Background: The NH2-terminal titin segment is firmly anchored to the Z-disk through the Z1 and Z2 Ig domains.
Results: Z1Z2 domains alone induce spontaneous dimerization in otherwise monomeric proteins.
Conclusion: A mechanical force of 700 pN is required to induce dimer rupture.
Significance: Such extremely high mechanical stability is likely to be a natural protective mechanism that guarantees muscle integrity.
Keywords: Muscle, Protein Conformation, Single Molecule Biophysics, Spectroscopy, Titin, Titin Z1Z2 Domains, Elastomeric Proteins, Force Spectroscopy AFM, Nanomechanical Anchoring, Single Molecule
Abstract
Muscle elasticity strongly relies on the mechanical anchoring of the giant protein titin to both the sarcomere M-band and the Z-disk. Such strong attachment ensures the reversible dynamics of the stretching-relaxing cycles determining the muscle passive elasticity. Similarly, the design of biomaterials with enhanced elastic function requires experimental strategies able to secure the constituent molecules to avoid mechanical failure. Here we show that an engineered titin-mimicking protein is able to spontaneously dimerize in solution. Our observations reveal that the titin Z1Z2 domains are key to induce dimerization over a long-range distance in proteins that would otherwise remain in their monomeric form. Using single molecule force spectroscopy, we measure the threshold force that triggers the noncovalent transition from protein dimer to monomer, occurring at ∼700 piconewtons. Such extremely high mechanical stability is likely to be a natural protective mechanism that guarantees muscle integrity. We propose a simple molecular model to understand the force-induced dimer-to-monomer transition based on the geometric distribution of forces occurring within a dimeric protein under mechanical tension.
Introduction
Elucidating the molecular-level determinants that underlie the specific mechanisms of adhesion remains a crucial goal in surface and materials sciences, especially in those approaches aiming to explore the nanometer realm. In this vein, outstanding lessons of highly specific and resilient adhesion can be taken from biological processes (1). For example, cell-cell adhesion in vertebrates and invertebrates is mediated by the interaction of cadherin proteins (2). Although the main players involving specific adhesive contact between cells were identified more than a decade ago through crystallization techniques (2), the molecular mechanisms by which cadherin extracellular domains form adhesive contacts have remained controversial for a long time (3). The current accepted view suggests that adhesive binding by these proteins occurs via a subtle strand-swapped interface (4–7). Similarly, the integrity of vertebrate striated muscle represents yet another natural example of a macromolecular assembly in biology, with precisely assigned localizations for their constituent proteins (8). This ordered assembly is likely to be orchestrated by the giant protein titin, which is perhaps the most paradigmatic example of a natural protein that conducts its function under mechanical stress (9, 10). Such a micrometer-long protein is known as a true molecular spring in muscle cells and actively participates as a scaffold protein aiding myofibrillar assembly (11). Titin is composed of a string of folded immunoglobulin domains, which act as shock absorbers, combined with unique sequences of unstructured regions that function as entropic springs (12). The NH2-terminal titin segment is firmly anchored to the Z-disk, and the protein expands all the way to the center of the sarcomere, the M-band (11, 13). These tight attachments ensure the functioning of titin as a molecular ruler for sarcomere assembly, ultimately responsible for the reversible resting elasticity of the muscle (14). At the titin N terminus, the Ig domains Z1 and Z2 of two parallel molecules interact in a palindromic way, mediated by the telethonin (Tcap) protein (15, 16). Such a molecular complex is thought to withstand unusually high mechanical forces (17, 18). However, recent experiments on telethonin knock-out mice and zebrafish surprisingly demonstrated that telethonin-depleted animals are viable, showing no dramatic defects in their muscle properties (13, 19, 20). Therefore, it is plausible that other elusive molecular mechanisms are involved in the mechanical gluing process of two interacting titin molecules. Single molecule force spectroscopy with AFM3 has provided a new vista on the mechanical stability of a wide variety of individual proteins with mechanical function (21–24). In particular, the titin protein has become the prima donna in the single molecule studies, revealing with unprecedented detail the diverse mechanical properties of the distinct building blocks that compose the largest protein in the human body (21, 25–27).
EXPERIMENTAL PROCEDURES
Protein Engineering
PolyZ1 (Z1)8 and PolyZ2 (Z2)8 were constructed by consecutive subcloning of the respective monomers using the BamHI, KpnI, and BglII restriction sites (28). The eight-domain polyproteins were cloned into the pQE80L (Qiagen) expression vector and transformed into the BLR DE3 Escherichia coli expression strain. Each polyprotein construct was finally purified by histidine metal affinity chromatography with Talon resin (Clontech) and by gel filtration using Superdex 200 HR column (GE Biosciences). The proteins were stored in PBS buffer. In the case of the (I27)4Z1Z2 protein and the (I27)4Z1Z2(I27)4 protein, the polyproteins were constructed with one or two handles, respectively, composed of four repeats of the I27 modules of human cardiac titin. We followed a multistep cloning procedure to construct the I27 handle polyproteins and subsequent insertion of the Z1Z2 dimer in between the handles. Cloning and purification were conducted as in the case of the (Z1)8 and (Z2)8 polyproteins. Finally, the control (I27)4Z1Z2 protein (supplemental Fig. S2) without terminal cysteines was cloned into the pQE16 (Qiagen) expression vector while keeping the rest of the cloning and purification steps invariant.
Single Molecule Force Spectroscopy
The details of our custom-made AFM apparatus have been described elsewhere (22). The control of the AFM head is carried out by data acquisition cards (6052E and 6703) from National Instruments (Austin, TX). Each protein sample was prepared by depositing 1–10 μl of protein in PBS solution (at a concentration of 0.2–1 mg ml−1) onto a freshly evaporated gold cover slide. Each cantilever (Si3N4 Veeco MLCT-AUHW) was individually calibrated using the equipartition theorem, which gave a typical spring constant of ∼20 or ∼50 pN/nm. Single proteins were picked up from the surface by pushing the cantilever onto the surface with a contact force of 500–1,000 pN to promote the nonspecific adhesion of the proteins on the cantilever surface. The pulling speed was set for all the experiments at 400 nm/s. All data were recorded and analyzed using custom software written in Igor Pro 6.0 (WaveMetrics, Lake Oswego, OR).
