Background: Photoreceptor PDE6 is the central enzyme in vision, but conformational changes during visual transduction are not understood.
Results: Binding of cGMP or regulatory proteins to PDE6 induces conformational changes detected by analytical ultracentrifugation.
Conclusion: Allosteric communication may contribute to regulating PDE6.
Significance: Understanding PDE6 molecular organization will aid understanding of how defects in PDE6 can result in retinal disease.
Keywords: Allosteric Regulation, Analytical Ultracentrifugation, Cyclic GMP (cGMP), Cyclic Nucleotides, Phosphodiesterases, Photoreceptors, Protein Conformation, Signal Transduction, PDE6, Transducin, Visual Transduction, Prenyl-binding Protein, Analytical Ultracentrifugation
Abstract
As the central effector of visual transduction, the regulation of photoreceptor phosphodiesterase (PDE6) is controlled by both allosteric mechanisms and extrinsic binding partners. However, the conformational changes and interactions of PDE6 with known interacting proteins are poorly understood. Using a fluorescence detection system for the analytical ultracentrifuge, we examined allosteric changes in PDE6 structure and protein-protein interactions with its inhibitory γ-subunit, the prenyl-binding protein (PrBP/δ), and activated transducin. In solution, the PDE6 catalytic dimer (Pαβ) exhibits a more asymmetric shape (axial ratio of 6.6) than reported previously. The inhibitory Pγ subunit behaves as an intrinsically disordered protein in solution but binds with high affinity to the catalytic dimer to reconstitute the holoenzyme without a detectable change in shape. Whereas the closely related PDE5 homodimer undergoes a significant change in its sedimentation properties upon cGMP binding to its regulatory cGMP binding site, no such change was detected upon ligand binding to the PDE6 catalytic dimer. However, when Pαβ was reconstituted with Pγ truncation mutants lacking the C-terminal inhibitory region, cGMP-dependent allosteric changes were observed. PrBP/δ bound to the PDE6 holoenzyme with high affinity (KD = 6.2 nm) and induced elongation of the protein complex. Binding of activated transducin to PDE6 holoenzyme resulted in a concentration-dependent increase in the sedimentation coefficient, reflecting a dynamic equilibrium between transducin and PDE6. We conclude that allosteric regulation of PDE6 is more complex than for PDE5 and is dependent on interactions of regions of Pγ with the catalytic dimer.
Introduction
Photoreceptor phosphodiesterase (PDE6) is the central effector enzyme of phototransduction. The PDE6 activation mechanism and its catalytic efficiency distinguish it from the other mammalian phosphodiesterase families (1–3) and underlie the ability of photoreceptor cells to rapidly respond to light stimuli with changes in cGMP levels on the millisecond time scale (4, 5). During the first steps in vision, photoisomerized rhodopsin activates the heterotrimeric G-protein, transducin, which binds GTP and releases its activated α-subunit (Tα-GTP)2 to activate membrane-associated rod PDE6 holoenzyme by displacing the inhibitory γ-subunit (Pγ) from the enzyme active site. The drop in cGMP that results from PDE6 activation causes cGMP-gated ion channels to close, resulting in membrane hyperpolarization that is transmitted to second-order retinal neurons (6, 7).
Considering the wealth of electrophysiological, biochemical, and genetic information about the activation and deactivation of the visual transduction pathway (8), it is surprising that fundamental aspects of PDE6 structure and regulation remain obscure. The rod PDE6 holoenzyme is composed of two non-identical catalytic subunits (α and β) to which two inhibitory Pγ subunits bind. Each catalytic subunit consists of two regulatory GAF domains (one of which, GAFa, allosterically binds cGMP), a catalytic domain, and an isoprenylated C terminus that causes PDE6 to be membrane-associated (4). Although the molecular organization of the catalytic subunits have been revealed at low resolution using electron microscopy and image analysis of rod PDE6 (9–11), very little is known about the conformational changes likely to occur to the Pαβ catalytic dimer upon binding of the Pγ subunit to Pαβ or upon binding of cGMP to the GAFa domain of the protein. Furthermore, several proteins besides Tα-GTP have been shown to interact with PDE6 in rod outer segments (e.g. prenyl-binding protein/δ (PrBP/δ), regulator of G-protein signaling-9 (RGS9), glutamic acid-rich protein-2 (GARP2), and aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1); reviewed in Ref 4), but the changes in molecular organization of PDE6 and (with the exception of transducin) the regulatory significance of these interactions remains unclear.
Previous enzymological studies of PDE6 have demonstrated that catalytic activity is regulated by its tight binding of its Pγ inhibitory subunit. Furthermore, the strength of the interaction of Pγ with Pαβ is modulated by cGMP binding to the GAFa domain. This allosteric regulation of Pγ affinity for Pαβ by cGMP is reciprocal in that cGMP binding affinity for the GAFa domain is enhanced when Pγ is associated with Pαβ (12–14). Unlike the structurally related PDE5 enzyme in which cGMP binding to the GAFa domain induces a conformational change that leads to allosteric enzyme activation (3), direct allosteric activation of PDE6 catalysis has not been observed (14–16). The underlying reason for the absence of direct allosteric activation of PDE6 is not understood; nor is the structural basis for the reciprocal allosteric effect between cGMP binding and Pγ binding to the PDE6 catalytic dimer.
Analytical ultracentrifugation is a well established biophysical method that can provide hydrodynamic information concerning the size, shape, and interactions of proteins in their native state under physiological solution conditions (17). Early hydrodynamic studies of PDE6 (18, 19) were limited by issues of protein purity and stability as well as the inability to distinguish molecular size and surface charge from potential asymmetry of the protein complex. The standard optical systems for analytical ultracentrifugation (i.e. interference and absorbance) have impeded progress due to their low sensitivity and the need for large quantities of purified proteins. The recent development of a fluorescence detection system for the analytical ultracentrifuge (AU-FDS) enables the study of a fluorescently labeled protein with high sensitivity (i.e. subnanomolar detection levels) and in complex, concentrated solutions that permit study of protein-protein and protein-ligand interactions (20, 21). In this paper, we utilize AU-FDS to examine the structure/function relationships of PDE6 subunits and to characterize conformational changes occurring when PDE6 interacts with ligands and with regulatory binding partners.
EXPERIMENTAL PROCEDURES
Materials
Bovine retinas were purchased from W.L. Lawson, Inc. The Superdex 200 and Mono-Q columns and the SP-Sepharose chromatography media were from GE Healthcare. The C4 reversed-phase column (Vydac 214TP, 22 × 250 mm) was from Grace Davison Discovery Sciences. Affinity chromatography media and HRV3c protease were from Qiagen or ThermoFisher Scientific. Filtration and ultrafiltration products were from Millipore. 5-Iodoacetamidofluorescein (IAF) and protein assay reagents were from Thermo Scientific/Pierce. The full-length open reading frame sequence for the human PDE5A gene (BAA28945) and the modified pFastBac Htb dual vector were a gift of Dr. Peter Rae (Bayer Healthcare AG). Full-length recombinant human PDE5, bovine PrBP/δ, and bovine rod Pγ truncation mutants were kindly provided by Dr. Karyn Cahill, Hannah Gitschier, and Dr. Xiujun Zhang, respectively (University of New Hampshire). All other chemicals were obtained from Sigma.
Preparation of PDE6 Holoenzyme and PDE6 Heterodimer
Bovine rod outer segments (ROS) were prepared from frozen bovine retinas under dark-adapted conditions on a discontinuous sucrose gradient. Rod PDE6 holoenzyme (Pαβγγ) was then extracted with a hypotonic buffer from illuminated ROS homogenates and purified by Mono-Q anion exchange chromatography and Superdex 200 gel filtration chromatography. The purified PDE6 was concentrated by ultrafiltration and stored with 50% glycerol at −20 °C (22). Hydrolytic activity was assayed with a colorimetric method (23).
The PDE6 catalytic dimer (Pαβ) was prepared from the PDE6 holoenzyme by removing the inhibitory Pγ subunits through limited trypsin proteolysis (24). A time course of proteolytic activation of PDE6 was determined to ensure that >90% of the Pγ subunit was destroyed without altering the apparent molecular weight of the catalytic subunits (22). To deplete endogenous cGMP bound to the noncatalytic cGMP binding sites on PDE6, purified Pαβ was incubated at 30 °C for 4 h, and the removal of 60–80% of the endogenous cGMP was verified by a membrane filtration assay of [3H]cGMP binding (25). The PDE6 concentration was determined as described previously (26).
Preparation of Recombinant Full-length PDE5 and Catalytic Domain of PDE5
The expression and purification of His-tagged human PDE5 in Sf9 insect cells was performed essentially as described by Corbin et al. (27). Briefly, bacmid DNA was generated by transforming competent DH10Bac cells (Invitrogen) using the manufacturer's protocol. Bacterial colonies were screened by PCR for the presence and correct orientation of the human PDE5 sequence within the viral DNA. Sf9 cells were transfected with the bacmid DNA using SuperFect (Qiagen). The virus-containing cell supernatant was used in viral plaque assays to screen viral isolates for protein expression and to generate high titer viral stocks.
To purify recombinant, His-tagged PDE5, suspension cultures of Sf9 cells were infected (multiplicity of infection = 5 pfu/ml) for 48–60 h at 27 °C. Harvested cells (∼2 × 108 cells) were disrupted by sonication in 20 mm Tris, pH 8, 100 mm NaCl, supplemented with Sigma protease inhibitor mixture lacking EDTA. Following centrifugation, the supernatant was affinity-purified on a 2.5-ml nickel-nitrilotriacetic acid-agarose column (Qiagen). After washing, His-tagged PDE5 was eluted with 20 mm Tris, pH 8, 100 mm NaCl, and 250 mm imidazole. The purified protein was immediately buffer-exchanged into 20 mm Tris, pH 7.5, 100 mm NaCl, 2 mm MgCl2, and 2 mm DTT; concentrated by ultrafiltration; and stored in 45% glycerol at −20 °C. Purified PDE5 (>95% as judged by SDS-PAGE) was stable (as judged by catalytic activity and [3H]cGMP binding measurements) under these conditions for up to a year.
