Background: NaD1 is a potent antifungal plant defensin from Nicotiana alata flowers.
Results: NaD1 forms dimers as determined by x-ray crystallographic, biophysical, and biochemical approaches.
Conclusion: Dimerization of NaD1 enhances its fungal cell killing.
Significance: Understanding the molecular basis of NaD1 antifungal activity helps define defensin function and has potential application for improving plant resistance against agronomically important fungal pathogens.
Keywords: Antimicrobial Peptides, Defensins, Innate Immunity, Plant Defense, X-ray Crystallography, Cell Permeabilization, Fungi, NaD1, Peptide Interactions, Small Angle X-ray Scattering
Abstract
The plant defensin, NaD1, from the flowers of Nicotiana alata, is a member of a family of cationic peptides that displays growth inhibitory activity against several filamentous fungi, including Fusarium oxysporum. The antifungal activity of NaD1 has been attributed to its ability to permeabilize membranes; however, the molecular basis of this function remains poorly defined. In this study, we have solved the structure of NaD1 from two crystal forms to high resolution (1.4 and 1.58 Å, respectively), both of which contain NaD1 in a dimeric configuration. Using protein cross-linking experiments as well as small angle x-ray scattering analysis and analytical ultracentrifugation, we show that NaD1 forms dimers in solution. The structural studies identified Lys4 as critical in formation of the NaD1 dimer. This was confirmed by site-directed mutagenesis of Lys4 that resulted in substantially reduced dimer formation. Significantly, the reduced ability of the Lys4 mutant to dimerize correlated with diminished antifungal activity. These data demonstrate the importance of dimerization in NaD1 function and have implications for the use of defensins in agribiotechnology applications such as enhancing plant crop protection against fungal pathogens.
Introduction
Plants are continually exposed to a myriad of potentially damaging microorganisms, phytophagous insects, and environmental stresses. As a counter-measure, plants have developed sophisticated innate defense mechanisms for protection. These range from preformed physical barriers such as the cell wall to the constitutive or inducible expression of chemical compounds, including secondary metabolites and defensive proteins.
Of the defense proteins that are produced by plants, the defensins represent one of the largest families. They are small (∼5 kDa), basic, and cysteine-rich and are abundantly expressed in various plant tissues. As a result of recent advances in whole-genome sequencing and proteomics, coupled with improved bioinformatic analyses, there has been a huge expansion in the number of described defensins, many of which are encoded by multigene families (1–5).
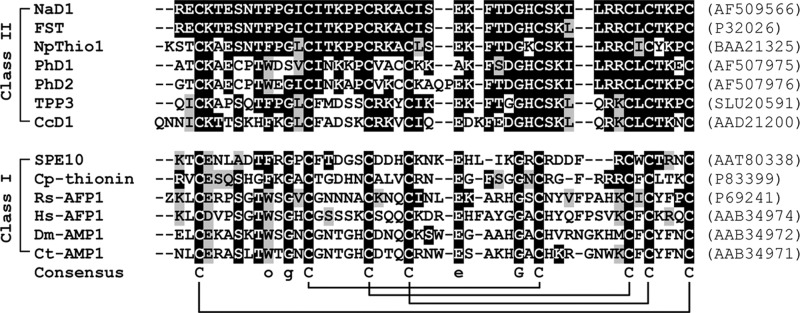
Plant defensins are distinguished from other protein families based on the number and arrangement of cysteines, the configuration of the disulfide bridges (CI–CVIII, CII–CV, CIII–CVI, and CIV–CVII), and a distinct structural fold consisting of a triple-stranded antiparallel β-sheet tethered via three disulfide bonds to an α-helix in a configuration known as the cysteine-stabilized αβ (CSαβ) motif. The fourth disulfide bond links the N- and C-terminal regions, producing a pseudocyclic protein that renders the protein highly stable to chemical and thermal denaturation (6, 7). Interestingly, the Petunia hybrida floral defensins PhD1 and PhD2 have a fifth disulfide bond (8–10). Apart from the cysteines, only a limited number of other residues are conserved among members of this family. They include a serine (position 8), two glycines (position 13 and 34), an aromatic residue (position 11), and a glutamic acid (position 29) (numbering relative to the radish defensin Rs-AFP2) (7). An amino acid sequence alignment of select representative examples of plant defensins is given in Fig. 1.
FIGURE 1.
Amino acid sequence alignment of representative mature plant defensins from class I and II. The cysteine residues that are invariant in all members of the plant defensin family are highlighted and are included in the consensus sequence, together with a serine, an aromatic residue (denoted as o), two glycines, and a glutamic acid that are conserved in most of the sequences. The disulfide bonds are shown below the consensus sequence by connecting lines. Z is the one-letter code for pyroglutamic acid. The amino acids are shaded in black or gray to indicate identity or similarity to NaD1, respectively. Spaces have been introduced to maximize the alignment. The accession numbers for each protein are given in parentheses. Sources of protein are as follows: NaD1 (N. alata), FST (Nicotiana tabacum), NpThio1 (Nicotiana paniculata), PhD1 and PhD2 (P. hybrida), TPP3 (Solanum lycopersicum), CcD1 (Capsicum chinense), SPE10 (Pachyrrhizus erosus), Cp-thionin (Vigna unguiculata), Rs-AFP1 (Raphanus sativus), Hs-AFP1 (Heuchera sanguinea), Dm-AMP1 (Dahlia merckii), and Ct-AMP1 (Clitoria ternatea).
Despite the low primary sequence identity between different plant defensins, their three-dimensional structures are remarkably similar. To date, the structures of 13 defensins have been elucidated (8, 10–20). All of these defensin structures, with the exception of one, were solved by solution 1H NMR spectroscopy as monomeric proteins (7). More recently, the structure of the SPE10 defensin from Pachyrrhizus erosus seeds was solved by x-ray crystallography and presented as a dimer in the asymmetric unit (20).
Importantly, plant defensins display a variety of biological functions despite their conserved three-dimensional structures. Best characterized is their ability to inhibit fungal pathogens (9, 21–25). A number of defensins have also been described with antibacterial activity (26–28) or can act as inhibitors of protein synthesis (29–32), α-amylases (33, 34), proteases (35, 36), or ion channels (37, 38). It is interesting to note that the latter functions are more consistent with a role in defense against insect pests. Furthermore, some defensins have been implicated in plant tolerance to environmental stresses such as high levels of zinc (39), salt (40, 41), or cold (42). Hence, the varied biological functions of plant defensins make them attractive candidates for exploitation in agribiotechnological applications to confer enhanced resistances to pathogens, insect herbivory, or environmental stresses in commercially important crops.
In solanaceous plants, defensins are divided into two classes. Class I defensins encode a precursor protein with an endoplasmic reticulum signal peptide and a mature defensin domain, whereas class II defensins encode a precursor with an additional C-terminal propeptide (7, 9). At least for three class II defensins, namely NaD1, PhD1, and PhD2 (where the protein has been characterized), the C-terminal propeptide is removed from the mature biologically active protein (8–10).
NaD1 is a defensin from the flowers of the ornamental tobacco, Nicotiana alata (9). It has been suggested that its primary function is to protect the reproductive tissues against damage from potential fungal pathogens (9, 24, 25). This is corroborated by in situ hybridization studies on N. alata flowers demonstrating accumulation of NaD1 transcripts in tissues surrounding the reproductive organs. Immunoblot analysis with NaD1-specific antibodies also revealed high levels of the protein in floral tissues (9). Furthermore, NaD1 exhibits potent in vitro antifungal activity against a number of pathogenic filamentous fungi, including Fusarium oxysporum f. sp. dianthi (Race 2), F. oxysporum f. sp. vasinfectum, and Botrytis cinerea (9, 24, 25). Recent studies have demonstrated that the antifungal activity of NaD1 involves specific interactions with the fungal cell wall, followed by permeabilization of the plasma membrane and entry of NaD1 into the cytoplasm. It has been proposed that NaD1-mediated membrane permeabilization, together with the recognition of intracellular targets, leads to fungal cell death (24, 25). However, the molecular basis of this process has not been defined.
Previously, the structure of monomeric NaD1 was determined by solution 1H NMR spectroscopy (10). Here, we report the high resolution crystal structures of NaD1 from two crystal forms, both of which contain NaD1 in a dimeric configuration. Using cross-linking experiments as well as small angle x-ray scattering analysis and analytical ultracentrifugation, we demonstrate that NaD1 forms dimers in solution. Dimerization of NaD1 is critical for antifungal activity as a mutant that does not dimerize has a greatly reduced ability to kill the filamentous fungus F. oxysporum.
