Background: Gene expression is regulated by DNA elements that often lie far apart along genomic sequences.
Results: Novel computations and experiments provide new structural insights into long-range communication on chromatin.
Conclusion: Efficient long-range association of transcriptional elements requires intact tails on the core histones.
Significance: The understanding of action-at-a-distance in three dimensions helps to connect nucleosome structure/positioning to chromatin dynamics and gene regulation.
Keywords: Chromatin, Chromatin Structure, Computer Modeling, Gene Regulation, Histones, Nucleosome, Action-at-a-Distance, Enhancer-Promoter Communication, Histone Tails, Looping
Abstract
Action across long distances on chromatin is a hallmark of eukaryotic transcriptional regulation. Although chromatin structure per se can support long-range interactions, the mechanisms of efficient communication between widely spaced DNA modules in chromatin remain a mystery. The molecular simulations described herein suggest that transient binary internucleosomal interactions can mediate distant communication in chromatin. Electrostatic interactions between the N-terminal tails of the core histones and DNA enhance the computed probability of juxtaposition of sites that lie far apart along the DNA sequence. Experimental analysis of the rates of communication in chromatin constructs confirms that long-distance communication occurs efficiently and independently of distance on tail-containing, but not on tailless, chromatin. Taken together, our data suggest that internucleosomal interactions involving the histone tails are essential for highly efficient, long-range communication between regulatory elements and their targets in eukaryotic genomes.
Introduction
The action of protein molecules over large distances on DNA requires special facilitating mechanisms. Indeed, sequences separated by more than a thousand base pairs (1 kb) along linear DNA do not communicate efficiently in vitro (1, 2). Prokaryotic proteins bound to more closely spaced regulatory elements can take advantage of the natural looping properties of DNA in forming the long-range contacts needed to control gene expression. The protein-mediated DNA loops implicated in the expression of eukaryotic genes, however, are much longer than the closed (∼0.5 kb) DNA structures formed most easily in solution (3–5). Most eukaryotic genes are regulated by short DNA sequences called transcriptional enhancers that, after binding of proteins, can activate transcription over distances up to hundreds of kb. The action of these enhancers involves the direct association of the proteins bound at the enhancer site and at its target (promoter) site, with accompanying formation of a large loop of the intervening chromatin-covered DNA (6–8).
Although both kinetic and equilibrium aspects of enhancer-promoter communication over long distances on histone-free DNA have been extensively investigated (3, 9–12), the mechanisms of communication in chromatin remain poorly understood. New experimental approaches that yield quantitative data on the rate of enhancer-promoter communication on nucleosome-bound DNA constructs in vitro (13, 14) provide unique information for deciphering the mechanisms of eukaryotic gene expression in chromatin. The levels of RNA detected upon association of regulatory proteins that are bound to the ends of these constructs reveal surprising new information about the intervening DNA. In particular, the presence of nucleosomes along the DNA strongly facilitates distant communication between the bacterial nitrogen regulatory protein C (NtrC)3 and the RNA polymerase-σ54 assembly (a convenient prokaryotic system for monitoring long-range association) and suggests that the observed enhancement of gene expression is more than simple DNA compaction (15).
Understanding long-range communication in chromatin requires knowledge of the contributions of the nucleosomes and the interspersed DNA linkers to the enhancer-promoter search. Early molecular simulations (16, 17) focused on a few of the many facets of the complex protein-DNA assembly that determine the degree of chromatin compaction, such as the interplay between the length of linker DNA and the entry-exit angle of the DNA wrapped on the histone octamer. More recent studies (18–20) have begun to look at the effects of other structural features, such as the histone tails (effective electrostatic charge, tail modifications, etc.), on the equilibrium configuration and dynamics of short fragments of chromatin. None of the aforementioned work has looked at the contribution of chromatin structure to long-range enhancer-promoter interactions or explored the chromatin-folding problem from a multiscale perspective.
In this study, to identify the features of chromatin that are important for long-range communication, we have performed Monte Carlo simulations of different coarse-grained representations of chromatin and explored the relative likelihood of enhancer-promoter contacts in nucleosome-bound and protein-free DNA chains. Our computations suggest that transient electrostatic internucleosomal interactions mediated by the N-terminal tails of the core histones could strongly facilitate enhancer-promoter communication between widely separated sites on DNA and contribute to enhancer action. Our measurements of enhancer-promoter communication on physiologically relevant, saturated arrays of regularly spaced and precisely positioned nucleosomes, obtained with the techniques noted above, corroborate the predicted looping propensities. The fully saturated arrays support highly efficient, enhancer-promoter communication over DNA distances from 0.7 to at least 4.5 kb and further show that the core histone N-terminal tails become progressively more important as the distance between elements is increased. Taken together, the data suggest that transient, internucleosomal electrostatic interactions mediated by the histone tails give rise to highly efficient, long-range communication between regulatory elements and their targets in eukaryotic chromatin.