RESULTS
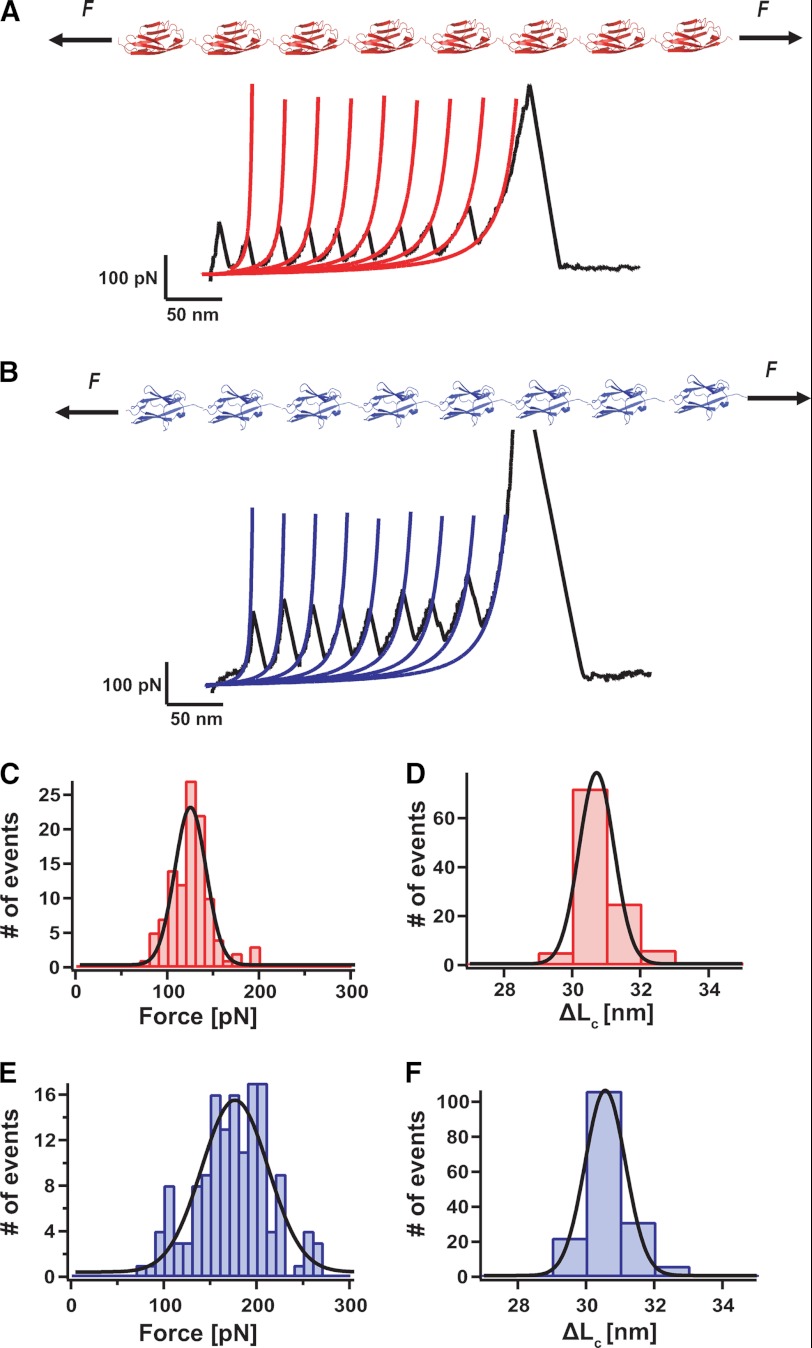
Using the polyprotein approach (Fig. 1), which provides unmistakable molecular fingerprints (28, 29), we individually characterized the mechanical properties of the Z1 and Z2 titin domains. Fig. 1A shows a force-extension trajectory of a (Z1)8 polyprotein, resulting in a typical saw-tooth pattern, where each individual peak corresponds to the unfolding of a single monomer in the polyprotein chain. Fig. 1C shows the histogram corresponding to the distribution of unfolding forces, yielding an average unfolding force of 125 ± 23 pN, n = 108. By fitting the worm-like chain (WLC) model of polymer elasticity (30) to each individual unfolding trajectory (in red), we measured an increment in contour length of ΔL = 30.8 ± 0.6 nm upon unfolding of each individual monomer in the chain (Fig. 1D). A similar approach was used to characterize the mechanical stability of the (Z2)8 polyprotein (Fig. 1B), yielding an average unfolding force of 174 ± 40 pN, n = 148, and an associated ΔL = 30.8 ± 0.7 nm (Fig. 1, E and F). The different mechanical stability of both domains agrees well with the results obtained by molecular dynamics simulations (17), showing a hierarchy in the mechanical unfolding of he Z1Z2 complex whereby the Z1 domain unfolded first, followed by the unfolding of the Z2 domain at longer pulling times.
FIGURE 1.
The Z1 and Z2 domains of titin exhibit distinct mechanical stability. A and B, diagram of the polyprotein (Z1)8 and (Z2)8 constructs and typical force-extension trajectories, where all the monomers in the chain unfold. C, distribution of unfolding forces for the (Z1)8 construct, yielding an average force 125 ± 23 pN, n = 108. D, fitting the data to the WLC model of polymer elasticity (red lines) yields an average increment in contour length of ΔL = 30.8 ± 0.6 nm. E, distribution of unfolding forces for the (Z2)8 construct, yielding an average force of 174 ± 40 pN, n = 148 (F) and an associated contour length increase of ΔL 30.8 ± 0.7 nm (F).
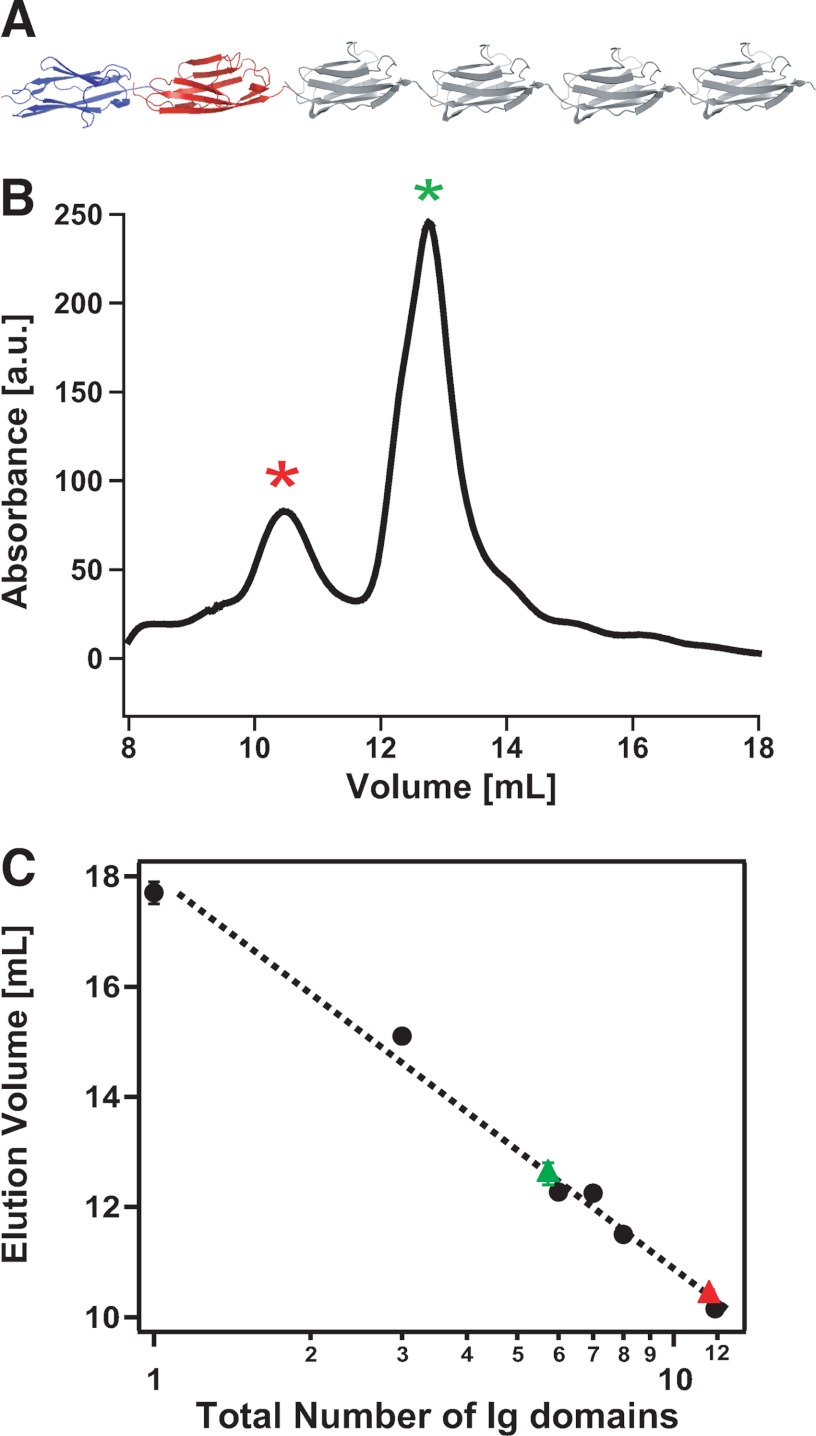
To study the mechanical properties of a protein mimicking the titin protein, we constructed the (I27)4Z1Z2 polyprotein (Fig. 2A). Surprisingly, the size-exclusion chromatogram resulting from this protein (Fig. 2B) showed two well defined peaks: the first one occurring at ∼10.4 ml (red asterisk) and the second one occurring at a higher elution volume of ∼12.6 ml (green asterisk). A typical representation relating the elution volume with the size of the protein (31) was used to elucidate the nature of the protein fraction contained in each elution peak. For calibration purposes, we used I27 polyproteins prepared using the same expression system but composed of a varying number of monomers, ranging from 1 to 12. Remarkably, the two peaks of the chromatogram corresponding to the elution of the (I27)4Z1Z2 polyprotein fall onto the same master curve. The elution time of the first peak corresponds to a polyprotein made up of 12 Ig folded domains, ∼11 kDa/domain (red triangle), whereas the second peak (green triangle) is consistent with the elution of a protein composed of six Ig domains. Hence, these results indicate that two different molecular species are detected in solution: a first monomeric species consistent with the expected size of the polyprotein (I27)4Z1Z2 and a second dimeric species consistent with a 2((I27)4Z1Z2) structure exhibiting lower retention times in size-exclusion chromatography.
