Background: The yeast Shu complex promotes repair through inhibition of the Srs2 anti-recombinase.
Results: Psy3 and Csm2 share a similar structure that binds to DNA through a Rad51-like motif.
Conclusion: The DNA binding activity of the Shu complex is essential for in vivo function.
Significance: The structural and biochemical studies on the Shu proteins provide a good foundation for further understanding of the Shu complex in eukaryotic cells.
Keywords: DNA-binding Protein, DNA Damage, DNA Damage Response, Protein Complexes, Protein-DNA Interaction, X-ray Crystallography
Abstract
The yeast Shu complex, consisting of the proteins Shu1, Shu2, Psy3, and Csm2, maintains genomic stability by coupling post-replication repair to homologous recombination. However, a lack of biochemical and structural information on the Shu proteins precludes revealing their precise roles within the pathway. Here, we report on the 1.9-Å crystal structure of the Psy3-Csm2 complex. The crystal structure shows that Psy3 forms a heterodimer with Csm2 mainly through a hydrophobic core. Unexpectedly, Psy3 and Csm2 share a similar architecture that closely resembles the ATPase core domain of Rad51. The L2 loop present in Psy3 and Csm2 is similar to that of Rad51 and confers the DNA binding activity of the Shu complex. As with Rad51, the Shu complex appears to form a nucleoprotein filament by binding nonspecifically to DNA. Structure-based mutagenesis studies have demonstrated that the DNA binding activity of the Shu complex is essential for repair of the methyl methanesulfonate-induced DNA damage. Our findings provide good foundations for the understanding of the Srs2 regulation by the Shu complex.
Introduction
All living cells possess the ability to repair DNA lesions caused by toxic chemicals and radiation or that which arise during the process of DNA replication. Several different mechanisms such as homologous recombination repair (HRR),4 nucleotide excision repair, and post-replication repair (PRR) are employed to deal with such damage (1, 2). In Saccharomyces cerevisiae, monoubiquitination of the proliferating cell nuclear antigen by Rad6-Rad18 activates the error-prone translesion synthesis (TLS) pathway of PRR (3–5). Further ubiquitination of the monoubiquitinated proliferating cell nuclear antigen by Rad5-MMS2-Ubc13 switches the damage response into another PRR branch, error-free PRR (6–9). The yeast Shu complex consisting of Shu1, Shu2, Psy3, and Csm2, originally identified through a genetic screen for mutants that suppress the growth defects of the Top3 mutant (10), was proposed to maintain genome stability by linking error-free PRR to homologous recombination (HR) (9). All the single Shu mutants exhibit moderate sensitivity to the DNA-alkylating agent methyl methanesulfonate (MMS),and mutation of all four does not cause any additive effects (9–11). Taken together with the two-hybrid data showing the interactive relationship of the Shu proteins (9, 10), it was suggested that the Shu proteins form a stable entity to resist MMS-induced DNA damage (9, 11). Further studies have revealed that the Shu mutants also suppress the slow growth of RMI1 and the hyper-recombination phenotype associated with the loss of TOP3 or SGS1 (11). It was proposed that the Shu complex functions in Rad51-, Rad52-, and Rad54-dependent HRR by promoting the formation of homologous intermediates that are ultimately processed by Sgs1-Rmi1-Top3 (9–11). Sgs1, a RecQ-type DNA helicase, is thought to function as an anti-recombinase by dissolving HR intermediates such as Holliday junctions into the noncrossover product. In addition to the Sgs1, the Srs2 anti-recombinase also acts to antagonize HR by disrupting the Rad51 nucleoprotein filament (12–14). These anti-recombinases play an important role in precluding inappropriate or excessive HR, which is exemplified by the BLM gene (human homologue of the Sgs1) mutation causing Blood syndrome (15). Recently, it was demonstrated that the Shu complex was a regulator of Srs2 (16). By inhibiting the disassembly reaction of Srs2, the Shu complex shifted the balance of damage repair toward Rad51 filament stabilization (16).
Notably, a study using Schizosaccharomyces pombe led to the discovery of an HR-related gene, Sws1, whose mutation would cause sensitivity to MMS and suppress the poor growth of rqh1Δ and srs2Δ, similar to the observation of Shu mutants in S. cerevisiae (17). Sws1, also present in humans, is identified as the homologue of Shu2 (17). Moreover, Shu1 shares homology with the fission yeast protein Rlp1 and the human protein XRCC2; Psy3 is homologous to Rdl1 from fission yeast and RAD51D in humans, and the Walker B motif included in the latter two proteins is also observed in Psy3 (17). These data reveal that the Shu complex is conserved from yeast to humans and plays a significant role in DNA damage repair. However, the underlying mechanism of the pathway remains largely unknown.
Here, we report on the crystal structure of the Psy3-Csm2 complex that shows Psy3 and Csm2 share a similar architecture that closely resembles the ATPase core domain of Rad51. The L2 loops in Psy3 and Csm2 confer the DNA binding activity of the Shu complex. The structural and biochemical studies on the Shu complex provide further insights into the mechanism by which the conserved Shu complex participates in the repair of DNA damage.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The full-length DNA fragments of Shu1 and Csm2 were cloned into pET-22b vector, respectively. Full-length fragments of Psy3 and Shu2 were inserted into a modified pET-28a with a His-MBP-TEV cleavage site tag in the N terminus of the recombinant protein. All the mutants were generated using PCRs and subcloned, overexpressed, and purified in the same way as that of wild-type proteins. For protein coexpression, the reading frame of each recombinant protein described above was inserted into pETDuet and RSFDuet (Novagen). Overexpression of recombinant protein was induced in Escherichia coli Rosetta (DE3) by 0.5 mm isopropyl β-d-thiogalactoside when the cell density reached an A600 nm of 0.6–0.8. After additional growth for about 20 h at 16 °C, the cells were collected. The recombinant proteins were purified using Ni2+-nitrilotriacetate affinity resin (Qiagen) in 20 mm Tris-HCl, pH 8.0, and 300 mm NaCl. Then the His-MBP tag in the target protein was removed by tobacco etch virus protease digestion in 4 °C overnight. The proteins were further purified using HiTrap Q FF (5 ml) or HiTrap SP FF (5 ml) and HiLoad 16/60 Superdex 200 (GE Healthcare). The final proteins in buffer containing 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, and 1 mm DTT were concentrated to 30–40 mg/ml for crystallization trials. For the DNA binding assay, Shu protein complexes were purified without tobacco etch virus protease digestion. All the Shu protein complexes were concentrated to 30–40 mg/ml in buffer containing 20 mm Tris-HCl, pH 7.5, 50 mm NaCl except the holo-Shu complex, which was in buffer containing 20 mm Tris-HCl, pH 7.5, 100 mm NaCl.
