and, finally, lead to poor developmental capability from morula to blastocyst stage (Fig. 11).
The qPCR data of blastocyst stage showed almost no significant differences among the NT groups, which indicated that those embryos that successfully reaching the blastocyst stage might have a similar gene expression profile. However, all the NT groups had significant higher 47 S rRNA expression levels than that of the ICSI group. This phenomenon has been reported previously between mouse in vitro fertilization and tail tip cell NT embryos. The author believed that, in NT blastocyst, the accumulation unprocessed 47 S rRNA and rRNA intermediates failed to be processed into mature rRNAs (31). Because no significant differences were found in FBL and B23 expressions between ICSI and NT groups in our study, the exact mechanism might be more complicated and need further investigations.
rDNA reactivation plays a basic but very important role in preimplantation development. Insufficient reactivation may lead to poor ribosomal biogenesis and impair developmental competence. Moreover, in human blastocyst, half NORs could be inactivated post-implantation (13). If the same mechanism is also adopted in mice, the embryos reconstructed from donor cells with low rDNA activity, like MEFNT embryos in our study, may face ever more problems after implantation. Thus, according to our results, choosing a cell line that has a lower rDNA methylation level and/or more active NORs as donor cell will be a good strategy for NT experiments. Artificially improving the rDNA epigenetic status before or immediately after NT is another option. Recently, several histone deacetylase inhibitors were used to treat the NT embryos to achieve better clone efficiency (52–55). The histone deacetylase inhibitor can increase the overall histone acetylation level of donor cell chromatins, then make the genes more reachable to maternal factors, therefore, promoting reprogramming. In a recent study, scriptaid (a kind of histone deacetylase inhibitor) helped NT embryos overcome the asynchronous rRNA transcription at the 2-cell stage by activation of rDNA and promotion of nucleolar protein allocation (56). This study revealed how histone deacetylase inhibitor could improve NT embryo development in rDNA activation levels. We may also expect that, for those terminally differentiated cells, if the inactive rDNA could be activated before or after entering the oocyte appropriately, better NT outcomes can be achieved thereafter.
This work was supported by grants from the State Key Development Program of Basic Research of China (2012CBA01303) and the Innovative Fund of Harbin Medical University Graduate Student (HCXB2010005).

This article contains supplemental Tables 1 and 2 and Figs. 1–6.
- NOR
- nucleolar organizer region
- UBF
- upstream biding factor
- RPI
- RNA polymerase I
- FBL
- fibrillarin
- B23
- nucleophosmin
- ESC
- embryonic stem cell
- CC
- cumulus cell
- MEF
- mouse embryonic fibroblast
- NT
- nuclear transfer
- ESNT
- embryonic cell nuclear transfer
- CCNT
- cumulus cell nuclear transfer
- MEFNT
- mouse embryonic fibroblast nuclear transfer
- ICSI
- Intracytoplasmic Sperm Injection
- hpa
- hours post-activation
- qPCR
- quantitative PCR.
REFERENCES
- 1. Grummt I. (2007) Different epigenetic layers engage in complex cross-talk to define the epigenetic state of mammalian rRNA genes. Hum. Mol. Genet. 16, R21-R27 [DOI] [PubMed] [Google Scholar]
- 2. Roussel P., André C., Comai L., Hernandez-Verdun D. (1996) The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol. 133, 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fair T., Hyttel P., Lonergan P., Boland M.P. (2001) Immunolocalization of nucleolar proteins during bovine oocyte growth, meiotic maturation, and fertilization. Biol. Reprod. 64, 1516–1525 [DOI] [PubMed] [Google Scholar]