EXPERIMENTAL PROCEDURES
Protein Purification
NaD1 was purified from whole N. alata flowers up to the petal coloration stage of flower development as described in van der Weerden et al. (25).
Crystallization and Data Collection
NaD1 crystals were obtained by the sitting drop vapor diffusion method in 20% PEG 1500 and 10% succinate/phosphate/glycine buffer, pH 9, and native data were collected from two different crystal forms, A and B, as described previously (43).
Heavy Atom Derivatization and Derivative Data Collection
Initial attempts to solve the structure of NaD1 by molecular replacement using either the NMR structure of NaD1 (PDB4 code 1MR4) or other plant defensin structures failed. Consequently, NaD1 crystals from crystal form A were soaked in mother liquor supplemented with 0.2 m 5-amino-2,4,6-triiodoisophthalic acid (44) and 10% ethylene glycol for 5 min. Derivative diffraction data were collected at 100 K using a wavelength of 1.5 Å at the Australian Synchrotron (beamline 3ID1) and processed using XDS (45).
Model Building and Refinement
5-Amino-2,4,6-triiodoisophthalic acid sites were found with ShelX (46) and refined using Phenix (47). Clear and continuous electron density was obtained for all residues of the entire molecule. Two chains of NaD1 were built for crystal form A. NaD1 crystal form B was solved by molecular replacement using monomeric NaD1 from crystal form A as a search model with PHASER (48). The final models for both crystal forms were built with Coot (49) and refined with Phenix to a resolution of 1.4 Å (form A) and 1.58 Å (form B). All data collection and refinement statistics are summarized in Table 1. Refinement yielded Rwork and Rfree values of 11.9 and 13.7% for crystal form A, and 20.1 and 23.2% for crystal form B, respectively. The coordinates have been deposited in the Protein Data Bank (accession codes 4aaz and 4ab0, respectively). Figures were prepared using PyMol (DeLano Scientific).
TABLE 1.
X-ray crystallographic data collection and refinement statistics
I3C is 5-amino-2,4,6-triiodoisophthalic acid.
| Crystal form A | I3C derivative | Crystal form B | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | P3221 |
| Cell dimensions | |||
| a, b, c (Å) | 32.70, 32.69, 41.98 | 32.64, 32.57, 42.06 | 33.09, 33.09, 128.77 Å |
| α, β, γ (°) | 90.00, 100.83, 90.00° | 90.00, 100.81, 90.00 | 90.00, 90.00, 120.00 |
| Wavelength (Å) | 0.9537 | 1.5 | 0.9537 |
| Resolution (Å)a | 41.23 to 1.4 (1.47 to 1.40) | 41.32 to 1.58 (1.66 to 1.58) | 64.39 to 1.64 (1.72 to 1.64) |
| Rsym or Rmergea | 0.089 (0.404) | 0.079 (0.366) | 0.081 (0.479) |
| I/σIa | 18.1 (4.6) | 16.6 (5.0) | 16.4 (3.1) |
| Completenessa (%) | 98.6 (90.5) | 99.0 (93.3) | 99.1 (93.7) |
| Redundancya | 6.9 (4.8) | 6.7 (5.0) | 9.4 (5.5) |
| Refinement | |||
| Resolution (Å) | 41.23 to 1.4 | 42.94 to 1.64 | |
| No. of reflections | 17,174 | 10,549 | |
| Rwork/Rfree | 0.119/0.137 | 0.201/0.232 | |
| No. atoms | |||
| Protein | 1474 | 1400 | |
| Ligand/ion | 1 PO4, 6 EDO | 2 PO4 | |
| Water | 369 | 78 | |
| B-factors | |||
| Protein | 8.9 | 20.9 | |
| Ligand/ion | 19.5 | 36.3 | |
| Water | 21.0 | 32.6 | |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.011 | 0.006 | |
| Bond angles (°) | 1.543 | 1.3° | |
a Values in parentheses are for the highest resolution shell.
Expression of NaD1 and NaD1 Mutants in Pichia pastoris
NaD1 and NaD1 mutants were recombinantly expressed in the methylotrophic yeast P. pastoris essentially as described by Cabral et al. (50). Briefly, DNA encoding the mature region of NaD1 was cloned into the pPIC9 expression vector (Invitrogen) directly in-frame with the α-mating factor secretion signal using the restriction enzymes XhoI and NotI. An alanine was added to the N terminus of the NaD1 sequence to ensure efficient cleavage of the signal at the Kex2 cleavage site. The glutamic acid at position 2 and the lysine at position 4 of the mature NaD1 sequence were mutated to alanine using the Phusion site-directed mutagenesis kit (Finnzymes) to generate mutants rNaD1(E2A) and rNaD1(K4A), respectively. After transformation into Escherichia coli TOP10 cells, the pPIC9-NaD1, pPIC9-NaD1(E2A), and pPIC9-NaD1(K4A) plasmids were isolated and linearized using SalI to allow integration at the his4 locus of the P. pastoris genome. Linearized DNA was transformed into electrocompetent yeast as described by Chang et al. (51), and His+ transformants were selected for by plating onto MD agar (13.4 g/liter yeast nitrogen base (YNB), 400 μg/liter biotin, 10 g/liter dextrose, and 15 g/liter agar). A single His+ colony was used to inoculate 200 ml of BMG (100 mm potassium phosphate, pH 6.0, 13.4 g/liter YNB, 400 μg/liter biotin, 1% (v/v) glycerol) and incubated with constant shaking at 30 °C until the A600 reached ∼3.0. The cell mass was collected by centrifugation (1,500 × g, 10 min) and resuspended into 1 liter of BMM (100 mm potassium phosphate, pH 6.0, 13.4 g/liter YNB, 400 μg/liter biotin, 0.5% (v/v) methanol) to induce expression. Expression was continued for 72 h with constant shaking at 30 °C after which time the cell mass was removed by centrifugation (10,000 × g, 10 min) and the supernatant collected. rNaD1, rNaD1(E2A), and rNaD1(K4A) were isolated from the supernatant using ion-exchange chromatography using SP-Sepharose as described for isolation of NaD1 from plant tissue.
Fungal Growth Inhibition Assays
The ability of NaD1, rNaD1, rNaD1(E2A), and rNaD1(K4A) to inhibit the growth of F. oxysporum f. sp. vasinfectum was assessed as described previously (25).
Protein Cross-linking
NaD1 at 125–500 μg/ml was cross-linked through primary amino groups by the addition of 6.25 mm bis[sulfosuccinimidyl] suberate (BS3) (Thermo Scientific) in a buffer containing 20 mm sodium phosphate and 150 mm NaCl, pH 7.1, at room temperature for 30 min. The samples were reduced with dithiothreitol, denatured, and subjected to SDS-PAGE prior to Coomassie Brilliant Blue staining. NaD1, rNaD1, rNaD1(E2A), and rNaD1(K4A) were also cross-linked with 6.25 mm BS3 or with 10 mm bis-N-succinimidyl-(pentaethylene glycol) ester (BS(PEG)5) (Thermo Scientific) at final protein concentrations of 250 or 375 μg/ml, respectively, as described above.
Small Angle X-ray Scattering
SAXS data were collected at the SAXS/WAXS beamline at the Australian Synchrotron. NaD1 at 0.125, 0.25, 0.5, 1, and 2 mg/ml in distilled water at pH 7.0 was measured in a Q range between 0.035 and 0.6 Å−1 at 12 keV and with a 1.6-meter camera length. Normalization was achieved via an integrating beam stop. Data were measured on a Pilatus 1 M camera (Dectris) and absolute scaled using distilled water. To control for radiation damage, the samples were measured in a 1.5-mm quartz capillary and flowed past the beam while 10 1-s exposures were measured on samples and blanks. The exposures were compared for agreement before being averaged. The radius of gyration was calculated from the high resolution models using the program Moleman2 (Uppsala Software Factory).
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted in a Beckman model XL-A analytical ultracentrifuge at a temperature of 20 °C. Samples of rNaD1 and rNaD1(K4A) were studied at a concentration of 0.5 mg/ml (94 μm) solubilized in PBS, pH 7.4. 380 μl of sample and 400 μl of reference solution were loaded into a conventional double sector quartz cell, which were subsequently mounted in a Beckman 4-hole An-60 Ti rotor, and centrifuged at a rotor speed of 40,000 rpm overnight. Data were collected at a single wavelength (237 nm) in continuous mode at 15-min intervals using a step size of 0.003 cm without averaging. Solvent density (1.0053 g/ml at 20 °C), viscosity (1.0184 centipoise), and an estimate of the partial specific volume of rNaD1 (0.727 ml/g) and rNaD1(K4A) (0.725 ml/g) were computed using the program SEDNTERP (52). Sedimentation velocity data at multiple time points were fitted to a continuous sedimentation coefficient (c(s)) distribution and a continuous mass (c(M)) distribution model (53–55) using the program SEDFIT, which is available on-line.
