Background: Cell-specific expression of mammalian genes is typically controlled by transcription via dedicated promoters.
Results: The GPR41 gene lacks a dedicated promoter; it is transcribed from the promoter of the upstream GPR40 gene.
Conclusion: The locus is a rare example of a mammalian operon.
Significance: Mammalian operons may be more common than appreciated and participate in coordinated gene expression.
Keywords: 7-Helix Receptor, Insulin, Pancreas, Transcriptional Regulation, Translational Regulation
Abstract
GPR41 is a G protein-coupled receptor activated by short chain fatty acids. The gene encoding GPR41 is located immediately downstream of a related gene encoding GPR40, a receptor for long chain fatty acids. Expression of GPR41 has been reported in a small number of cell types, including gut enteroendocrine cells and sympathetic ganglia, where it may play a role in the maintenance of metabolic homeostasis. We now demonstrate that GPR41, like GPR40, is expressed in pancreatic beta cells. Surprisingly, we found no evidence for transcriptional control elements or transcriptional initiation in the intergenic GPR40-GPR41 region. Rather, using 5′-rapid amplification of cDNA ends analysis, we demonstrated that GPR41 is transcribed from the promoter of the GPR40 gene. We confirmed this finding by generating bicistronic luciferase reporter plasmids, and we were able to map a potential internal ribosome entry site-containing region to a 2474-nucleotide region of the intergenic sequence. Consistent with this, we observed m7G cap-independent reporter gene expression upon transfection of RNA containing this region. Thus, GPR41 expression is mediated via an internal ribosome entry site located in the intergenic region of a bicistronic mRNA. This novel sequence organization may be utilized to permit coordinated regulation of the fatty acid receptors GPR40 and GPR41.
Introduction
Pancreatic beta cells play a central role in maintaining metabolic homeostasis through their unique capacity to produce the hormone insulin (1, 2). Insulin secretion by beta cells is tightly regulated according to precise physiological needs. The primary trigger for insulin secretion is elevated blood glucose concentration; however, numerous additional stimuli, including a wide range of nutrients, fine-tune insulin output to precisely match metabolic requirements (3, 4). Among these, long chain fatty acids (LCFA)3 are well documented regulators of glucose-stimulated insulin secretion. LCFA have complex and paradoxical effects on insulin secretion; acute exposure of beta cells to LCFA leads to augmented glucose-stimulated insulin secretion, whereas prolonged treatment leads to severely impaired beta cell function or “lipotoxicity” (5, 6).
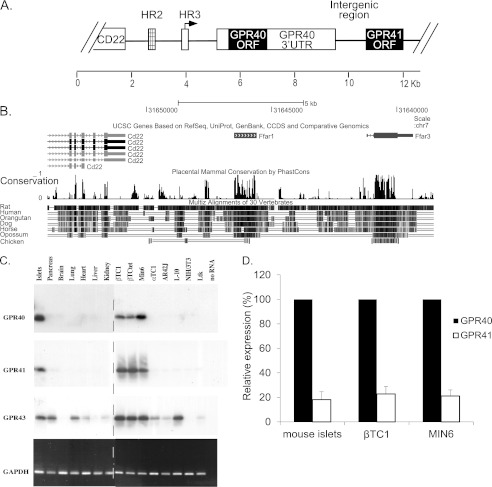
The G protein-coupled receptor GPR40 (FFAR1) is expressed selectively in beta cells (7, 8) and gut enteroendocrine cells (9). It is activated by a range of medium and long chain fatty acids (7, 8). Several studies indicate that GPR40 plays an important role in mediating the effects of LCFA on glucose-stimulated insulin secretion (10–12). Immediately downstream of GPR40 in the mammalian genome is the gene encoding GPR41 (FFAR3) (Fig. 1, A and B) (13), a related G protein-coupled receptor that is activated by short chain fatty acids (SCFA) (14). Approximately 40 kb downstream of the GPR41 gene is the GPR43 (FFAR2) gene (13), encoding another SCFA receptor (14). SCFA are generated primarily in the gastrointestinal tract as a result of microflora-mediated metabolism of ingested food (15). SCFA constitute an important caloric source in mammals (16), yet the physiological role of SCFA is incompletely understood. GPR41 expression has been reported in the peripheral nervous system, gut enteroendocrine cells, and adipose tissue (17–20). Genetic studies in mice have indicated that these receptors may play an important role in mediating the responsiveness of target cells to fatty acids (17, 19).
FIGURE 1.
Genome structure and gene expression pattern of GPR40/41 locus. A, representation of the GPR40/41 locus. GPR40 and GPR41 ORFs are shown as black boxes. White boxes flanking the GPR40 ORF indicate 5′- and 3′UTRs. Boxes upstream of the GPR40 5′UTR show the transcriptional control regions HR2 (hatched) and HR3 (white). The arrow above HR3 indicates the GPR40 transcription start site. B, comparative genomic analysis using the UCSC genome browser showing high conservation of GPR40 and GPR41 ORFs. C, RT-PCR analysis of RNA from mouse tissues and cell lines. Total RNA was subjected to RT-PCR followed by Southern blot analysis using specific probes of GPR40, GPR41, and GPR43. The specific activity of probes used was 107 to 108 cpm/μg. The lower panel shows ethidium bromide staining of RT-PCR using GAPDH primers. βTC1, βTCtet, and MIN6, mouse pancreatic β cells; αTC1, mouse pancreatic α cell; AR4-2J, rat pancreatic exocrine cell; L-10, mouse lymphoid cell; NIH-3T3, mouse embryonic cell; Ltk, mouse fibroblast cell. The dotted line indicates the separation point between two blots processed under identical conditions. D, endogenous expression of GPR40 and GPR41. Expression was measured by qRT-PCR and presented relative to GPR40. The results are the mean ± S.E. (n ≥5).
Studies from our laboratory have shown that selective expression of GPR40 in beta cells is controlled by transcriptional mechanisms (21). Of particular importance is HR2, a beta cell-specific transcriptional enhancer located 1.1 kb upstream of the gene (Fig. 1A). We now report that GPR41 is also expressed in beta cells. Unexpectedly, the GPR41 gene lacks a dedicated promoter; rather, transcription of GPR41 initiates from the GPR40 promoter, located 6619 bp upstream of the GPR41 gene, generating a bicistronic transcript encoding both GPR40 and GPR41. Translation of GPR41 is mediated by an internal ribosome entry site (IRES) located in the intergenic region.
EXPERIMENTAL PROCEDURES
Total RNA Extraction
Total RNA was extracted from cell cultures using the TRI Reagent procedure (Molecular Research Center Inc.) according to the manufacturer's instructions. Where indicated, RNA was treated with DNase to remove traces of genomic or plasmid DNA. Ten μg of total RNA were incubated with 1 unit of DNase I (RQ1, Promega), 1 mm DTT, 40 units of RNasin, and 1× RQ1 buffer (Promega) in a final volume of 20 μl at 37 °C for 20 min. Following phenol/chloroform extraction, RNA was ethanol-precipitated and dissolved in diethyl pyrocarbonate-treated H2O. For quantitative reverse transcriptase-PCR (qRT-PCR) experiments, the extracted RNA was loaded on Direct-zol RNA MiniPrep columns (Zymo Research Corp.) and treated with on-column DNase according to the manufacturer's instructions.
RT-PCR
RT-PCR was performed using the Access RT-PCR system (Promega) with 20 ng of total RNA and 20 pmol each of the forward and reverse primers of GPR40, GPR41, GPR43, and GAPDH (supplemental Table 1). RT-PCR conditions were as follows: 48 °C for 45 min, 94 °C for 2 min, 35 cycles: 94 °C for 30 s, 56 °C for 1 min, 68 °C for 2 min, and a final extension step at 68 °C for 7 min. RT-PCR products were resolved on a 1% agarose gel, transferred to a nylon membrane (Nytran N, Schleicher & Schuell), and hybridized in hybridization solution (50% formamide, 2× SSC, 5× Denhardt's, 1% SDS, 100 μg/ml boiled salmon sperm DNA) at 42 °C with radioactively labeled probes generated by random primed labeling (Roche Applied Science). For qRT-PCR experiments, first strand cDNA synthesis was performed by SuperScript II reverse transcriptase (Invitrogen). RNA was hybridized with hexamer random primers (Fermentas) at 65 °C for 5 min. RT-PCR conditions were 25 °C for 10 min, 42 °C for 50 min, and inactivation at 70 °C for 15 min. qRT-PCR was performed with power SYBR Green (Applied Biosystems) using cDNA derived from 20 ng of RNA samples according to the manufacturer's instructions. Primer details are shown in supplemental Table 1. The results were normalized to expression of β-actin and hypoxanthine-guanine phosphoribosyltransferase. The results were quantitated by the ΔΔCt method, relative to control (22).