To prepare the SeMet derivative protein, Psy3 and Csm2 were expressed in E. coli strain B834 (Novagen) using M9 medium supplemented with SeMet and six amino acids, including leucine, isoleucine, valine, phenylalanine, lysine, and threonine, respectively. The SeMet derivative Psy3-Csm2 complex was then copurified by a procedure similar to that described above.
Crystallization and Data Collection
Crystals of both native and SeMet derivative Psy3-Csm2 complex were grown using the hanging drop vapor diffusion method at 285 K and grew to maximum size in about 72 h in buffer containing 100 mm Tris-HCl, pH 8.0, 10% glycerol, 14% ethanol. Both the native and multiple wavelength anomalous dispersion data were collected at 100 K with cryoprotectant (100 mm Tris-HCl, pH 8.0, 30% glycerol, 14% ethanol). The multiple wavelength anomalous dispersion data were collected from a single SeMet derivative crystal at beamline 3W1A of the Beijing Synchrotron Radiation Facility at the Institute of High Energy Physics, Chinese Academy of Sciences. The native data were collected at beamline BL17U of Shanghai Synchrotron Radiation Facility. All the data were processed using HKL2000 (18) and programs in the CCP4 package (19).
Structure Determination and Refinement
Selenium positions were determined by SOLVE (20) using Bijvoet differences of the multiple wavelength anomalous dispersion data. The initial phases were calculated by RESOLVE (21) with the resolution ranging from 50 to 3.0 Å, and an initial model was autobuilt by RESOLVE. The model was further built and refined against the native data at 1.9 Å resolution using Refmac5 (22) and COOT (23). Further cycles of refinement and model building were carried out using PHENIX.refine (24). At last, all the density in the map could be interpreted well, and all residues had acceptable chemical conformation. TLS restraint was used in the last several cycles of refinement. The final model was checked by MolProbity (25). Details about data collection and processing were presented in Table 1. Figures were prepared using PyMOL (DeLano Scientific LLC).
TABLE 1.
Data collection and refinement statistics for Psy3-Csm2
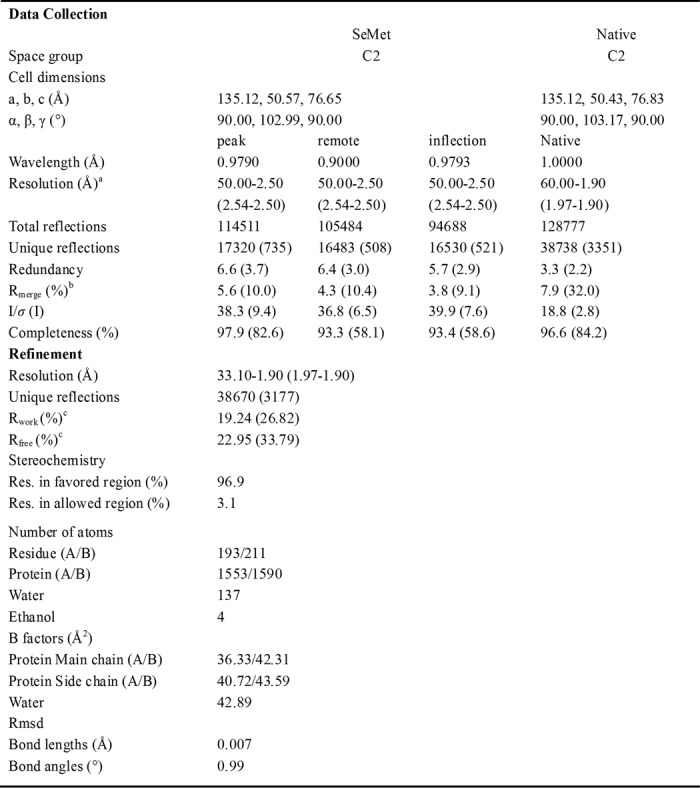
a High resolution shell is shown in parentheses.
b Rmerge = Σ|Ii−〈I〉|/Σ|I|, where Ii is the intensity of an individual reflection and 〈I〉 is the average intensity of that reflection.
c Rwork = Σ‖Fo|−|Fc‖/Σ|Fo| for all reflections and Rfree = Σ‖Fo|−|Fc‖/Σ|Fo|, calculated on the 5% of data excluded from refinement.
Pulldown Assays
A similar amount of E. coli cells expressing the wild-type or mutant Shu proteins were lysed alone or together with the indicated partners in binding buffer containing 20 mm Tris-HCl, pH 8.0, and 300 mm NaCl. The supernatant was incubated with 100 μl of Ni2+-nitrilotriacetate beads for 30 min at room temperature. Then the beads were washed two times with the binding buffer. Finally, the bound proteins were eluted from beads with 100 μl of binding buffer supplemented with 500 mm imidazole. All the samples were resolved by SDS-PAGE (15%) and stained with Coomassie Brilliant Blue.
Size-exclusion Chromatography Assay
The size-exclusion chromatography assays were performed with a Superdex 200 column (10/300 GL) (GE Healthcare). The protein sample or molecular mass standards were applied to the Superdex 200 column (10/300 GL) and eluted with 50 mm Tris-HCl, pH 7.5, and 300 mm NaCl. Standard proteins (GE Healthcare) were thyroglobulin (669.0 kDa), ferritin (440.0 kDa), albumin (69.0 kDa), ovalbumin (43.0 kDa), and ribonuclease A (13.7 kDa). The void volume was determined with blue dextran (GE Healthcare).
Electrophoretic Mobility Shift Assay (EMSA)
DNA-binding reactions (10 μl) were carried out for 1 h at 4 °C in binding buffer (20 mm Tris-HCl, pH 7.5, 50 mm NaCl for the Shu-protein subcomplexes and 100 mm NaCl for the holo-Shu complex and the same binding buffer were used in the fluorescence polarization assays) with the indicated concentration of protein and 0.6 μm dsDNA substrate. The used DNA probe is described in supplemental Table S1. The dsDNA probe was formed by annealing the two complementary ssDNAs. After the addition of 3 μl of gel loading buffer (50% glycerol, 0.02% bromphenol blue), the reaction mixtures were resolved in 5% native polyacrylamide gel in 0.5× TBE buffer (45 mm Tris borate, 1 mm EDTA, pH 8.0) at 4 °C for 50–65 min and visualized by Gel-Red staining.