RESULTS
Crystal Structure of NaD1
The structure of NaD1 (crystal form A) was solved by single heavy atom isomorphous replacement with anomalous signal and refined to a resolution of 1.4 Å. A second crystal form (form B) was solved by molecular replacement using monomeric NaD1 from the previous experimentally determined structure of form A. As expected, NaD1 adopts the cysteine-stabilized αβ (CSαβ) motif formed by a triple-stranded antiparallel β-sheet and a single α-helix that is tethered to the sheet via three disulfide bonds (Fig. 2A). The fourth disulfide bond reinforces the N- and C-terminal regions of the molecule. Superimpositions of monomeric NaD1 from crystal form A with chain 1 or all 20 chains within the ensemble of lowest energy NMR structures for NaD1 (PDB code 1MR4) are shown in Fig. 2, B and C, respectively. As evident from Fig. 2C, there are no appreciable differences between the crystal and NMR structures for NaD1, with the crystal structure fitting well within the NMR ensemble. Moreover, superimposition of the monomers from crystal form A or B with the NMR structure of NaD1 yields root mean square deviation values of 1.0–1.3 Å over all 47 Cα backbone atoms.
FIGURE 2.
Crystal structure of NaD1. A, schematic representation of NaD1 from crystal form A. Disulfide bonds are shown as ball and stick. B, superimposition of the NaD1 NMR structure (gray, PDB code 1MR4) with NaD1 from crystal form A (green). C, superimposition of a single NaD1 monomer (green) from crystal from A onto the NMR ensemble of PDB code 1MR4 (gray).
Two Alternative Dimeric Forms
NaD1 is present as a dimer in the asymmetric unit of both crystal forms (Fig. 3). In crystal form A, the α1-β2 loop of one monomer faces the α-helix of the other monomer (Fig. 3A). However, there are no protein-protein contacts at the dimer interface, with all interactions being mediated by ordered water molecules. Consequently, it is possible that this dimeric form only exists in the crystal and is not sufficiently stable to be observed in solution. In contrast, the dimeric configuration of crystal form B is formed by the association of the β1-strands from the two participating monomers (Fig. 3B). This creates an extended β-sheet leading to the formation of a more compact and symmetrical dimer. The dimer interface has a combined buried surface area of 678 Å2 and is stabilized by three hydrogen bonds, two from Lys4–Lys4 and one from Ser35–Cys47. Additionally, a salt bridge is found between Cys47–Arg40 (Fig. 3C).
FIGURE 3.
Two alternative dimeric configurations for NaD1. A and B, schematic representations of NaD1 crystal forms A and B. Residues involved in the dimer interface for dimer form B are shown as sticks, and hydrogen bonds and salt bridges are shown in black as dotted lines. C and D, electrostatic surface representation of NaD1 crystal forms A and B. The views are conserved from A and B. E, electrostatic surface representation of NaD1 crystal form A. The view is rotated by 180° through the horizontal axis from C. F, electrostatic surface representation of NaD1 crystal form B. The view is rotated by 180° through the horizontal axis from D. G, surface representation of NaD1 dimer form B. The view is as in F. The NaD1 surface is shown in gray; Lys4 residues are shown in magenta and cyan from each monomeric chain.
Electrostatic Surfaces of the NaD1 Dimers
The biological activity of NaD1 has been linked to an activity at the fungal cell wall (24, 25). To investigate the possibility of NaD1 surface interactions with components of the fungal cell wall, we examined the electrostatic surface potential of the two different dimer configurations (Fig. 3, C–F).
The NaD1 dimer in crystal form A has two distinct negatively charged pockets at the ends of the dimer, with two small areas of local positive charge (Fig. 3, C and E). In contrast, the charge distribution on the surface of the NaD1 dimer from crystal form B is markedly different. A continuous surface of positive charge is present on one side of the dimer that begins on the first monomer and continues to the end of the second monomer (Fig. 3F). The two Lys4 residues make key contributions to the middle of the positively charged face of the dimer (Fig. 3G). Flanking either side of the positive charge are two well separated negatively charged pockets (Fig. 3F). Furthermore, the opposite side of this dimer presents a weaker acidic and hydrophobic surface (Fig. 3D).
Protein Cross-linking Studies
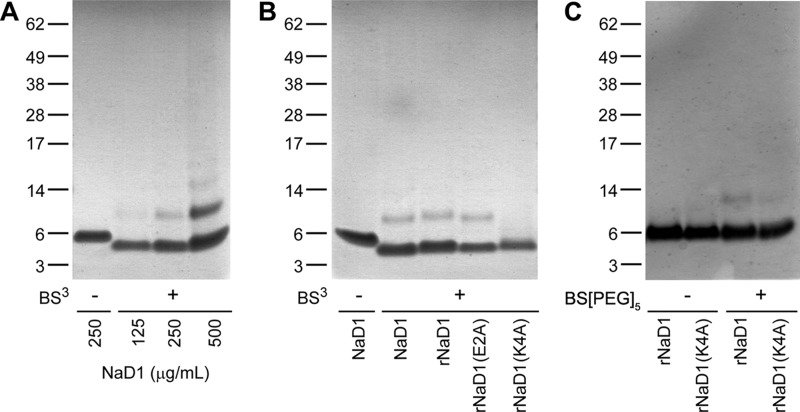
To investigate the possibility that NaD1 forms a dimer in solution, we utilized chemical cross-linking studies to directly assess the configuration of the dimer in solution using BS3. BS3 contains an amine-reactive N-hydroxysulfosuccinimide ester at each end of an 8-carbon (11.4 Å) spacer arm. Once the N-hydroxysulfosuccinimide esters react with primary amines (present in the side chain of lysine residues and the N terminus of each polypeptide) at pH 7–9, it forms stable amide bonds, along with release of the N-hydroxysulfosuccinimide leaving group.
As illustrated in Fig. 4A, NaD1 migrates as a single protein band at ∼5 kDa (monomer) on reducing SDS-PAGE in the absence of BS3. When the BS3 cross-linker is added, an additional protein band appears with increased relative molecular mass that is consistent with that of a NaD1 dimer (∼10 kDa) (Fig. 4A). The protein band corresponding to the dimer becomes more pronounced with increasing amounts of NaD1 being used in the cross-linking reaction. At the higher concentrations (e.g. 500 μg/ml NaD1 + BS3), multimers of 15 and 20 kDa are also apparent, consistent with the expected sizes of a trimer and tetramer of NaD1, respectively.
FIGURE 4.
Chemical cross-linking of NaD1 in solution. A, ability of NaD1 to form dimers in solution as determined by protein-protein cross-linking with BS3. NaD1 at 125, 250, and 500 μg/ml were incubated at room temperature for 30 min in the absence or presence of BS3 (6.25 mm) prior to SDS-PAGE analysis under reducing and denaturing conditions and staining with Coomassie Brilliant Blue. B, NaD1, rNaD1, rNaD1(E2A), and rNaD1(K4A) at 250 μg/ml cross-linked with BS3. C, rNaD1 and rNaD1(K4A) at 375 μg/ml in the absence or presence of the cross-linker BS(PEG)5. Samples in B and C were subsequently treated as per A.
Based on our structural studies that suggested a pivotal role for Lys4 in NaD1 dimerization, we generated a K4A mutant of NaD1 in P. pastoris to examine whether dimerization would be affected. As controls, rNaD1 and rNaD1(E2A) (glutamic acid at position 2 is not involved at the dimer interface) were also produced. These proteins were subjected to BS3 cross-linking. As shown in Fig. 4B, rNaD1 or rNaD1(E2A) was able to form dimers. In contrast, dimerization was abolished in the rNaD1(K4A) mutant.