5′-Rapid Amplification of cDNA Ends (5′-RACE)
5′-RACE was performed as described previously (21) with the following modifications. Method A, first strand cDNA was synthesized from 5 μg of DNase-treated βTC1 RNA, using reverse transcriptase (SuperScript II; Invitrogen) according to the manufacturer's instructions. The reaction mixture was incubated at 42 °C for 1 h. Primers used were complementary to GPR40–41 intergenic region, GPR40 3′-UTR or GPR41 ORF. A second procedure (method B) was used to characterize particularly long transcripts. Method B, first strand cDNA was synthesized from 10 μg of DNase-treated βTC1 RNA, using reverse transcriptase (StrataScript 5.0; Stratagene) according to the manufacturer's instructions. The reaction mixture was incubated at 55 °C for 1.5 h. The rest of the experiment was identical in the two methods. The cDNA was purified using RBC HiYieldTM Gel/PCR DNA extraction kit (Real Biotech Corp.), and eluted in 40 μl of H2O. A poly(dG) tail was added to the cDNA 3′ end using terminal deoxynucleotidyltransferase (Promega). Following a second purification of the cDNA using RBC purification kit, cDNA was eluted in 100 μl of DDW. PCR was performed using the Accuzyme PCR system (Bioline), with 10 μl of cDNA, 30 pmol of a reverse primer complementary to GPR40/41 sequences located nested to the primer used for reverse transcription, and 30 pmol forward primer GAATTC(C)24. PCR conditions were as follows: 94 °C for 2 min, followed by 25 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 2 min, followed by an additional 72 °C for 5 min. A fraction of the PCR product was resolved on a 1% agarose gel. When a band appeared, it was excised from the gel, purified, and sequenced. Primer details are shown in supplemental Table 1.
3′-RACE
First strand cDNA was synthesized from 5 μg of DNase-treated βTC1 RNA, using reverse transcriptase (Super Script II; Invitrogen) according to the manufacturer's instructions. The reaction mixture was incubated at 42 °C for 1 h. The primer used was 5′-GCGAGCACAGAATTAATACGACTCACTATAGG(T)12. PCR was performed using the Expand high Fidelity PCR system (Roche Applied Science), with one-third of the cDNA, 30 pmol of a reverse primer 5′-GCGAGCACAGAATTAATACGAC (complementary to the 5′ flank of the primer used for reverse transcription), and a forward gene-specific primer complementary to GPR40 or GPR41 sequence. PCR conditions were 94 °C for 2 min, followed by 25 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 2 min, followed by an additional 72 °C for 5 min. A fraction of the PCR product was resolved on a 1% agarose gel. When a band appeared, it was excised from the gel, purified, and sequenced.
Plasmid Constructions
Plasmids for GPR41 promoter activity analysis were constructed using pGL3-basic vector (Promega). Fragments from the region upstream to the GPR41 gene were generated by digestion of a genomic library clone described previously (21) using the following restriction enzymes SacI and NcoI (construct 4106), SacI (construct 3450), SacI and EcoRV (construct 2054), NcoI (construct 1427), and BamHI and HindIII (construct 1691). Construct 5054 was generated by insertion of fragment 1691 into construct 4106 using the HindIII site.
Islet Isolation
Islets were isolated from 1.5 to 3-month-old mice by in situ pancreatic perfusion with collagenase (23). After isolation, islets were incubated overnight in culture media (RPMI, 10% FCS, 1% l-glutamine, 1% penicillin/streptomycin, and 1% gentamycin), and RNA was isolated as described above.
Cell Culture
The following established cell lines were used in this study: βTC1, βTCtet, and MIN6 (mouse beta cells); HIT M2.2.2 (hamster beta cells); αTC1 (mouse alpha cells), and HeLa and NIH-3T3 (mouse fibroblast cells). βTC1, βTCtet HIT, αTC1, HeLa, and NIH-3T3 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), penicillin (200 IU/ml), and streptomycin (100 μg/ml). MIN6 cells were grown on Falcon tissue culture plates in DMEM containing 10 mm d-glucose, supplemented with 15% FCS, penicillin (200 IU/ml), streptomycin (100 μg/ml), 2 mm l-glutamine, and 72 mm β-mercaptoethanol. All experiments using MIN6 cells were performed with passages not higher than 33.
Transient Transfections
Transfections were carried out by the calcium phosphate coprecipitation technique, as described previously (24), or using the transfection reagent JetPEITM (PolyPlus transfection) according to the manufacturer's instructions. Forty eight h after transfection, cells were harvested by passive lysis buffer (Promega), and extracts were subjected to assays to determine the activity of reporter enzymes.
siRNA Transfection
Cells were plated 24 h before transfection to 12-well plates (7 × 105 cells/well). Four microliters of Dharmafect 4 (Dharmacon) were used to transfect 1.25 μl of ON-TARGET PLUS pool siRNA (Dharmacon) (final concentration 25 nm). Each siRNA pool contained four different siRNAs targeting coding regions of GPR40 or GPR41. A nontarget siRNA pool was used as control. After 48 h, cells were harvested for RNA purification. Transfection efficiency was determined using siGLO positive control (Dharmacon) transfected under the same conditions. Transfection efficiency was ∼85% as evaluated by fluorescence microscopy.
Luciferase Assays
Firefly luciferase and Renilla luciferase assays were carried out as follows: whole cell extracts containing 5–50 μg (1–5 μl) of protein were added to 100 μl of either firefly luciferase assay buffer (20 mm Tricine, 0.1 mm EDTA, 1.07 mm (MgCO3)4Mg(OH)2·5H2O, 2.67 mm MgSO4, 3.3 mm DTT, 270 μm coenzyme A, 470 μm luciferin (Promega E1602) and 530 μm ATP, pH 7.8.) or Renilla luciferase assay buffer (0.1 m K2HPO4 and 0.1 m KH2PO4, pH 7.4, and 0.5 μm coelenterazine (Calbiochem)). The samples were placed in a luminometer (LUMAC Biocounter M2500 or Modulus microplate, Turner Biosystems), and the light output was determined over a 10-s interval. Firefly luciferase activity was normalized to the activity of Renilla luciferase.
In Vitro Transcription and RNA Transfections
In vitro transcription of pcDNA3-RL, pcDNA3-2474-RL, and pc-FL was performed with mMESSAGE mMACHINE kit (Ambion) according to the manufacturer's instructions with the following modifications: all the plasmids were linearized by incubation for 2 h with enzyme XbaI, which digests downstream of the end of the RL and FL coding sequences. Following linearization, the plasmids were subjected to ethanol precipitation and treated with proteinase K for removal of remaining enzyme. Then the DNA was purified by phenol/chloroform extraction and ethanol precipitation and dissolved in diethyl pyrocarbonate-treated H2O. The in vitro transcriptional reaction was performed in a 10-μl reaction volume using 0.5 μg of DNA template in the presence of ARCA m7G cap analog (Ambion) or unmethylated cap analog (Epicenter) and incubated for 2 h. The resulting transcripts were purified using MEGAclear kit (Ambion) and analyzed by agarose gel electrophoresis to verify size and integrity of the RNAs. Transfection was carried into HeLa cells using TransMessenger transfection reagent (Qiagen) according to the manufacturer's instruction with the following modifications: 0.8 μg of methylated/unmethylated cap-RL or methylated/unmethylated cap-2474-RL, and 0.2 μg of methylated cap-FL RNA was transfected at a ratio of 4:1 (μl of transfection reagent to μg of RNA). Twenty h after transfection, cells were harvested using passive lysis buffer (Promega), and extracts were subjected to luciferase assay.