Fluorescence Polarization Assays (FPAs)
Fluorescence polarization assays were performed at 298 K using a SpectraMax M5 microplate reader system. The wavelengths of fluorescence excitation and emission were 490 and 522 nm, respectively. Each well of a 384-well plate contained 100 nm fluorescent-labeled (5′-6-carboxyfluorescein) DNA probe and different amounts of protein complexes with a final volume of 80 μl. For each assay, DNA-free controls were included. The fluorescence polarization (in mP units) was calculated with Equation 1,
The fluorescence polarization change ΔP (in mP units) was fit to Equation 2,
The DNA probe used is 5′-6-carboxyfluorescein-DNA-1 or 5′-6-carboxyfluorescein-ssDNA-1.
Dynamic Light Scattering (DLS)
All DLS measurements were done with DynaPro-MS800 instrument (DynaPro). The Shu1/MBP-Shu2/MBP-Psy3/Csm2 (MBP-Shu complex) complex at a concentration of about 1 mg/ml (20 mm Tris-HCl, pH 7.5, 100 mm NaCl) was mixed with the various DNA substrates in 1:1 molar ratio at 298 K. After an incubation of 30 min, the protein-DNA sample was subjected to DLS analysis. Data analysis was performed using Dynamics Version 6. The hydrodynamic radius and size distribution were determined by the means of at least three DLS measurements.
Semi-quantitative Growth Assay
The pRS316-Csm2 (Csm2-mt1 or Csm2-mt2) plasmids were constructed by cloning fragments containing the Csm2 coding region with or without the indicated mutations and its upstream 500-bp and downstream 500-bp regions into the BamHI-XhoI sites of pRS316. The plasmid was confirmed by DNA sequencing. The pRS315-Psy3 (Psy3-mt2) plasmids were cloned in a similar way as that of pRS316-Csm2. Psy3Δ and Csm2Δ strains and wild-type BY4742 (MAT his3Δ1 leu2Δ0 lys2Δ0 ura3Δ) strain were used. Phenotypic analysis was performed using serial dilutions of yeast cells grown at 30 °C to early mid-log phase (A600 of 1.0–1.5). Serial dilutions were carried out by spotting cells and diluting 5-fold for each spot onto the indicated plates. Plates were grown at 30 °C for 2–3 days and imaged.
RESULTS
Purification and Structure Determination of the Shu Protein Complexes
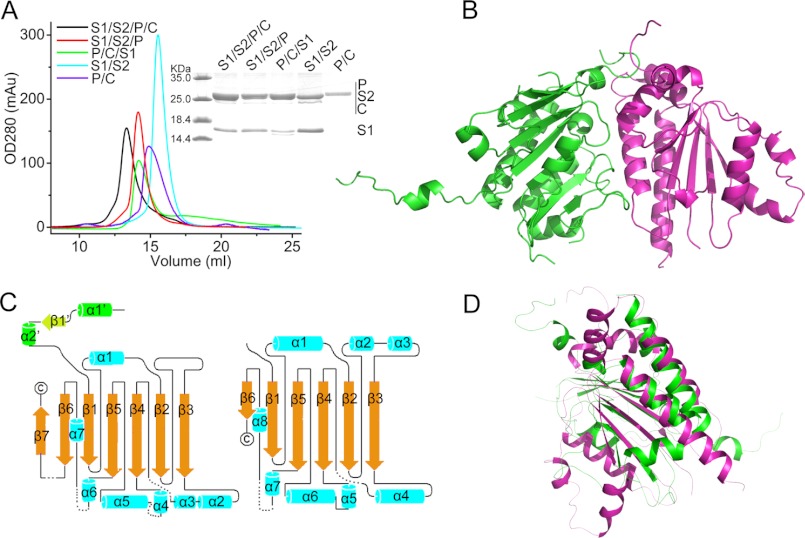
Based on the previous two-hybrid data, the Shu proteins were coexpressed in E. coli with the following combinations: Shu1/MBP-Shu2, MBP-Psy3/Csm2, Shu1/MBP-Shu2/MBP-Psy3, Shu1/MBP-Psy3/Csm2 and Shu1/MBP-Shu2/MBP-Psy3/Csm2. After removal of the maltose-binding protein tags in the recombinant proteins, the different protein complexes were further purified by ion exchange and gel filtration chromatography. During the purification process, each Shu protein complex eluted as a single peak (Fig. 1A), and the presence of each component in that complex was confirmed by mass spectrometry (data not shown), suggesting the different assembled Shu protein complexes were stable in vitro.
FIGURE 1.
Purification of the Shu complex and overall structure of the Psy3-Csm2 heterodimer. A, gel filtration analysis of the purified Shu protein complexes. “S1/S2/P/C” stands for Shu1-Shu2-Psy3-Csm2 complex. B, ribbon representation of the Psy3-Csm2 heterodimer, with Psy3 in green and Csm2 in magenta. C, Psy3 and Csm2 share a similar topological structure. Invisible residues are modeled with black dashes. D, overlay of the structures of Psy3 and Csm2.
To improve our understanding of the Shu complex, we performed structure analysis on the Shu proteins, of which only the Psy3-Csm2 complex yielded diffractable crystals. The 1.9-Å crystal structure showed that one Psy3-Csm2 heterodimer formed the asymmetric unit, consistent with the observation of Psy3 and Csm2 eluting as a heterodimer during the size exclusion chromatography. Unexpectedly, although Psy3 and Csm2 have a low sequence identity (18%), they share a similar architecture (3.0 Å root mean square deviation over 125 Cα atoms) consisting mainly of a parallel, twisted β-sheet flanked by α-helices on both sides (Fig. 1, B–D). In addition to the core domain, Psy3 was shown to contain an N-terminal protruding element that folded into two small helices and irregular coils with residues Lys4–Leu41 (Fig. 1, B and C).