EXPERIMENTAL PROCEDURES
Bead-spring Model
In the bead-spring model of chromatin, the spring is modeled by a finitely extensible nonlinear elastic potential, which captures the resistance of chromatin to stretching. An energy cost to bending is introduced to capture the bending rigidity of chromatin, and phantom beads are introduced so that the springs and beads are self-avoiding and the model polymer is non-self-intersecting.
Modeling the screened electrostatic interaction between nucleosomes requires some experimentation. We argue that the histone tails, because of their flexibility, are available for temporary electrostatic interactions with nucleosomes. The possibility of nucleosome-nucleosome interaction has been raised in this and many other studies (21–23). Such interactions have been detected, and participating molecular surfaces have been identified experimentally (24, 25). In this picture, the histone tails are either extended and available for electrostatic interaction with other nucleosomes or wrapped on the nucleosome surface, and unable to mediate interactions with other nucleosomes. We have simplified this picture further and introduced a single effective tail at each nucleosome that is in two discrete states, available or unavailable, that it switches between stochastically.
We ignored the orientational dependence of nucleosomal interactions, which should not matter in a coarse-grained description. The screened attractive potential between beads available for bonding is a Gaussian well potential, which is harmonic in bead separation within the screening length and assigned a depth (attained when beads are touching) of a few kBT, where kB is the Boltzmann constant, and T is the temperature in Kelvin. The interaction state of a histone tail was modeled by an internal variable, which is either available or unavailable. This internal variable encountered a uniform potential barrier to flipping. We tuned the height of this barrier and the depth of the potential well for comparisons in our simulations. We simulated a ring of 200 beads using standard Metropolis-Monte Carlo methods. All beads were identical, and the data generated were the set of temporary bonds formed between beads at fixed sampling time. We collected the order of 5 × 106 accepted Monte Carlo steps for each bead, where each step is 5% of the bead size.
Mesoscale Chromatin Model
Our mesoscale treatment of chromatin represents DNA in terms of the set of rigid-body base-pair-step parameters that specify the orientation and displacement of successive base pairs. These details, excluded in earlier models of fluctuating chromatin fibers (17, 18), are needed to keep precise account of the spatial disposition of interacting proteins. The protein-free linker DNA is governed by an ideal elastic potential that allows for deformations in local structure consistent with the solution properties of DNA (26), and the nucleosomes are treated as rigid bodies. The atoms of proteins are stored in the frames of the nucleosomal base pairs and thereby included in the model (27). Each nucleosome also carries a superhelical reference frame, which is used in the detection of excluded volume and the characterization of multinucleosomal arrangements. The enhancer-binding protein NtrC was represented as a cylinder (12.4 nm diameter × 4 nm high), and the RNA polymerase-σ54 assembly, on the promoter, was represented as a prolate ellipsoid (15 × 11 × 11.5 nm) consistent with the observed dimensions (28, 29) (supplemental Fig. S1).
The electrostatic interactions that govern nucleosomal formation and interaction (30–32) were treated approximately in terms of charge clusters identified on the basis of the spatial locations and chemical identities of protein and DNA atoms in the core-particle structure. Our treatment ignored the charge-compensated cationic and anionic residues in the protein interior and placed the negative charges on DNA into groups comparable in size with those identified for protein side groups (supplemental Fig. S2). The positions of charges on DNA and bound nucleosomes thus varied with change in linker conformation. The charges on protein also moved in accordance with a Gaussian distribution based on the positions of the amino acid atoms comprising the clusters. These small moves very crudely mimicks the dynamics of the histone tails, which adopt fairly defined structures in solution (33, 34). The model thus included structural features more realistic than those in typical physics-based models (17) but simpler and easier to decipher and control at the local level than those fitted to all-atom treatments of electrostatic properties (18).
We simulated the chromatin system, given its complexity, in multiple stages (see supplemental Methods). The dense packing of nucleosomes on the system made the computations challenging in that internucleosomal contacts often trap the system in small, deep energy wells. Samples of 106-108 configurations were generated to access the available configuration space.
We computed the distances and angles that specify the relative spatial disposition of the proteins at the ends of the simulated chains and a loop-closure probability term analogous to the classic Jacobson-Stockmayer cyclization factor (35). The latter quantity depends upon the fraction of configurations that meet the selected chain-closure criteria (27), here the probability that the distance between the bound proteins terminated in the desired range.