Northern Blot
Poly(A)+ mRNA was purified using mRNA isolation kit (Roche Applied Science) according to the manufacturer's instructions. The mRNA (5 μg) was mixed with sample buffer (50% formamide, 16% formaldehyde, 1× MOPS buffer (50 mm MOPS, pH 7.0, 5 mm NaAc, pH 5.5, 1 mm EDTA, pH 7.2)), heated to 60 °C for 10 min, cooled on ice, and 5 μl of sample dye (50% glycerol, 1 mm EDTA, 0.25% bromphenol blue, 0.25% xylene cyanol) were added. Samples were resolved on 0.7% agarose, 17.5% formaldehyde gel in 1× MOPS buffer at ∼70 V for 6–10 h. Following electrophoresis, the gel was UV-irradiated for 1.5 min, washed with H2O, and then with 20× SSC (1× SSC = 0.015 m sodium citrate, 0.15 m NaCl). RNA was capillary-blotted overnight to a GeneScreen Plus membrane (PerkinElmer Life Sciences) using 10× SSC as the transfer solution. The membrane was rinsed for 30 min with 2× SSC, 0.1% SDS, and RNA was fixed to the membrane using UV cross-linking (UV Stratalinker 1800, wavelength 254 nm, optimal cross-linking setting). The membrane was pre-hybridized for 3 h at 42 °C with hybridization solution (50% formamide, 2× SSC, 5× Denhardt's, 1% SDS, and 100 μg/ml boiled salmon sperm DNA), and then hybridization was performed overnight at 42 °C with hybridization solution containing 106 cpm/ml radiolabeled probe. Following hybridization, the membrane was washed four times with 2× SSC, 0.1% SDS solution for 5 min at RT, and once with 0.2× SSC, 0.1% SDS solution for 15 min at 50 °C. Radioactive probe was prepared using a random priming procedure as follows: 30 ng of digested DNA were heated at 100 °C for 5 min and quickly chilled on ice. Then it was incubated in the presence of 0.05 mm dATP, dGTP, and dTTP, Hexamix buffer, 50 μCi of [α-32P]dCTP (3,000 Ci/mmol), and 2 units of DNA polymerase I (Klenow fragment, Roche Applied Science) in a final volume of 20 μl for 3 h at 37 °C. The radiolabeled probe was separated from free nucleotides using ProbeQuantTM G-50 micro columns (Amersham Biosciences) according to the manufacturer's instructions.
Bioinformatics
EST and sequence conservation data were from the UCSC genome browser.
RESULTS
Expression Pattern of the GPR40 Gene Family
To compare the expression patterns of the GPR40, GPR41, and GPR43 genes, we prepared RNA from a range of mouse tissues and cell lines. RT-PCR analysis was performed using sets of primers specific for each ORF. We observed GPR40 and GPR41 expression in pancreatic islets and in the beta cell lines βTC1, βTCtet, and MIN6 but not in any of the other tissues examined (Fig. 1C). GPR43, however, was more widely expressed. It was found in islets, pancreas, lung, and in the beta cell lines βTC1, βTCtet, MIN6, and in L-10 (a lymphoid cell line), and to a lesser extent in heart, kidney, αTC1, and AR4-2J. Because this analysis was performed using radioactively labeled RNA probes of unequal specific activity, it can provide information regarding the relative expression of each gene in different tissues and cells, but it cannot permit comparison of expression of the different genes. To compare relative expression of GPR40 and GPR41 in mouse islets and beta cell lines, we used qRT-PCR. This analysis showed that GPR40 mRNA is expressed at ∼5 times higher levels as compared with GPR41 mRNA in MIN6 and βTC1 cell lines and in mouse pancreatic islets (Fig. 1D).
Mapping of GPR41 Gene Boundaries
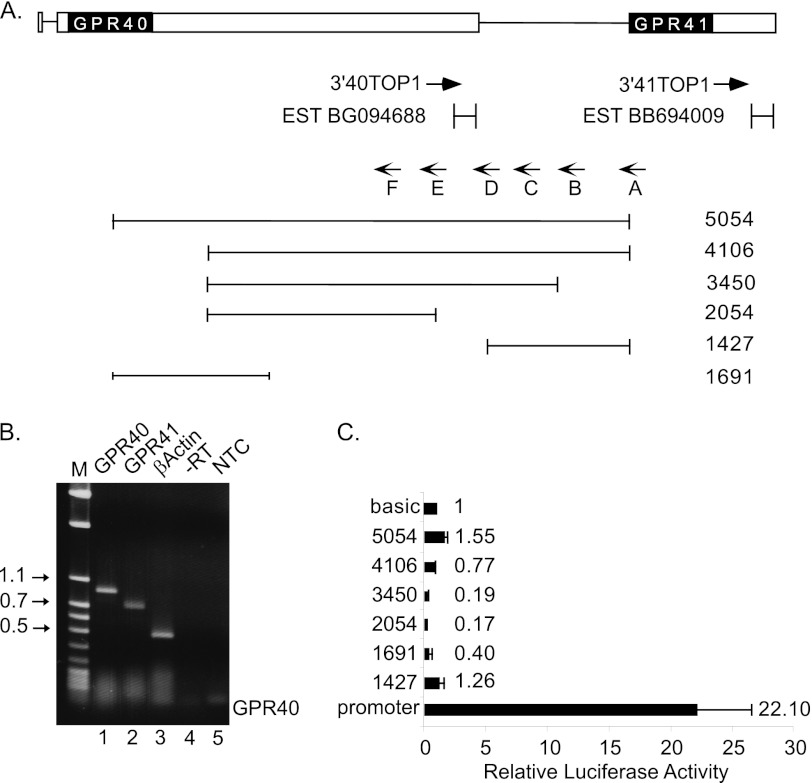
To study the regulatory mechanisms responsible for the cell-specific expression of the GPR41 gene, we mapped the transcriptional boundaries of GPR41. To identify the 3′ end of GPR41 mRNA, we performed 3′-RACE using primers complementary to the ORF of GPR41 (Fig. 2). A major product of 780 bp was generated and sequenced using a primer from the GPR41 ORF (Fig. 2B, lane 2). The sequence defined the 3′ end of GPR41 mRNA, which is positioned 611 nt downstream to the GPR41 stop codon. An EST (BB694009) found in databases from mouse sympathetic ganglion is consistent with these findings. In addition, sequence analysis of the region revealed a poly(A) signal (AATAAA) 20 nt upstream to the 3′ end. In parallel, the 3′ end of GPR40 mRNA was determined using this method (Fig. 2B, lane 1); the result is consistent with EST information present in databases.
FIGURE 2.
Analysis of GPR41 gene expression by 5′-RACE and reporter gene assay. A, map illustrating GPR40/41 locus. GPR40 and GPR41 coding regions are shown as black boxes. White boxes indicate 5′- and 3′UTRs of the GPR40 gene and 3′UTR of the GPR41 gene. Primer 3′41top1 and primer 3′40top1 used for 3′-RACE are indicated. ESTs from this region are also shown. Primers A–F were used in 5′-RACE experiment (method A). Six different analyses were conducted, and the primers shown are those used for reverse transcription. Also indicated are the fragments from the 5′ region of the GPR41 gene used to generate reporter plasmids. B, 3′-RACE analysis of cDNA from βTC1 cells using a forward gene-specific primer (3′40top1 or 3′41top1) and a reverse primer complementary to the flank region of 3′ poly(dT) primer (lanes 1 and 2, respectively). As a positive control, the cDNA was amplified using a specific primer from β-actin coding region (lane 3). As a negative control, PCR was carried out with RNA that was not reverse transcribed (−RT, lane 4) or without cDNA (NTC, lane 5). M, DNA size markers. C, functional analysis of promoter activity. Fragments from 5′-flanking region of GPR41 were ligated upstream to the firefly luciferase reporter gene in the promoter-less vector pGL3-basic. As a positive control, the GPR40 promoter fragment (21) was used. Promoter activity of each construct was determined in the β cell line HIT. Values are normalized for transfection efficiency according to the activity of a co-transfected Renilla luciferase plasmid and expressed relative to the activity of pGL3-basic vector. Values shown are the mean ± S.E. (n ≥3).
To define the 5′ end of the GPR41 mRNA, we performed 5′-RACE (method A, see under “Experimental Procedures”). We used six different primers complementary to the sequence between GPR40 and GPR41 ORFs with intervals of ∼600 nt between them (Fig. 2A, primers A–F). Surprisingly, none of the analyses produced a discrete band (data not shown), indicating that the 5′ end of the GPR41 mRNA may be highly heterogeneous or that it may be upstream of the primers used.
Transcriptional Regulation of GPR41 Gene Expression
To determine whether the GPR40-GPR41 intergenic region contains a functional promoter, we generated luciferase reporter plasmids containing several fragments spanning the GPR41 upstream region and transfected them into beta cells (Fig. 2A, lower panel). The construct 5054-GPR41P-LUC covers the entire region between GPR40 and GPR41 coding regions, and all other fragments are derived from this full fragment. As a positive control, a GPR40 promoter fragment was fused upstream to the firefly luciferase reporter gene. None of the GPR41 constructs exhibited transcriptional activity significantly higher than the parental promoter-less vector pGL3-basic upon transfection of the beta cell line HIT (Fig. 2C). This is consistent with the results of the above 5′-RACE experiments and indicates that the GPR41 promoter may lie upstream of the GPR40 ORF.