The catalytic domain of PDE5 (amino acids 503–875) was generated by excising a 1130-nucleotide fragment from the full-length human PDE5 sequence using the BamHI and NotI restriction enzyme recognition sites. The catalytic domain fragment was then inserted into the pET47b vector (Millipore), and the plasmid was transformed into the Rosetta strain of Escherichia coli BL21(DE3) cells for protein expression. The His-tagged catalytic domain was affinity-purified as described above, followed by concentration and storage at −20 °C. For both the full-length and the catalytic domain proteins, fluorescent labeling (see below) had no discernible effect on the catalytic properties of the recombinant proteins.
Preparation and Purification of Pγ and Pγ Fragments
Recombinant full-length Pγ was expressed in E. coli BL21(DE3) cells using a pET11 vector containing the nucleotide sequence for bovine rod Pγ. Cation exchange chromatography (SP-Sepharose) followed by reverse-phase chromatography (28) yielded Pγ that was >90% pure, as judged by SDS-PAGE. The activity of purified Pγ was assessed by its ability to stoichiometrically inhibit Pαβ, and it was determined to be >95% active. Truncation mutants of Pγ used in this study were generated, purified, and characterized as described previously (26).
Purification of Recombinant PrBP/δ
The coding sequence for bovine PrBP/δ was inserted into the pGEX6p1 vector to generate a pGEX6p1-PrBP/δ construct with a glutathione S-transferase (GST) fusion tag. Recombinant protein was expressed in E. coli BL21(DE3) cells following induction with 0.5 mm isopropylthiogalactoside at 37 °C for 3 h. The expressed protein was purified by affinity chromatography on a glutathione-agarose column, and the GST tag was removed by incubation with HRV3C protease followed by repurification of the PrBP/δ and storage at 4 °C until use.
Purification of Persistently Activated Transducin α-Subunit (Tα-GTPγS)
Transducin α-subunits were extracted from PDE6-depleted ROS membranes by adding 50 μm GTPγS to the ROS membranes and recovering the solubilized Tα-GTPγS by centrifugation. The extracted Tα-GTPγS was purified on a blue Sepharose column (29, 30) and, in some experiments, further purified by gel filtration chromatography on a Superdex 200 column. The concentration of Tα-GTPγS was determined by a colorimetric protein assay (31). Purified Tα-GTPγS was stored in 50% glycerol at −20 °C with 50 μm GTPγS to ensure persistent activation.
Protein Labeling with IAF
Proteins were concentrated, and the buffer was exchanged using Centricon ultrafiltration devices. The solution used for the labeling reaction was as follows: 10 mm Tris, pH 7.5, 100 mm NaCl, and 2 mm MgCl2. Protein disulfide bonds were reduced with a 10-fold molar excess of Tris (2-carboxyethyl) phosphine-HCl. After the pH was adjusted to 7.5–8.0 with 100 mm Tris base, reduced proteins were incubated overnight at 4 °C in darkness with a 20-fold excess of IAF suspended in DMSO. Excess label was removed by gel filtration chromatography, ultrafiltration, or dialysis, depending on the protein. Labeled proteins were stored in 50% glycerol. The degree of label incorporation was determined spectrophotometrically (21).
Analytical Ultracentrifugation and Data Analysis
Experiments were performed in an Optima XLI analytical ultracentrifuge (Beckman-Coulter) equipped with a fluorescence detection system (AVIV Biomedical) (32). Sedimentation velocity studies were typically run at 50,000 rpm at 20 °C (or 55,000 rpm at 20 °C for Pγ) using a Beckman An-50 Ti rotor with double-sector cells with sapphire windows (Spin Analytical). Fluorescence scans (reported in arbitrary units) were acquired at 2-min intervals for all samples simultaneously. Analysis of 250–500 scans of each sample was performed according to a c(s) distribution model using the program Sedfit (33) to determine sedimentation coefficients of each protein or complex. In one instance, the absorbance optical system of an Optima XLA/XLI centrifuge was used for comparison with the fluorescent detection system results. In some cases, green fluorescent protein (GFP) (2.6 S) served as an internal standard.
The Stokes radius (RS) and prolate axial ratio (a/b) were calculated with the program SEDNTERP (34) using the observed sedimentation coefficient, composition molecular weight, and partial specific volume (assuming solvation = 0.4 g of H2O/g of protein). The predicted hydrodynamic behavior for those proteins for which crystallographic data were available was determined using Solution Modeler (SOMO) (35, 36).
All experimental measurements were performed at least three times, and data are reported as the mean ± S.E. Curve fitting and statistical tests of significance (unpaired, two-tailed t test) were performed using SigmaPlot.
RESULTS AND DISCUSSION
Hydrodynamic Analysis of PDE6 Catalytic Dimer and Inhibitory Subunit
Because there is a lack of detailed information about the structural organization of PDE6 holoenzyme and its constituent subunits, we began our study by examining the hydrodynamic properties of PDE6 subunits using analytical ultracentrifugation. We relied on the recent development of AU-FDS to enable us to detect ∼100-fold lower concentrations of protein compared with the traditional optical systems previously used for sedimentation velocity studies. This approach requires labeling the protein of interest with a fluorescent dye to enable detection during sedimentation velocity analysis.
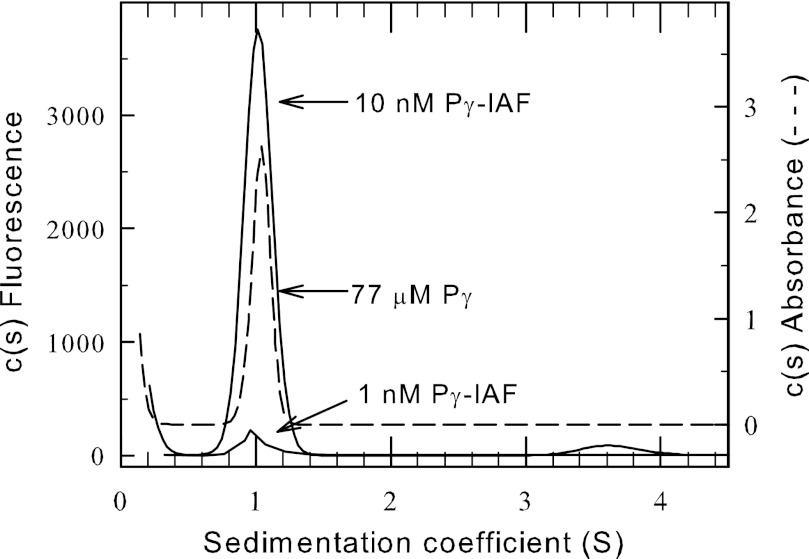
To study the hydrodynamic properties of Pγ using AU-FDS, we covalently attached IAF to the 87-amino acid bovine rod Pγ molecule at its single cysteine residue (Cys-68) and verified stoichiometric label incorporation. We ascertained that covalent modification of Cys-68 with IAF did not alter its ability to inhibit the PDE6 catalytic heterodimer (Pαβ) compared with unlabeled Pγ (data not shown). We also compared the sedimentation properties of Pγ-IAF with those of unlabeled Pγ (using an absorbance detection system). Fig. 1 shows that Pγ-IAF and unlabeled Pγ have identical sedimentation properties, with a sedimentation coefficient of 1.0 ± 0.02 S for Pγ-IAF and 1.0 ± 0.06 S for the unlabeled Pγ. We conclude that covalently attaching a fluorescein molecule at Cys-68 of the Pγ sequence does not affect either the shape or the inhibitory activity of this protein.
FIGURE 1.
Hydrodynamic properties of the purified Pγ subunit. Pγ was purified, and a portion was labeled with IAF (see “Experimental Procedures”). Sedimentation velocity studies were carried out using either the absorbance optical system (77 μm Pγ; 55,000 rpm at 20 °C; dashed line) or the fluorescence optical system (1 or 10 nm Pγ-IAF; 50,000 rpm at 20 °C; solid line). The resulting data were analyzed with Sedfit to determine a sedimentation coefficient of 1.0 S for all samples.
Based on the mass of Pγ (9.6 kDa) and its observed s value, an RS of 2.2 × 10−7 cm was calculated, corresponding to an axial ratio of 5.9, indicating that the Pγ solution structure is asymmetric compared with a tightly folded protein of identical molecular mass. This result is consistent with other measurements of Pγ solution structure that indicate that Pγ can be considered an intrinsically disordered protein with only limited amounts of secondary structure (37–39).
We next fluorescently labeled PDE6 catalytic heterodimer (Pαβ) with IAF to study its sedimentation properties. We consistently were able to incorporate 5–6 molecules of IAF per Pαβ, indicating that only a small fraction of the 36 total cysteines in the α- and β-subunits were accessible to and reactive with the dye. Fluorescently labeled PDE6 exhibited catalytic and cGMP-binding properties that were indistinguishable from those of unlabeled enzyme. Comparison of the sedimentation properties of the unlabeled versus labeled PDE6 catalytic dimer (reconstituted with labeled or unlabeled Pγ, respectively) revealed no statistically significant differences in PDE6 conformation upon labeling (p = 0.3, two-tailed t test).
Sedimentation velocity analysis of Pαβ-IAF yielded a major species with a sedimentation coefficient of 7.5 ± 0.02 S (Fig. 2). Taking into consideration the molecular mass of Pαβ, the hydrodynamic behavior of the catalytic dimer is consistent with an elongated shape in solution (axial ratio (a/b) of 6.6, assuming a prolate ellipsoid shape; see Table 1. A minor species (typically less than 10% of the total fluorescent signal) was routinely observed with a sedimentation coefficient of ∼5 S and most likely represents proteolytic fragments of Pαβ or a minor contaminating protein co-purifying with PDE6. Based on hydrodynamic modeling of the solution behavior of the structurally related PDE2 crystal structure (Table 1), monomeric PDE6 catalytic subunits would be expected to sediment at 4.3 S, not 5.0 S. Thus, it is unlikely that the 5 S material represents monomeric α- and/or β-subunits of PDE6.