Plasmids, Proteins, and Chromatin Assembly
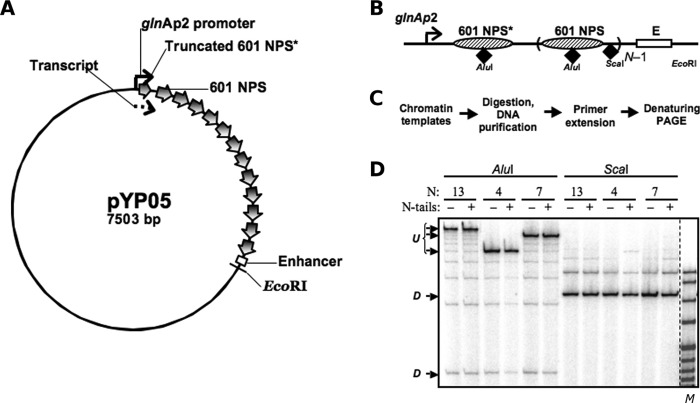
Plasmids were constructed with the approaches given in supplemental Table S1. All templates for in vitro transcription used in this work are derivatives of the pLY10 plasmid, which has been described before (2) and used in previous studies (15). The pYP05 plasmid (Fig. 1A) containing an NtrC-dependent enhancer and a glnAp2 promoter separated by a spacer DNA of 0.7–4.5 kb, including specific numbers of strong 601 nucleosome-positioning sequences, was cloned from pLY10 in several consecutive steps. In the first nucleosome-positioning sequence (NPS), 35 bp were truncated from the promoter-distal end and replaced by the bacteriophage T7 transcription terminator site, thereby limiting the length of the transcript by 176 nucleotides (Fig. 1B). The lengths of the spacers between all NPSs within the pYP05 plasmid were the same (29 bp). The plasmids were constructed using PCR amplification of the desired fragments followed by restriction digestion of the fragments and the target vectors and ligation.
FIGURE 1.
Nucleosome positions and occupancies on saturated 601 arrays. A, schematic diagram of the pYP05 template. The 601 nucleosome-positioning sequences are indicated by wide arrows. The glnAp2 promoter and the enhancer are positioned over 0.7–4.5-kb distances from each other (2.5 kb in the image). The first 601 nucleosome-positioning sequence (601 NPS*) is truncated by 35 bp from the 3′ end and overlaps with the 176-nucleotide transcript (dashed arrow). B, schematic map of the arrays containing different numbers N of 601 nucleosome-positioning sequences (NPS) between the glnAp2 promoter and the enhancer (E). AluI sites for restriction endonucleases are located close to the middle of the positioning sequences, and ScaI sites are in the middle of the 30-bp DNA linkers. C, experimental approach: a restriction enzyme sensitivity assay with primer extension. Chromatin was assembled on a negatively supercoiled pYP05 plasmid and digested with an excess of restriction enzymes AluI or ScaI. Purified DNA was next digested with EcoRI restriction endonuclease to create one DNA end and then subjected to primer extension using a promoter-proximal end-labeled primer. D, characterization of the 601177×13 saturated nucleosomal arrays assembled on pYP05 using the restriction digestion sensitivity assay. Analysis of primer end-labeled DNA by denaturing PAGE was performed. Undigested (U) and fully digested (D) DNA fragments are indicated. Note that the increase in the chromatin assembly level results in almost quantitative protection of the templates from AluI, but not ScaI restriction enzyme. M denotes the MspI digest of pBR322 plasmid.
Established methods were used to purify all transcriptional machinery proteins (12), to prepare H1/H5-depleted chicken erythrocyte donor chromatin (36), to prepare tailless donor chromatin (by digestion of long H1/H5-depleted chromatin with trypsin) (37), and to reconstitute chromatin on supercoiled templates in vitro (36). Nucleosome positioning within the assembled arrays was verified using a restriction enzyme sensitivity assay. After chromatin assembly, the samples were digested with an excess of restriction enzymes, either AluI or ScaI. Purified DNA was then digested with EcoRI restriction endonuclease to set one DNA end and subjected to primer extension with Taq DNA polymerase using a radioactively labeled primer localized immediately upstream of the promoter.
Transcription
Conditions for in vitro transcription were optimized for maximal utilization of the chromatin templates. Transcription was conducted as described previously (13). In short, all templates were linearized, the closed initiation complexes were formed, and the enhancer-binding protein (NtrC) was prebound to DNA in the absence of ATP. Next, ATP was added for a short time (1 min) to initiate enhancer-promoter communication and formation of the open initiation complex, which is competent to begin elongation. A mixture of all four NTPs (containing [α-32P]GTP and heparin) was then added to the reaction to start transcript elongation and to limit it to a single round. The reaction was continued for 15 min, and labeled RNA was purified and analyzed by denaturing PAGE. The gel was dried, exposed, and scanned on a Cyclone imager (PerkinElmer Life Sciences). The data were analyzed using the OptiQuant software. Further details will be presented elsewhere.