- 4. Boisvert F.M., van Koningsbruggen S., Navascués J., Lamond A.I. (2007) The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 [DOI] [PubMed] [Google Scholar]
- 5. Maddox-Hyttel P., Bjerregaard B., Laurincik J. (2005) Meiosis and embryo technology. Renaissance of the nucleolus. Reprod. Fertil. Dev. 17, 3–14 [DOI] [PubMed] [Google Scholar]
- 6. Baran V., Pavlok A., Bjerregaard B., Wrenzycki C., Hermann D., Philimonenko V.V., Lapathitis G., Hozak P., Niemann H., Motlik J. (2004) Immunolocalization of upstream binding factor and pocket protein p130 during final stages of bovine oocyte growth. Biol. Reprod. 70, 877–886 [DOI] [PubMed] [Google Scholar]
- 7. Bjerregaard B., Wrenzycki C., Philimonenko V.V., Hozak P., Laurincik J., Niemann H., Motlik J., Maddox-Hyttel P. (2004) Regulation of ribosomal RNA synthesis during the final phases of porcine oocyte growth. Biol. Reprod. 70, 925–935 [DOI] [PubMed] [Google Scholar]
- 8. Paynton B.V., Rempel R., Bachvarova R. (1988) Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev. Biol. 129, 304–314 [DOI] [PubMed] [Google Scholar]
- 9. Schultz R.M., Wassarman P.M. (1977) Biochemical studies of mammalian oogenesis. Protein synthesis during oocyte growth and meiotic maturation in the mouse. J. Cell Sci. 24, 167–194 [DOI] [PubMed] [Google Scholar]
- 10. Zeng F., Schultz R.M. (2005) RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev. Biol. 283, 40–57 [DOI] [PubMed] [Google Scholar]
- 11. Geuskens M., Alexandre H. (1984) Ultrastructural and autoradiographic studies of nucleolar development and rDNA transcription in preimplantation mouse embryos. Cell Differ. 14, 125–134 [DOI] [PubMed] [Google Scholar]
- 12. Santoro R., Grummt I. (2001) Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell 8, 719–725 [DOI] [PubMed] [Google Scholar]
- 13. Schlesinger S., Selig S., Bergman Y., Cedar H. (2009) Allelic inactivation of rDNA loci. Genes Dev. 23, 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baran V., Vignon X., LeBourhis D., Renard J.P., Fléchon J.E. (2002) Nucleolar changes in bovine nucleotransferred embryos. Biol. Reprod. 66, 534–543 [DOI] [PubMed] [Google Scholar]
- 15. Baran V., Fabian D., Rehak P., Koppel J. (2003) Nucleolus in apoptosis-induced mouse preimplantation embryos. Zygote 11, 271–283 [DOI] [PubMed] [Google Scholar]
- 16. Laurincik J., Zakhartchenko V., Stojkovic M., Brem G., Wolf E., Müller M., Ochs R.L., Maddox-Hyttel P. (2002) Nucleolar protein allocation and ultrastructure in bovine embryos produced by nuclear transfer from granulosa cells. Mol. Reprod. Dev. 61, 477–487 [DOI] [PubMed] [Google Scholar]
- 17. Bjerregaard B., Pedersen H.G., Jakobsen A.S., Rickords L.F., Lai L., Cheong H.T., Samuel M., Prather R.S., Strejcek F., Rasmussen Z.R., Laurincik J., Niemann H., Maddox-Hyttel P., Thomsen P.D. (2007) Activation of ribosomal RNA genes in porcine embryos produced in vitro or by somatic cell nuclear transfer. Mol. Reprod. Dev. 74, 35–41 [DOI] [PubMed] [Google Scholar]
- 18. Svarcova O., Dinnyes A., Polgar Z., Bodo S., Adorjan M., Meng Q., Maddox-Hyttel P. (2009) Nucleolar reactivation is delayed in mouse embryos cloned from two different cell lines. Mol. Reprod. Dev. 76, 132–141 [DOI] [PubMed] [Google Scholar]
- 19. Wakayama T., Perry A.C., Zuccotti M., Johnson K.R., Yanagimachi R. (1998) Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394, 369–374 [DOI] [PubMed] [Google Scholar]
- 20. Kato Y., Tani T., Tsunoda Y. (2000) Cloning of calves from various somatic cell types of male and female adult, newborn and fetal cows. J. Reprod. Fertil. 120, 231–237 [PubMed] [Google Scholar]
- 21. Yang F., Hao R., Kessler B., Brem G., Wolf E., Zakhartchenko V. (2007) Rabbit somatic cell cloning. Effects of donor cell type, histone acetylation status, and chimeric embryo complementation. Reproduction 133, 219–230 [DOI] [PubMed] [Google Scholar]