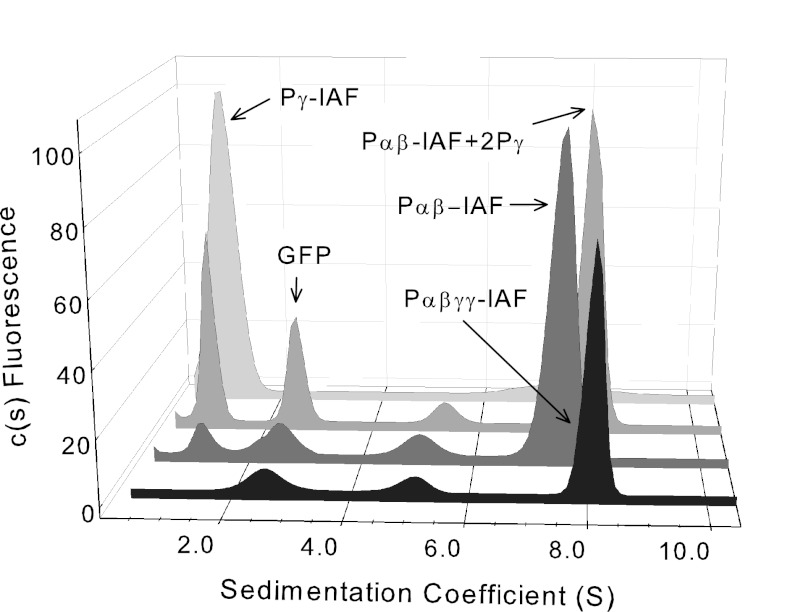
FIGURE 2.
Hydrodynamic properties of PDE6 holoenzyme and catalytic dimer reconstituted with Pγ. Sedimentation velocity experiments (50,000 rpm at 20 °C) were conducted with fluorescently labeled PDE6 holoenzyme (Pαβγγ-IAF; black), PDE6 catalytic dimer (Pαβ-IAF; dark gray), PDE6 catalytic dimer reconstituted with stoichiometric amounts of unlabeled Pγ (Pαβ-IAF + 2 Pγ; medium gray), and purified Pγ (light gray). As an internal standard, GFP (2.6 S) was added to samples containing catalytic subunits. The sedimentation coefficients determined in this representative experiment were as follows: PDE6 holoenzyme, 7.9 S; catalytic dimer, 7.5 S; reconstituted PDE6 holoenzyme, 7.9 S; purified Pγ, 1.0 S. See Table 1 for averaged data and statistical significance for this and subsequent figures.
TABLE 1.
Summary of sedimentation properties
Sedimentation velocity analysis using the fluorescence detection system (see “Experimental Procedures”) was carried out on the indicated protein samples. The data were analyzed using Sedfit with the following parameters: partial specific volume = 0.7300, buffer density = 0.99823 g/ml, and viscosity = 0.01002 Poise. The sedimentation coefficient (s) values are the mean ± S.E. for n separate determinations.
| Protein sample | n | sa | RSb | Axial ratio (a/b)c | Predicted sd |
|---|---|---|---|---|---|
| cm | |||||
| Pαβγγ | 32 | 7.9 ± 0.02 | 6.3 × 10−7 | 6.8 | |
| Pαβ | 24 | 7.5 ± 0.02* | 6.1 × 10−7 | 6.6 | |
| Pαβ + 2γ | 12 | 7.9 ± 0.03 | 6.3 × 10−7 | 6.8 | |
| Pαβ + cGMP | 20 | 7.5 ± 0.04* | 6.1 × 10−7 | 6.7 | |
| Pγ | 13 | 1.0 ± 0.02 | 2.2 × 10−7 | 5.9 | |
| PrBP/δ | 24 | 2.0 ± 0.04 | 2.0 × 10−7 | 1.3 | 1.9 |
| 2PrBP/δ + Pαβγγ | 20 | 8.1 ± 0.05 | 7.2 × 10−7 | 8.6 | |
| Tα/GTPγS | 13 | 3.1 ± 0.04 | 3.1 × 10−7 | 3.7 | 3.2 |
| Tα/GTPγS + Pγ | 26 | 3.5 ± 0.08** | 3.4 × 10−7 | 4.1 | |
| PDE5 dimer | 16 | 7.7 ± 0.04 | 6.4 × 10−7 | 7.2 | 4.3/7.1 |
| PDE5 dimer + cGMP | 12 | 7.4 ± 0.04* | 6.7 × 10−7 | 8.4 | |
| PDE5-CAT monomer | 6 | 3.4 ± 0.12 | 3.1 × 10−7 | 3.2 | 3.1 |
| PDE5-CAT + cGMP | 6 | 3.3 ± 0.04 | 3.2 × 10−7 | 3.6 |
a To determine statistical significance referenced to the boldface entry in each section, a two-tailed t test was performed, and the degree of significance noted by an asterisk (p < 0.001), a double asterisk (p < 0.01), or no asterisk (not statistically significant).
b RS was calculated using SEDNTERP, a knowledge of the composition molecular weight, and the partial specific volume (assuming solvation = (0.4 g of H2O/g of protein)).
c Axial ratio was calculated assuming a prolate ellipsoid model of the hydrodynamic properties, assuming the monomeric state except for PDE5 and PDE6 catalytic dimers.
d Predicted sedimentation coefficient is based on hydrodynamic modeling using SOMO (35) with the following Protein Data Bank structure files: PrBP/δ (1KSH), Tα-GTPγS (1TND), PDE5 monomer/dimer (modeled using the dimeric PDE2 crystal structure, 3IBJ), PDE5 catalytic domain (no ligand bound, 1T9R).
Binding of Pγ to Pαβ Does Not Alter Conformation of Resulting Holoenzyme
The catalytic subunits of PDE6 consist of three folding domains: two N-terminal tandem GAF domains (GAFa and GAFb) and the C-terminal catalytic domain. Biochemical evidence indicates that the N-terminal portion of Pγ interacts with the GAFa domain of the catalytic subunits, whereas the C-terminal portion of Pγ interacts with the catalytic domains of Pαβ (40). To test the hypothesis that binding of Pγ to Pαβ might induce a structural change in conformation of the domain organization of PDE6, we examined the effects of mixing Pγ with Pαβ prior to determining the hydrodynamic properties of the reconstituted holoenzyme.
Previous studies have documented that the addition of stoichiometric amounts of unlabeled Pγ to Pαβ can quantitatively inhibit cGMP hydrolysis (41, 42). This titration behavior of Pγ to inhibit Pαβ was also observed for the IAF-labeled catalytic dimer (data not shown). Upon reconstituting the PDE6 holoenzyme by adding back stoichiometric amounts of Pγ to our Pαβ-IAF preparation, we found an increase of the sedimentation coefficient from 7.5 ± 0.02 to 7.9 ± 0.03 S (Fig. 2). Highly purified PDE6 holoenzyme also was labeled with IAF, which resulted in labeling of both the Pγ subunits and the Pαβ catalytic dimer (3–5 molecules of IAF per holoenzyme). The resulting sedimentation behavior of the native holoenzyme (s = 7.9 ± 0.02 S; Table 1) was indistinguishable from that of reconstituted PDE6-IAF. These results support the idea that the limited proteolysis of PDE6 used to prepare purified Pαβ did not adversely affect the dimerization of the catalytic subunits or their ability to bind Pγ to reform the αβγγ holoenzyme. At all concentrations tested, neither Pαβ nor PDE6 holoenzyme revealed any evidence for the existence of monomeric catalytic subunits.
Calculation of the Stokes radius based on the observed sedimentation coefficient leads to the conclusion that the Pαβ catalytic dimer (RS = 6.1 × 10−7 cm) does not undergo a significant change in shape upon binding of Pγ subunits (RS = 6.3 × 10−7 cm; see Table 1). The simplest interpretation is that the intrinsically disordered Pγ subunit (39) binds to the elongated Pαβ by conforming to overall shape of the catalytic dimer without causing detectable alterations in the conformation of the GAF or catalytic domains.
Comparison of the hydrodynamic properties of PDE6 holoenzyme (Fig. 2) with available structural information for PDE6 holoenzyme (examined by electron microscopy and image analysis (9, 10)) suggests that the solution structure of PDE6 is more asymmetric than the molecular organization described by electron microscopy. Our estimate of the molecular volume of PDE6 (785 nm3, based on its Stokes radius) is 20–40% smaller than the two published values for the molecular dimensions of purified PDE6 holoenzyme examined by electron microscopy (9, 10). In addition, PDE6 in solution appears more elongated than what is seen for PDE6 particles viewed by electron microscopy. These differences may reflect an overestimate of the molecular dimensions of negatively stained particles used for the electron microscopic analysis and/or distortion of the molecular organization of PDE6 during sample preparation for electron microscopy.
cGMP Binding Induces Large Conformational Changes in PDE5 but Not in PDE6
Because AU-FDS is well suited to detect conformational changes in proteins upon ligand binding, we sought to determine whether cGMP binding to the GAF domains of PDE6 could alter its overall shape. Support for this idea comes from studies of the closely related enzyme, PDE5, which undergoes changes in electrophoretic mobility (43, 44) and NMR spectral properties (45) upon binding of cGMP. Because PDE5 and PDE6 are closely related structurally and biochemically (see Introduction), we examined whether ligand-induced conformational changes could be observed for both the PDE5 and PDE6 enzyme families.
We first tested fluorescently labeled recombinant human PDE5 (1–2 IAF molecules per dimer) and observed a sedimentation coefficient of 7.7 ± 0.04 S (n = 16) in the absence of added cGMP (Fig. 3A). The calculated Stokes radius (RS = 6.4 × 10−7 cm) for PDE5 is somewhat larger than for the PDE6 catalytic dimer, indicating a more elongated molecular structure (axial ratio of 7.2). Upon cGMP binding, the sedimentation coefficient is lowered from 7.7 to 7.4 ± 0.04 S (n = 12; Fig. 3A), indicative of a statistically significant (p < 0.0001) elongation of PDE5 structure (e.g. increase in axial ratio to 8.4) upon cGMP binding. This finding agrees with a cGMP-dependent decrease in PDE5 electrophoretic mobility on native gels and a shift in elution during gel filtration chromatography (44, 46). This cGMP-dependent conformational change in PDE5 holoenzyme was not observed when we tested the isolated catalytic domain of PDE5; in this instance, the catalytic domain behaves as a monomeric, mildly asymmetric protein (3.4 S), and cGMP addition caused no statistically significant change in sedimentation coefficient (p = 0.46). For both the full-length PDE5 homodimer and the monomeric catalytic domain fragment, the predicted hydrodynamic properties (based on modeling the solved crystal structures) were in good agreement with the observed sedimentation behavior of these two proteins (Table 1).