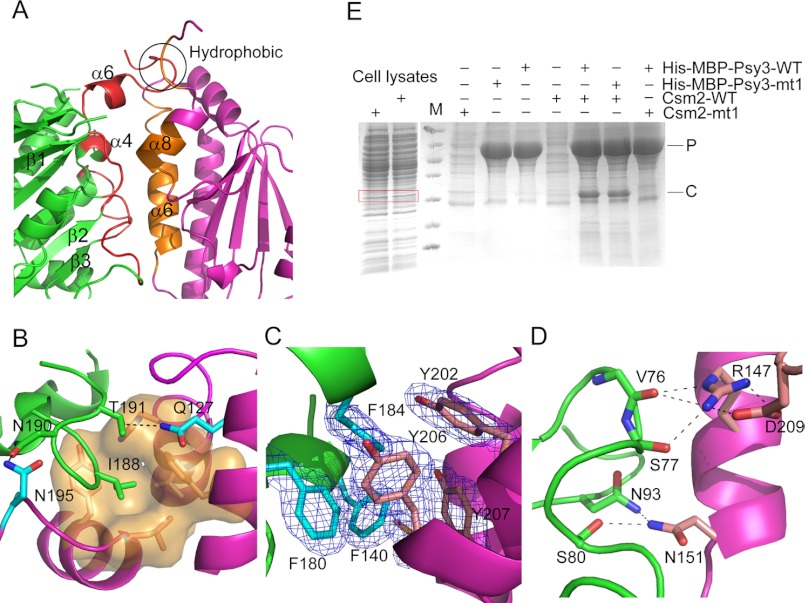
Interface of the Psy3-Csm2 Complex
The α6- and α8-helices of Csm2 and loops following β1-, β2-, β3-, α4-, and α6-helices of Psy3 are the main structural elements involved in heterodimer formation that buries a pairwise surface area of 1115 Å2 (Fig. 2A). As observed in the Rad51 polymerization motif (Rad51-PM)-mediated interactions (26, 27), the side chain of Ile188 in Psy3 is placed into a hydrophobic cavity formed by residues Phe123, His136, Leu139, Leu199, and Tyr202 from Csm2, displaying a “ball-and-socket” module (Fig. 2B). Following the ball-and-socket, the residues Phe140, Phe180, and Phe184 from Psy3 form a complementary hydrophobic core with the residues Tyr202, Tyr206, and Tyr207 from Csm2 through staggering the aromatic rings (Fig. 2C). Csm2 residues Leu144 and Val148 also make hydrophobic contacts with residues in Psy3 (supplemental Fig. S1).
FIGURE 2.
Interface of the Psy3-Csm2 heterodimer. A, overall view of the Psy3-Csm2 interface. Structural elements constructing the interface are shown in red (Psy3) and orange (Csm2), respectively. B, ball and socket mode of hydrophobic interaction. Csm2 residues forming the “socket” are presented as orange sticks and surface and the following residues forming the two hydrogen bonds are shown as cyan sticks. C, hydrophobic core formed by Psy3 residues Phe140, Phe180, and Phe184 and Csm2 residues Tyr202, Tyr206, and Tyr207. D, polar contacts within the interface. E, “hydrophobic core” maintains the heterodimer formation. The red box indicates the Csm2 (mt1) inputs. P and C represent Psy3 (mt1) and Csm2 (mt1), respectively.
Polar contacts occur mainly at the two ends of the interface. The Asn151 in Csm2 forms two hydrogen bonds with Psy3 residues Ser80 and Asn93 through its side chain (Fig. 2D). The residue Arg147 of Csm2 is hydrogen-bonded to the main chain carbonyls of Val76 and Ser77 in Psy3. It also interacts electrostatically with Csm2 Asp209 (Fig. 2D), which keeps the two side chains in position. On the other side, Psy3 residues Asn190 and Thr191 form two hydrogen bonds with residues Asn195 and Gln127 on Csm2, respectively (Fig. 2B). These two hydrogen interactions stabilize the local conformation of Ile188 in Psy3, conferring additional affinity between Psy3 and Csm2.
To further validate the interface and to understand the determinant interaction elements, we generated two mutants to disrupt the observed hydrophobic contacts, Psy3-mt1 (Psy3I188D) and Csm2-mt1 (Csm2Y206E/Y207E). Pulldown assays showed that although His-MBP-Psy3-mt1 still retained Csm2 on the nickel column, Csm2-mt1 failed to bind to His-MBP-Psy3 (Fig. 2E), indicating the “hydrophobic core” dominates the heterodimer formation, consistent with the instability of Psy3-Csm2 complex at low salt concentrations.
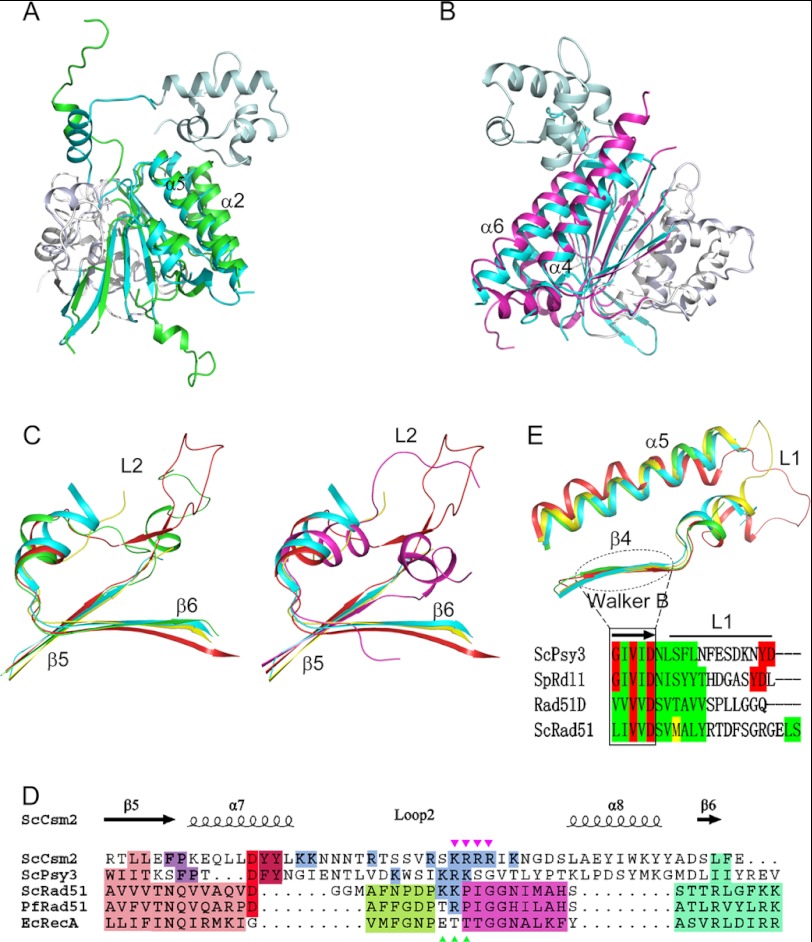
Comparison of Psy3 and Csm2 with Rad51
Structure-based search of the Protein Data Bank using Dali reveals that Psy3 and Csm2 closely resemble the ATPase core domain of the RecA family recombinase, which plays a crucial role in the maintenance of genome stability through HR (27–29). The identified structural similarity strengthens the previous results showing the Shu complex functions in HR-related DNA damage repair. Both Psy3 and Csm2 lack the N-terminal subdomain and C-terminal β-strands (β8/β9 for Psy3 and β7/β8/β9 for Csm2), present in Rad51 (Fig. 1C and supplemental Fig. S2). Structural superposition of Psy3-Rad51 (Protein Data Bank code 1SZP, sequence identity 13%) and Csm2-Rad51 (sequence identity 10%) gives a respective root mean square deviation of 2.6 Å (153 Cα) and 2.8 Å (153 Cα). The structural resemblance of Psy3 (Csm2) and Rad51 mainly resides in the central β-sheet as well as α2- (α4 for Csm2) and