- 22. Bryja V., Bonilla S., Arenas E. (2006) Derivation of mouse embryonic stem cells. Nat. Protoc. 1, 2082–2087 [DOI] [PubMed] [Google Scholar]
- 23. Gao S., McGarry M., Latham K.E., Wilmut I. (2003) Cloning of mice by nuclear transfer. Cloning Stem Cells 5, 287–294 [DOI] [PubMed] [Google Scholar]
- 24. Wakayama T., Rodriguez I., Perry A.C., Yanagimachi R., Mombaerts P. (1999) Mice cloned from embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 96, 14984–14989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conner D.A. (2001) Mouse embryo fibroblast (MEF) feeder cell preparation. Curr. Protoc. Mol. Biol. Chapter 23, Unit 23.2 [DOI] [PubMed] [Google Scholar]
- 26. Henegariu O., Heerema N.A., Lowe Wright L., Bray-Ward P., Ward D.C., Vance G.H. (2001) Improvements in cytogenetic slide preparation. Controlled chromosome spreading, chemical aging, and gradual denaturing. Cytometry 43, 101–109 [PubMed] [Google Scholar]
- 27. Tesarik J., Rienzi L., Ubaldi F., Mendoza C., Greco E. (2002) Use of a modified intracytoplasmic sperm injection technique to overcome sperm-borne and oocyte-borne oocyte activation failures. Fertil. Steril. 78, 619–624 [DOI] [PubMed] [Google Scholar]
- 28. Araki Y., Yoshizawa M., Abe H., Murase Y., Araki Y. (2004) Use of mouse oocytes to evaluate the ability of human sperm to activate oocytes after failure of activation by intracytoplasmic sperm injection. Zygote 12, 111–116 [DOI] [PubMed] [Google Scholar]
- 29. Hu L.L., Shen X.H., Zheng Z., Wang Z.D., Liu Z.H., Jin L.H., Lei L. (August 15, 2011) Cytochalasin B treatment of mouse oocytes during intracytoplasmic sperm injection (ICSI) increases embryo survival without impairment of development. Zygote doi: 10.1017/S0967199411000438 [DOI] [PubMed] [Google Scholar]
- 30. Zhou Q., Renard J.P., Le Friec G., Brochard V., Beaujean N., Cherifi Y., Fraichard A., Cozzi J. (2003) Generation of fertile cloned rats by regulating oocyte activation. Science 302, 1179. [DOI] [PubMed] [Google Scholar]
- 31. Suzuki T., Minami N., Kono T., Imai H. (2007) Comparison of the RNA polymerase I-, II-, and III-dependent transcript levels between nuclear transfer and in vitro fertilized embryos at the blastocyst stage. J. Reprod. Dev. 53, 663–671 [DOI] [PubMed] [Google Scholar]
- 32. Mamo S., Gal A.B., Bodo S., Dinnyes A. (2007) Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev. Biol. 7, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maddox-Hyttel P., Svarcova O., Laurincik J. (2007) Ribosomal RNA and nucleolar proteins from the oocyte are to some degree used for embryonic nucleolar formation in cattle and pig. Theriogenology 68, S63–S70 [DOI] [PubMed] [Google Scholar]
- 34. Leung A.K., Gerlich D., Miller G., Lyon C., Lam Y.W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A.I. (2004) Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 166, 787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simonsson S., Gurdon J. (2004) DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat. Cell Biol. 6, 984–990 [DOI] [PubMed] [Google Scholar]
- 36. Tsunoda Y., Kato Y. (2003) Nuclear transfer and reprogramming mechanism. Nihon Rinsho 61, 406–410 [PubMed] [Google Scholar]
- 37. Eckardt S., McLaughlin K.J. (2004) Interpretation of reprogramming to predict the success of somatic cell cloning. Anim. Reprod. Sci. 82, 97–108 [DOI] [PubMed] [Google Scholar]
- 38. King W.A., Niar A., Chartrain I., Betteridge K.J., Guay P. (1988) Nucleolus organizer regions and nucleoli in preattachment bovine embryos. J. Reprod. Fertil. 82, 87–95 [DOI] [PubMed] [Google Scholar]
- 39. Kopecný V., Fléchon J. E., Camous S., Fulka J., Jr. (1989) Nucleologenesis and the onset of transcription in the eight-cell bovine embryo. Fine-structural autoradiographic study. Mol. Reprod. Dev. 1, 79–90 [DOI] [PubMed] [Google Scholar]