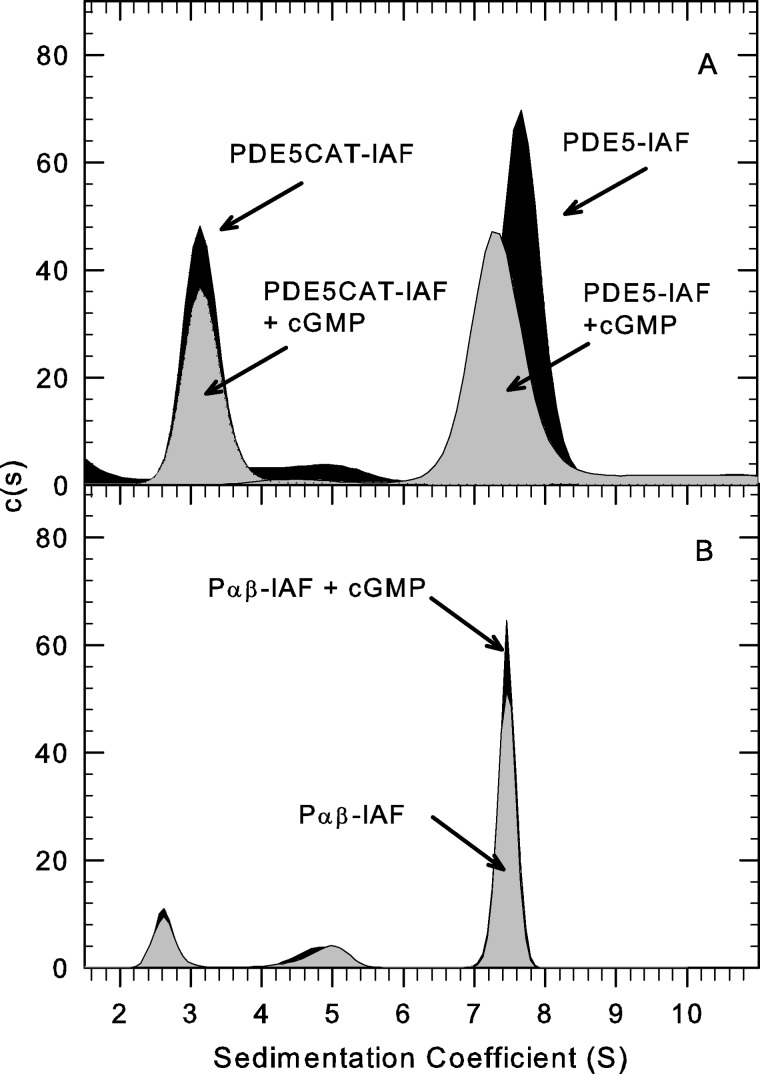
FIGURE 3.
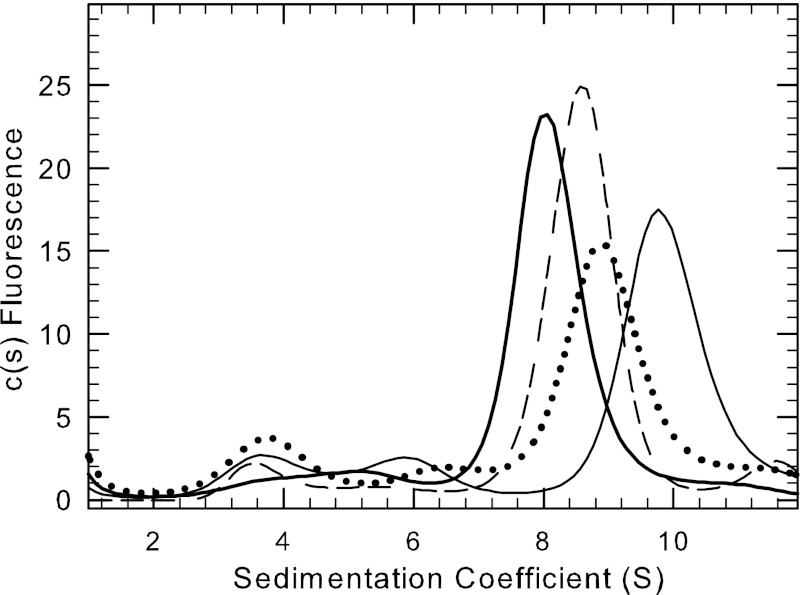
Evidence for ligand-induced conformational changes in PDE5 but not PDE6. AU-FDS Sedimentation velocity studies (50,000 rpm at 20 °C) were carried out to examine conformational changes induced by cGMP. A, His-tagged, recombinant PDE5, either full-length or catalytic domain, were fluorescently labeled and repurified by gel filtration chromatography prior to sedimentation velocity analysis of 10 nm protein in the presence or absence of 10 mm cGMP. The sedimentation coefficients determined in this experiment were as follows: PDE5-IAF alone, 7.7 S; PDE5-IAF plus cGMP, 7.4 S; PDE5CAT-IAF alone, 3.3 S; PDE5CAT-IAF + cGMP, 3.2 S. B, purified PDE6-IAF was activated by limited trypsin proteolysis, repurified by gel filtration chromatography, and depleted of endogenous nucleotides (see “Experimental Procedures”). 10 nm Pαβ-IAF (plus 1 mm zaprinast or 0.1 mm vardenafil) was analyzed in the presence or absence of 10 mm cGMP. The resulting sedimentation coefficients were as follows: Pαβ-IAF, 7.5 S; Pαβ-IAF plus cGMP, 7.5 S.
To examine whether similar allosteric regulation of PDE6 by cGMP could be detected, we compared the hydrodynamic properties of Pαβ-IAF in the absence of ligands or in the presence of 10 mm cGMP plus a PDE5/6 catalytic site inhibitor (either 1 mm zaprinast or 0.1 mm vardenafil) to prevent destruction of cGMP. (cGMP binding assays confirmed earlier findings (14, 42) that, under these conditions, all high affinity cGMP binding sites on Pαβ would remain occupied during centrifugation.) No significant change in sedimentation coefficient was observed for PDE6 catalytic dimer in the absence or presence of cGMP (Fig. 3B). (We were unable to test the effects of cGMP in the absence of a cyclic nucleotide phosphodiesterase inhibitor because the high catalytic rate of Pαβ will degrade all of the added cGMP during the time needed to conduct the sedimentation velocity experiment.) We also examined the ability of various PDE5/6 inhibitors to alter the sedimentation properties of Pαβ but found no effects of zaprinast or vardenafil on the hydrodynamic behavior of the PDE6 catalytic dimer (data not shown). We conclude that, unlike with PDE5, no significant conformational changes can be detected upon binding of cGMP (or other small ligands) to the PDE6 catalytic dimer.
cGMP-dependent Conformational Changes in PDE6 Can Be Observed upon Stabilization of cGMP Binding by Pγ Fragments
The failure to observe structural rearrangement of Pαβ upon Pγ binding (Fig. 2) or cGMP binding (Fig. 3) was unexpected. Previous work has documented positive cooperativity of cGMP and Pγ binding to Pαβ and allosteric effects of N-terminal fragments of Pγ on cGMP binding and catalytic activity (13, 14, 26, 47, 48). This led us to consider whether the combination of cGMP occupancy plus interaction of Pγ might be necessary to observe structural changes in PDE6 conformation.
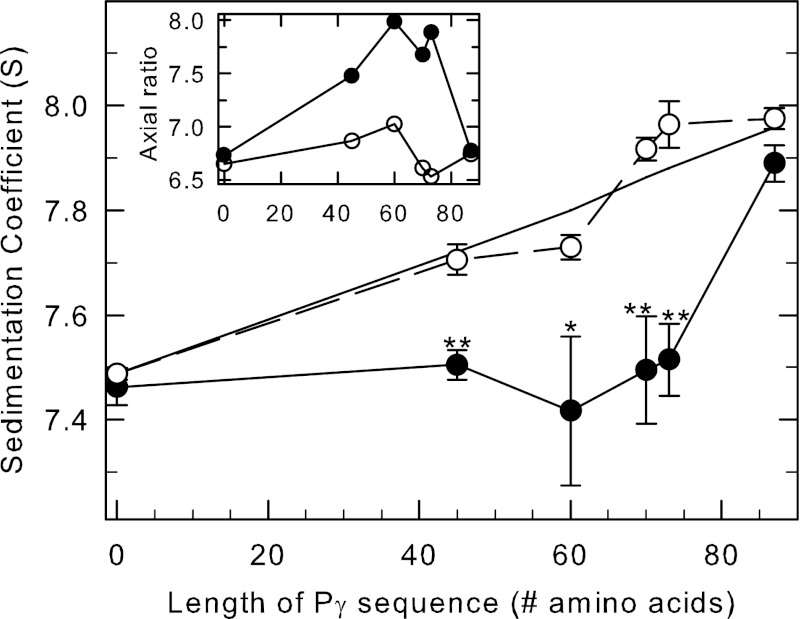
To test this, we compared the hydrodynamic behavior of Pαβ when incubated with various Pγ fragments in the absence or presence of cGMP. Fig. 4 shows that the sedimentation coefficient of the PDE6 catalytic dimer in the absence of cGMP (open circles) linearly increased as a function of the length (i.e. mass) of the Pγ fragment that was added. This behavior is consistent with a change in size of the reconstituted PDE6 holoenzyme without a change in shape (Fig. 4, inset, open circles).
FIGURE 4.
cGMP-induced conformational changes in PDE6 observed when Pγ truncation mutants are bound to the catalytic dimer. Purified, nucleotide-depleted Pαβ-IAF (10 nm) was incubated with 1 μm concentrations of the following: nothing (control), full-length Pγ (Pγ(1–87)), and the C-terminal truncation mutants Pγ(1–45), Pγ(1–60), Pγ(1–70), and Pγ(1–73). One portion of each preparation was incubated with 10 mm cGMP, and samples were centrifuged as soon as possible thereafter to minimize cGMP hydrolysis. The x axis represents the number of amino acids in each of the C-terminal truncation mutants. Data points represent the mean ± S.E. of 4–9 separate experiments. The asterisks indicate statistical difference at p < 0.05 (*) p < 0.005 (**) level of significance. The straight line is the predicted s value for each sample, calculated using the additional mass contributed by each Pγ truncation mutant, assuming a binding stoichiometry of 2 Pγ per Pαβ and no change in shape of the reconstituted protein. Inset, the axial ratio was calculated with SEDNTERP based on the composition molecular weight and the s value for each condition in the absence (open circles) and presence (filled circles) of cGMP.