α5 (α6 for Csm2)-helices (Fig. 3, A and B). Notably, the superimposed structures reveal that β5/β6 of Psy3 and Csm2 overlay perfectly with the Rad51 elements flanking the L2 loop (Fig. 3C), which is implicated in DNA binding (27, 29, 30). As occurs for the Rad51 L2 loop, the loops connecting the Psy3 and Csm2 β5/β6 are flexible and are not included in the final model. Moreover, structure-based sequence alignment shows that both of them are rich in positively charged amino acids (Fig. 3D). Therefore, we termed the loops L2. The relative orientation of Psy3 and Csm2 is similar to that of hDMC1 dimer (31), bringing the two L2 loops close to each other in a three-dimensional space. In addition to the L2 loop, Psy3 appears to contain a flexible L1 loop connecting the β4/α5 (Fig. 3E). The previously identified Walker B, which is intermediately N-terminal to the L1 loop, aligns well with those of Rad51 proteins from primary sequence to tertiary structure (Fig. 3E).
FIGURE 3.
Psy3 and Csm2 resemble Rad51. A, superposition of Psy3 with yeast Rad51 (Protein Data Bank code 1SZP). The overlaid parts are colored in green for Psy3 and cyan for Rad51. B, superposition of Csm2 with yeast Rad51 (1SZP). The overlaid parts are colored in magenta for Csm2. C, L2 loop observed in Psy3 (left) and Csm2 (right). Green indicates Psy3; magenta indicates Csm2; red indicates E. coli RecA (Protein Data Bank code 3CMW); cyan indicates yeast Rad51 (1SZP); and yellow indicates Pyrococcus furiosus Rad51 (Protein Data Bank code 1PZN). D, structure-based sequence alignment of the L2 loop present in Psy3, Csm2, and Rad51 proteins. Magenta triangle indicates the residues responsible for DNA binding of Csm2 and green triangle for those of Psy3 (see below). E, structure and sequence alignment of the Psy3 Walker B motif and L1 loop. S. pombe Rdl1, human Rad51D. and Saccharomyces cerevisiae Rad51 are included. Diagram are colored as before.
Psy3 lacks the “FXXA” motif in the N-terminal protruding coil, corresponding to the Rad51-PM that mediates the formation of the helical filament through hydrophobic and hydrogen contacts (27, 31). However, the Psy3 residues Arg8–Leu12 interact with the β3 of the neighboring Csm2 in a similar way in the crystal lattice (supplemental Fig. S3). The observed interactions may not be strong enough to support polymer formation in solution. Nevertheless, it is possible that the protruding coil directs oligomer formation when a cofactor such as DNA is present. Taken together, our structural findings reveal that Psy3, as a Rad51 paralogue protein, shares great similarity with Rad51, and Csm2 may also be a member of Rad51 paralogues.
Interaction between the Shu Protein Complex and DNA
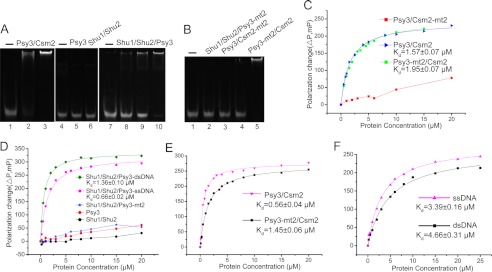
Structural analysis strongly suggests that the Shu protein complex interacts with DNA. Therefore, we used the electrophoretic mobility shift assay (EMSA) and FPA to query whether purified Shu proteins and their complexes interacted with DNA in vitro. During the investigation, maltose-binding protein-tagged Psy3 and Shu2 were used to avoid protein precipitation at low ionic strength.
Initially we investigated two subcomplexes, Shu1-Shu2 and Psy3-Csm2. Both the EMSA and FPA results indicated that the Shu1-Shu2 complex did not bind to the DNA substrates used in the tests (see “Experimental Procedures”) (Fig. 4, A and D). By contrast, the Psy3-Csm2 complex exhibited an appreciable affinity toward ssDNA and dsDNA (Fig. 4, A, C and E). Psy3 alone lacked DNA binding activity. Interestingly, Shu1-Shu2 in complex with Psy3 showed high affinity toward ssDNA and dsDNA (Fig. 4, A and D). Considering the above results, we speculate that a DNA-binding interface is formed when Shu1-Shu2 is complexed with Psy3.
FIGURE 4.