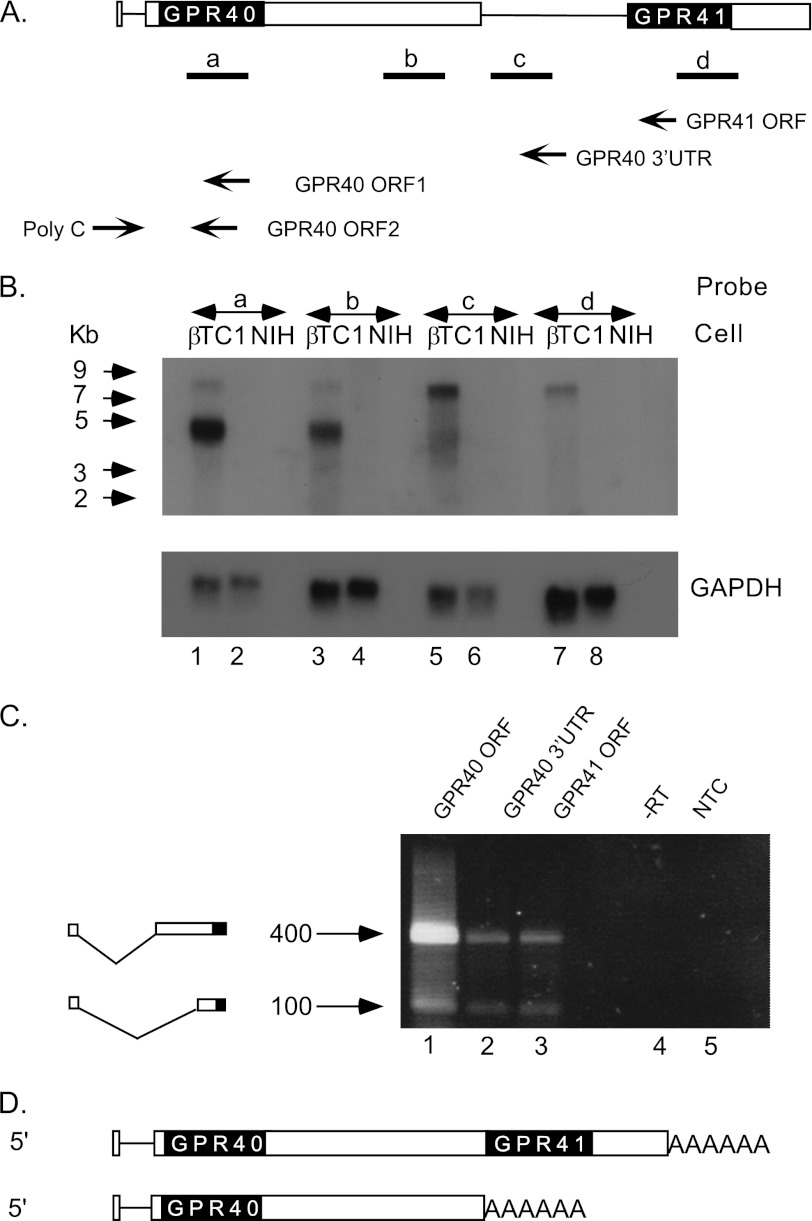
Because reporter gene experiments could not identify the GPR41 promoter, we performed Northern blot analysis to estimate the length of GPR41 mRNA (Fig. 3). DNA probes spanning the GPR40 ORF (Fig. 3B, lane 1) or GPR40 3′UTR (lane 3) gave a pattern of two bands, and the stronger band corresponds in mobility to the expected size of GPR40 mRNA (4.4 kb), according to 5′-RACE (21) and 3′-RACE (see above). The second weaker band is larger (>7 kb), and its structure was not previously characterized. Quantitation of autoradiograms indicated that the abundance of this band is ∼30% relative to the stronger band. Probes spanning the intergenic region (Fig. 3B, lane 5) or GPR41 ORF (lane 7) gave a single band, corresponding in migration to the weaker band obtained with GPR40 probes (lanes 1 and 3). These results indicate that GPR40 mRNA appears in two forms, a shorter 4.4 kb and a longer form of >7 kb (Fig. 3D). GPR41 mRNA exists in only one form (>7 kb). Because the distance from the GPR40 ATG to the 3′ end of GPR41 RNA is 7 kb, whereas the length of the major band corresponding to GPR41 mRNA is larger than that, we surmised that the 5′ end of GPR41 mRNA is located upstream to the GPR40 ORF.
FIGURE 3.
Analysis of GPR40 and GPR41 transcripts by Northern blot and 5′-RACE. A, probes a–d correspond to GPR40 ORF, GPR40 3′UTR, intergenic region, and GPR41 ORF, respectively. The location of primers used for 5′-RACE is indicated by arrows. B, Northern blot analysis of 5 μg of poly(A)+ RNA from βTC1 cells (lanes 1, 3, 5, and 7) or non-beta cells (NIH-3T3) (lanes 2, 4, 6, and 8) using probe a (lanes 1 and 2), probe b (lanes 3 and 4), probe c (lanes 5 and 6), or probe d (lanes 7 and 8). Migration of RNA molecular weight markers is indicated by arrows. Gel loading control (lower panel) was GAPDH probe that spans GAPDH ORF. C, analysis of cDNA from βTC1 cells by modified 5′-RACE. Primers used for reverse transcriptase reaction were complementary to GPR40 ORF1 (lane 1), GPR40 3′UTR (lane 2), or GPR41 ORF (lane 3). The cDNA was poly(dG)-tailed and amplified by PCR using a poly(dC) primer and primer GPR40 ORF2. As a negative control, the PCR was carried out with RNA that was not reverse-transcribed (−RT, lane 4) or without cDNA (NTC, lane 5). D, proposed structure of transcripts indicating the bicistronic GPR40/41 transcript and the monocistronic GPR40 transcript.
To examine this possibility, we utilized a second 5′-RACE procedure that permits characterization of long transcripts (method B, see under “Experimental Procedures”). Three different primers were used (Fig. 3A) as follows: complementary to the GPR40 ORF (Fig. 3C, lane 1), GPR40 3′UTR (Fig. 3C, lane 2), or GPR41 ORF (Fig. 3C, lane 3). All three primers generated the same pattern of two major products (Fig. 3C, lanes 1–3). Sequence analysis of the larger (400 bp) band derived from all three lanes was consistent with the previously defined transcription start site of GPR40 (21), including a 24-base exon, 698 base intron, and a 4.4-kb second exon that includes a 321-base 5′UTR. Sequence analysis of the smaller (100 base) band revealed a novel mRNA splice variant containing the same first short exon linked to exon 2 truncated by 265 nt at the 5′ end (Fig. 3C). Taken together, our analysis shows that the GPR40 and GPR41 genes share the same promoter and transcription start site and that GPR41 and GPR40 are encoded on the same bicistronic mRNA (Fig. 3D). In addition, this experiment revealed a new splice variant of the GPR40 gene that is present in both the monocistronic and bicistronic forms of RNA (Fig. 3C).