RESULTS
Internucleosomal Interactions Increase Long-range Chromatin Looping
Many models of chromatin, incorporating varying degrees of structural detail, have been explored for different goals (23, 38–41). Here we focused on the effects of electrostatic interactions between nucleosomes, likely mediated by flexible histone tails, in enhancer-promoter communication. The existing models are insufficient for our purposes as we explain below. We initially modeled chromatin minimally as a bead-spring polymer, where the beads represent a few nucleosomes and the spring connecting them represents the linker DNA and overall flexibility of packed nucleosomes. We also introduced a discrete variable for each bead, which indicates whether the flexible histone tails are available for interaction with other beads. Two nearby beads that are available for interaction could form a temporary “bond.” Two key parameters of this model are the free energy barrier to the tails becoming available and the free energy loss when the temporary bond is broken.
The key quantity of interest is the contact probability distribution as a function of the separation of beads. For a self-avoiding, semiflexible polymer, this distribution exhibits a maximum for separations on the order of the persistence length. Below the persistence length, contacts are unlikely because of the cost of bending the polymer, and above this reference point, the contact probability falls off as a power law of the separation. This fall-off stems from the entropic cost of bringing two monomers, separated along the polymer, in proximity in three-dimensional space. We observed qualitative departures from this functional form induced by the internucleosomal interactions within the polymer.
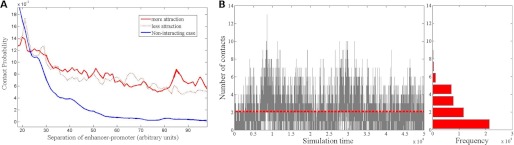
At intermediate interaction strength (1.25 or 1.5 kBT, Fig. 2A), the probability distribution displays a distinct profile with a much slower fall-off at longer separation of beads when compared with that of the noninteracting case. This flattened tail of the distribution implies enhancement of long-range communication between nucleosomes, mediated by histone-tail interactions, and is not simply an overall enhancement of contact frequency. The time evolution of the number of contacts (Fig. 2B) shows that the system intermittently goes through states with a relatively high number of contacts. We conclude that the interactions lead to a novel mechanism of long-range action, which avoids a collapsed state but enhances long-range interactions.
FIGURE 2.
Internucleosomal interactions increase probability of long-range chromatin looping through intermittent high-contact configurations. A, probabilities of contact of varying strength as a function of site separation along chromatin modeled as a ring of beads subject to elastic deformations and transient electrostatic interactions. The typical contact probability is peaked around a separation determined by the choice of persistence length, and that peak is excluded from this plot. As expected, the tail of the distribution falls off as a power law for the noninteracting case (blue line), but has a much slower fall-off for our interaction model (dotted and solid red lines correspond to two different interaction strengths). B, number of contacts against simulation time (arbitrary units). The timeline plot (simulation steps) shows the number of transient bonds formed in a window of a typical run. Notice the occasional peaks in the number of contacts, where the value fluctuates significantly above the mean. The side panel shows the frequencies of the contacts.
Histone Tails Introduce Intrinsic Compaction and Curvature That Enhance Long-range Chromatin Interactions
A closer look at chromatin structure at the level of specifically positioned nucleosomes and fluctuating linker DNA segments shows how the presence of the histone tails enhances the likelihood of long-range intramolecular contacts when compared with that in both free DNA of the same chain length and chromatin fragments modeled with tailless nucleosomes in the same positions. We simulated saturated arrays of regularly spaced nucleosomes with spacings known to form higher-order chromatin structures in vitro (42, 43), i.e. 147-bp nucleosomal DNA segments separated by 30-bp linkers. We treated DNA at the base-pair level and incorporated, in the bound nucleosomes, the precise DNA pathway and the positions of charged amino acid atoms found in the currently best-resolved core-particle structure (44). We let the elastic properties of the linker DNA, the steric overlap of nucleosome cylinders, and the electrostatic interactions of the DNA polyanion and histone polycations guide the spatial arrangements of the mesoscale assembly (see “Experimental Procedures”).