In contrast, adding cGMP to occupy all of the cGMP binding sites on Pαβ reconstituted with various Pγ fragments suppressed this linear increase in s value because the size of the Pγ fragments was increased (Fig. 4, filled circles). Pγ fragments ranging in size from the first 45 to the first 73 residues all showed highly significant differences in s value when cGMP was present compared with identical samples lacking cGMP during the experiment. This cGMP-dependent change in sedimentation properties of the reconstituted enzyme is accompanied by a previously documented enhancement of cGMP binding to low affinity sites of Pαβ by Pγ (42) or N-terminal Pγ fragments (14).
Only in the case of full-length Pγ does this cGMP-induced change in PDE6 holoenzyme conformation revert to an axial ratio similar to that seen in the absence of cGMP (Fig. 4, inset). It should also be noted that another study has reported a cGMP-dependent shift in the elution profile of partially activated PDE6 (αβγ) studied using gel filtration chromatography (49).
Taken together, the results of Figs. 3 and 4 suggest that both PDE5 and PDE6 undergo a cGMP-induced elongation of the catalytic dimer but that in the case of PDE6, this allosteric mechanism requires the participation of the GAF-interacting region of Pγ (for discussion, see below).
Binding of PrBP/δ to PDE6 Holoenzyme Causes Elongation of Protein Complex
As mentioned in the Introduction, PrBP/δ is a small prenyl-binding protein that co-purifies with a soluble form of PDE6 when isolated from retinal extracts and has been hypothesized to play a role in PDE6 transport and/or regulation of its catalytic activity (50–52). To examine this interaction, we first purified recombinantly expressed PrBP/δ, labeled it with IAF (∼2 IAF molecules per protein), and determined its hydrodynamic properties using AU-FDS. PrBP/δ exhibits a sedimentation coefficient of 2.0 ± 0.04 S (Table 1). This corresponds to a Stokes radius of 2.0 × 10−7 cm and an axial ratio of 1.3, indicating that the protein is essentially a spherical globular protein in solution. Our results are consistent with the predicted hydrodynamic properties of PrBP/δ based on modeling the x-ray crystal structure (Protein Data Bank entry 1KSH, chain B) using SOMO (35, 36), supporting the characterization of PrBP/δ as a tightly organized, immunoglobulin-like fold protein (53).
Following incubation of fluorescently labeled PrBP/δ with PDE6 holoenzyme, a PrBP/δ-PDE6 complex was detected with a sedimentation coefficient of 8.1 ± 0.05 S (Fig. 5A). To evaluate the affinity of the interaction between PrBP/δ and PDE6, we preincubated PDE6 holoenzyme with a large excess of PrBP/δ-IAF, performed gel filtration to remove unbound PrBP/δ-IAF, and then examined the sedimentation properties of a series of dilutions of the protein complex. As shown in Fig. 5B, the extent to which PrBP/δ-IAF remained bound to PDE6 was concentration-dependent (K½ = 6.1 nm), indicating a strong affinity of PrBP/δ-IAF for PDE6 holoenzyme. However, the above experiments are unable to determine the binding stoichiometry of this interaction, specifically whether one or both of the isoprenylated catalytic subunits bind PrBP/δ-IAF.
FIGURE 5.
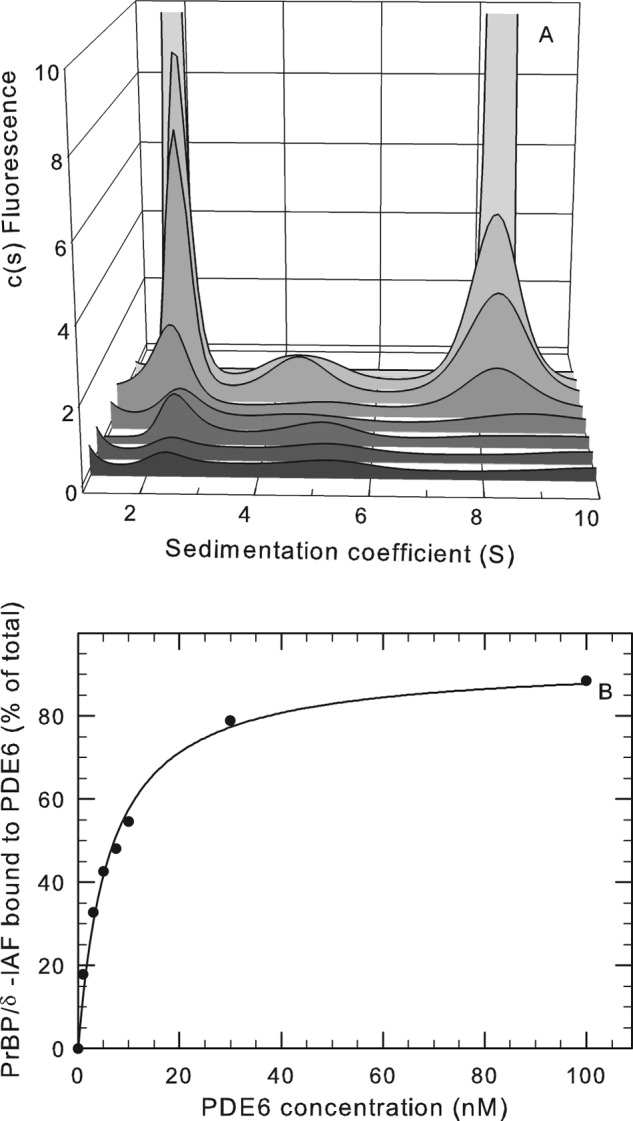
Binding of PDE6-interacting protein PrBP/δ with PDE6 holoenzyme. A, purified PrBP/δ-IAF (1 μm) was incubated overnight at 4 °C with 420 nm PDE6 holoenzyme, and then unbound PrBP/δ-IAF was separated from PDE6 by gel filtration chromatography. The recovered protein complex was then diluted to the following PDE6 concentrations (listed from front to back): 1, 3, 5, 7.5, 10, 30, and 100 nm. Sedimentation velocity analysis of the dilution series was then performed, along with a control sample (not shown) consisting of purified PrBP/δ-IAF, which sediments at 2.0 S. At 100 nm, the amplitude of the fluorescent signal (2 S peak = 29.4 units and 8 S peak = 113 units) is off scale so that the results at lower concentrations can be seen. B, the percentage of the total PrBP/δ fluorescence associated with PDE6 was determined for each PDE6 concentration by integrating the areas under the observed peaks and calculating the percentage of the total fluorescence migrating at 8.1 S. The curve represents the fit of the data to a two-parameter hyperbolic function (K½ = 6.1 nm).
To estimate the binding stoichiometry of PrBP/δ to PDE6, we quantitated the amount of PrBP/δ-IAF bound to purified PDE6 holoenzyme or to PDE6 solubilized from ROS membranes with PrBP/δ following gel filtration chromatography to remove unbound PrBP/δ. The amount of PrBP/δ bound was quantified by either spectrofluorimetric analysis (with reference to a set of PrBP/δ-IAF standards of known concentration) or following SDS-PAGE and quantitation of Coomassie-stained gel bands. Both approaches yielded similar results with a maximum binding stoichiometry of 2.2 ± 0.4 (n = 12), consistent with a recent study (11). With this information, we could now interpret the modest shift in sedimentation coefficient from 7.9 to 8.1 S upon PrBP/δ binding to reflect a significant elongation of the protein complex (axial ratio increased from 6.8 to 8.6) upon PrBP/δ-IAF binding to the isoprenyl group at the C terminus of each PDE6 catalytic subunit.
Interactions of Activated Transducin α-Subunit with PDE6
The heterotrimeric G-protein, transducin, plays a critical role in the activation of membrane-associated rod PDE6 during visual excitation (see Introduction). We utilized AU-FDS to study the hydrodynamic properties of the persistently activated transducin α-subunit (Tα-GTPγS) and its interactions with the inhibitory Pγ subunit and with the PDE6 holoenzyme. Purified Tα-GTPγS fluorescently labeled with IAF (0.2–0.4 molecules of IAF per Tα-GTPγS) was found to have a sedimentation coefficient of 3.1 ± 0.04 S, consistent with a slightly elongated globular protein (axial ratio 3.7; Table 1). The predicted asymmetry (axial ratio of 3.0) based on computer modeling of the hydrodynamic properties from the x-ray crystal structure (Protein Data Bank entry 1TND) is comparable with the observed solution properties of Tα-GTPγS (Table 1).
Upon binding of Pγ to transducin α-subunit, the sedimentation coefficient increases from 3.1 to 3.5 S (Table 1). This increase in s value can be predominantly accounted for by the increased mass of the complex of Tα-GTPγS with Pγ (1:1 stoichiometry), with only a modest increase in the axial ratio (from 3.7 to 4.1). Similar results are obtained regardless of how the complex of Tα-GTPγS with Pγ is formed or which protein was fluorescently labeled (not shown).
When purified Tα-GTPγS is incubated with fluorescently labeled PDE6 holoenzyme, the sedimentation coefficient of the labeled species increases in a concentration-dependent manner (Fig. 6), from 7.9 S (no added Tα-GTPγS) to 9.4 S (10,000-fold molar excess of Tα-GTPγS over PDE6). (The efficiency with which Tα-GTPγS activates PDE6 catalysis in solution occurs over the same concentration range as this observed shift in s value (54).) This progressive increase in s value as the Tα-GTPγS concentration is increased can be explained by either (a) two molecules of Tα-GTPγS binding to the PDE6 holoenzyme with an insignificant change in shape or (b) one Tα-GTPγS binding to PDE6 that induces a reduction in asymmetry of the protein complex. The presence of smaller s value peaks complicates the interpretation of these data. In particular, the several peaks between 2 and 7 S do not exhibit systematic shifts as the transducin concentration increases. Because purified Tα-GTPγS has a sedimentation coefficient of 3.1 S and Tα-GTPγS complexed with Pγ is shifted to 3.5 (Table 1), it is likely that increased quantities of material sedimenting approximately at 3–4 S may result from a portion of the Pγ subunit completely dissociating from the PDE6 holoenzyme following Tα-GTPγS binding the enzyme. Also notable is the shape of the leading edge of the 8 S PDE6 peak in the presence of higher concentrations of Tα-GTPγS. The skewing toward higher s values may be indicative of multiple equilibria between transducin-PDE6 complexes consisting of one or two Tα-GTPγS molecules binding to the PDE6 holoenzyme.