FIGURE 2.
The (I27)4Z1Z2 protein spontaneously forms dimers in solution. A, diagram of the engineered (I27)4Z1Z2 polyprotein. B, size-exclusion chromatogram of the (I27)4Z1Z2 protein, exhibiting two well defined peaks, the first one occurring at ∼10.4 ml (red asterisk) and the second one occurring at a higher elution volume of ∼12.6 ml (green asterisk). a. u., arbitrary units. C, representation of the elution volume as a function of the size of the protein (in logarithmic scale). The Z1 and Z2 Ig domains of titin have approximately the same size (99 and 100 amino acids, respectively) as the I27 Ig module of titin (89 amino acids). With calibration purposes, we used I27 polyproteins prepared using the same expression system but composed of a varying number of monomers: (I27)n. Specifically, for calibration of the column, we used I27 polyproteins composed of n = 1, 3, 6, 7, 8, or 12 I27 domains. Interpolation of the elution volumes corresponding to the two peaks indicate that the first peak corresponds to the size of a 12-mer (∼11 kDa/domain), thus consistent with a dimeric 2((I27)4Z1Z2) protein, and the second peak corresponds to a 6-mer, suggestive of a monomeric (I27)4Z1Z2 protein.
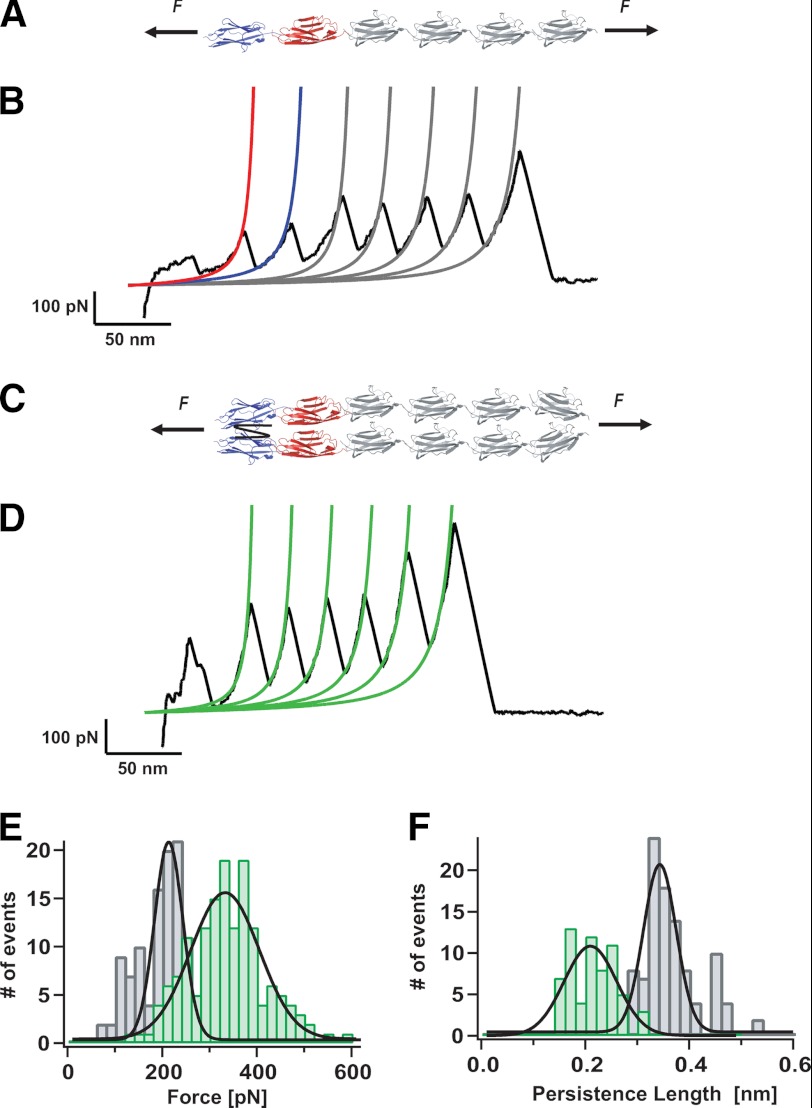
The mechanical characterization of the (I27)4Z1Z2 polyprotein using our force spectroscopy approach (Fig. 3A) resulted in individual unfolding trajectories that showed four distinctive and well defined patterns. Interestingly, all four unfolding patterns can be observed when protein fractions from both peaks in Fig. 2 are studied in the single molecule mechanical experiments, suggestive of a dynamic equilibrium between both species in solution. Fig. 3B shows the unfolding trajectory corresponding to a single (I27)4Z1Z2 protein monomer (∼20% occurrence). In this trajectory, the two first unfolding peaks correspond to the unfolding of the Z1 and Z2 modules (red and blue fits, respectively) as marked by their characteristic ∼31 nm increase in contour length and unfolding forces in the range of 120–180 pN (Fig. 1, C–F). The remaining four peaks correspond to the unfolding of the well characterized I27 module, occurring at a force value of ∼200 pN with a concomitant increase of 28.6 nm in length (28) (gray WLC fits). The distribution of forces corresponding to the unfolding of the complete protein are shown in the histogram of Fig. 3E (gray bars), resulting in an overall average unfolding force of 189 ± 48 pN, n = 107. Fitting of the WLC model to each individual unfolding trajectory of such monomeric proteins yielded a distribution of persistence length values that is shown in Fig. 3F (gray bars) with an average value of P = 0.36 ± 0.06 nm (where P indicates persistence length).