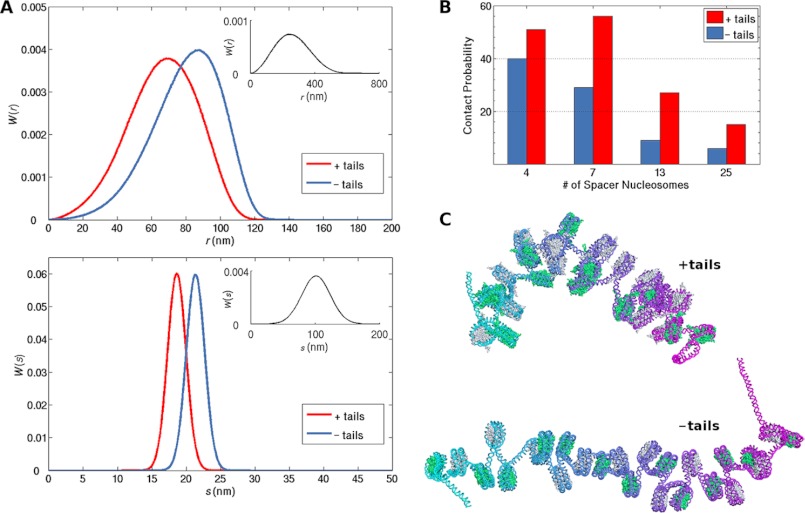
The interactions of the histone tails have a profound effect on the configurations of the simulated chains. The charges on the surfaces of modeled intact nucleosomes shift the distributions of both the end-to-end distance and the radius of gyration toward lower values than those of the corresponding DNA with bound tailless nucleosomes (Fig. 3A, supplemental Fig. S3). Both nucleosome-bound systems are globally stiffer and substantially more compact than free DNA and thus more prone to looping mediated by other proteins. For example, the simulated likelihood of close contact between a cylindrically shaped NtrC activator protein complex located on an enhancer site at the 3′ end of the DNA and a prolate ellipsoidal RNA polymerase-σ54 complex bound at a promoter site on the 5′ end is more than an order of magnitude greater for duplexes with 4–25 intervening nucleosomes than for ideal, protein-free DNA of the same chain length (Fig. 3B). Moreover, the computed enhancement of looping for fragments with nucleosomes that contain intact histone tails is 1.3–3.0 times greater than that for the corresponding tailless arrays. Remarkably, the positive impact of the histone tails on the efficiency of looping in chromatin becomes larger as the size of the loop increases (Fig. 3B). In other words, in the presence of histone tails, the likelihood of chromatin looping is less distance-dependent, the same result found at longer chain lengths with the bead-spring polymer model. The combined data suggest that the histone tails are essential for distant communication within a chromatin fiber.
FIGURE 3.
Histone tails induce a natural compaction and curvature in chromatin that enhance long-range intramolecular contacts. A, simulated distributions of the end-to-end distances r (top) and radii of gyration s (bottom) of DNA chains containing 13 nucleosomes, evenly spaced by 30-bp DNA linkers and with either fully intact (native) or tailless histones (red and blue curves, respectively) when compared with those of protein-free DNA chains of the same length (black curves in insets). See supplemental Fig. S3 for the corresponding distributions with different numbers of nucleosomes. B, estimated likelihood of close (≤15 nm) contacts between NtrC and RNA polymerase-σ54 complexes bound respectively at the 3′ and 5′ ends of DNA chains with 4, 7, 13, or 25 intervening nucleosomes. Red and blue bars: native and tailless histones, respectively. C, representative configurations of chromosomal fragments bearing 25 intact (top) or tailless (bottom) nucleosomes. The DNA is represented by a color-coded backbone (progression from 5′ to 3′ ends of the DNA is depicted by the aqua to violet to magenta color sequence), and the histone cores are represented by alternating green and silver space-filled atomic representations. Images are rendered with PyMOL. See Figs. S1 and S4 for other images and curvature values.
The estimated enhancement of loop formation in nucleosome-bound versus free DNA can be altered if constraints are placed on both the distance and the orientation of the terminal proteins. Thus the precise enhancement level depends upon the assumed spatial arrangement of the NtrC-polymerase-σ54 (enhancer-promoter) complex. For example, an arrangement that orients the cylindrical axis of the NtrC complex roughly parallel to the long axis of the polymerase-σ54 ellipsoid reduces the likelihood of complex formation by ∼80% in chains with 13 nucleosomes when compared with free DNA, i.e. reduction of the loop-closure enhancement factors plotted for chains with intact and tailless nucleosomes in Fig. 3B by ∼80%. Further restrictions on the “twisting” of the polymerase ellipsoid with respect to the cylindrical axis of the NtrC reduce the simulated looping propensities even further.
The detailed depiction of individual nucleosomes and linker base pairs incorporated in the mesoscale representation of chromosomal DNA reveals an intrinsic compaction and curvature in the fragments bearing intact nucleosomes when compared with those that bind tailless nucleosomes (Fig. 3C, supplemental Figs. S1 and S4). This difference in chain configuration underlies the increased levels of end-to-end contact detected in the simulations of nucleosomal arrays.