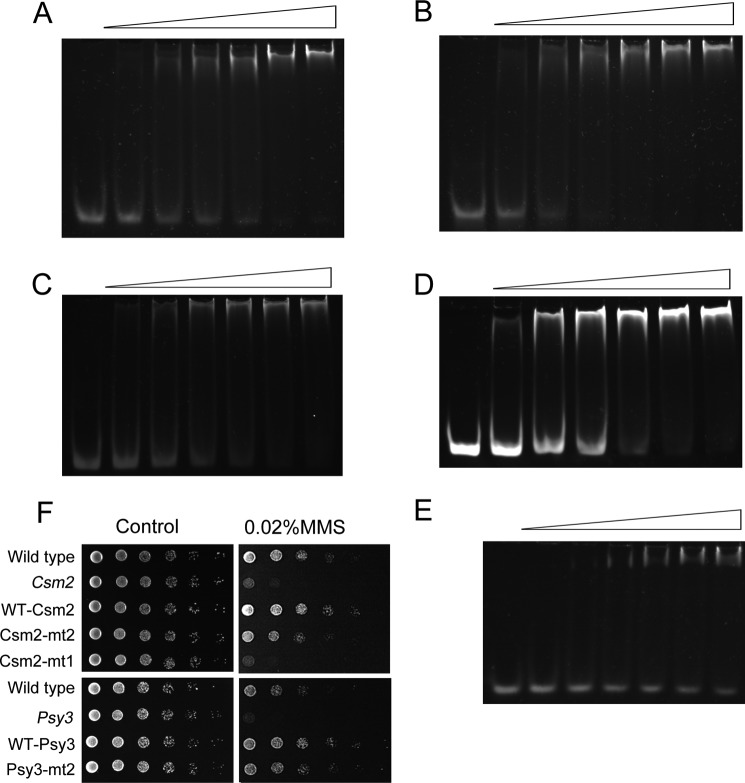
Shu protein complexes interact with DNA. A, EMSA results of Shu protein complexes with dsDNA. 0.6 μm dsDNA-5 substrate; protein at lanes 1–3 (Psy3/Csm2): 0, 2, and 6 μm, respectively; lanes 4 and 5 (Psy3): 0 and 20 μm, respectively; lane 6 (Shu1/Shu2): 20 μm; lanes 7–10 (Shu1/Shu2/Psy3): 0, 5, 10, and 20 μm, respectively. B, EMSA results of Psy3 and Csm2 mutants. 0.6 μm dsDNA-5; proteins at lanes 1 and 2 (Shu1/Shu2/Psy3-mt2): 0 and 20 μm, respectively; lanes 3 and 4 (Psy3/Csm2-mt2): 5 and 20 μm, respectively; lane 5 (Psy3-mt2/Csm2): 5 μm. C, FPAs of Psy3/Csm2 and the mutants with dsDNA-1. The data were fitted according to Equation 2 (see “Experimental Procedures”). D, FPAs of Shu1/Shu2/Psy3 and the mutants with DNA-1. The data were fitted according to Equation 2. The dsDNA-1 substrate was used for Shu1/Shu2/Psy3, Shu1/Shu2/Psy3-mt2, Psy3, and Shu1/Shu2, and ssDNA-1 substrate was also used for Shu1/Shu2/Psy3. E, FPAs of Psy3/Csm2 and the mutant with ssDNA-1. The data were fitted according to Equation 2. F, FPAs of the holo-Shu complex with DNA-1. The ssDNA-1 and dsDNA-1 substrates were used. The data were fitted according to Equation 2.
We then investigated whether the DNA binding activity was conferred by the L2 loops of Psy3 and Csm2. To this end, two mutants were prepared by substituting the consecutive basic residues in the L2 loop with alanine: Psy3-mt2 (K199A/R200A/R201A) and Csm2-mt2 (K189A/R190A/R191A/R192A) (Fig. 3D). The assembled mutant complexes behaved well in solution, indicating these mutations did not affect protein folding and stability (supplemental Fig. S4). Although the mutations in Csm2 completely abolished the ssDNA and dsDNA binding activity of the Psy3-Csm2 complex, those in Psy3 did not affect its affinity to bind dsDNA, reflected by the small increase in dissociation constant (Kd) (from 1.57 to 1.95 μm) (Fig. 4, B and C). However, a 2.6-fold decrease in affinity toward ssDNA was observed (Fig. 4E). In contrast to the Psy3-Csm2 complex, the mutations in Psy3 totally eliminated the ssDNA and dsDNA binding activity of the Shu1-Shu2-Psy3 complex (Fig. 4, B and D), suggesting the L2 loop of Psy3 has a role in DNA binding in the Shu1-Shu2-Psy3 complex.
These investigations reveal that the L2 loops of both Psy3 and Csm2 are the DNA-binding sites of the Shu protein complexes. Thus, the holo-Shu complex seems to possess two separate DNA-binding regions, one created by the Csm2 L2 loop and the other created by the Psy3 L2 loop and Shu1-Shu2 complex. We then used FPA to characterize the DNA-binding property of the holo-Shu complex. Because of the instability of the holo-Shu complex, we performed the FPA at a higher salt concentration (see “Experimental Procedures”), which may justify the increase of the determined Kd value of holo-Shu complex toward ssDNA and dsDNA relative to the subcomplexes (Fig. 4F). Notably, the experimental data could only be fitted using a one-site binding mode, instead of a two-site binding mode, indicating that the predicted two DNA-binding regions are not independent and act together to form a single DNA-binding site. Considering the Psy3-Csm2 structure, this finding is not surprising as the Csm2 L2 loop is located in the vicinity of the Psy3 L2 loop in the three-dimensional space. The existence of redundant DNA-binding motifs in one site may ensure the stability of the formed protein-DNA complex and direct the binding efficiency toward different types of DNA substrates such as ssDNA and dsDNA.
DNA-binding Properties of the Shu Complex
We next tested if Shu protein complexes had a sequence preference. Several dsDNA substrates with random sequences were selected (supplemental Table S1). As shown in Fig. 5, A–D, the Psy3-Csm2 complex showed no obvious sequence preference to the different substrates. Although the Psy3-Csm2 complex bound to dsDNA of different lengths ranging from 30 to 80 bp with similar affinity, it showed a weaker binding toward the 11-bp dsDNA, suggesting that a dsDNA molecule longer than one turn is required for efficient binding (Fig. 5E). The Shu1-Shu2-Psy3 complex also exhibited a similar result (data not shown).
FIGURE 5.
Nonspecific DNA binding is required for resisting MMS-induced damage. EMSA results of the Psy3-Csm2 complex toward various DNA substrates. 0.6 μm dsDNA-2 (30 bp) (A), dsDNA-5 (50 bp) (B), dsDNA-4 (59 bp) (C), dsDNA-3 (80 bp) (D), and dsDNA-6 (11 bp) (E) were incubated with increasing proteins: 0–6 μm, respectively. F, sensitivity to MMS of Shu mutants. The growth defects of Psy3 and Csm2 mutants could be rescued by the wild-type plasmids but not the indicated mutants.