FIGURE 3.
Mechanical fingerprint of a parallel dimeric 2((I27)4Z1Z2) polyprotein. A, scheme of the engineered (I27)4Z1Z2 polyprotein under mechanical unfolding conditions. B, typical unfolding trajectory obtained form pulling a (I27)4Z1Z2 monomer. Fitting the WLC model of polymer elasticity to the traces (red fit for Z1, blue fit for Z2, and gray fits for I27 modules) measures the increment in contour length, ΔL, and the persistence length, P, for the monomeric species. C, diagram of the protein dimer 2((I27)4Z1Z2). D, typical unfolding trace corresponding to in-register unfolding of the 2((I27)4Z1Z2) dimer. Fittings to the WLC (green fits) measure the ΔL and P values associated to each unfolding peak. E, histogram corresponding to the unfolding forces measured for the protein monomer trajectories (gray bars) and for the protein dimers (green bars). The measured average unfolding for the dimers (338 ± 85 pN) is almost double that obtained for the monomers, 189 ± 48 pN, n = 107. F, by contrast, the persistence length is almost halved for the protein dimers (0.21 ± 0.04 nm, n = 63) with respect to that obtained for the protein monomers (P = 0.36 ± 0.06 nm).
A typical individual trajectory corresponding to the second distinctive pattern (∼10% occurrence) observed for the unfolding trajectories of the (I27)4Z1Z2 polyprotein is shown in Fig. 3D. In this case, the unfolding peaks occur at a much higher force of 338 ± 85 pN (Fig. 3E, green bars). Fitting these trajectories to the WLC model (green lines) yields a distribution of persistence lengths (Fig. 3F, green bars) that is much lower (0.21 ± 0.04 nm, n = 63) than the one observed for the monomeric species. Thus, in these trajectories, the unfolding force measured for the monomeric form is almost doubled and the persistence length is halved, whereas the contour length increase is kept unchanged. Taken together, these mechanical features are the fingerprint for the unfolding of a 2((I27)4Z1Z2) dimer that extends in perfect register (32, 33). It is noteworthy that these mechanical features corresponding to the unfolding of a dimer protein are very rarely observed when pulling on the extensively well characterized I27 polyproteins, devoid of Z1Z2 domains.
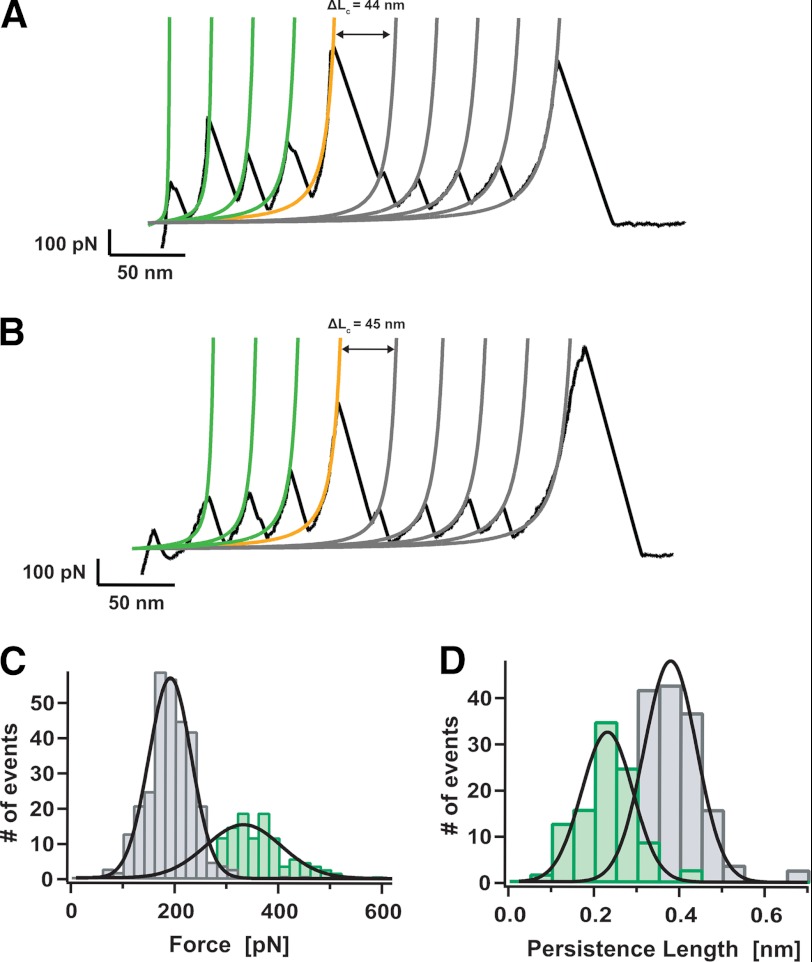
Fig. 4, A and B, show two characteristic examples of a third group of individual unfolding trajectories of the (I27)4Z1Z2 polyprotein (∼20% occurrence). The most striking feature of both trajectories is the presence of a high-force peak of ∼700 pN. In both traces, such a high-force peak takes place concomitant with an increase in length of ∼45 nm (orange fit). Prior to such a high-force peak, the observed unfolding peaks exhibit a mechanical stability of 337 ± 84 pN, n = 152 (Fig. 4C, green bars), and a persistence length of 0.22 ± 0.07 nm (Fig. 4D, green bars in histogram). Hence, these mechanical features directly correlate with the in-register unfolding of a protein dimer (32). By contrast, after the high-force peak, the remaining unfolding events (gray fits) feature an unfolding force of 192 ± 48 pN, n = 303 (Fig. 4C, gray bars) and a persistence length of 0.40 ± 0.09 nm (Fig. 4D, gray bars), thus consistent with the unfolding of a monomeric polyprotein. In light of these results, in the individual trajectories shown in Fig. 4, A and B, we suggest that initially three domains of each protein chain unfold in perfect register (green peaks). Subsequently, a dimer-to-monomer transition occurs, which is hallmarked by the presence of the high-force peak. Once the dimer is broken, the four remaining individual monomers unfold (gray peaks).