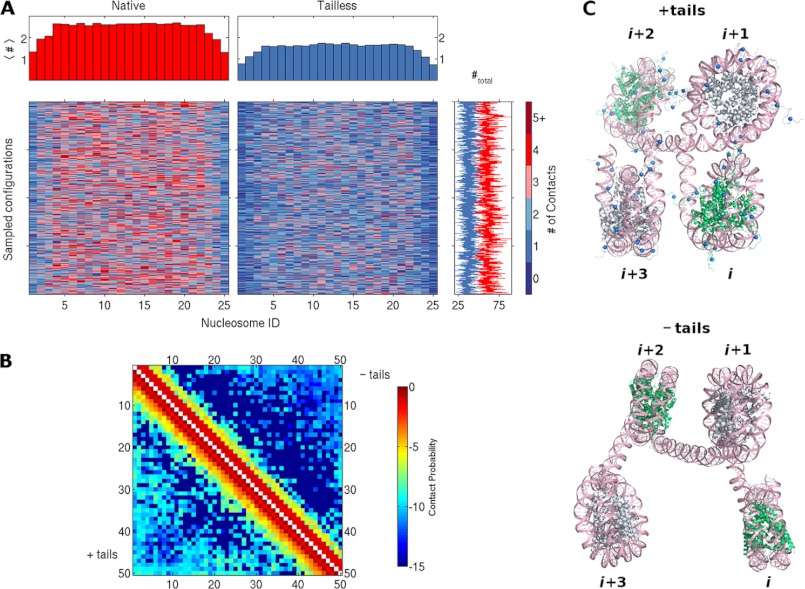
The changes in average global structure reflect slight differences in the local arrangements of nucleosomes, which arise, in turn, from the modeled interactions of the histone tails. These interactions affect both the number and the type of intramolecular contacts within the simulated chromatin fragments. For example, there are more close contacts of nucleosomes in the modeled chromatin systems with histone tails than in those without tails (Fig. 4A, supplemental Fig. S5). There are also more such contacts in the middle than at the ends of the nucleosome-bearing DNA. The three nucleosomes at either end of the chains make substantially fewer contacts on average with other nucleosomes than those in the middle. The central tail-bearing nucleosomes make as many as eight and on average 2.6 contacts with other nucleosomes in the simulated structures. These contacts tend to occur at close range, involving the three sequentially closest nucleosomes, i.e. nucleosomes in positions ±1, ±2, or ±3 with respect to that of the nucleosome of interest (Fig. 4B). Moreover, there is a much greater likelihood that a nucleosome in these chains will contact a nucleosome at position ±2, the nucleosome known to be spatially closest in a perfect zigzag model of chromatin, than at positions ±1 or ±3 (supplemental Fig. S6). By contrast, the nucleosomes in the middle of the chains that bind tailless histones contact 1.7 other nucleosomes on average and associate with sequentially close nucleosomes with slightly different propensities. These subtle changes in number and type of contacts lead to the differences in mean internucleosomal structure that give rise to the overall compaction and curvature noted above. For example, both the angle between the cylindrical axes of nearby nucleosomes and the displacement of the centers of the nucleosomes tend to be greater when the tails are removed (Fig. 4C). The few longer-range contacts captured in the simulations, such as the (n, n + 24) contact illustrated on a 50-nucleosome construct in Fig. 5, occur in concert with numerous shorter-range contacts and appreciable bending of the intervening DNA. The formation of such interactions is much less frequent in chains with tailless histones than in those with intact nucleosomes (Fig. 4B).
FIGURE 4.
Histone tails affect contacts and spatial arrangements of nucleosomes in simulated chromatin fragments. A, color-coded mosaic of the numbers and locations of close (≤15 nm) internucleosomal contacts detected in simulated chains bearing 25 evenly spaced nucleosomes with intact (left) or tailless (right) histones. The positions of nucleosomes are denoted by the numbers at the bottom of the images, and the number of contacts (#) are denoted by the scale on the right. The total number of contacts per simulated configuration, plotted to the right of each row, fluctuates around an average value of 60 for intact (red line) and 38 for tailless (blue line) nucleosomes. See supplemental Fig. S5 for mosaics of 4-, 7-, and 13-mers. B, color-coded map of the frequency of close contacts between all pairs of nucleosomes in simulated chains binding 50 regularly spaced nucleosomes. The color coding is based on a logarithmic scale to pinpoint the locations of rare, longer-range interactions, e.g. −5 denotes a frequency of 10−5. Note the distinctly different frequencies (colors) for the intact (+tails) nucleosomes plotted below the diagonal versus those for the tailless (−tails) nucleosomes plotted above the diagonal. See supplemental Fig. S6 for frequency counts. C, representative configurations of four-nucleosome chromatin fragments in simulated chains with intact (top) and tailless (bottom) histones. The DNA is shown in pink, with the phosphorus atoms on complementary strands depicted by ribbons and the bases drawn as stick figures. The histones of successive nucleosomes are shown alternately in green and silver; the blue spheres depict charge cluster sites on the histone tails (see supplemental Fig. S2).