FIGURE 6.
Binding of the G-protein, transducin, to PDE6 holoenzyme. Purified PDE6 holoenzyme (2.5 or 6 nm) covalently labeled with IAF was incubated with various concentrations of purified, activated Tα-GTPγS. Sedimentation velocity analysis was then carried out (50,000 rpm at 20 °C), and the data were analyzed to determine the sedimentation coefficient for each concentration: no added Tα-GTPγS (thick solid line), 3000-fold molar excess of Tα-GTPγS (dashed line), 5000-fold excess (dotted line), and 10,000-fold excess (thin solid line). The traces shown were normalized based on the total protein fluorescence in each sample.
Conclusions
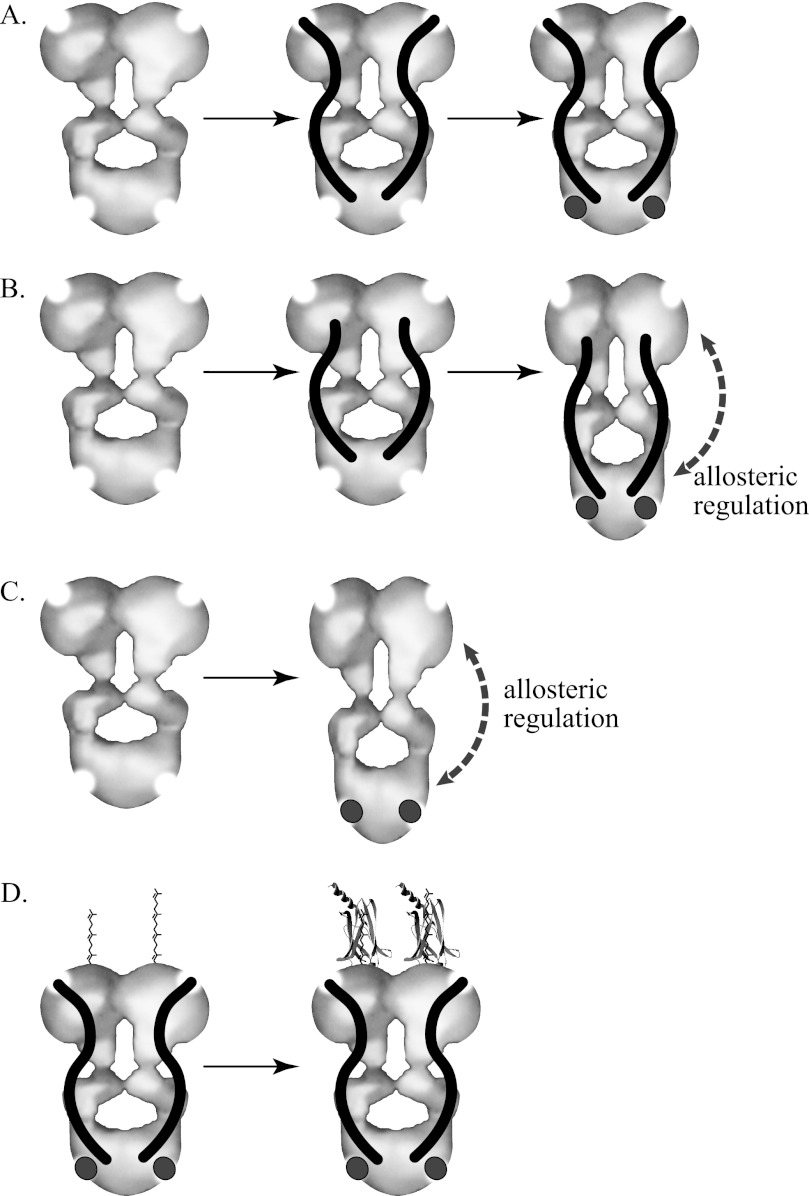
The photoreceptor-specific PDE6 is one of five members of the mammalian phosphodiesterase superfamily that contain tandem GAF domains. In contrast to the other GAF-containing phosphodiesterases (PDE2, PDE5, PDE10, and PDE11) in which ligand binding to the GAF domains is allosterically communicated to the enzyme active site to enhance catalysis (reviewed in Refs. 55 and 56), there is no biochemical evidence supporting direct allosteric regulation of the PDE6 catalytic dimer (Pαβ) by cGMP binding (14–16). Comparison of the hydrodynamic behavior of PDE5 and Pαβ in the absence and presence of cGMP (Fig. 3) demonstrates that PDE5, but not PDE6, undergoes a large scale conformational change of the entire catalytic dimer upon cGMP binding to the GAFa domain (Fig. 7C) that probably represents the structural basis for PDE5 allosteric activation.
FIGURE 7.
Changes in molecular organization of PDE6 and PDE5. The depicted catalytic dimers of PDE6 and PDE5 are based on electron microscopic image analysis of the molecular organization of the enzymes reported by Kameni et al. (9); the enzyme active sites (top) and GAFa regulatory cGMP binding site (bottom) are represented by semicircular areas deleted from the molecular model. The inhibitory Pγ subunit of PDE6 is represented as an S-shaped rod, and cGMP is shown as a dark circle. A, the PDE6 catalytic dimer (Pαβ) fails to undergo a detectable change in conformation upon binding of Pγ and/or cGMP. B, although full-length Pγ fails to support allosteric changes upon cGMP binding, C-terminal truncations of Pγ allow detection of cGMP-induced elongation of the catalytic dimer. C, similarly, an elongation of the PDE5 homodimer is observed upon cGMP binding, consistent with previous hydrodynamic studies (44, 46). D, post-translational farnesylation (left subunit) or geranylgeranylation (right subunit) of the C terminus of the PDE6 α- and β-subunits permit PrBP/δ (crystal structure derived from Protein Data Bank entry 1KSH) to associate with the catalytic subunits, causing an overall elongation of the protein complex.
The absence of direct allosteric regulation of PDE6 by cGMP binding (Fig. 3B) reflects a failure of the GAF domains to communicate to the catalytic domain upon cGMP occupancy because the isolated GAFa domain of cone PDE6 undergoes a similar ligand-dependent stabilization of secondary structure (57) that has been observed for cGMP binding to PDE2 and PDE5 GAF domains (45, 58). The ability of Pγ1–45, known to bind with high affinity to the GAF domains of Pαβ (14, 59, 60), to restore a cGMP-dependent conformational change to the PDE6 catalytic dimer (Fig. 4) dramatically illustrates that allosteric communication from the tandem GAF domains of PDE6 requires participation of the GAF-interacting region of Pγ (Fig. 7B). In this context, the N-terminal half of Pγ may play a role in PDE6 regulation not unlike the N-terminal regions of other GAF-containing cyclic nucleotide phosphodiesterase catalytic subunits, which exert regulatory control over catalytic activity (61, 62).
This ligand-induced conformational change of PDE6 also has important implications for the mechanism of transducin activation of PDE6 during visual excitation. It is generally accepted that that activated transducin α-subunit (Tα*-GTP) binds to the C-terminal region of Pγ to displace the inhibitory constraint of Pγ at the enzyme active site (63–65). A structural study of the complex of Pγ(70–87) with a chimeric PDE5/PDE6 catalytic domain (66) suggests a mechanism in which Tα*-GTP may interact with Pγ residues 71–77 and induce a hingelike movement of the last 10 residues away from the enzyme active site without the Tα*-GTP-Pγ complex dissociating completely from the PDE6 holoenzyme. Our experiments revealing a cGMP-dependent elongation of Pαβ-Pγ(1–60) or Pαβ-Pγ(1–73) (Figs. 4 and 7B) may in fact mimic the transducin-activated state of the PDE6 holoenzyme in which activated transducin has displaced the C-terminal third of Pγ from Pαβ to Tα*-GTP. The state of occupancy of the GAF domains of Pαβ could regulate the conformational transition from an elongated (cGMP-bound) conformation to a more compact (empty site) conformation of the activated enzyme; this may be relevant under physiological conditions of rod photoreceptor light adaptation (i.e. persistent PDE6 activation and sustained lowering of cytoplasmic cGMP levels), where negative feedback regulation of the lifetime of PDE6 activation is expected (67). The failure to detect cGMP-dependent changes in the nonactivated PDE6 holoenzyme conformation (Fig. 7A) may be indicative of the fact that tight binding of the C-terminal region of full-length Pγ to the catalytic domains of Pαβ prevents a cGMP-induced conformational change that would otherwise occur.
Our analysis of the hydrodynamic behavior of Pγ in aqueous solution using AU-FDS agrees well with structural data using other methods (38, 40) and lends support to the conclusion that Pγ is a disordered, extended structure in solution and binds at multiple sites along the surface of the PDE6 catalytic dimer (consistent with chemical cross-linking studies, reviewed in Ref 40). The observation that binding of Pγ to Pαβ occurs without a significant change in overall shape of the resulting heterotetramer (Fig. 7A) supports the idea that Pγ conforms to the overall shape of the catalytic dimer upon binding (Table 1). The function of these multiple sites of interactions between the extended, partially unfolded structure of Pγ and the highly organized three-domain structure of the PDE6 catalytic subunits remains a subject of inquiry and is probably fundamental to the multiple roles Pγ plays in visual excitation and recovery (40).
AU-FDS has been successfully used to quantify the high affinity protein-protein interactions between PrBP/δ and PDE6 holoenzyme (Fig. 5). In conjunction with independently determining the stoichiometry of two molecules of PrBP/δ binding per catalytic dimer, the sedimentation coefficient of the protein complex indicates that significant elongation results from PrBP/δ binding to the isoprenyl groups attached to the catalytic domain of PDE6 (Fig. 7D). This transition to a more elongated structure for the PDE6-PrBP/δ complex in solution is consistent with electron microscopic observations of negatively stained preparations of a GST-PrBP/δ fusion protein mixed with purified PDE6 holoenzyme (11).