When using BS3, three potential cross-links can be formed in dimer form A (Lys17–Lys22, Lys28–Lys45, and Lys17–Lys28) and one in dimer form B (Lys4–Lys4). Because mutation of Lys4 could prevent cross-linking with BS3 in dimer form B and thus lead to an apparent loss of cross-linking that is not correlated with loss of dimerization, we also performed chemical cross-linking using BS(PEG)5, which has a spacer arm of 21.7 Å (Fig. 4C). BS(PEG)5 enables additional cross-links in dimer form B between Lys28–Lys45 and Lys45–Lys45. When using BS(PEG)5, rNaD1 again revealed cross-linked dimers on SDS-PAGE, whereas rNaD1(K4A) showed substantially reduced amounts of dimer. Overall, our chemical cross-linking results suggest that rNaD1(K4A) has a significantly lower propensity to form dimers compared with the wild-type rNaD1.
Circular dichroism analysis of NaD1, rNaD1, rNaD1(E2A), and rNaD1(K4A) indicated that there was no major spectral differences between the proteins suggesting that they were similarly folded (data not shown). Therefore, the reduced dimerization of rNaD1(K4A) is not a result of incorrect folding and provides evidence to support the importance of Lys4 in NaD1 dimerization.
Antifungal Activity of NaD1, rNaD1, and rNaD1 Mutants
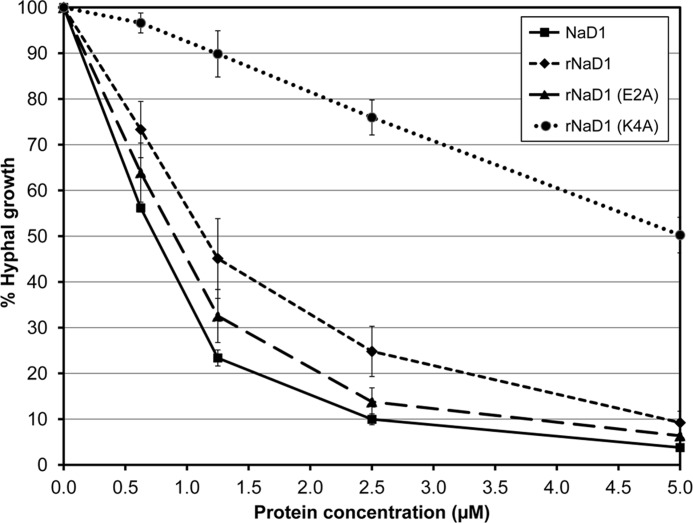
To determine whether dimerization of NaD1 contributes to its antifungal activity, the ability of rNaD1(K4A) to inhibit the growth of F. oxysporum f. sp. vasinfectum was assessed and compared with rNaD1 and rNaD1(E2A) (Fig. 5). The inhibitory activity of rNaD1 and rNaD1(E2A) was similar to native NaD1 (IC50 value of <1.15 μm). In contrast, there was a significant decrease in the ability of rNaD1(K4A) to inhibit fungal growth, with an approximate 5-fold increase in its IC50 value (5 μm) relative to rNaD1 and rNaD1(E2A).
FIGURE 5.
Antifungal activity of NaD1, rNaD1, and rNaD1 variants E2A and K4A. Growth inhibition of F. oxysporum f. sp. vasinfectum by NaD1 compared with rNaD1 and rNaD1 variants E2A and K4A. Hyphal growth is plotted relative to a no protein control. Error bars represent mean ± S.E. (three independent experiments performed at least in triplicate).
SAXS Analysis
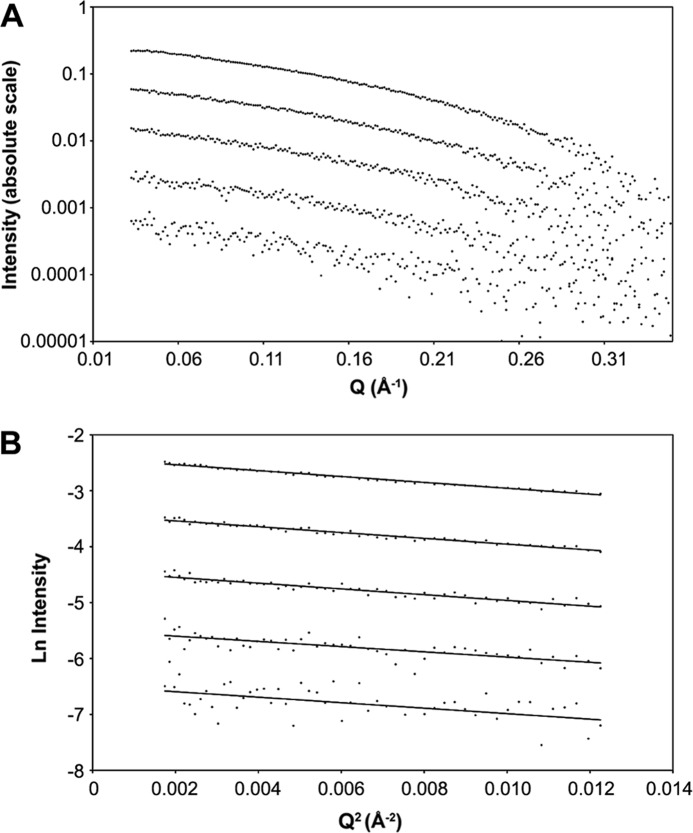
As the solution NMR structure of NaD1 indicated that the molecule was present as a monomer during the spectra acquisition (at pH 4) (10), we sought to establish if NaD1 could be present as a dimer in solution under more physiologically relevant conditions. For this purpose, we utilized SAXS analysis. The NaD1 protein was measured at five concentrations, 0.125, 0.25, 0.5, 1, and 2 mg/ml. The scattering curves show a similar shape at each concentration (Fig. 6A). The lowest angle regions of the scattering curves conform to a straight line on a Guinier plot (Fig. 6B), and the calculated radius of gyration does not vary significantly across the concentration range, indicating an absence of significant concentration effects (Table 2).
FIGURE 6.
SAXS analysis of NaD1. A, log-log plot showing raw SAXS data for NaD1 at five concentrations (0.125, 0.25, 0.5, 1, and 2 mg/ml; from bottom to top). B, Guinier plots of NaD1 SAXS data at 0.125, 0.25, 0.5, 1, and 2 mg/ml (from bottom to top) with straight line fits.
TABLE 2.
Small angle x-ray scattering data
| Concentration | I(0) | Rg | Guinier range | Mass | Oligomerization (monomer is 5.3 kDa) |
|---|---|---|---|---|---|
| mg/ml | absolute calibration | Å | Å | kDa | |
| 0.125 | 0.00071 | 12.6 | 0.015–0.11 | 9.02 | 1.70 |
| 0.25 | 0.0014 | 12.9 | 0.015–0.11 | 8.81 | 1.66 |
| 0.5 | 0.0029 | 14.1 | 0.015–0.11 | 9.23 | 1.74 |
| 1 | 0.0056 | 13.4 | 0.015–0.11 | 8.87 | 1.67 |
| 2 | 0.011 | 12.9 | 0.015–0.11 | 8.56 | 1.61 |
Two elements of the scattering data indicate that NaD1 was largely a dimer in solution. First, the molecular mass, calculated from I(0) on the absolute scattering scale across the concentration range, was around 8.9 kDa. This corresponded to an oligomerization state around 1.7, suggesting that more of the protein is in a dimeric state than a monomeric state. In addition, the radius of gyration was calculated from the high resolution structures of monomer and dimer. The Rg of the monomeric structure was calculated to be 10 Å, although the dimer was 15 Å. The measured Rg from the data was 13.2 Å or somewhere between monomer and dimer.
Sedimentation Velocity Analysis of rNaD1 and rNaD1(K4A)
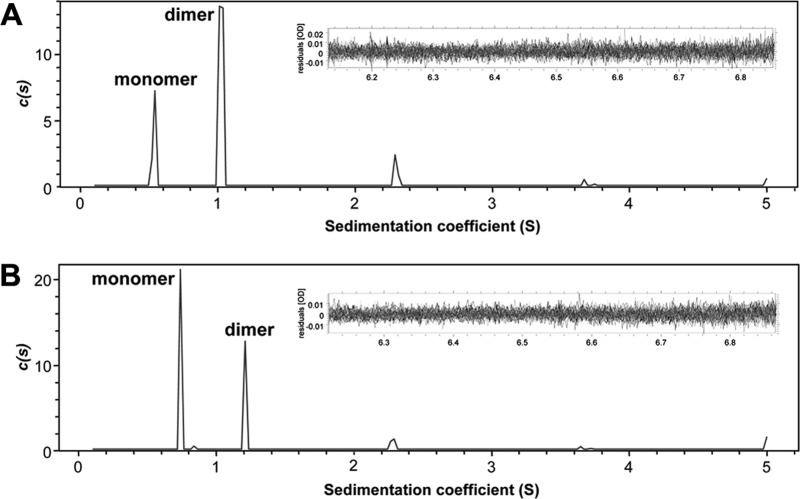
Sedimentation velocity studies of rNaD1 and rNaD1(K4A) were employed in the analytical ultracentrifuge to further validate the observations from our chemical cross-linking experiments and SAXS analysis. The absorbance versus radial position profiles at different time points were initially fitted to a continuous c(s) distribution model (Fig. 7). The c(s) distribution analyses yielded excellent fits to the data as demonstrated by the low root mean square deviation values of 0.00464 for rNaD1 and 0.00410 for rNaD1(K4A), as well as the low Runs test Z values of 1.50 for rNaD1 and 0.86 for rNaD1(K4A). The resulting c(s) distributions show that both rNaD1 and rNaD1(K4A) exist as a mixture of monomers (s ∼0.57–0.77 S) and dimers (s ∼1.05–1.26 S); however, rNaD1(K4A) shows significantly less propensity to dimerize relative to rNaD1, which predominantly exists as a dimer.