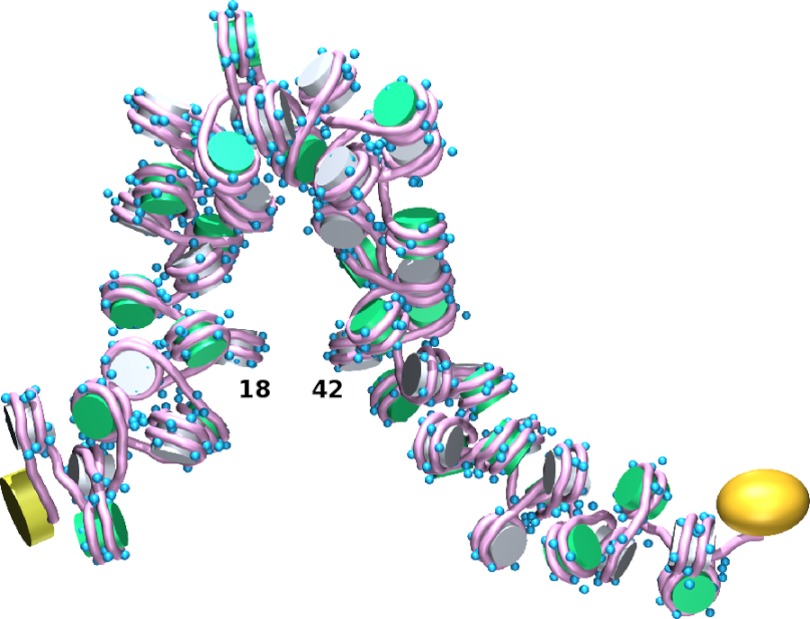
FIGURE 5.
Long-range contacts of nucleosome-decorated DNA occur in concert with numerous short-range interactions and appreciable bending of intervening chain. Simulated hairpin configuration of chromosomal DNA bearing 50 intact, evenly spaced nucleosomes is shown. In this example, one pair of nucleosomes, at positions 18 and 42, comes within 21 nm of each other, and almost every nucleosome pair separated by 1–3 intervening nucleosomes lies within 20 nm of each other. The NtrC assembly bound to the enhancer at the 3′ end of the chain is represented by a yellow cylinder, and the polymerase-σ54 complex bound to the promoter at the 5′ end is represented by a gold prolate ellipsoid. The blue spheres depict charged clusters on the histone tails. Images were created with MathematicaTM and rendered with Blender.
Histone N-tails Mediate Long-range Enhancer-Promoter Communication in Vitro
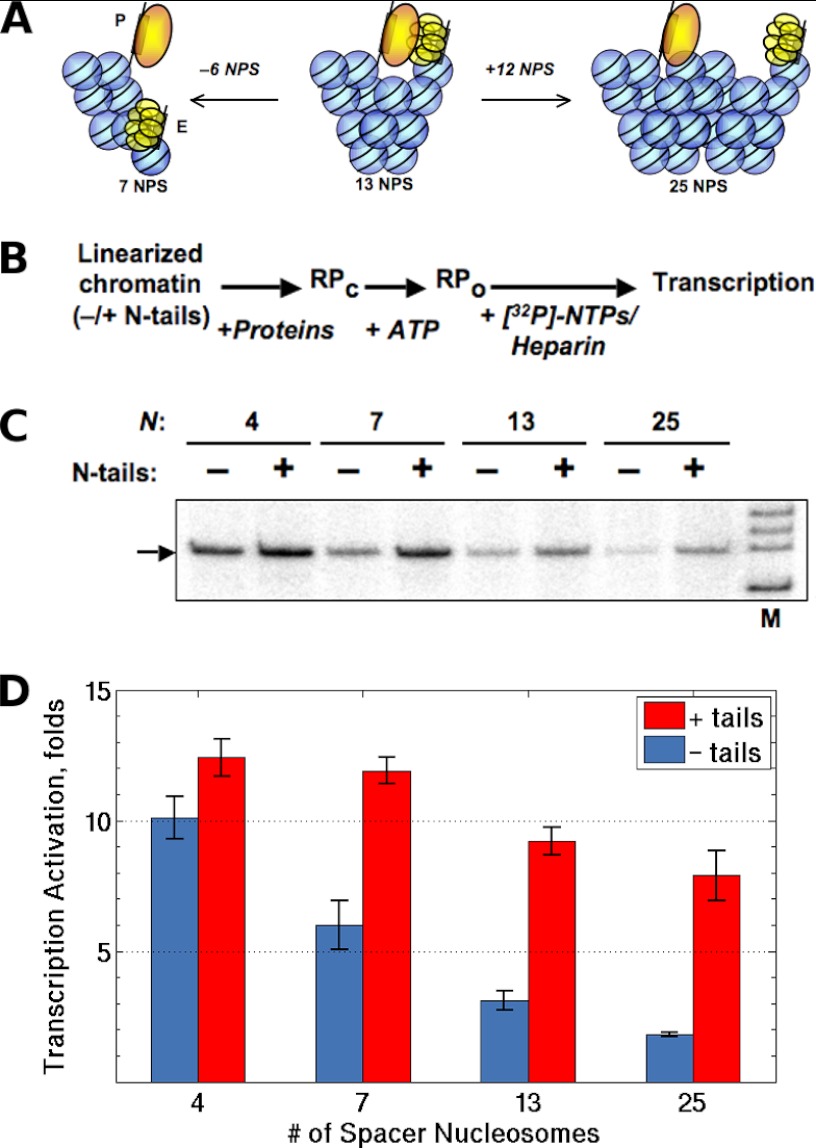
To evaluate the primary prediction of the computational studies (the effect of histone tails on distant communication in chromatin), we constructed a series of 601177×N plasmid derivatives with different numbers of 601 nucleosome-positioning sequences (45) between enhancer and promoter sites (n = 4, 7, 13, or 25) (Fig. 1, A and B). Nucleosomal arrays were reconstituted by transfer methods by use of continuous dialysis from 1 m NaCl (14) with intact or tailless core histones (Fig. 1C). Each 177-bp long nucleosomal repeat contained 147 bp of nucleosome-covered DNA and a 30-bp spacer between successive positioning sites. These constructs place the promoter and enhancer on the same or opposite sides of a fiber with the 6- and 12-nucleosome repeats per turn deduced from chemical cross-linking and low-resolution structural studies (46, 47) (Fig. 6A).
FIGURE 6.
Histone N-tails mediate long-range enhancer-promoter communication. A, relative positions of the enhancer (E) and promoter (P) on nucleosomal arrays of different lengths. The structure of the fiber is represented by a two-start, zigzag model with 12 nucleosomes per turn, roughly to scale. If the chromatin fiber were rigid, shortening (left) or lengthening (right) of the 13-mer nucleosomal