Studies on the mechanism of transducin activation of PDE6 are hampered by the fact that in vivo, this process occurs with both proteins confined to the photoreceptor outer segment membrane and at high local concentrations compared with what can be achieved with purified proteins in solution (5). Our hydrodynamic results reveal that binding of transducin to PDE6 in solution can be demonstrated using sedimentation velocity analysis, but the complexity of the data preclude straightforward analysis of the binding affinity or stoichiometry. Future efforts will be directed at reconstituting transducin and PDE6 on buoyant membranes (e.g. nanodiscs (68)) that are amenable to sedimentation velocity analysis.
Mutations in each of the catalytic subunit genes (rod PDE6A, rod PDE6B, and cone PDE6C) as well as the rod inhibitory Pγ subunit (PDE6G) have all been associated with retinal degenerative diseases, such as retinitis pigmentosa, congenital stationary night blindness, and cone-rod dystrophy (69). Indeed, mutations in the rod catalytic subunits are the second most common identifiable cause of autosomal recessive retinitis pigmentosa (70). In addition, it is increasingly clear that genetic defects in proteins known to interact with PDE6 subunits (e.g. transducin, RGS9-1, and GARP2) are also capable of causing retinal dystrophy in humans and in animal models (69), perhaps due to impaired regulation of PDE6 activity. Our biophysical approaches to studying PDE6 and its regulatory binding partners will provide the mechanistic basis for understanding how mutations in phototransduction genes alter their activity, allosteric regulation, and ability to form signal transducing complexes required for normal vision.
Acknowledgment
We thank Susan Chase for providing the GFP used in this study.
This work was supported, in whole or in part, by National Institutes of Health Grant EY-05798. Partial funding was provided by the New Hampshire Agricultural Experiment Station. This is Scientific Contribution Number 2473.
- Tα
- transducin α-subunit
- Tα*
- activated transducin α-subunit
- GAF
- a protein domain named for some of the proteins in which it is found, including cGMP-binding phosphodiesterases, adenylyl cyclases, and the bacterial FhlA protein
- Pαβ
- catalytic dimer of PDE6 α- and β-subunits
- Pγ
- inhibitory γ-subunit of PDE6
- ROS
- rod outer segment(s)
- IAF
- 5-iodoacetamidofluorescein
- PrBP/δ
- prenyl-binding protein-δ
- GTPγS
- guanosine 5′-O-(γ-thio)triphosphate
- AU-FDS
- analytical ultracentrifugation using a fluorescence detection system
- RS
- Stokes radius.
REFERENCES
- 1. Cote R. H. (2004) Characteristics of photoreceptor PDE (PDE6). Similarities and differences to PDE5. Int. J. Impot. Res. 16, S28–S33 [DOI] [PubMed] [Google Scholar]
- 2. Conti M., Beavo J. (2007) Biochemistry and physiology of cyclic nucleotide phosphodiesterases. Essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 76, 481–511 [DOI] [PubMed] [Google Scholar]
- 3. Francis S. H., Blount M. A., Corbin J. D. (2011) Mammalian cyclic nucleotide phosphodiesterases. Molecular mechanisms and physiological functions. Physiol. Rev. 91, 651–690 [DOI] [PubMed] [Google Scholar]
- 4. Cote R. H. (2006) Photoreceptor phosphodiesterase (PDE6). A G-protein-activated PDE regulating visual excitation in rod and cone photoreceptor cells. in Cyclic Nucleotide Phosphodiesterases in Health and Disease (Beavo J. A., Francis S. H., Houslay M. D., eds) pp. 165–193, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 5. Wensel T. G. (2008) Signal transducing membrane complexes of photoreceptor outer segments. Vision Res. 48, 2052–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arshavsky V. Y., Lamb T. D., Pugh E. N., Jr. (2002) G proteins and phototransduction. Annu. Rev. Physiol. 64, 153–187 [DOI] [PubMed] [Google Scholar]
- 7. Cote R. H. (2008) The cGMP signaling pathway in retinal photoreceptors and the central role of photoreceptor phosphodiesterase (PDE6). In Visual Transduction and Non-Visual Light Perception (Tombran-Tink J., Barnstable C. J., eds) pp. 141–169, Humana Press, Totawa, NJ [Google Scholar]
- 8. Burns M. E., Pugh E. N., Jr. (2010) Lessons from photoreceptors. Turning off G-protein signaling in living cells. Physiology 25, 72–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kameni Tcheudji J. F., Lebeau L., Virmaux N., Maftei C. G., Cote R. H., Lugnier C., Schultz P. (2001) Molecular organization of bovine rod cGMP-phosphodiesterase 6. J. Mol. Biol. 310, 781–791 [DOI] [PubMed] [Google Scholar]
- 10. Kajimura N., Yamazaki M., Morikawa K., Yamazaki A., Mayanagi K. (2002) Three-dimensional structure of non-activated cGMP phosphodiesterase 6 and comparison of its image with those of activated forms. J. Struct. Biol. 139, 27–38 [DOI] [PubMed] [Google Scholar]
- 11. Goc A., Chami M., Lodowski D. T., Bosshart P., Moiseenkova-Bell V., Baehr W., Engel A., Palczewski K. (2010) Structural characterization of the rod cGMP phosphodiesterase 6. J. Mol. Biol. 401, 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yamazaki A., Hayashi F., Tatsumi M., Bitensky M. W., George J. S. (1990) Interactions between the subunits of transducin and cyclic GMP phosphodiesterase in Rana catesbiana rod photoreceptors. J. Biol. Chem. 265, 11539–11548 [PubMed] [Google Scholar]
- 13. Cote R. H., Bownds M. D., Arshavsky V. Y. (1994) cGMP binding sites on photoreceptor phosphodiesterase. Role in feedback regulation of visual transduction. Proc. Natl. Acad. Sci. U.S.A. 91, 4845–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mou H., Cote R. H. (2001) The catalytic and GAF domains of the rod cGMP phosphodiesterase (PDE6) heterodimer are regulated by distinct regions of its inhibitory γ subunit. J. Biol. Chem. 276, 27527–27534 [DOI] [PubMed] [Google Scholar]
- 15. Arshavsky V. Y., Dumke C. L., Bownds M. D. (1992) Noncatalytic cGMP-binding sites of amphibian rod cGMP phosphodiesterase control interaction with its inhibitory γ-subunits. A putative regulatory mechanism of the rod photoresponse. J. Biol. Chem. 267, 24501–24507 [PubMed] [Google Scholar]
- 16. D'Amours M. R., Cote R. H. (1999) Regulation of photoreceptor phosphodiesterase catalysis by its non-catalytic cGMP-binding sites. Biochem. J. 340, 863–869 [PMC free article] [PubMed] [Google Scholar]
- 17. Howlett G. J., Minton A. P., Rivas G. (2006) Analytical ultracentrifugation for the study of protein association and assembly. Curr. Opin. Chem. Biol. 10, 430–436 [DOI] [PubMed] [Google Scholar]
- 18. Baehr W., Devlin M. J., Applebury M. L. (1979) Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J. Biol. Chem. 254, 11669–11677 [PubMed] [Google Scholar]
- 19. Kohnken R. E., Eadie D. M., Revzin A., McConnell D. G. (1981) The light-activated GTP-dependent cyclic GMP phosphodiesterase complex of bovine retinal rod outer segments. Dark resolution of the catalytic and regulatory proteins. J. Biol. Chem. 256, 12502–12509 [PubMed] [Google Scholar]
- 20. Kroe R. R., Laue T. M. (2009) NUTS and BOLTS. Applications of fluorescence-detected sedimentation. Anal. Biochem. 390, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kingsbury J. S., Laue T. M. (2011) Fluorescence-detected sedimentation in dilute and highly concentrated solutions. Methods Enzymol. 492, 283–304 [DOI] [PubMed] [Google Scholar]
- 22. Pentia D. C., Hosier S., Collupy R. A., Valeriani B. A., Cote R. H. (2005) Purification of PDE6 isozymes from mammalian retina. Methods Mol. Biol. 307, 125–140 [DOI] [PubMed] [Google Scholar]
- 23. Cote R. H. (2000) Kinetics and regulation of cGMP binding to noncatalytic binding sites on photoreceptor phosphodiesterase. Methods Enzymol. 315, 646–672 [DOI] [PubMed] [Google Scholar]
- 24. Hurley J. B., Stryer L. (1982) Purification and characterization of the gamma regulatory subunit of the cyclic GMP phosphodiesterase from retinal rod outer segments. J. Biol. Chem. 257, 11094–11099 [PubMed] [Google Scholar]
- 25. Cote R. H. (2005) Cyclic guanosine 5′-monophosphate binding to regulatory GAF domains of photoreceptor phosphodiesterase. Methods Mol. Biol. 307, 141–154 [DOI] [PubMed] [Google Scholar]
- 26. Zhang X. J., Skiba N. P., Cote R. H. (2010) Structural requirements of the photoreceptor phosphodiesterase γ-subunit for inhibition of rod PDE6 holoenzyme and for its activation by transducin. J. Biol. Chem. 285, 4455–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Corbin J. D., Blount M. A., Weeks J. L., 2nd, Beasley A., Kuhn K. P., Ho Y. S., Saidi L. F., Hurley J. H., Kotera J., Francis S. H. (2003) [3H]Sildenafil binding to phosphodiesterase-5 is specific, kinetically heterogeneous, and stimulated by cGMP. Mol. Pharmacol. 63, 1364–1372 [DOI] [PubMed] [Google Scholar]
- 28. Artemyev N. O., Arshavsky V. Y., Cote R. H. (1998) Photoreceptor phosphodiesterase. Interaction of inhibitory γ subunit and cyclic GMP with specific binding sites on catalytic subunits. Methods 14, 93–104 [DOI] [PubMed] [Google Scholar]
- 29. Kleuss C., Pallast M., Brendel S., Rosenthal W., Schultz G. (1987) Resolution of transducin subunits by chromatography on blue Sepharose. J. Chromatogr. 407, 281–289 [DOI] [PubMed] [Google Scholar]
- 30. Wensel T. G., He F., Malinski J. A. (2005) Purification, reconstitution on lipid vesicles, and assays of PDE6 and its activator G protein, transducin. Methods Mol. Biol. 307, 289–313 [DOI] [PubMed] [Google Scholar]
- 31. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 32. MacGregor I. K., Anderson A. L., Laue T. M. (2004) Fluorescence detection for the XLI analytical ultracentrifuge. Biophys. Chem. 108, 165–185 [DOI] [PubMed] [Google Scholar]