- 40. Dev V.G., Tantravahi R., Miller D.A., Miller O.J. (1977) Nucleolus organizers in Mus musculus subspecies and in the RAG mouse cell line. Genetics 86, 389–398 [PMC free article] [PubMed] [Google Scholar]
- 41. Kurihara Y., Suh D.S., Suzuki H., Moriwaki K. (1994) Chromosomal locations of Ag-NORs and clusters of ribosomal DNA in laboratory strains of mice. Mamm. Genome 5, 225–228 [DOI] [PubMed] [Google Scholar]
- 42. Korobova F.V., Romanova L.G., Noniashvili E.M., Dyban A.P., Zatsepina O.V. (2004) Localization of chromosomal nucleus organizing regions in one-cell mouse embryos and oocytes by fluorescence in situ hybridization. Ontogenez 35, 336–345 [PubMed] [Google Scholar]
- 43. Mamaeva S.E., Tsvileneva N.N. (1985) A study of chromosome content of Friend virus-induced mouse erythroleukemia cells (clone M2) via karyotype reconstruction. Cancer Genet. Cytogenet. 16, 199–205 [DOI] [PubMed] [Google Scholar]
- 44. Jean P., Hartung M., Mirre C., Stahl A. (1983) Association of centromeric heterochromatin with the nucleolus in mouse Sertoli cells. Anat. Rec. 205, 375–380 [DOI] [PubMed] [Google Scholar]
- 45. Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D., Buetow K.H., Gingeras T.R., Misteli T., Meshorer E. (2008) Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Conconi A., Widmer R.M., Koller T., Sogo J.M. (1989) Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57, 753–761 [DOI] [PubMed] [Google Scholar]
- 47. Weisenberger D., Scheer U. (1995) A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J. Cell Biol. 129, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dyban A.P., Severova E.L., Zatsepina O.V., Chentsov Y.S. (1990) The silver-stained NOR and argentophilic nuclear proteins in early mouse embryogenesis. A cytological study. Cell Differ. Dev. 29, 165–179 [DOI] [PubMed] [Google Scholar]
- 49. Roussel P., André C., Masson C., Géraud G., Hernandez-Verdun D. (1993) Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci. 104, 327–337 [DOI] [PubMed] [Google Scholar]
- 50. Kato Y., Tsunoda Y. (2010) Role of the donor nuclei in cloning efficiency. Can the ooplasm reprogram any nucleus? Int. J. Dev. Biol. 54, 1623–1629 [DOI] [PubMed] [Google Scholar]
- 51. Schneider D.A., Michel A., Sikes M.L., Vu L., Dodd J.A., Salgia S., Osheim Y.N., Beyer A.L., Nomura M. (2007) Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell 26, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kishigami S., Bui H.T., Wakayama S., Tokunaga K., Van Thuan N., Hikichi T., Mizutani E., Ohta H., Suetsugu R., Sata T., Wakayama T. (2007) Successful mouse cloning of an outbred strain by trichostatin A treatment after somatic nuclear transfer. J. Reprod. Dev. 53, 165–170 [DOI] [PubMed] [Google Scholar]
- 53. Su G.H., Sohn T.A., Ryu B., Kern S.E. (2000) A novel histone deacetylase inhibitor identified by high-throughput transcriptional screening of a compound library. Cancer Res. 60, 3137–3142 [PubMed] [Google Scholar]
- 54. Zhao J., Ross J.W., Hao Y., Spate L.D., Walters E.M., Samuel M.S., Rieke A., Murphy C.N., Prather R.S. (2009) Significant improvement in cloning efficiency of an inbred miniature pig by histone deacetylase inhibitor treatment after somatic cell nuclear transfer. Biol. Reprod. 81, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ono T., Li C., Mizutani E., Terashita Y., Yamagata K., Wakayama T. (2010) Inhibition of class IIb histone deacetylase significantly improves cloning efficiency in mice. Biol. Reprod. 83, 929–937 [DOI] [PubMed] [Google Scholar]
- 56. Bui H.T., Seo H.J., Park M.R., Park J.Y., Thuan N.V., Wakayama T., Kim J.H. (2011) Histone deacetylase inhibition improves activation of ribosomal RNA genes and embryonic nucleolar reprogramming in cloned mouse embryos. Biol. Reprod. 85, 1048–1056 [DOI] [PubMed] [Google Scholar]