FIGURE 4.
Disruption of protein dimer results in a peak of extremely high mechanical resistance. A and B, individual unfolding trajectories showing the rupture of a protein dimer under force. WLC fits to the data measure the increase in contour length before (green fits) and after (gray fits) the peak of high force (orange fit) takes place. C, histogram of the unfolding forces for the force peaks occurring before (green bars, 337 ± 84 pN, n = 152) or after (gray bars, 192 ± 48 pN, n = 303) the high-force peak. D, histogram of the measured persistence length for the force peaks occurring before (green bars, 0.22 ± 0.07 nm) or after (gray bars, 0.40 ± 0.09 nm) the high-force peak. These results demonstrate that before the high-force peak, the protein is in its dimeric form, converting into a protein monomer once after the high-force event occurs. The high-force event thus fingerprints the dimer-to-monomer transition.
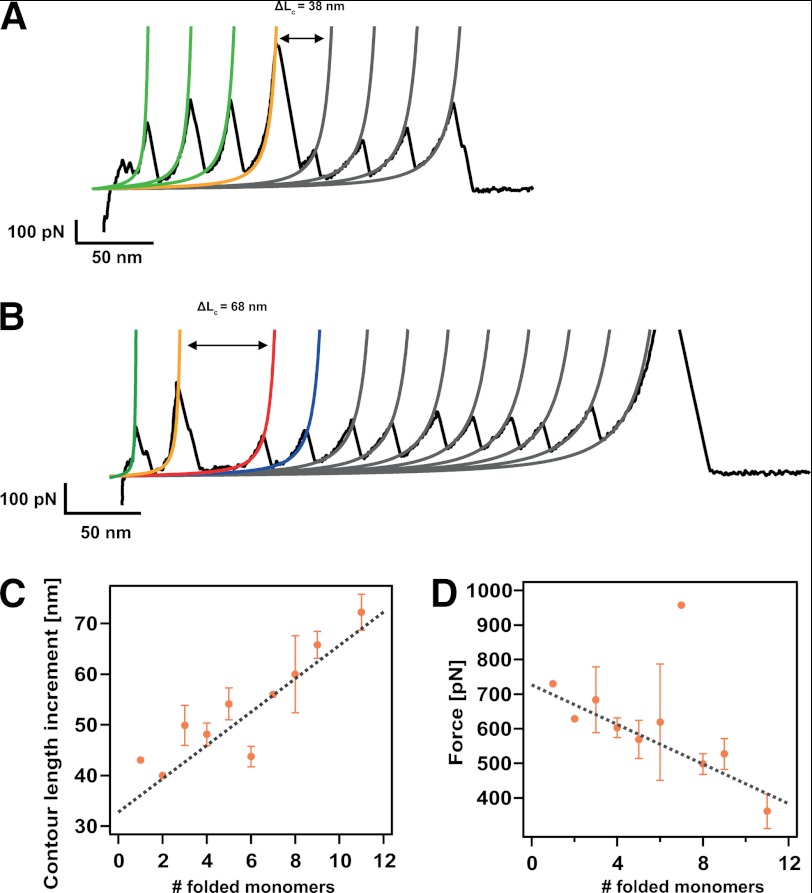
Although the presence of such a remarkable high-force peak seems to be a clear fingerprint for a dimer-to-monomer transition, the molecular mechanisms involving such force-activated dimer disruption remain unclear. In the trajectories shown in Fig. 4, A and B, disrupting the protein dimer elicits a similar extension of ∼45 nm. By contrast, in the shorter trace shown in Fig. 5A, the ΔL corresponding to the disruption of the protein dimer is smaller (38 nm). For those trajectories where a larger number of protein monomers unfold after the occurrence of the high-force peak, the associated ΔL increases (68 nm in the case of the trajectory shown in Fig. 5B). Fig. 5C shows the dependence of the ΔL associated to the dimer disruption as a function of the number of protein monomers that remained folded once the dimer rupture event occurs. These data demonstrates a linear increase of the ΔL with the number of folded monomers. Linear fit to the data results in a slope of 3.3 nm/folded monomer and an intercept of 32.8 nm. Strikingly, the slope of the fit closely coincides with the size of a folded Ig domain. Moreover, the measured intercept (32.8 nm) nearly corresponds to the contour length increment of a single immunoglobulin domain upon unfolding (Fig. 1, D–F). Hence, these results demonstrate that such dimer-to-monomer transition involves the unfolding of a protein monomer along with the extension of the remaining folded protein monomers. Interestingly, we measure a negative linear correlation between the force at which this transition occurs and the number of modules that are still folded once the dimer-to-monomer transition takes place (Fig. 5D). This suggests that those dimeric proteins for which their transition to a monomeric species involves a higher number of folded domains are, on average, mechanically less stable (Fig. 5B). Such an unexpected result is entirely compatible with the induction of distal ordering by the Z1Z2 domains. The efficiency of such an ordering mechanism dies out with distance (Fig. 5D), probably reflecting an increased level of conformational flexibility of the long chain.
FIGURE 5.
The molecular mechanism of dimer disruption involves unfolding of one protein domain and extension of the domains that still remained folded. A, typical unfolding trajectory where three domains are still folded once the forced-induce rupture occurs, eliciting an increase in length of 38 nm. B, unfolding trajectory where the dimer-to-monomer transition occurs when nine monomers remained folded. In this case, such disruption entails a longer increment of released length of ΔL = 68 nm. C, dependence of the increment in contour length, ΔL, associated with the dimer-to-monomer transition as a function of the folded monomers, showing a linear dependence. Linear fit to the data (gray discontinuous line) yielded a slope of 3.3 nm (close to the size of a folded monomer) and an intercept of 32.8 nm (corresponding to the unfolding of one monomer in the chain). D, dependence of the force at which the dimer-to-monomer transition occurs with the number of folded modules remaining in the chain. The negative linear correlation indicates that on average, the presence of each extra monomer in the chain decreases the mechanical stability of the complex by ∼29 pN (linear fit, gray line). Error bars stand for S.E. in each case. The individual data points devoid of error bars correspond to measurements stemming from an individual unfolding trajectory.