- 33. Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. (2002) Size-distribution analysis of proteins by analytical ultracentrifugation. Strategies and application to model systems. Biophys. J. 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Laue T. M., Shah B., Ridgeway T. M., Pelletier S. L. (1992) Computer-aided interpretation of sedimentation data for proteins. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (Harding S. E., Rowe A. J., Horton J. C., eds) Royal Society of Chemistry, Cambridge [Google Scholar]
- 35. Rai N., Nöllmann M., Spotorno B., Tassara G., Byron O., Rocco M. (2005) SOMO (SOlution MOdeler) differences between x-ray- and NMR-derived bead models suggest a role for side chain flexibility in protein hydrodynamics. Structure 13, 723–734 [DOI] [PubMed] [Google Scholar]
- 36. Brookes E., Demeler B., Rosano C., Rocco M. (2010) The implementation of SOMO (SOlution MOdeller) in the UltraScan analytical ultracentrifugation data analysis suite. Enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur. Biophys. J. 39, 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berger A. L., Cerione R. A., Erickson J. W. (1997) Real time conformational changes in the retinal phosphodiesterase γ subunit monitored by resonance energy transfer. J. Biol. Chem. 272, 2714–2721 [DOI] [PubMed] [Google Scholar]
- 38. Uversky V. N., Permyakov S. E., Zagranichny V. E., Rodionov I. L., Fink A. L., Cherskaya A. M., Wasserman L. A., Permyakov E. A. (2002) Effect of zinc and temperature on the conformation of the γ subunit of retinal phosphodiesterase. A natively unfolded protein. J. Proteome Res. 1, 149–159 [DOI] [PubMed] [Google Scholar]
- 39. Song J., Guo L. W., Muradov H., Artemyev N. O., Ruoho A. E., Markley J. L. (2008) Intrinsically disordered γ-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. Proc. Natl. Acad. Sci. U.S.A. 105, 1505–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo L. W., Ruoho A. E. (2008) The retinal cGMP phosphodiesterase γ-subunit. A chameleon. Curr. Protein Pept. Sci. 9, 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wensel T. G., Stryer L. (1986) Reciprocal control of retinal rod cyclic GMP phosphodiesterase by its γ subunit and transducin. Proteins 1, 90–99 [DOI] [PubMed] [Google Scholar]
- 42. Mou H., Grazio H. J., 3rd, Cook T. A., Beavo J. A., Cote R. H. (1999) cGMP binding to noncatalytic sites on mammalian rod photoreceptor phosphodiesterase is regulated by binding of its γ and δ subunits. J. Biol. Chem. 274, 18813–18820 [DOI] [PubMed] [Google Scholar]
- 43. Francis S. H., Bessay E. P., Kotera J., Grimes K. A., Liu L., Thompson W. J., Corbin J. D. (2002) Phosphorylation of isolated human phosphodiesterase-5 regulatory domain induces an apparent conformational change and increases cGMP binding affinity. J. Biol. Chem. 277, 47581–47587 [DOI] [PubMed] [Google Scholar]
- 44. Corbin J. D., Zoraghi R., Francis S. H. (2009) Allosteric site and catalytic site ligand effects on PDE5 functions are associated with distinct changes in physical form of the enzyme. Cell. Signal. 21, 1768–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heikaus C. C., Stout J. R., Sekharan M. R., Eakin C. M., Rajagopal P., Brzovic P. S., Beavo J. A., Klevit R. E. (2008) Solution structure of the cGMP binding GAF domain from phosphodiesterase 5. Insights into nucleotide specificity, dimerization, and cGMP-dependent conformational change. J. Biol. Chem. 283, 22749–22759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Francis S. H., Chu D. M., Thomas M. K., Beasley A., Grimes K., Busch J. L., Turko I. V., Haik T. L., Corbin J. D. (1998) Ligand-induced conformational changes in cyclic nucleotide phosphodiesterases and cyclic nucleotide-dependent protein kinases. Methods 14, 81–92 [DOI] [PubMed] [Google Scholar]
- 47. Yamazaki A., Bartucca F., Ting A., Bitensky M. W. (1982) Reciprocal effects of an inhibitory factor on catalytic activity and noncatalytic cGMP binding sites of rod phosphodiesterase. Proc. Natl. Acad. Sci. U.S.A. 79, 3702–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X. J., Cahill K. B., Elfenbein A., Arshavsky V. Y., Cote R. H. (2008) Direct allosteric regulation between the GAF domain and catalytic domain of photoreceptor phosphodiesterase PDE6. J. Biol. Chem. 283, 29699–29705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamazaki A., Hayashi F., Matsuura I., Bondarenko V. A. (2011) Binding of cGMP to the transducin-activated cGMP phosphodiesterase, PDE6, initiates a large conformational change involved in its deactivation. FEBS J. 278, 1854–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Norton A. W., Hosier S., Terew J. M., Li N., Dhingra A., Vardi N., Baehr W., Cote R. H. (2005) Evaluation of the 17-kDa prenyl-binding protein as a regulatory protein for phototransduction in retinal photoreceptors. J. Biol. Chem. 280, 1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Karan S., Zhang H., Li S., Frederick J. M., Baehr W. (2008) A model for transport of membrane-associated phototransduction polypeptides in rod and cone photoreceptor inner segments. Vision Res. 48, 442–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gitschier H. J., Cote R. H. (2011) Phosphodiesterase 6D, cGMP-specific rod δ, UCSD-Nature Molecule Pages doi: 10.1038/mp.a001757.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hanzal-Bayer M., Renault L., Roversi P., Wittinghofer A., Hillig R. C. (2002) The complex of Arl2-GTP and PDEδ. From structure to function. EMBO J. 21, 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Y. T., Matte S. L., Corbin J. D., Francis S. H., Cote R. H. (2009) Probing the catalytic sites and activation mechanism of photoreceptor phosphodiesterase using radiolabeled phosphodiesterase inhibitors. J. Biol. Chem. 284, 31541–31547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heikaus C. C., Pandit J., Klevit R. E. (2009) Cyclic nucleotide binding GAF domains from phosphodiesterases. Structural and mechanistic insights. Structure 17, 1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jäger R., Russwurm C., Schwede F., Genieser H. G., Koesling D., Russwurm M. (2012) Activation of PDE10 and PDE11 phosphodiesterases. J. Biol. Chem. 287, 1210–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martinez S. E., Heikaus C. C., Klevit R. E., Beavo J. A. (2008) The structure of the GAF A domain from phosphodiesterase 6C reveals determinants of cGMP binding, a conserved binding surface, and a large cGMP-dependent conformational change. J. Biol. Chem. 283, 25913–25919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pandit J., Forman M. D., Fennell K. F., Dillman K. S., Menniti F. S. (2009) Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc. Natl. Acad. Sci. U.S.A. 106, 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Muradov K. G., Granovsky A. E., Schey K. L., Artemyev N. O. (2002) Direct interaction of the inhibitory γ-subunit of rod cGMP phosphodiesterase (PDE6) with the PDE6 GAFa domains. Biochemistry 41, 3884–3890 [DOI] [PubMed] [Google Scholar]
- 60. Guo L. W., Muradov H., Hajipour A. R., Sievert M. K., Artemyev N. O., Ruoho A. E. (2006) The inhibitory γ subunit of the rod cGMP phosphodiesterase binds the catalytic subunits in an extended linear structure. J. Biol. Chem. 281, 15412–15422 [DOI] [PubMed] [Google Scholar]
- 61. Bruder S., Schultz A., Schultz J. E. (2006) Characterization of the tandem GAF domain of human phosphodiesterase 5 using a cyanobacterial adenylyl cyclase as a reporter enzyme. J. Biol. Chem. 281, 19969–19976 [DOI] [PubMed] [Google Scholar]
- 62. Gross-Langenhoff M., Stenzl A., Altenberend F., Schultz A., Schultz J. E. (2008) The properties of phosphodiesterase 11A4 GAF domains are regulated by modifications in its N-terminal domain. FEBS J. 275, 1643–1650 [DOI] [PubMed] [Google Scholar]
- 63. Slepak V. Z., Artemyev N. O., Zhu Y., Dumke C. L., Sabacan L., Sondek J., Hamm H. E., Bownds M. D., Arshavsky V. Y. (1995) An effector site that stimulates G-protein GTPase in photoreceptors. J. Biol. Chem. 270, 14319–14324 [DOI] [PubMed] [Google Scholar]
- 64. Slep K. C., Kercher M. A., He W., Cowan C. W., Wensel T. G., Sigler P. B. (2001) Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 Å. Nature 409, 1071–1077 [DOI] [PubMed] [Google Scholar]
- 65. Granovsky A. E., Artemyev N. O. (2001) A conformational switch in the inhibitory γ-subunit of PDE6 upon enzyme activation by transducin. Biochemistry 40, 13209–13215 [DOI] [PubMed] [Google Scholar]
- 66. Barren B., Gakhar L., Muradov H., Boyd K. K., Ramaswamy S., Artemyev N. O. (2009) Structural basis of phosphodiesterase 6 inhibition by the C-terminal region of the γ-subunit. EMBO J. 28, 3613–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Burns M. E., Arshavsky V. Y. (2005) Beyond counting photons. Trials and trends in vertebrate visual transduction. Neuron 48, 387–401 [DOI] [PubMed] [Google Scholar]
- 68. Bayburt T. H., Sligar S. G. (2010) Membrane protein assembly into Nanodiscs. FEBS Lett. 584, 1721–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baehr W., Frederick J. M. (2009) Naturally occurring animal models with outer retina phenotypes. Vision Res. 49, 2636–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ferrari S., Di Iorio E., Barbaro V., Ponzin D., Sorrentino F. S., Parmeggiani F. (2011) Retinitis pigmentosa. Genes and disease mechanisms. Curr. Genomics 12, 238–249 [DOI] [PMC free article] [PubMed] [Google Scholar]