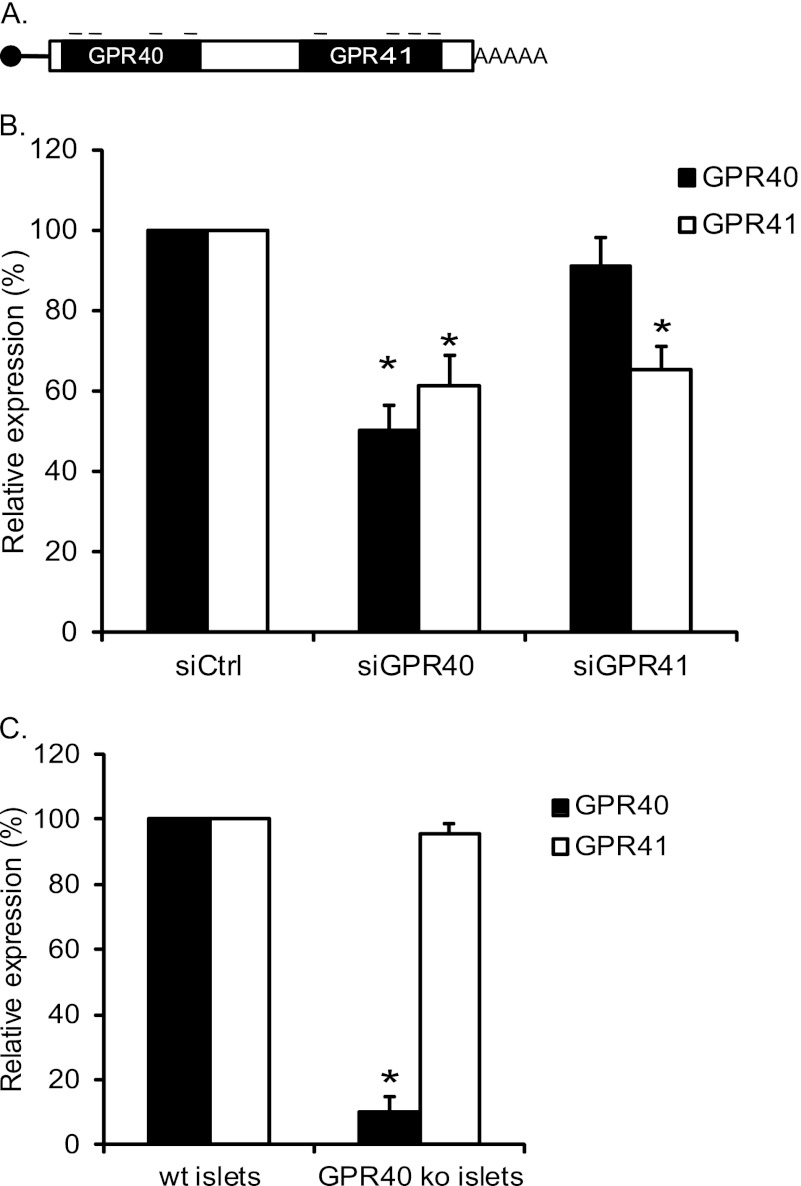
To verify the proposed bicistronic structure, we used siRNA-mediated knockdown of GPR40 and GPR41. Because the bicistronic RNA is present at ∼3-fold lower concentrations than the monocistronic RNA encoding GPR40, we predicted that knockdown of GPR40 would lead to reduced levels of RNA encoding both GPR40 and GPR41, whereas knockdown of GPR41 would reduce RNA encoding GPR41 without substantially reducing the expression of GPR40. MIN6 cells were transfected using siRNA pools against GPR40 or GPR41 (Fig. 4A). We observed 30–50% knockdown of GPR40 and GPR41 RNA by their homologous siRNAs (Fig. 4B). As predicted, siRNA targeting GPR40 reduced significantly RNA encoding GPR41, whereas siRNA targeting GPR41 did not significantly reduce RNA encoding GPR40 (Fig. 4B). Similar results upon siRNA knockdown were also observed using the cell line βTC1 (data not shown). An alternative explanation of these results would be a requirement of the GPR40 protein for the expression of GPR41. To test this possibility, we analyzed expression of GPR40 and GPR41 in islets from GPR40 knock-out mice (lacking the GPR40 coding region but containing an intact GPR40 promoter region (11)). As expected, these mice showed very low levels of GPR40 RNA; importantly, they showed no significant reduction of GPR41 RNA as compared with wild type mice, demonstrating that the GPR40 protein is not required for expression of the GPR41 gene. Taken together, the results strongly support the proposed bicistronic mode of expression of the locus.
FIGURE 4.
Validation of bicistronic mRNA expression. A, illustration of siRNA system. The small bars represent targeted areas of GPR40/41. Each pool consists of four different siRNAs targeting the coding region of GPR40 or GPR41 as indicated. B, knockdown of GPR40 or GPR41. MIN6 cells were transfected with siCtrl, siGPR40, or siGPR41, and gene expression was measured 48 h after transfection. The results were normalized to β-actin and hypoxanthine-guanine phosphoribosyltransferase and are expressed relative to siCtrl. The values indicated are mean ± S.E. (n = 5; *, p < 0.05). C, GPR40 and GPR41 expression in GPR40 knock-out mice. Islet RNA was isolated from 2- to 3-month-old WT and GPR40 knock-out mice. GPR40 and GPR41 RNA were measured by qRT-PCR, and expression levels were normalized to β-actin. Results are presented relative to expression of each gene in WT mice. The results are the mean ± S.E. (n ≥3).
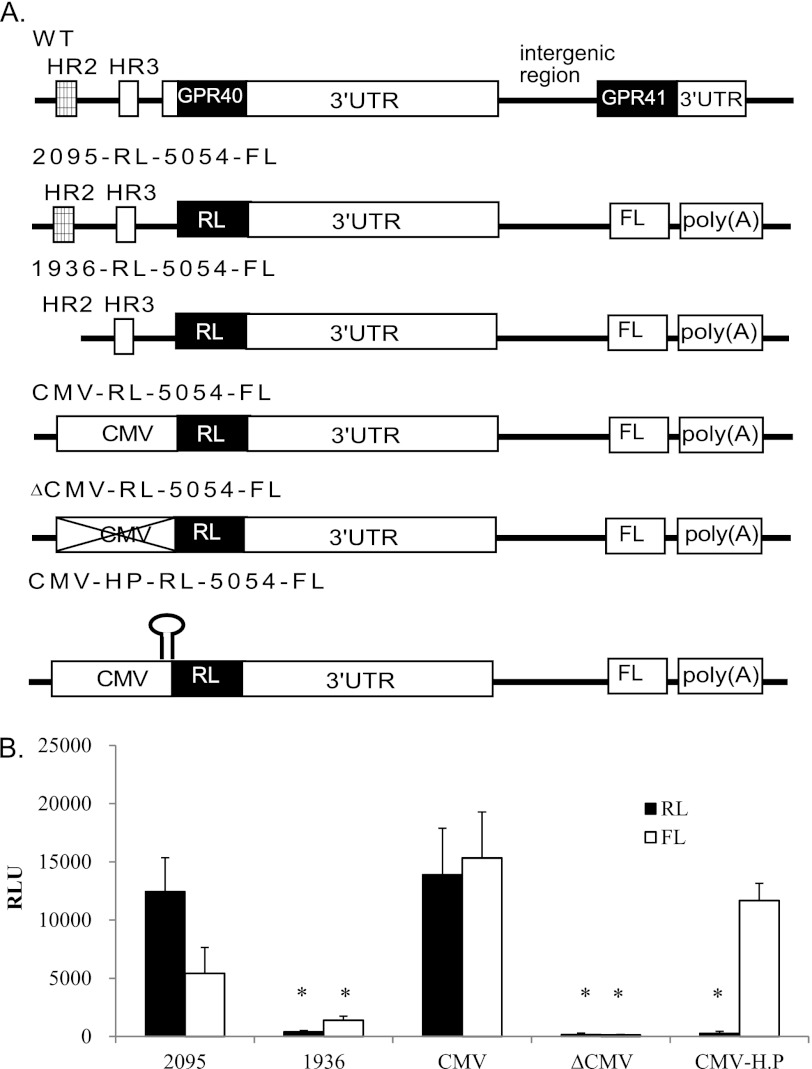
To determine whether GPR40 and GPR41 share the same transcriptional control elements, we constructed several bicistronic reporter vectors (Fig. 5A) based on the structure of the native GPR40/GPR41 locus; the vector 2095-RL-5054-FL contains the RL reporter gene replacing GPR40 ORF under the control of the intact GPR40 promoter (containing both the HR2 enhancer region and the HR3 proximal promoter region) and the FL reporter gene replacing GPR41 ORF. Downstream to RL and upstream to FL, the construct includes the intergenic region between GPR40 and GPR41 containing the intact GPR40 3′UTR and the region immediately upstream to the GPR41 ORF. The vector 1936-RL-5054-FL contains all of these sequences except the HR2 enhancer. The vector CMV-RL-5054-FL contains the CMV promoter replacing the full GPR40 endogenous promoter, and the vector ΔCMV contains the same sequences except for the CMV promoter. The constructs were transfected into the beta cell line MIN6, and RL and FL activity was measured (Fig. 5B). Deletion of either the HR2 enhancer region or the CMV promoter abolished the activity of both RL and FL, confirming that transcription of both upstream and downstream reporters are completely dependent on the presence of the promoter controlling the upstream gene.
FIGURE 5.