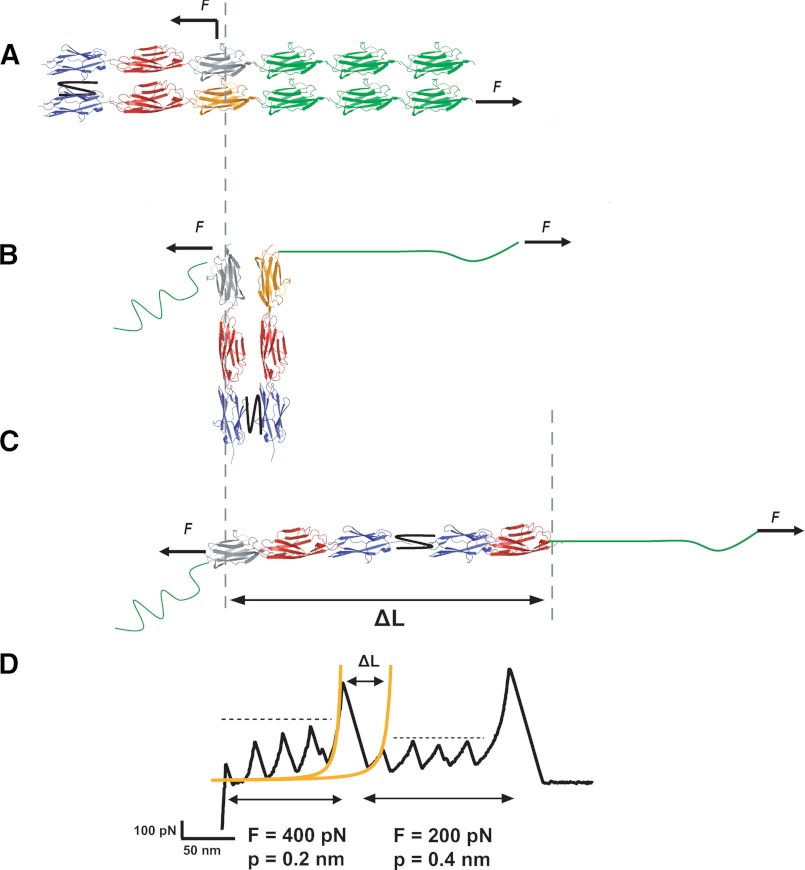
A simple molecular model able to account for this dimer-to-monomer transition mechanism is shown in Fig. 6. The AFM cantilever tip can pick a protein from random positions within the structure. In the case of a dimeric protein, it is likely that the stretching force is applied from the two different protein chains in the dimer. Assuming that the strong mechanical link (black connecting line) between the two chains occurs within the Z2 domains, pulling from the positions in Fig. 6A marked with arrows would result first in the parallel in-register unfolding of three I27 modules (in green). During this process, the protein will rotate, forming an angle of up to 90° with the pulling direction (Fig. 6B). Further application of force will result in the breakage of the protein interface, which triggers a rotation until the protein aligns with the pulling force (Fig. 6C). This process increases the contour length of the protein, ΔL, by an amount equal to the length released upon unfolding of one domain (orange module) plus the length gain corresponding to the number of monomers that remained folded. Therefore, this scenario predicts that changing the force application point (Fig. 6A) would result in a different increase in contour length (Fig. 6C) because a different number of modules would remain folded, consistent with the data shown in Fig. 5C. Finally, further stretching the protein will result in the sequential unfolding of the remaining monomers in the large polyprotein chain.
FIGURE 6.
Schematic model of force-induced dimer-to-monomer mechanism. A, pulling the protein dimer from random positions along a different pulling direction results in the parallel in-register unfolding of three I27 modules (green). B, during this process, the protein will rotate, forming an angle up to 90° with the pulling direction. C, the pulling force will then disrupt the interface between the two parallel polyprotein chains forming the dimer, forcing the protein to rotate until it aligns with the pulling force. D, this process increases the contour length of the protein, ΔL, by an amount equal to the length released upon unfolding of one domain (orange monomer in panel B) plus the length gain corresponding to the number of monomers that remained folded.
A striking observation in our mechanical experiments is that dimerization occurs in more than 50% of the cases. It is likely that the two additional cysteine residues included in the C terminus of our engineered polyproteins (see “Experimental Procedures”) enhance the probability of protein dimerization. Indeed, in the more frequent fourth class of typical trajectories that we observe (supplemental Fig. S1), where we do not typically identify the presence of the high-force peak, we measure a number of unfolding peaks ranging from 7 up to 12, which corresponds to the complete unfolding of a protein dimer (supplemental Fig. S1b). In sharp contrast with these observations, typical unfolding trajectories of extremely well characterized polyproteins such as the (I27)8 (28) or the (ubiquitin)9 (34), which also contain the two cysteines in their termini, hardly ever give rise to protein dimers (probability lower than 0.002), as is shown in the schematic representation and typical force-extension unfolding trajectory of the (I27)8 polyprotein in supplemental Fig. S1c. The capture of the full unfolding trajectories of protein dimers featuring 12 peaks (supplemental Fig. S1b) most probably implies that a disulfide bond has been formed between the termini of the two adjacent proteins, thus giving rise to a head-to-head linkage conformation (supplemental Fig. S1a). The large proportion of these long dimeric trajectories (50%) when compared with other proteins expressed in the same vector suggests that the interactions between the Z1Z2 domains brings the two proteins closer, such that the formation of the disulfide bond is greatly enhanced. In this case, the monomer-dimer equilibrium is rapidly shifted toward the irreversible formation of protein dimers. As a control experiment, we expressed the same (I27)4Z1Z2 polyprotein in the pQE16 vector, depleted of cysteines in the protein termini. In this case (supplemental Fig. S2), the formation of long head-to-head dimers such as those shown in supplemental Fig. S1 is completely hindered because there is no covalent link between both protein monomers. Instead, the two other main unfolding scenarios, i.e. the unfolding of the protein monomer (supplemental Fig. S2b) and the unfolding pattern exhibiting the disruption of the protein dimer (supplemental Fig. S2c), are retained. In this case, the probability of unfolding the monomeric form is higher (∼80% occurrence), although this probability shifts toward the dimeric species with time (timescale of days after purification of the protein). Hence, these results further demonstrate the ability of the Z1Z2 domains to trigger dimerization in proteins that would otherwise remain in their monomeric form. Indeed, pulling on the protein (I27)4Z1Z2(I27)4, where two (I27)4 handles rather than one are inserted, also exhibits force-extension curves consistent with the monomeric (∼80%) and dimeric (∼20%) forms, the latter also exhibiting the high-force peak (supplemental Fig. S3). Finally, it is noteworthy that protein samples collected from both peaks in the chromatogram shown in Fig. 2 resulted in unfolding trajectories corresponding to the unfolding of both monomeric and dimeric species, albeit with different proportion. The unfolding trajectories corresponding to the protein fraction contained within the first elution peak in Fig. 2B displayed a larger population of unfolding traces exhibiting more than six unfolding events, thus corresponding to protein dimers. Similarly, protein samples obtained from the second elution peak in Fig. 2B mainly gave rise to unfolding trajectories such as those shown in Fig. 3B, corresponding to the mechanical unfolding of a single polyprotein chain. However, a non-negligible number of traces exhibited mechanical features corresponding to the unfolding of a protein dimer, such as those shown in Fig. 2D, Fig. 4, A and B, and supplemental Fig. S1b. In that case, the proportion of trajectories corresponding to the mechanical unfolding of protein dimers readily increased with time (in the scale of days/weeks from the moment the protein was first purified). Hence, these observations altogether suggest a dynamic equilibrium between the monomeric and dimeric species, and that is the main reason why both species could not be perfectly separated in our mechanical experiments. Undoubtedly, the disulfide bond formation irreversibly shifts the equilibrium toward the dimeric species, although with a slow timescale of days. As a further proof of such dynamic equilibrium, we injected a protein fraction collected from the second elution peak (thus corresponding to the monomeric species) into our chromatography column a week after the first purification. The resulting chromatogram revealed a second elution peak, at ∼10.1 ml, corresponding to the dimeric species.