FIGURE 7.
Sedimentation velocity analysis of rNaD1 and rNaD1(K4A). Continuous sedimentation coefficient, c(s) (Svedberg−1), distribution plotted as a function of sedimentation coefficient (Svedberg) for rNaD1 (A) and rNaD1(K4A) (B). The c(s) analysis was performed using the program SEDFIT (53–55) at a resolution of 200, with smin = 0.1 S, smax = 5 S, and a confidence level (p value) = 0.5. Insets, residuals plotted as a function of radial position (cm) for the c(s) distribution best fits shown in each respective panel.
DISCUSSION
The formation of dimers and/or higher order oligomers in antimicrobial peptides has been proposed as a contributing factor to their ability to disrupt biological membranes (56–61). In this study, we employed a number of biophysical approaches to examine the three-dimensional and quaternary structures of NaD1, both in solid state (i.e. crystals) and in solution. In all examined environments, NaD1 formed dimers and smaller amounts of higher order oligomers. This is consistent with previous observations made for various other plant defensins. For example, Terras et al. (22) demonstrated using SDS-PAGE analysis that unreduced radish defensins Rs-AFP1 and Rs-AFP2 had apparent molecular masses of 15 (trimer) and/or 20 kDa (tetramer). This was in contrast to the reduced and S-pyridylethylated derivatives of both proteins that migrated as single bands with a relative molecular mass of ∼5 kDa. This was similarly demonstrated for other plant defensins, namely Dm-AMP1, Dm-AMP2, Hs-AFP1, Ah-AMP1, and Ct-AMP1 (21). Melo et al. (35) also demonstrated that the cowpea defensin, Cp-thionin, could exist as dimers along with smaller quantities of other multimers by MALDI-TOF mass spectrometry.
More recently, Song and colleagues (20, 62, 63) provided experimental evidence, including a crystal structure to support a dimeric configuration for the plant defensin SPE10. Similar to our study, the authors observed a dimer of SPE10 in the asymmetric unit (20). However, unlike the β-sheet to β-sheet configuration of NaD1 crystal form B (Fig. 8A), the two SPE10 monomers are arranged in a side by side manner with the α-helix of one monomer contacting the β-sheet of the second monomer (Fig. 8B), resulting in an extended and slightly twisted molecular surface. Dimerization buries 606 Å2 of solvent accessible surface area and results in the formation of intermolecular hydrogen bonds involving Arg36–Asp21, Arg40–Asp22, Glu4–Lys25, Asn5–Asn26, Asn17–Asp37, and Gly18–Asp37 (20).
FIGURE 8.
Dimer assemblies for NaD1 compared with SPE10 plant defensin and human defensins HNP-3 and HBD-2. All views are down the dimer interfaces. The PDB accession numbers for SPE10, HNP-3, and HBD-2 are 3PSM, 1DFN, and 1FD3, respectively.
To date, the relevance of dimer formation in plant defensins has not been examined. However, it has been suggested that for some cationic antimicrobial proteins, such as the mammalian defensins, dimeric configurations represent the functional biological units (56–61). Furthermore, the dimeric protein unit may undergo orchestrated oligomerization to form larger assemblies that are pertinent to the ability of the given cationic antimicrobial protein to kill. This hypothesis is certainly central to models in which cationic antimicrobial proteins operate via the creation of oligomeric pores.
Considering that oligomerization of defensins may play a key role for their biological function and that their initial site of activity is either a cell wall or plasma membrane, our observation of a dimer coupled with an extended positively charged surface area on one side of the NaD1 dimer is significant. Fungal cell walls are decorated with negatively charged glycoproteins (64, 65), and dimeric NaD1 may be important for initial binding to these cell wall components as a precursor step for oligomerization.
An important functional link between NaD1 dimerization and antifungal activity was strongly suggested by site-directed mutagenesis of Lys4. As described, the solved structure of NaD1 crystal form B indicated that Lys4 plays a crucial role in the monomer-monomer interaction to form the dimeric conformation. Indeed, the substitution of Lys4 with alanine resulted in a markedly reduced ability of NaD1 to form dimers, as shown by chemical cross-linking and sedimentation velocity analyses. The importance of the NaD1 dimeric configuration for its antifungal activity was demonstrated as the K4A mutant displayed a greatly reduced capacity to kill the filamentous fungus F. oxysporum with an approximate 5-fold increase in its IC50 value compared with native and recombinant NaD1. Examination of the amino acid sequences of all class II defensins described to date (7) indicates that Lys4 is totally conserved. As such, it is tempting to speculate that all class II defensins may have the ability to dimerize and that dimerization may be important for their respective functions (e.g. by enhancing their antifungal activity).
It is interesting to compare the findings described herein for NaD1 dimerization with defensins from other species such as those from mammals. The mammalian defensins are defined under three distinct structural subfamilies, α, β, or θ, based on their differences in size, the placement and connectivity of their (six) cysteines, the nature of their precursors, and their sites of expression (66–71). It is worth noting here that plant defensins do not share primary or three-dimensional structures with these mammalian namesakes except that they all have an implicated role in innate host immunity. Indeed, it was in the context of their defense role that the name “defensin” was originally coined (72, 73).
Human α-defensins or human neutrophil peptides exhibit broad antimicrobial activity against Gram-negative and Gram-positive bacteria, fungi, and enveloped viruses (74) and have also been linked with adaptive immunity as immunostimulating agents (67, 68). The three-dimensional structures of several α- and β-defensins have been solved (57–59, 61, 75–82). Interestingly, the structures of several human defensins, including HNP-1, HNP-3, and HBD-2, indicated the presence of dimers and/or multimers in the asymmetric units. For HNP-1, Wei et al. (61) recently suggested a functional link between multimerization and antibacterial activity as mutants with reduced capacity to self-associate exhibited poor bactericidal activity.
HNP-3 (PDB code 1DFN) consists of a triple-stranded antiparallel β-sheet that forms a six-stranded sheet as part of a symmetrical dimer (57). The dimer is stabilized by four direct hydrogen bonds between the β2-strands of the monomers, three hydrogen bonds mediated through water molecules, and several hydrophobic interactions (Fig. 8C). This is somewhat reminiscent of the β-sheet packing observed for NaD1 crystal form B. Moreover, HNP-3 was also able to form dimers and higher oligomers by equilibrium centrifugation in solution conditions that matched that used for the crystallization (except that PEG8000 was omitted) (57).
HBD-2 consists of a triple-stranded antiparallel β-sheet and an α-helix but in an αβββ arrangement (βαββ for NaD1) (59), which was observed in two crystal forms (PDB codes 1FD3 and 1FD4). The orthorhombic form contained four monomers in the asymmetric unit, formed by two topologically identical dimers (Fig. 8D). Its dimer interface is also mediated by two hydrogen bonds and several hydrophobic (van der Waals) interactions from the β1-strands of the monomers, resulting in a six-stranded β-sheet for the dimer as in NaD1 and HNP-3. The monoclinic form contained two octameric assemblies within the asymmetric unit, which were proposed to be the stable, native quaternary structure of HBD-2 for activity (59), although no additional evidence for the presence of such an oligomer in solution was presented. Intriguingly, subsequent dynamic light scattering experiments revealed that concentrated solutions of HBD-2 mainly formed dimers with other aggregated forms (59). This led Hoover et al. (59) to suggest that additional stabilizing interactions such as those provided by the negatively charged target (bacterial) membrane could be needed to drive octamer formation. As NaD1 is known to interact with components (as yet undetermined) on the fungal cell wall and membrane, together with the observation that cations (e.g. Ca2+) can abrogate antifungal activity (25), it would be plausible to suggest that electrostatic attraction mediated by anionic surface components (such as phospholipids, sphingolipids, or glycoproteins), may be important.