Analysis of transcriptional and translational control of GPR40 and GPR41 gene expression using bicistronic luciferase constructs. A, bicistronic vectors used in this analysis. WT represents the genomic organization of the locus; open reading frames are shown as black boxes; 5′ and 3′ UTR as white boxes, and the conserved region HR2 as a hatched box. The fragment 2095 represents the full GPR40 promoter containing both HR2 (enhancer) and HR3 (transcription start site) regions; 1936 represents a truncated promoter fragment without the HR2 enhancer region. In the three constructs CMV-RL-5054-FL, ΔCMV-RL-5054-FL, and CMV-HP-RL-5054-FL, the CMV promoter replaces the endogenous promoter of GPR40. In all constructs, the reporter genes Renilla luciferase (RL, black) and firefly luciferase (FL, white) replace GPR40 and GPR41 coding regions, respectively. B, constructs were transfected into the beta cell line MIN6, and Renilla and firefly luciferase reporter activity was determined. RLU, relative light units. Each data point represents the mean ± S.E. (n ≥3; *, p < 0.05).
Translational Regulation of GPR41 Gene
In eukaryotes virtually all mRNAs are monocistronic. Initiation of translation typically starts with binding of the ribosome to the 5′ m7G cap and scanning downstream on the mRNA until it reaches the translational initiation codon AUG. The existence of a bicistronic mRNA raises the question of how the downstream ORF of GPR41 is translated. A possible mechanism for translation of the downstream ORF is via an IRES-based pathway (25). Because such a mechanism would be expected to be m7G cap-independent, we tested whether in fact GPR41 translation is m7G cap-independent. For this, we inserted a hairpin structure in the reporter plasmid between the CMV promoter and the reporter gene RL in a bicistronic plasmid (CMV-HP-RL-5054-FL, see Fig. 5A). The construct was transfected into the beta cell line MIN6, and RL and FL activity was measured (Fig. 5B); we observed that the hairpin led to a complete loss of RL activity, confirming that RL translation is fully m7G cap-dependent. The activity of FL was reduced much less dramatically (∼24%), indicating that FL activity (representing GPR41 translation) is relatively m7G cap-independent and thus may be IRES-dependent.
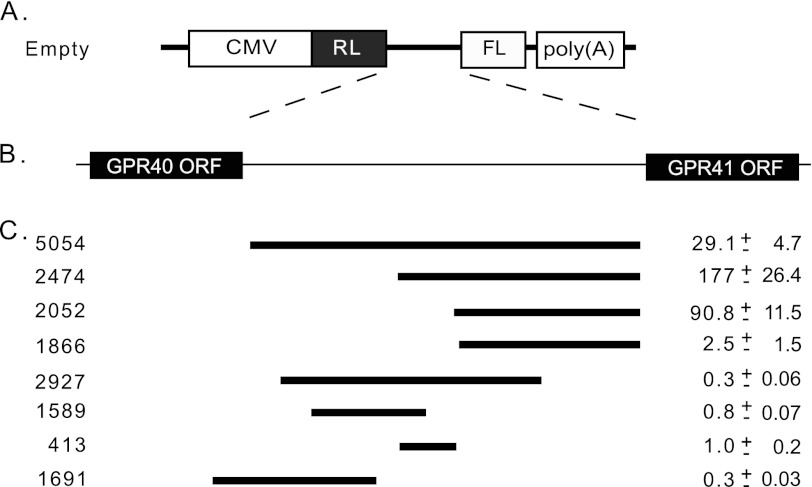
To map the sequence that is required for activity of the putative IRES, we generated a series of bicistronic plasmids containing variable lengths of DNA from the intergenic region between GPR40 and GPR41 (Fig. 6). The fragment 5054 represents the entire sequence between the GPR40 and GPR41 ORFs. The constructs were transfected into the beta cell line MIN6. The ratio of FL to RL was determined and compared with that of a control plasmid lacking the intergenic region. Several fragments demonstrated significantly elevated FL/RL ratios indicating the presence of IRES activity (Fig. 6C). Fragment 2474 is the shortest sequence identified that can direct full IRES activity. This fragment is located 2474 bases upstream to the GPR41 ATG; its activity is 177-fold higher than that of the empty vector (Fig. 6C). A positive control sequence of an established IRES (from the apoptosis inhibitor gene XIAP (26)) also showed high activity compared with the empty vector (407-fold, data not shown).
FIGURE 6.
Mapping of IRES region. A, bicistronic vector used in this analysis. The empty vector contains the CMV promoter and the two reporter genes RL and FL. B, genomic region from which the intergenic region fragments were taken. C, fragments indicated were inserted to the empty vector between the RL to FL reporter genes. 5054 represents the entire sequence between the GPR40 and GPR41 ORFs. Bicistronic vectors were transfected into the beta cell line MIN6, and Renilla and firefly luciferase reporter activity was determined. FL/RL activity is expressed relative to the empty vector. The values indicated are the mean ± S.E. (n ≥4).
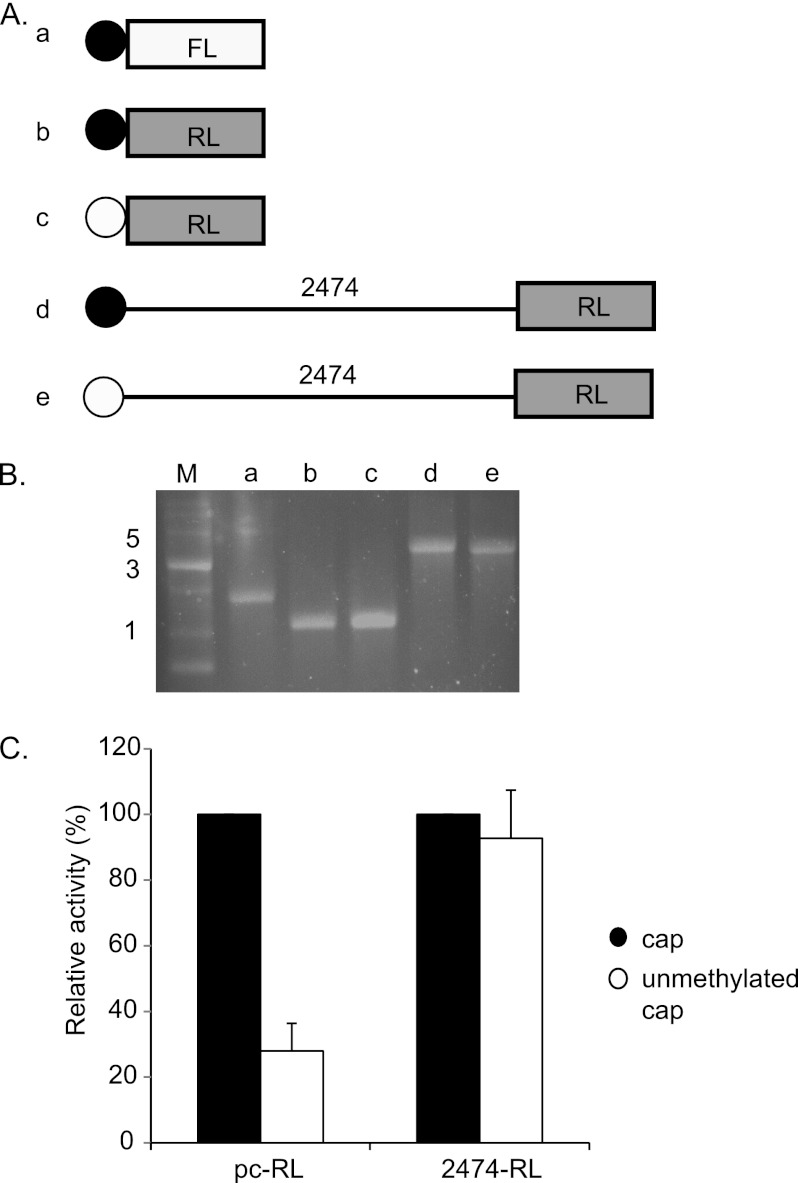
To further test the notion that the fragment 2474 contains an IRES element, and to exclude the possibility that the observed activity results from an alternate mechanism (such as cryptic promoter or splice sequences, frameshifting, ribosome hopping, or leaky ribosome scanning), we performed RNA transfection experiments. For this, we generated RL reporter plasmids with or without the 2474-bp fragment of the GPR40–41 intergenic region located downstream of a T7 RNA polymerase-dependent promoter (plasmids pCDNA3–2474-RL and pCDNA3-RL). The linearized plasmids were transcribed with either standard m7G cap analog (black circle) or unmethylated cap analog (which does not support binding of the ribosome, white circle) (Fig. 7A). The resulting RNA molecules (Fig. 7A, panels a–e) migrated as expected on agarose gel electrophoresis and showed minimal degradation (Fig. 7B). The methylated and unmethylated transcripts were transfected into HeLa cells together with m7G-capped transcript derived from an FL plasmid lacking the putative IRES sequence (Fig. 7, A and B) as an internal control, and the activity of FL and RL was measured. As expected, the presence of nonmethylated cap analog in the short RL RNA molecule led to a dramatic reduction (∼75%) in translation efficiency of RL compared with the standard m7G cap analog (Fig. 7C) indicating that initiation of translation in this short RNA is mainly controlled by binding of the ribosome to the m7G cap. By contrast, RNA containing the 2474 fragment upstream to the RL ORF shows no significant reduction in the translation efficiency of RL with the nonmethylated cap analog as compared with the standard m7G cap analog. We are currently unable to accurately predict the intracellular ratio of GPR40 protein to GPR41 protein. The ratio of monocistronic to bicistronic RNAs combined with the observed relative efficiency of IRES-dependent as compared with m7G cap-dependent translation (Fig. 7 and data not shown) would suggest a relative rate of translation initiation of GPR40 to GPR41 of ∼50–100-fold. However, this ratio might be substantially affected by post-translational parameters, including protein stability and stress conditions. Taken together, our data show that the 2474 sequence confers on the transcript the ability to undergo m7G cap-independent translation, strongly supporting the view that this sequence contains an IRES that permits translation of the GPR41 gene in the bicistronic RNA.