DISCUSSION
Altogether, the experiments presented here demonstrate the exceptional role of the Z1Z2 titin modules in triggering the formation of a mechanically ultrastable protein dimer. Remarkably, the breakage of such strong interaction occurs at forces as high as ∼700 pN, which is the strongest rupture force measured in force spectroscopy involving the breakage of noncovalent bonds. Indeed, such a rupture force is even higher than the force required to unfold scaffoldin, taking place at ∼560 pN, which is the mechanically strongest protein measured to date with AFM (35). Because the pulling geometry in our single molecule experiments closely resembles that experienced in vivo by titin molecules in the muscle sarcomere, we speculate that the measured force value for the dimer-to-monomer transition accurately represents the mechanical resistance of an individual Z1Z2 attachment in the Z-line. Incidentally, the distribution of forces at which such a high-force peak occurs in our experiments closely coincides with the force required to break the Z1Z2 interface in the presence of telethonin (18). Remarkably, our experiments demonstrate such extremely high mechanical stability in the absence of Tcap. In light of the low fraction of measured superstable dimers (∼10%), it is plausible that telethonin enhances the probability of creating these highly mechanical resistant dimers under in vivo conditions. Despite much effort, we could not directly test such a hypothesis because we did not succeed in expressing the telethonin protein.
In light of our experimental data, a new plausible and simpler molecular mechanism for efficient titin oligomerization emerges whereby two neighboring titin chains would spontaneously dimerize. It is admittedly intriguing that the isolated Ig Z1 and Z2 tandem crystallizes with no indication of tandem formation (36). It is therefore likely that the (I27)4 adaptor promotes the dimerization process. Although a precise picture of the Z1Z2 interaction is missing, we hypothesize that a putative domain-swapping mechanism between the two Z1Z2 moieties can explain the molecular anchoring process described here. Further future molecular dynamics simulations on the Z1Z2 titin domains will certainly help elucidate the atomic details of such interaction. Indeed, domain swapping has been also described in nature as an effective mechanism to promote adhesion between partner molecules under mechanical tension. In the case of type I cadherins, adhesive binding occurs via a strand-swapped interface (6) in which anchoring occurs via the insertion of a side chain of a conserved Trp-2 residue into a complementary hydrophobic pocket in the partner molecule (37). Such an interface mediates binding between cadherins presented from opposing cells.
From the perspective of the organization of titin in the sarcomere structure, the results presented here might help shed light onto the existing riddle regarding the oligomeric state of titin through the I-band (11). In this vein, it is still an open discussion in the field how the titin molecule satisfies at the same time the three-fold symmetry of the A band while keeping the two-fold symmetry of the Z-disk (11, 38, 39). Our experiments unambiguously demonstrate the capacity of the Z1 and Z2 modules to induce dimerization to proteins that would otherwise remain in their monomeric form. In this respect, Z1Z2 functions in a similar way as the 33-amino acid α-helical coiled-coil domain GCN4, which readily assembles into dimers (40). Most interestingly, the GCN4 domain bundles together proteins of interest such as the I27 protein (32), inducing directionality to the dimeric bundle. It is possible that the Z1Z2 moiety also induces a similar quaternary helicoidal structure with improved mechanical behavior to the titin-like protein, reminiscent of a mechanical nanorope. Finally, the extraordinary high mechanical stability that we measure can serve as a platform toward the design of biomaterials with tailored mechanical stability. Indeed, many artificial scaffolds composed of naturally occurring building blocks such as extracellular matrix components rely on the tight and reversible attachment of their components (1, 41, 42). Here we propose that the Z1Z2 domains can be used in the design of biomimetic materials as an analog of the muscle anchoring system efficiently devised by nature.
Acknowledgment
We thank Prof. Mathias Gautel (King's College London, United Kingdom) for useful comments on the manuscript.
This work was supported by the Fundación IberCaja (to S. G. M. and J. A. C.), by the Fundación Martín Escudero (J. A. C.), and by National Institute of Health Grants HL66030 and HL61228 (to J. M. F.).

This article contains supplemental Figs. S1–S3.
- AFM
- atomic force microscopy
- pN
- piconewtons
- WLC
- worm-like chain
- ΔL
- contour length
- P
- persistence length.
REFERENCES
- 1. Langer R., Tirrell D. A. (2004) Designing materials for biology and medicine. Nature 428, 487–492 [DOI] [PubMed] [Google Scholar]
- 2. Shapiro L., Fannon A. M., Kwong P. D., Thompson A., Lehmann M. S., Grübel G., Legrand J. F., Als-Nielsen J., Colman D. R., Hendrickson W. A. (1995) Structural basis of cell-cell adhesion by cadherins. Nature 374, 327–337 [DOI] [PubMed] [Google Scholar]
- 3. Harrison O. J., Corps E. M., Berge T., Kilshaw P. J. (2005) The mechanism of cell adhesion by classical cadherins: the role of domain 1. J. Cell Sci. 118, 711–721 [DOI] [PubMed] [Google Scholar]
- 4. Vendome J., Posy S., Jin X., Bahna F., Ahlsen G., Shapiro L., Honig B. (2011) Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins. Nat. Struct. Mol. Biol. 18, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ciatto C., Bahna F., Zampieri N., VanSteenhouse H. C., Katsamba P. S., Ahlsen G., Harrison O. J., Brasch J., Jin X., Posy S., Vendome J., Ranscht B., Jessell T. M., Honig B., Shapiro L. (2010) T-cadherin structures reveal a novel adhesive binding mechanism. Nat. Struct. Mol. Biol. 17, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrison O. J., Bahna F., Katsamba P. S., Jin X., Brasch J., Vendome J., Ahlsen G., Carroll K. J., Price S. R., Honig B., Shapiro L. (2010) Two-step adhesive binding by classical cadherins. Nat. Struct. Mol. Biol. 17, 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Posy S., Shapiro L., Honig B. (2008) Sequence and structural determinants of strand swapping in cadherin domains: do all cadherins bind through the same adhesive interface? J. Mol. Biol. 378, 954–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lange S., Ehler E., Gautel M. (2006) From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 16, 11–18 [DOI] [PubMed] [Google Scholar]