It is known that for the seed defensins from radish (Rs-AFP2) and dahlia (Dm-AMP1), interactions with specific sphingolipids (glucosylceramide and mannose-(inositol phosphate)2-ceramide, respectively) on fungal plasma membranes is required for their antifungal activity (83, 84). They are able to permeabilize the fungal membrane and induce Ca2+ influx and K+ efflux. Given that fungi grow from the tip and require the maintenance of an intracellular Ca2+ concentration gradient to drive polarized growth (85, 86), Thevissen et al. (87) suggested that the growth inhibition may be due to dissipation of this gradient. Interestingly, other plant defensins from alfalfa and corn have been reported to block ion channels, although interaction with fungal Ca2+ channels has not yet been demonstrated (37, 38). It is noteworthy that scorpion toxins, which are bone fide ion channel blockers, also display the CSαβ motif that is conserved in the plant defensins (88–90).
Numerous models have been proposed in the literature to account for the potential molecular mechanisms of action for various cationic antimicrobial peptides on target membranes (91). Based on the crystal structure of HNP-3, Hill et al. (57) proposed three possible models for HNP-3-membrane interaction as follows: (i) a wedge model, (ii) a dimer pore model, and (iii) a general pore model. In all proposed models, the dimer is the protomeric unit, building from a single dimer, to two stacked dimers onto a large annular pore involving multiple dimers. Intriguingly, these models (or variations thereof) are conceivable for HNP-3 and other dimer-forming cationic antimicrobial proteins such as NaD1 and may indeed represent transition states. As such, factors including protein concentration, the local environment of the target membrane (e.g. lipid composition), the volume of the molecule, its structure, and its oligomeric state in solution and in membranes would make important contributions to determining which model operated.
In summary, NaD1 adopts a dimeric configuration in solution under physiological conditions that enhances its antifungal activity. The dimerization of NaD1 results in the formation of a large continuous positively charged surface. This region of the NaD1 dimer may be important for mediating interactions with target cell surfaces such as the fungal cell wall and plasma membrane, thereby promoting cell permeabilization and entry of NaD1 into the cell to reach intracellular targets.
Acknowledgments
We thank the staff at the Bio21 Collaborative Crystallization Centre for the setup of crystal screens. Diffraction and scattering data were measured at the MX and SAXS/WAXS beamlines at the Australian Synchrotron. We thank the staff from the MX team for assistance with x-ray diffraction data collection.
This work was supported by Balmoral Australia Pty Ltd., Hexima Ltd., Australian Research Council Project Grant DP0881732 (to M. A. A.), and the National Health and Medical Research Council of Australia Fellowship 637372 (to M. K).
The atomic coordinates and structure factors (codes 4aaz and 4ab0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PDB
- Protein Data Bank
- BS3
- bis(sulfosuccinimidyl)suberate
- BS(PEG)5
- bis-N-succinimidyl-(pentaethylene glycol) ester
- SAXS
- small angle x-ray scattering.
REFERENCES
- 1. De-Paula V. S., Razzera G., Medeiros L., Miyamoto C. A., Almeida M. S., Kurtenbach E., Almeida F. C., Valente A. P. (2008) Evolutionary relationship between defensins in the Poaceae family strengthened by the characterization of new sugarcane defensins. Plant Mol. Biol. 68, 321–335 [DOI] [PubMed] [Google Scholar]
- 2. Graham M. A., Silverstein K. A., Cannon S. B., VandenBosch K. A. (2004) Computational identification and characterization of novel genes from legumes. Plant Physiol. 135, 1179–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mergaert P., Nikovics K., Kelemen Z., Maunoury N., Vaubert D., Kondorosi A., Kondorosi E. (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132, 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Silverstein K. A., Graham M. A., Paape T. D., VandenBosch K. A. (2005) Genome organization of more than 300 defensin-like genes in Arabidopsis. Plant Physiol. 138, 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Silverstein K. A., Moskal W. A., Jr., Wu H. C., Underwood B. A., Graham M. A., Town C. D., VandenBosch K. A. (2007) Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51, 262–280 [DOI] [PubMed] [Google Scholar]
- 6. Broekaert W. F., Terras F. R., Cammue B. P., Osborn R. W. (1995) Plant defensins. Novel antimicrobial peptides as components of the host defense system. Plant Physiol. 108, 1353–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lay F. T., Anderson M. A. (2005) Defensins-Components of the innate immune system in plants. Curr. Protein Pept. Sci. 6, 85–101 [DOI] [PubMed] [Google Scholar]
- 8. Janssen B. J., Schirra H. J., Lay F. T., Anderson M. A., Craik D. J. (2003) Structure of Petunia hybrida defensin 1, a novel plant defensin with five disulfide bonds. Biochemistry 42, 8214–8222 [DOI] [PubMed] [Google Scholar]
- 9. Lay F. T., Brugliera F., Anderson M. A. (2003) Isolation and properties of floral defensins from ornamental tobacco and petunia. Plant Physiol. 131, 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lay F. T., Schirra H. J., Scanlon M. J., Anderson M. A., Craik D. J. (2003) The three-dimensional solution structure of NaD1, a new floral defensin from Nicotiana alata and its application to a homology model of the crop defense protein alfAFP. J. Mol. Biol. 325, 175–188 [DOI] [PubMed] [Google Scholar]
- 11. Almeida M. S., Cabral K. M., Kurtenbach E., Almeida F. C., Valente A. P. (2002) Solution structure of Pisum sativum defensin 1 by high resolution NMR. Plant defensins, identical backbone with different mechanisms of action. J. Mol. Biol. 315, 749–757 [DOI] [PubMed] [Google Scholar]
- 12. Bloch C., Jr., Patel S. U., Baud F., Zvelebil M. J., Carr M. D., Sadler P. J., Thornton J. M. (1998) 1H NMR structure of an antifungal γ-thionin protein SIα1. Similarity to scorpion toxins. Proteins 32, 334–349 [DOI] [PubMed] [Google Scholar]
- 13. Bruix M., González C., Santoro J., Soriano F., Rocher A., Méndez E., Rico M. (1995) 1H NMR studies on the structure of a new thionin from barley endosperm. Biopolymers 36, 751–763 [DOI] [PubMed] [Google Scholar]
- 14. Bruix M., Jiménez M. A., Santoro J., González C., Colilla F. J., Méndez E., Rico M. (1993) Solution structure of γ1-H and γ1-P thionins from barley and wheat endosperm determined by 1H NM. A structural motif common to toxic arthropod proteins. Biochemistry 32, 715–724 [DOI] [PubMed] [Google Scholar]
- 15. de Paula V. S., Razzera G., Barreto-Bergter E., Almeida F. C., Valente A. P. (2011) Portrayal of complex dynamic properties of sugarcane defensin 5 by NMR. Multiple motions associated with membrane interaction. Structure 19, 26–36 [DOI] [PubMed] [Google Scholar]
- 16. Fant F., Vranken W., Broekaert W., Borremans F. (1998) Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1H NMR. J. Mol. Biol. 279, 257–270 [DOI] [PubMed] [Google Scholar]
- 17. Fant F., Vranken W. F., Borremans F. A. (1999) The three-dimensional solution structure of Aesculus hippocastanum antimicrobial protein 1 determined by 1H nuclear magnetic resonance. Proteins 37, 388–403 [DOI] [PubMed] [Google Scholar]
- 18. Lin K. F., Lee T. R., Tsai P. H., Hsu M. P., Chen C. S., Lyu P. C. (2007) Structure-based protein engineering for α-amylase inhibitory activity of plant defensin. Proteins 68, 530–540 [DOI] [PubMed] [Google Scholar]
- 19. Liu Y. J., Cheng C. S., Lai S. M., Hsu M. P., Chen C. S., Lyu P. C. (2006) Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids. Proteins 63, 777–786 [DOI] [PubMed] [Google Scholar]