FIGURE 7.
RNA transfection. A, illustration of in vitro transcribed RNAs used in this analysis. Two reporter genes were used as follows: Renilla luciferase (RL, gray) and firefly luciferase (FL, white). Filled circles represent methylated (active) cap, and empty circles represent unmethylated (nonactive) cap. Panel a, RNA derived from pCDNA3-FL. Panels b and c, RNA derived from pCDNA3-RL. Panels d and e, RNA derived from pCDNA3-2474-RL. B, analysis by agarose gel electrophoresis of the in vitro transcribed RNA molecules used for the RNA transfections. M, RNA marker; panels a–e, the RNAs shown in A. C, in vitro transcribed RNA was transfected into HeLa cells, and Renilla and firefly luciferase reporter activity was determined. RL/FL ratio of unmethylated transcript is expressed relative to the methylated transcript. Values shown are the mean ± S.E. (n ≥3; *, p < 0.005).
DISCUSSION
To perform their sophisticated role in production and regulated secretion of insulin, beta cells express a characteristic repertoire of genes, including transcription factors, enzymes, receptors, ion channels, and secretory granule proteins (3). Expression of these genes is typically mediated through transcriptional control mechanisms acting on dedicated promoter elements for each gene (27). The best studied example of this is the insulin gene itself, which is controlled by a transcriptional regulatory region located in the 350 bp upstream of the transcription start site (28, 29); several transcription factors expressed preferentially in beta cells are required, including PDX1, BETA2, E2A, and MAFA (27). Recently, we analyzed GPR40 gene expression demonstrating that it is indeed regulated at the transcription level via transcriptional control elements located within the ∼1.5-kb region upstream from the transcription start site (21).
In this study, we have extended our analysis of the GPR40 family to examine the mechanisms controlling expression of the GPR41 gene. Of the mouse tissues and cell types surveyed, we observed the highest expression in cultured beta cells and pancreatic islets, in a pattern closely similar to that seen for GPR40 and distinctly different from GPR43. This is consistent with reports from the literature indicating that GPR41 is expressed in a highly selective pattern (19, 20, 30), whereas GPR43 expression is much more general (31). Because the GPR40 gene is located in close proximity to the GPR41 gene, we anticipated that GPR41 would be controlled transcriptionally via control elements residing in the intergenic region. Unexpectedly, we were unable to find evidence for transcriptional control elements or a transcription start site within this region. Rather, through use of an optimized 5′-RACE procedure, we observed that transcription of the GPR41 gene initiated from the transcription start site of the GPR40 gene controlled by the GPR40 promoter.
Because antibodies capable of sensitive detection of GPR40 and GPR41 are not available, we used bicistronic luciferase reporter plasmids to confirm this finding: whereas expression of the downstream reporter was completely dependent on the presence of upstream promoter activity, it was only modestly inhibited by the insertion of a stem loop structure downstream of the promoter, consistent with the possibility that translation of the GPR41 gene is controlled by the presence of an IRES sequence in the intergenic region. Accordingly, we were able to map the putative IRES to a 2,474 nt region of the intervening sequence. To exclude other possibilities (e.g. cryptic promoter or splice sequences, frameshifting, ribosome hopping, and leaky ribosome scanning), we performed RNA transfection experiments. Indeed, we observed m7G cap-independent expression from reporter RNA molecules containing the putative IRES, supporting the notion that GPR41 translation is dependent on the presence of an IRES element in the intergenic region.
Taken together our results indicate that beta cell-specific expression of the GPR41 gene is controlled by an unusual mechanism that involves control at both the level of transcription and translation. Transcriptional control is mediated through the previously characterized GPR40 promoter that is active selectively in pancreatic beta cells. This promoter generates two distinct forms of RNA, a monocistronic form encoding exclusively GPR40 and a longer bicistronic transcript encoding both GPR40 and GPR41. The monocistronic transcript is present at significantly higher levels in both cultured beta cells and islets. Consistent with this, siRNA-mediated knockdown of GPR40 reduces the levels of GPR41 but not vice versa.
Whereas bicistronic and polycistronic genes are common in bacteria, they are extremely rare in mammal protein-coding genes (32, 33). Because polycistronic gene expression in bacteria often permits coordinated gene expression (34), we speculate that the operon-like structure may have evolved to permit coordinated regulation of these two genes at the transcriptional level. This is consistent with the physiological roles of these two proteins as fatty acid-activated receptors. Thus, coordinated expression of these two receptors may be required to enable them to function optimally as fatty acid sensors on the beta cell.
The bicistronic nature of the GPR40/41 transcript raises an interesting issue regarding translation of GPR41. Translation in the mammalian cell typically proceeds via a ribosome scanning mechanism involving attachment of the ribosome to the 5′ end in an m7G cap-dependent process followed by scanning to a consensus ATG initiation codon, translation of the open reading frame, and disengagement from the RNA (35, 36). Hence, translation of a downstream ORF in mammalian cells requires a noncanonical translational mechanism for translational initiation. Our results are consistent with the presence in the intergenic region of an IRES element. Initially observed in viral genes (37, 38), IRES elements have now been identified in an increasing number of 5′UTR regions of cellular genes, often in genes whose expression is up-regulated under conditions of cellular stress, when m7G cap-dependent translation is inhibited (39, 40). Future studies should investigate in more detail the physiological relevance of the unusual bicistronic structure of the GPR40/41 transcript and its regulation at the transcriptional and translation levels.
Acknowledgments
We thank Dr. Adi Kimchi and Noa Liberman for helpful advice, provision of the bicistronic vector, and instruction in RNA transfection procedures. We also thank Dr. Rivka Dikstein, Dr. Orna Elroy-Stein, Dr. Oded Meyuhas, and Dr. Reut Bartoov-Shifman for valuable discussions, and Sara Weiss for expert technical assistance.
This work was supported in part by grants from the Israel Science Foundation, the Benjamin and Seema Pulier Charitable Foundation, and the Florence Blau, Morris Blau, and Rose Patterson Fund.

This article contains supplemental Table 1.
- LCFA
- long chain fatty acid
- IRES
- internal ribosome entry site
- RACE
- rapid amplification of cDNA ends
- qRT-PCR
- quantitative reverse transcriptase-PCR
- SCFA
- short chain fatty acid
- FL
- firefly luciferase
- RL
- Renilla luciferase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- nt
- nucleotide.