The nonspecific DNA binding activity prompts us to speculate that the Shu-complex yields the nucleoprotein filament by binding to the DNA molecule. To address the question, we employed DLS to check the hydrodynamic radius (Rh) of the various assembled protein-DNA samples. The holo-Shu complex or DNA alone has a relatively small Rh of about 5.5 and 3.3 nm, respectively. As the DNA length increased, the Rh of the Shu-DNA complex increased correspondingly (Table 2 and supplemental Fig. S5). No obvious increase in Rh was observed for the 11-bp DNA, indicating the binding site of one Shu molecule is large enough to coat one 11-bp DNA molecule, consistent with the EMSA results. The nonglobular shape of nucleoprotein filament may bring some inaccuracy into the Rh calculation, but the dramatic changes in Rh of different samples strongly suggest that nucleoprotein is indeed formed.
TABLE 2.
The Rh of different protein-DNA samples
| Hydrodynamic radius (Rh) | |
|---|---|
| nm | |
| Shu | 5.5 |
| DNA-4 (59 bp) | 3.3 |
| Shu + DNA-6 (11 bp) | 5.7 |
| Shu + DNA-2 (30 bp) | 7.5 |
| Shu + DNA-4 (59 bp) | 7.9 |
| Shu + ssDNA-4 (59 bp) | 8.0 |
Disruption of Shu Complex Function by Mutations in Psy3 and Csm2
The holo-Shu complex is thought to protect the genome from damage during DNA replication. To investigate whether the direct interaction between Psy3 and Csm2 is essential in vivo, we employed the yeast phenotypic assay. Plasmids containing the wild-type Csm2 or Csm2-mt1 gene were introduced into a Csm2 knock-out yeast strain. As expected, the wild-type Csm2 (WT-Csm2) rescued the yeast growth defect in YPD containing 0.02% MMS; however, this was not observed in the Csm2-mt1 culture (Fig. 5F), implying the loss of function of the Shu complex. To investigate the importance of the two newly identified DNA-binding motifs, we transformed Psy3-mt2 and Csm2-mt2 into the Psy3 and Csm2 knock-out strains, respectively. For both of the mutants, modest growth defects were observed compared with the wild-type plasmids (Fig. 5F). Thus, we believe that both of the DNA-binding motifs in the Shu complex are required for the efficient functioning in response to DNA damage.
DISCUSSION
In this study, we presented the crystal structure of the Shu proteins, Psy3-Csm2, and demonstrated that they are structural homologues of Rad51. The notion of the integral Shu complex functioning to resist MMS-induced damage was confirmed by the Psy3-Csm2 interface mutation phenotypic assay. As for the Rad51 L2 loop, the L2 loops of Psy3 and Csm2 confer the DNA binding activity of the Shu complex. In contrast to the requirement of ATP for DNA binding of Rad51, the Shu complex was able to bind to DNA without the presence of the Walker A motif. Rather than acting as two separate DNA-binding regions, the two L2 loops in Psy3 and Csm2 are located close to each other in three dimensions and form one DNA-binding site. The reason for the existence of redundant DNA-binding motifs in one site remains unclear. However, they may contribute differently to binding to different types of DNA, consistent with the observation that mutations in the Psy3 L2 loop only slightly impacted the dsDNA binding activity of the Psy3-Csm2 complex but led to a decrease in the ssDNA binding. Nevertheless, the redundant DNA-binding motifs guard the binding efficiency and may explain why the DNA-binding mutations in either Psy3 or Csm2 alone did not cause a deleterious effect in vivo similar to null mutations or mutations in Csm2 precluding interaction with Psy3.
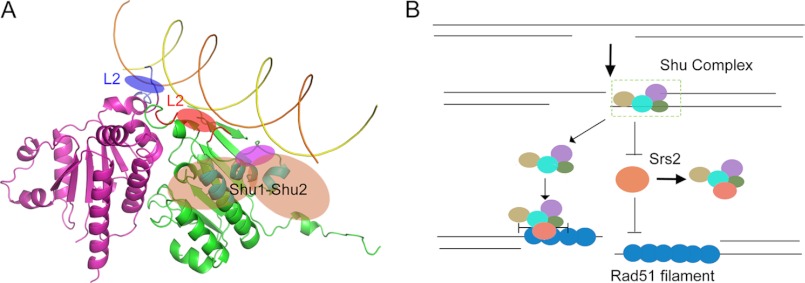
As a central part of the Shu complex, Psy3 not only simultaneously interacts with Shu1, Shu2, and Csm2, but it bridges the DNA-binding region in Shu1-Shu2 and Csm2. Based on the known data, we have proposed a model in which the DNA runs across the surface of the Shu complex with the Psy3 L2 loop in the center, the Csm2 L2 loop being located on one side and the Shu1-Shu2 complex on the other side (Fig. 6A). Considering the trajectory of bound DNA, Psy3 α2′-, α5-, and α7-helices may serve as the anchor site for Shu1-Shu2 binding (Fig. 6A).
FIGURE 6.
Model of Shu-DNA complex and the functional mechanism. A, proposed model of Shu-DNA complex. The blue shadow represents the Csm2 L2 loop, located in the distal side; the red shadow in the middle area indicates the Psy3 L2 loop; the magenta shadow stands for the binding motif from Shu1-Shu2. The orange shadow represents the Shu1-Shu2 complex bound to the Psy3 α2′-, α5-, and α7-helices shown in cyan. B, model of Shu complex in regulating DNA damage repair. Shu complex is recruited by binding to the DNA at the damage sites during the early stage of repair. Shu complex then associates with Srs2 to prevent its interaction with Rad51 filament (right). Shu complex may bind to the Rad51 nucleoprotein filament directly (left). When Srs2 moves to the site of Rad51-DNA-Shu filament, its translocase/helicase activity is inhibited, and thus a more stable Rad51 filament is ensured.