- 9. Granzier H. L., Irving T. C. (1995) Passive tension in cardiac muscle: contribution of collagen, titin, microtubules, and intermediate filaments. Biophys. J. 68, 1027–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tskhovrebova L., Trinick J. (2003) Titin: properties and family relationships. Nat. Rev. Mol. Cell Biol. 4, 679–689 [DOI] [PubMed] [Google Scholar]
- 11. Krüger M., Linke W. A. (2011) The giant protein titin: a regulatory node that integrates myocyte signaling pathways. J. Biol. Chem. 286, 9905–9912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Labeit S., Kolmerer B. (1995) Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science 270, 293–296 [DOI] [PubMed] [Google Scholar]
- 13. Gautel M. (2011) The sarcomeric cytoskeleton: who picks up the strain? Curr. Opin. Cell Biol. 23, 39–46 [DOI] [PubMed] [Google Scholar]
- 14. Voelkel T., Linke W. A. (2011) Conformation-regulated mechanosensory control via titin domains in cardiac muscle. Pflugers Arch. 462, 143–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zou P., Gautel M., Geerlof A., Wilmanns M., Koch M. H., Svergun D. I. (2003) Solution scattering suggests cross-linking function of telethonin in the complex with titin. J. Biol. Chem. 278, 2636–2644 [DOI] [PubMed] [Google Scholar]
- 16. Zou P., Pinotsis N., Lange S., Song Y. H., Popov A., Mavridis I., Mayans O. M., Gautel M., Wilmanns M. (2006) Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature 439, 229–233 [DOI] [PubMed] [Google Scholar]
- 17. Lee E. H., Gao M., Pinotsis N., Wilmanns M., Schulten K. (2006) Mechanical strength of the titin Z1Z2-telethonin complex. Structure 14, 497–509 [DOI] [PubMed] [Google Scholar]
- 18. Bertz M., Wilmanns M., Rief M. (2009) The titin-telethonin complex is a directed, superstable molecular bond in the muscle Z-disk. Proc. Natl. Acad. Sci. U.S.A. 106, 13307–133310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Markert C. D., Meaney M. P., Voelker K. A., Grange R. W., Dalley H. W., Cann J. K., Ahmed M., Bishwokarma B., Walker S. J., Yu S. X., Brown M., Lawlor M. W., Beggs A. H., Childers M. K. (2010) Functional muscle analysis of the Tcap knockout mouse. Hum. Mol. Genet 19, 2268–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang R., Yang J., Zhu J., Xu X. (2009) Depletion of zebrafish Tcap leads to muscular dystrophy via disrupting sarcomere-membrane interaction, not sarcomere assembly. Hum. Mol. Genet 18, 4130–4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rief M., Gautel M., Oesterhelt F., Fernandez J. M., Gaub H. E. (1997) Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 276, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 22. Oberhauser A. F., Marszalek P. E., Erickson H. P., Fernandez J. M. (1998) The molecular elasticity of the extracellular matrix protein tenascin. Nature 393, 181–185 [DOI] [PubMed] [Google Scholar]
- 23. Oberhauser A. F., Badilla-Fernandez C., Carrion-Vazquez M., Fernandez J. M. (2002) The mechanical hierarchies of fibronectin observed with single-molecule AFM. J. Mol. Biol. 319, 433–447 [DOI] [PubMed] [Google Scholar]
- 24. del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009) Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kellermayer M. S., Smith S. B., Granzier H. L., Bustamante C. (1997) Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science 276, 1112–1116 [DOI] [PubMed] [Google Scholar]
- 26. Li H., Linke W. A., Oberhauser A. F., Carrion-Vazquez M., Kerkvliet J. G., Lu H., Marszalek P. E., Fernandez J. M. (2002) Reverse engineering of the giant muscle protein titin. Nature 418, 998–1002 [DOI] [PubMed] [Google Scholar]
- 27. Li H., Oberhauser A. F., Redick S. D., Carrion-Vazquez M., Erickson H. P., Fernandez J. M. (2001) Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. U.S.A. 98, 10682–10686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carrion-Vazquez M., Oberhauser A. F., Fowler S. B., Marszalek P. E., Broedel S. E., Clarke J., Fernandez J. M. (1999) Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. U.S.A. 96, 3694–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garcia-Manyes S., Brujić J., Badilla C. L., Fernández J. M. (2007) Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin. Biophys. J. 93, 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bustamante C., Marko J. F., Siggia E. D., Smith S. (1994) Entropic elasticity of lambda-phage DNA. Science 265, 1599–1600 [DOI] [PubMed] [Google Scholar]
- 31. Hagel L. (2001) Gel filtration chromatography. Curr. Protoc. Protein Sci. Chapter 8, Unit 8.3 [DOI] [PubMed] [Google Scholar]
- 32. Sarkar A., Caamano S., Fernandez J. M. (2007) The mechanical fingerprint of a parallel polyprotein dimer. Biophys. J. 92, L36–L38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kellermayer M. S., Bustamante C., Granzier H. L. (2003) Mechanics and structure of titin oligomers explored with atomic force microscopy. Biochim. Biophys. Acta 1604, 105–114 [DOI] [PubMed] [Google Scholar]
- 34. Carrion-Vazquez M., Li H., Lu H., Marszalek P. E., Oberhauser A. F., Fernandez J. M. (2003) The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 10, 738–743 [DOI] [PubMed] [Google Scholar]
- 35. Valbuena A., Oroz J., Hervás R., Vera A. M., Rodríguez D., Menéndez M., Sulkowska J. I., Cieplak M., Carrión-Vázquez M. (2009) On the remarkable mechanostability of scaffoldins and the mechanical clamp motif. Proc. Natl. Acad. Sci. U.S.A. 106, 13791–13796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marino M., Zou P., Svergun D., Garcia P., Edlich C., Simon B., Wilmanns M., Muhle-Goll C., Mayans O. (2006) The Ig doublet Z1Z2: a model system for the hybrid analysis of conformational dynamics in Ig tandems from titin. Structure 14, 1437–1447 [DOI] [PubMed] [Google Scholar]
- 37. Patel S. D., Ciatto C., Chen C. P., Bahna F., Rajebhosale M., Arkus N., Schieren I., Jessell T. M., Honig B., Price S. R., Shapiro L. (2006) Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell 124, 1255–1268 [DOI] [PubMed] [Google Scholar]
- 38. Liversage A. D., Holmes D., Knight P. J., Tskhovrebova L., Trinick J. (2001) Titin and the sarcomere symmetry paradox. J. Mol. Biol. 305, 401–409 [DOI] [PubMed] [Google Scholar]
- 39. Tskhovrebova L., Trinick J. (2010) Roles of titin in the structure and elasticity of the sarcomere. J. Biomed. Biotechnol. 2010, 612482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harbury P. B., Zhang T., Kim P. S., Alber T. (1993) A switch between two-, three-, and four-stranded coiled-coils in GCN4 leucine zipper mutants. Science 262, 1401–1407 [DOI] [PubMed] [Google Scholar]
- 41. Lv S., Dudek D. M., Cao Y., Balamurali M. M., Gosline J., Li H. (2010) Designed biomaterials to mimic the mechanical properties of muscles. Nature 465, 69–73 [DOI] [PubMed] [Google Scholar]
- 42. Cao Y., Li H. (2008) Engineered elastomeric proteins with dual elasticity can be controlled by a molecular regulator. Nat. Nanotechnol. 3, 512–516 [DOI] [PubMed] [Google Scholar]