REFERENCES
- 1. Ohneda K., Ee H., German M. (2000) Regulation of insulin gene transcription. Semin. Cell Dev. Biol. 11, 227–233 [DOI] [PubMed] [Google Scholar]
- 2. Steiner D. F., Chan S. J., Welsh J. M., Kwok S. C. (1985) Structure and evolution of the insulin gene. Annu. Rev. Genet. 19, 463–484 [DOI] [PubMed] [Google Scholar]
- 3. Halban P. A., Kahn S. E., Lernmark A., Rhodes C. J. (2001) Gene and cell replacement therapy in the treatment of type 1 diabetes. How high must the standards be set? Diabetes 50, 2181–2191 [DOI] [PubMed] [Google Scholar]
- 4. Henquin J. C., Ravier M. A., Nenquin M., Jonas J. C., Gilon P. (2003) Hierarchy of the beta-cell signals controlling insulin secretion. Eur J. Clin. Invest. 33, 742–750 [DOI] [PubMed] [Google Scholar]
- 5. Yaney G. C., Corkey B. E. (2003) Fatty acid metabolism and insulin secretion in pancreatic beta cells. Diabetologia 46, 1297–1312 [DOI] [PubMed] [Google Scholar]
- 6. Zraika S., Dunlop M., Proietto J., Andrikopoulos S. (2002) Effects of free fatty acids on insulin secretion in obesity. Obes. Rev. 3, 103–112 [DOI] [PubMed] [Google Scholar]
- 7. Briscoe C. P., Tadayyon M., Andrews J. L., Benson W. G., Chambers J. K., Eilert M. M., Ellis C., Elshourbagy N. A., Goetz A. S., Minnick D. T., Murdock P. R., Sauls H. R., Jr., Shabon U., Spinage L. D., Strum J. C., Szekeres P. G., Tan K. B., Way J. M., Ignar D. M., Wilson S., Muir A. I. (2003) The orphan G protein-coupled receptor GPR40 is activated by medium and long chain fatty acids. J. Biol. Chem. 278, 11303–11311 [DOI] [PubMed] [Google Scholar]
- 8. Itoh Y., Kawamata Y., Harada M., Kobayashi M., Fujii R., Fukusumi S., Ogi K., Hosoya M., Tanaka Y., Uejima H., Tanaka H., Maruyama M., Satoh R., Okubo S., Kizawa H., Komatsu H., Matsumura F., Noguchi Y., Shinohara T., Hinuma S., Fujisawa Y., Fujino M. (2003) Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature 422, 173–176 [DOI] [PubMed] [Google Scholar]
- 9. Edfalk S., Steneberg P., Edlund H. (2008) Gpr40 is expressed in enteroendocrine cells and mediates free fatty acid stimulation of incretin secretion. Diabetes 57, 2280–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shapiro H., Shachar S., Sekler I., Hershfinkel M., Walker M. D. (2005) Role of GPR40 in fatty acid action on the beta cell line INS-1E. Biochem. Biophys. Res. Commun. 335, 97–104 [DOI] [PubMed] [Google Scholar]
- 11. Steneberg P., Rubins N., Bartoov-Shifman R., Walker M. D., Edlund H. (2005) The FFA receptor GPR40 links hyperinsulinemia, hepatic steatosis, and impaired glucose homeostasis in mouse. Cell Metab. 1, 245–258 [DOI] [PubMed] [Google Scholar]
- 12. Latour M. G., Alquier T., Oseid E., Tremblay C., Jetton T. L., Luo J., Lin D. C., Poitout V. (2007) GPR40 is necessary but not sufficient for fatty acid stimulation of insulin secretion in vivo. Diabetes 56, 1087–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sawzdargo M., George S. R., Nguyen T., Xu S., Kolakowski L. F., O'Dowd B. F. (1997) A cluster of four novel human G protein-coupled receptor genes occurring in close proximity to CD22 gene on chromosome 19q13.1. Biochem. Biophys. Res. Commun. 239, 543–547 [DOI] [PubMed] [Google Scholar]
- 14. Brown A. J., Goldsworthy S. M., Barnes A. A., Eilert M. M., Tcheang L., Daniels D., Muir A. I., Wigglesworth M. J., Kinghorn I., Fraser N. J., Pike N. B., Strum J. C., Steplewski K. M., Murdock P. R., Holder J. C., Marshall F. H., Szekeres P. G., Wilson S., Ignar D. M., Foord S. M., Wise A., Dowell S. J. (2003) The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 278, 11312–11319 [DOI] [PubMed] [Google Scholar]
- 15. Cummings J. H., Pomare E. W., Branch W. J., Naylor C. P., Macfarlane G. T. (1987) Short chain fatty acids in human large intestine, portal, hepatic, and venous blood. Gut 28, 1221–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bergman E. N. (1990) Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590 [DOI] [PubMed] [Google Scholar]
- 17. Kimura I., Inoue D., Maeda T., Hara T., Ichimura A., Miyauchi S., Kobayashi M., Hirasawa A., Tsujimoto G. (2011) Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41) Proc. Natl. Acad. Sci. U.S.A. 108, 8030–8035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tolhurst G., Heffron H., Lam Y. S., Parker H. E., Habib A. M., Diakogiannaki E., Cameron J., Grosse J., Reimann F., Gribble F. M. (2012) Short-chain fatty acids stimulate glucagon-like Peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 61, 364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Samuel B. S., Shaito A., Motoike T., Rey F. E., Backhed F., Manchester J. K., Hammer R. E., Williams S. C., Crowley J., Yanagisawa M., Gordon J. I. (2008) Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty acid-binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. U.S.A. 105, 16767–16772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiong Y., Miyamoto N., Shibata K., Valasek M. A., Motoike T., Kedzierski R. M., Yanagisawa M. (2004) Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc. Natl. Acad. Sci. U.S.A. 101, 1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bartoov-Shifman R., Ridner G., Bahar K., Rubins N., Walker M. D. (2007) Regulation of the gene encoding GPR40, a fatty acid receptor expressed selectively in pancreatic beta cells. J. Biol. Chem. 282, 23561–23571 [DOI] [PubMed] [Google Scholar]
- 22. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 23. Li D. S., Yuan Y. H., Tu H. J., Liang Q. L., Dai L. J. (2009) A protocol for islet isolation from mouse pancreas. Nat. Protoc. 4, 1649–1652 [DOI] [PubMed] [Google Scholar]
- 24. Wigler M., Pellicer A., Silverstein S., Axel R., Urlaub G., Chasin L. (1979) DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 76, 1373–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Plank T. D., Kieft J. S. (2012) The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip. Rev. RNA 3, 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henis-Korenblit S., Shani G., Sines T., Marash L., Shohat G., Kimchi A. (2002) The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl. Acad. Sci. U.S.A. 99, 5400–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chakrabarti S. K., Mirmira R. G. (2003) Transcription factors direct the development and function of pancreatic beta cells. Trends Endocrinol. Metab. 14, 78–84 [DOI] [PubMed] [Google Scholar]
- 28. German M., Ashcroft S., Docherty K., Edlund H., Edlund T., Goodison S., Imura H., Kennedy G., Madsen O., Melloul D., Moss L., Olson K., Permutt A., Philippe J., Robertson R. P., Rutter W. J., Serup P., Stein R., Steiner D., Tsai M. J., Walker M. D. (1995) The insulin gene promoter. A simplified nomenclature. Diabetes 44, 1002–1004 [DOI] [PubMed] [Google Scholar]
- 29. Walker M. D., Edlund T., Boulet A. M., Rutter W. J. (1983) Cell-specific expression controlled by the 5′-flanking region of insulin and chymotrypsin genes. Nature 306, 557–561 [DOI] [PubMed] [Google Scholar]
- 30. Kimura M., Mizukami Y., Miura T., Fujimoto K., Kobayashi S., Matsuzaki M. (2001) Orphan G protein-coupled receptor, GPR41, induces apoptosis via a p53/Bax pathway during ischemic hypoxia and reoxygenation. J. Biol. Chem. 276, 26453–26460 [DOI] [PubMed] [Google Scholar]
- 31. Karaki S., Mitsui R., Hayashi H., Kato I., Sugiya H., Iwanaga T., Furness J. B., Kuwahara A. (2006) Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res. 324, 353–360 [DOI] [PubMed] [Google Scholar]
- 32. Gray T. A., Saitoh S., Nicholls R. D. (1999) An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 5616–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hui D., Kumar K. N., Mach J. R., Srinivasan A., Pal R., Bao X., Agbas A., Höfner G., Wanner K. T., Michaelis E. K. (2009) A rat brain bicistronic gene with an internal ribosome entry site codes for a phencyclidine-binding protein with cytotoxic activity. J. Biol. Chem. 284, 2245–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jacob F., Monod J. (1961) Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 [DOI] [PubMed] [Google Scholar]
- 35. Kozak M. (1989) The scanning model for translocation. An update. J. Cell Biol. 108, 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234, 187–208 [DOI] [PubMed] [Google Scholar]
- 37. Jang S. K., Kräusslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., Wimmer E. (1988) A segment of the 5′-nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pelletier J., Sonenberg N. (1988) Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325 [DOI] [PubMed] [Google Scholar]
- 39. Elroy-Stein O., Merrick W. C. (2007) in Translational Control in Biology and Medicine (Mathews M., Sonenberg N., Hershey J. W. B., eds) pp. 155–172, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 40. Hellen C. U., Sarnow P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15, 1593–1612 [DOI] [PubMed] [Google Scholar]