- 20. Song X., Zhang M., Zhou Z., Gong W. (2011) Ultra-high resolution crystal structure of a dimeric defensin SPE10. FEBS Lett. 585, 300–306 [DOI] [PubMed] [Google Scholar]
- 21. Osborn R. W., De Samblanx G. W., Thevissen K., Goderis I., Torrekens S., Van Leuven F., Attenborough S., Rees S. B., Broekaert W. F. (1995) Isolation and characterization of plant defensins from seeds of Asteraceae, Fabaceae, Hippocastanaceae, and Saxifragaceae. FEBS Lett. 368, 257–262 [DOI] [PubMed] [Google Scholar]
- 22. Terras F. R., Schoofs H. M., De Bolle M. F., Van Leuven F., Rees S. B., Vanderleyden J., Cammue B. P., Broekaert W. F. (1992) Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J. Biol. Chem. 267, 15301–15309 [PubMed] [Google Scholar]
- 23. Thomma B. P., Cammue B. P., Thevissen K. (2002) Plant defensins. Planta 216, 193–202 [DOI] [PubMed] [Google Scholar]
- 24. van der Weerden N. L., Hancock R. E., Anderson M. A. (2010) Permeabilization of fungal hyphae by the plant defensin NaD1 occurs through a cell wall-dependent process. J. Biol. Chem. 285, 37513–37520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van der Weerden N. L., Lay F. T., Anderson M. A. (2008) The plant defensin, NaD1, enters the cytoplasm of Fusarium oxysporum hyphae. J. Biol. Chem. 283, 14445–14452 [DOI] [PubMed] [Google Scholar]
- 26. Franco O. L., Murad A. M., Leite J. R., Mendes P. A., Prates M. V., Bloch C., Jr. (2006) Identification of a cowpea γ-thionin with bactericidal activity. FEBS J. 273, 3489–3497 [DOI] [PubMed] [Google Scholar]
- 27. Segura A., Moreno M., Molina A., García-Olmedo F. (1998) Novel defensin subfamily from spinach (Spinacia oleracea). FEBS Lett. 435, 159–162 [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y., Lewis K. (1997) Fabatins. New antimicrobial plant peptides. FEMS Microbiol. Lett. 149, 59–64 [DOI] [PubMed] [Google Scholar]
- 29. Chen G. H., Hsu M. P., Tan C. H., Sung H. Y., Kuo C. G., Fan M. J., Chen H. M., Chen S., Chen C. S. (2005) Cloning and characterization of a plant defensin VaD1 from azuki bean. J. Agric. Food Chem. 53, 982–988 [DOI] [PubMed] [Google Scholar]
- 30. Colilla F. J., Rocher A., Mendez E. (1990) γ-Purothionins. Amino acid sequence of two polypeptides of a new family of thionins from wheat endosperm. FEBS Lett. 270, 191–194 [DOI] [PubMed] [Google Scholar]
- 31. Mendez E., Moreno A., Colilla F., Pelaez F., Limas G. G., Mendez R., Soriano F., Salinas M., de Haro C. (1990) Primary structure and inhibition of protein synthesis in eukaryotic cell-free system of a novel thionin, γ-hordothionin, from barley endosperm. Eur. J. Biochem. 194, 533–539 [DOI] [PubMed] [Google Scholar]
- 32. Méndez E., Rocher A., Calero M., Girbés T., Citores L., Soriano F. (1996) Primary structure of ω-hordothionin, a member of a novel family of thionins from barley endosperm, and its inhibition of protein synthesis in eukaryotic and prokaryotic cell-free systems. Eur. J. Biochem. 239, 67–73 [DOI] [PubMed] [Google Scholar]
- 33. Bloch C., Jr., Richardson M. (1991) A new family of small (5 kDa) protein inhibitors of insect α-amylases from seeds or sorghum (Sorghum bicolar (L) Moench) have sequence homologies with wheat γ-purothionins. FEBS Lett. 279, 101–104 [DOI] [PubMed] [Google Scholar]
- 34. Pelegrini P. B., Lay F. T., Murad A. M., Anderson M. A., Franco O. L. (2008) Novel insights on the mechanism of action of α-amylase inhibitors from the plant defensin family. Proteins 73, 719–729 [DOI] [PubMed] [Google Scholar]
- 35. Melo F. R., Rigden D. J., Franco O. L., Mello L. V., Ary M. B., Grossi de Sá M. F., Bloch C., Jr. (2002) Inhibition of trypsin by cowpea thionin. Characterization, molecular modeling, and docking. Proteins 48, 311–319 [DOI] [PubMed] [Google Scholar]
- 36. Wijaya R., Neumann G. M., Condron R., Hughes A. B., Polya G. M. (2000) Defense proteins from seed of Cassia fistula include a lipid transfer protein homologue and a protease inhibitory plant defensin. Plant Sci. 159, 243–255 [DOI] [PubMed] [Google Scholar]
- 37. Kushmerick C., de Souza Castro M., Santos Cruz J., Bloch C., Jr., Beirão P. S. (1998) Functional and structural features of γ-zeathionins, a new class of sodium channel blockers. FEBS Lett. 440, 302–306 [DOI] [PubMed] [Google Scholar]
- 38. Spelbrink R. G., Dilmac N., Allen A., Smith T. J., Shah D. M., Hockerman G. H. (2004) Differential antifungal and calcium channel-blocking activity among structurally related plant defensins. Plant Physiol. 135, 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mirouze M., Sels J., Richard O., Czernic P., Loubet S., Jacquier A., François I. E., Cammue B. P., Lebrun M., Berthomieu P., Marquès L. (2006) A putative novel role for plant defensins. A defensin from the zinc hyper-accumulating plant, Arabidopsis halleri, confers zinc tolerance. Plant J. 47, 329–342 [DOI] [PubMed] [Google Scholar]
- 40. Komori T., Yamada S., Imaseki H. (1997) cDNA cloning for γ-thionin from Nicotiana paniculata (accession number 005250) (PGR 97-132). Plant Physiol. 115, 314 [Google Scholar]
- 41. Yamada S., Komori T., Imaseki H. (1997) cDNA cloning for γ-thionin from Nicotiana paniculata (accession number AB005266) (PGR 97-131). Plant Physiol. 115, 314 [Google Scholar]
- 42. Koike M., Okamoto T., Tsuda S., Imai R. (2002) A novel plant defensin-like gene of winter wheat is specifically induced during cold acclimation. Biochem. Biophys. Res. Commun. 298, 46–53 [DOI] [PubMed] [Google Scholar]
- 43. Lay F. T., Mills G. D., Hulett M. D., Kvansakul M. (2012) Crystallization and preliminary x-ray crystallographic analysis of the plant defensin NaD1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beck T., Krasauskas A., Gruene T., Sheldrick G. M. (2008) A magic triangle for experimental phasing of macromolecules. Acta Crystallogr. D Biol. Crystallogr. 64, 1179–1182 [DOI] [PubMed] [Google Scholar]
- 45. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E. Combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Storoni L. C., McCoy A. J., Read R. J. (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 49. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 50. Cabral K. M., Almeida M. S., Valente A. P., Almeida F. C., Kurtenbach E. (2003) Production of the active antifungal Pisum sativum defensin 1 (Psd1) in Pichia pastoris. Overcoming the inefficiency of the STE13 protease. Protein Expr. Purif. 31, 115–122 [DOI] [PubMed] [Google Scholar]
- 51. Chang T., Schroder L. A., Thomson J. M., Klocman A. S., Tomasini A. J., Strømhaug P. E., Dunn W. A. (2005) PpATG9 encodes a novel membrane protein that traffics to vacuolar membranes, which sequester peroxisomes during pexophagy in Pichia pastoris. Mol. Biol. Cell 16, 4941–4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Laue T. M., Shah B. D., Ridgeway T. M., Pelletier S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., Horton J. C., eds) pp. 90–125, The Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 53. Perugini M. A., Schuck P., Howlett G. J. (2000) Self-association of human apolipoprotein E3 and E4 in the presence and absence of phospholipid. J. Biol. Chem. 275, 36758–36765 [DOI] [PubMed] [Google Scholar]
- 54. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. (2002) Size-distribution analysis of proteins by analytical ultracentrifugation. Strategies and application to model systems. Biophys. J. 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schibli D. J., Hunter H. N., Aseyev V., Starner T. D., Wiencek J. M., McCray P. B., Jr., Tack B. F., Vogel H. J. (2002) The solution structures of the human β-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J. Biol. Chem. 277, 8279–8289 [DOI] [PubMed] [Google Scholar]