To date, several studies have helped to explain where and how the Shu complex functions in vivo. Mankouri et al. (11) showed that the Shu complex promotes the formation of HRR intermediates that are then resolved by Sgs1-Rmi1-Top3. According to the study executed in S. pombe, the Shu complex seems to act as a mediator of HRR due to the protein homology (17). However, a recent result revealed that the Shu complex, acting as a regulator of Srs2 anti-recombinase, functioned to balance the productive error-free repair with the disassembly of nonproductive recombination intermediates (16). Our data support the latter model. When the replication fork is stalled, the Shu complex is recruited by binding to the processed DNA elements at the damaged site. Adopting a structure similar to that of Rad51, the Shu complex may compete with Rad51 in binding to Srs2, thus protecting the Rad51 filament from disassembly and promoting HRR (Fig. 6B).
While our paper was in preparation, Heyer and co-workers (32) reported that the yeast Rad51 paralogue proteins, Rad55-Rad57, also function to balance the Srs2 antirecombinase in Rad51 filament formation and further revealed that Rad55-Rad57 and Rad51 could bind to Srs2 simultaneously. Rad51 binding to DNA stimulates the Srs2 translocase/helicase activity; however, the stimulatory effect is suppressed by Rad55-Rad57 (32). Similarly, the Shu complex may also regulate the Srs2 activity by such a mechanism (Fig. 6B). Interestingly, the Shu complex is specific to MMS-induced DNA damage repair, although Rad55-Rad57 seems to be more effective in dealing with radiation-induced DNA damage (10, 32). It is possible that these proteins form a complementary group of proteins to repair different types of DNA damage through similar mechanisms.
In summary, through structural and biochemical studies, we have further characterized the yeast Shu complex and proposed its functional mechanism in vivo, providing a good foundation for understanding this conserved pathway in eukaryotic cells.
Acknowledgments
We thank Prof. Jianye Zang for helpful discussions and are grateful to the staff members in Beijing Synchrotron Radiation Facility and Shanghai Synchrotron Radiation Facility for the collection of diffraction data.
This work was supported by Fundamental Research Funds for the Central Universities, Chinese National Natural Science Foundation Grants 31130018, 30900224, and 10979039, Chinese Ministry of Science and Technology Grants 2009CB825500 and 2012CB917200, Science and Technological Fund of Anhui Province for Outstanding Youth Grant 10040606Y11.

This article contains supplemental Figs. S1–S5 and Table S1.
The atomic coordinates and structure factors (code 4DT1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HRR
- homologous recombination repair
- ssDNA
- single-stranded DNA
- DLS
- dynamic light scattering
- FPA
- fluorescence polarization assay
- PRR
- post-replication repair
- MMS
- methyl methanesulfonate
- HR
- homologous recombination
- SeMet
- selenomethionine.
REFERENCES
- 1. Jackson S. P., Bartek J. (2009) The DNA-damage response in human biology and disease. Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ciccia A., Elledge S. J. (2010) The DNA damage response. Making it safe to play with knives. Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Broomfield S., Hryciw T., Xiao W. (2001) DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486, 167–184 [DOI] [PubMed] [Google Scholar]
- 4. Barbour L., Xiao W. (2003) Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability. A yeast model. Mutat. Res. 532, 137–155 [DOI] [PubMed] [Google Scholar]
- 5. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 6. Hofmann R. M., Pickart C. M. (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 [DOI] [PubMed] [Google Scholar]
- 7. Brusky J., Zhu Y., Xiao W. (2000) UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet. 37, 168–174 [DOI] [PubMed] [Google Scholar]
- 8. Ulrich H. D., Jentsch S. (2000) Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19, 3388–3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ball L. G., Zhang K., Cobb J. A., Boone C., Xiao W. (2009) The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol. Microbiol. 73, 89–102 [DOI] [PubMed] [Google Scholar]
- 10. Shor E., Weinstein J., Rothstein R. (2005) A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3, and CSM2. Four genes involved in error-free DNA repair. Genetics 169, 1275–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mankouri H. W., Ngo H. P., Hickson I. D. (2007) Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell 18, 4062–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu L., Hickson I. D. (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 [DOI] [PubMed] [Google Scholar]
- 13. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., Sung P. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 [DOI] [PubMed] [Google Scholar]
- 14. Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., Fabre F. (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 [DOI] [PubMed] [Google Scholar]
- 15. German J. (1993) Bloom syndrome. A Mendelian prototype of somatic mutational disease. Medicine 72, 393–406 [PubMed] [Google Scholar]
- 16. Bernstein K. A., Reid R. J., Sunjevaric I., Demuth K., Burgess R. C., Rothstein R. (2011) The Shu complex, which contains Rad51 paralogues, promotes DNA repair through inhibition of the Srs2 anti-recombinase. Mol. Biol. Cell 22, 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martín V., Chahwan C., Gao H., Blais V., Wohlschlegel J., Yates J. R., 3rd, McGowan C. H., Russell P. (2006) Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 25, 2564–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19. Collaborative Computational Project No. 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 20. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Terwilliger T. C. (2003) Automated main chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pellegrini L., Yu D. S., Lo T., Anand S., Lee M., Blundell T. L., Venkitaraman A. R. (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420, 287–293 [DOI] [PubMed] [Google Scholar]
- 27. Shin D. S., Pellegrini L., Daniels D. S., Yelent B., Craig L., Bates D., Yu D. S., Shivji M. K., Hitomi C., Arvai A. S., Volkmann N., Tsuruta H., Blundell T. L., Venkitaraman A. R., Tainer J. A. (2003) Full-length archaeal Rad51 structure and mutants. Mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 22, 4566–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Story R. M., Weber I. T., Steitz T. A. (1992) The structure of the E. coli recA protein monomer and polymer. Nature 355, 318–325 [DOI] [PubMed] [Google Scholar]
- 29. Conway A. B., Lynch T. W., Zhang Y., Fortin G. S., Fung C. W., Symington L. S., Rice P. A. (2004) Crystal structure of a Rad51 filament. Nat. Struct Mol. Biol. 11, 791–796 [DOI] [PubMed] [Google Scholar]
- 30. Chen Z., Yang H., Pavletich N. P. (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494 [DOI] [PubMed] [Google Scholar]
- 31. Kinebuchi T., Kagawa W., Enomoto R., Tanaka K., Miyagawa K., Shibata T., Kurumizaka H., Yokoyama S. (2004) Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein Dmc1. Mol. Cell 14, 363–374 [DOI] [PubMed] [Google Scholar]
- 32. Liu J., Renault L., Veaute X., Fabre F., Stahlberg H., Heyer W. D. (2011) Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479, 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]