- 57. Hill C. P., Yee J., Selsted M. E., Eisenberg D. (1991) Crystal structure of defensin HNP-3, an amphiphilic dimer. Mechanisms of membrane permeabilization. Science 251, 1481–1485 [DOI] [PubMed] [Google Scholar]
- 58. Hoover D. M., Chertov O., Lubkowski J. (2001) The structure of human β-defensin-1. New insights into structural properties of β-defensins. J. Biol. Chem. 276, 39021–39026 [DOI] [PubMed] [Google Scholar]
- 59. Hoover D. M., Rajashankar K. R., Blumenthal R., Puri A., Oppenheim J. J., Chertov O., Lubkowski J. (2000) The structure of human β-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 275, 32911–32918 [DOI] [PubMed] [Google Scholar]
- 60. Zhang Y., Lu W., Hong M. (2010) The membrane-bound structure and topology of a human α-defensin indicate a dimer pore mechanism for membrane disruption. Biochemistry 49, 9770–9782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wei G., Pazgier M., de Leeuw E., Rajabi M., Li J., Zou G., Jung G., Yuan W., Lu W. Y., Lehrer R. I., Lu W. (2010) Trp-26 imparts functional versatility to human α-defensin HNP1. J. Biol. Chem. 285, 16275–16285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Song X., Zhou Z., Wang J., Wu F., Gong W. (2004) Purification, characterization, and preliminary crystallographic studies of a novel plant defensin from Pachyrrhizus erosus seeds. Acta Crystallogr. D Biol. Crystallogr. 60, 1121–1124 [DOI] [PubMed] [Google Scholar]
- 63. Song X., Wang J., Wu F., Li X., Teng M., Gong W. (2005) cDNA cloning, functional expression, and antifungal activities of a dimeric plant defensin SPE10 from Pachyrrhizus erosus seeds. Plant Mol. Biol. 57, 13–20 [DOI] [PubMed] [Google Scholar]
- 64. Bowman S. M., Free S. J. (2006) The structure and synthesis of the fungal cell wall. BioEssays 28, 799–808 [DOI] [PubMed] [Google Scholar]
- 65. Schoffelmeer E. A., Klis F. M., Sietsma J. H., Cornelissen B. J. (1999) The cell wall of Fusarium oxysporum. Fungal Genet. Biol. 27, 275–282 [DOI] [PubMed] [Google Scholar]
- 66. Tang Y. Q., Yuan J., Osapay G., Osapay K., Tran D., Miller C. J., Ouellette A. J., Selsted M. E. (1999) A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated α-defensins. Science 286, 498–502 [DOI] [PubMed] [Google Scholar]
- 67. Tang Y. Q., Yuan J., Miller C. J., Selsted M. E. (1999) Isolation, characterization, cDNA cloning, and antimicrobial properties of two distinct subfamilies of α-defensins from rhesus macaque leukocytes. Infect. Immun. 67, 6139–6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lehrer R. I., Ganz T. (2002) Defensins of vertebrate animals. Curr. Opin. Immunol. 14, 96–102 [DOI] [PubMed] [Google Scholar]
- 69. Selsted M. E., Tang Y. Q., Morris W. L., McGuire P. A., Novotny M. J., Smith W., Henschen A. H., Cullor J. S. (1993) Purification, primary structures, and antibacterial activities of β-defensins, a new family of antimicrobial peptides from bovine neutrophils. J. Biol. Chem. 268, 6641–6648 [PubMed] [Google Scholar]
- 70. Hancock R. E., Lehrer R. (1998) Cationic peptides. A new source of antibiotics. Trends Biotechnol. 16, 82–88 [DOI] [PubMed] [Google Scholar]
- 71. Ganz T. (2003) Defensins. Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 72. Ganz T., Selsted M. E., Szklarek D., Harwig S. S., Daher K., Bainton D. F., Lehrer R. I. (1985) Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Invest. 76, 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Selsted M. E., Harwig S. S., Ganz T., Schilling J. W., Lehrer R. I. (1985) Primary structures of three human neutrophil defensins. J. Clin. Invest. 76, 1436–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lehrer R. I., Ganz T. (1996) Endogenous vertebrate antibiotics. Defensins, protegrins, and other cysteine-rich antimicrobial peptides. Ann. N.Y. Acad. Sci. 797, 228–239 [DOI] [PubMed] [Google Scholar]
- 75. Sawai M. V., Jia H. P., Liu L., Aseyev V., Wiencek J. M., McCray P. B., Jr., Ganz T., Kearney W. R., Tack B. F. (2001) The NMR structure of human β-defensin-2 reveals a novel α-helical segment. Biochemistry 40, 3810–3816 [DOI] [PubMed] [Google Scholar]
- 76. Bach A. C., 2nd, Selsted M. E., Pardi A. (1987) Two-dimensional NMR studies of the antimicrobial peptide NP-5. Biochemistry 26, 4389–4397 [DOI] [PubMed] [Google Scholar]
- 77. Pardi A., Hare D. R., Selsted M. E., Morrison R. D., Bassolino D. A., Bach A. C., 2nd (1988) Solution structures of the rabbit neutrophil defensin NP-5. J. Mol. Biol. 201, 625–636 [DOI] [PubMed] [Google Scholar]
- 78. Pardi A., Zhang X. L., Selsted M. E., Skalicky J. J., Yip P. F. (1992) NMR studies of defensin antimicrobial peptides. 2. Three-dimensional structures of rabbit NP-2 and human HNP-1. Biochemistry 31, 11357–11364 [DOI] [PubMed] [Google Scholar]
- 79. Zhang X. L., Selsted M. E., Pardi A. (1992) NMR studies of defensin antimicrobial peptides. 1. Resonance assignment and secondary structure determination of rabbit NP-2 and human HNP-1. Biochemistry 31, 11348–11356 [DOI] [PubMed] [Google Scholar]
- 80. Zimmermann G. R., Legault P., Selsted M. E., Pardi A. (1995) Solution structure of bovine neutrophil β-defensin-12: the peptide fold of the β-defensins is identical to that of the classical defensins. Biochemistry 34, 13663–13671 [DOI] [PubMed] [Google Scholar]
- 81. Szyk A., Wu Z., Tucker K., Yang D., Lu W., Lubkowski J. (2006) Crystal structures of human α-defensins HNP4, HD5, and HD6. Protein Sci. 15, 2749–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pazgier M., Prahl A., Hoover D. M., Lubkowski J. (2007) Studies of the biological properties of human β-defensin 1. J. Biol. Chem. 282, 1819–1829 [DOI] [PubMed] [Google Scholar]
- 83. Thomma B. P., Cammue B. P., Thevissen K. (2003) Mode of action of plant defensins suggests therapeutic potential. Curr. Drug Targets Infect. Disord. 3, 1–8 [DOI] [PubMed] [Google Scholar]
- 84. Thevissen K., Cammue B. P., Lemaire K., Winderickx J., Dickson R. C., Lester R. L., Ferket K. K., Van Even F., Parret A. H., Broekaert W. F. (2000) A gene encoding a sphingolipid biosynthesis enzyme determines the sensitivity of Saccharomyces cerevisiae to an antifungal plant defensin from dahlia (Dahlia merckii). Proc. Natl. Acad. Sci. U.S.A. 97, 9531–9536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Robson G. D., Wiebe M. G., Trinci A. P. (1991) Involvement of Ca2+ in the regulation of hyphal extension and branching in Fusarium graminearum A 3/5. Exp. Mycol. 15, 263–272 [Google Scholar]
- 86. Garrill A., Jackson S. L., Lew R. R., Heath I. B. (1993) Ion channel activity and tip growth. Tip-localized stretch-activated channels generate an essential Ca2+ gradient in the oomycete Saprolegnia ferax. Eur. J. Cell Biol. 60, 358–365 [PubMed] [Google Scholar]
- 87. Thevissen K., Ghazi A., De Samblanx G. W., Brownlee C., Osborn R. W., Broekaert W. F. (1996) Fungal membrane responses induced by plant defensins and thionins. J. Biol. Chem. 271, 15018–15025 [DOI] [PubMed] [Google Scholar]
- 88. Possani L. D., Merino E., Corona M., Bolivar F., Becerril B. (2000) Peptides and genes coding for scorpion toxins that affect ion channels. Biochimie 82, 861–868 [DOI] [PubMed] [Google Scholar]
- 89. Bontems F., Gilquin B., Roumestand C., Ménez A., Toma F. (1992) Analysis of side-chain organization on a refined model of charybdotoxin. Structural and functional implications. Biochemistry 31, 7756–7764 [DOI] [PubMed] [Google Scholar]
- 90. Bontems F., Roumestand C., Gilquin B., Ménez A., Toma F. (1991) Refined structure of charybdotoxin: common motifs in scorpion toxins and insect defensins. Science 254, 1521–1523 [DOI] [PubMed] [Google Scholar]
- 91. Brogden K. A. (2005) Antimicrobial peptides. Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250 [DOI] [PubMed] [Google Scholar]