array would move the enhancer and promoter apart and result in a decrease in the efficiency of enhancer-promoter communication in chromatin. B, the primary experimental approach for the transcriptional analysis of the rate of enhancer-promoter communication in chromatin (61). Closed initiation complexes (RPc) were formed using intact or tailless nucleosomal arrays. The closed initiation complexes were converted to open complexes (RPo) after the addition of ATP for 1 min followed by the addition of NTPs and heparin to limit transcription to a single round and to remove the nucleosomal barrier to transcript elongation. C, transcription of the linear saturated arrays containing different numbers of intact or tailless positioned nucleosomes. Analysis of labeled specific transcripts (arrow) transcripts by denaturing PAGE was performed. D, quantitative analysis of the specific transcripts formed on templates containing different numbers of nucleosomes between the promoter and enhancer. Gels, similar to the ones shown in panel C, were quantified using a Cyclone Imager (Perkin Elmer). Error bars correspond to the S.D. of three independent experiments. The amounts of transcripts produced from the templates are directly proportional to the rate of enhancer-promoter communication (12).
We determined the rate of enhancer-promoter communication in the 601177×N nucleosomal arrays using the recently developed approach, outlined in Fig. 1C, which allows for the assembly of saturated nucleosomal arrays and quantitative characterization of the resulting chromatin system (13, 14). The DNA plasmids (Fig. 1B) contain, at one end, the NtrC-dependent enhancer, which can strongly activate the glnAp2 promoter placed at the other end (2, 12, 48).
Restriction digestion sensitivity assays (Fig. 1, B–D) show that the chromatin samples contain saturated nucleosomal arrays. The AluI restriction sites positioned within nucleosomes are fully protected from digestion by the core histones, whereas the ScaI sites positioned on the linker DNA are fully sensitive to restriction enzymes (Fig. 1, B–D). A different version of the restriction enzyme sensitivity assay shows that the majority of nucleosomes assemble specifically on the high-affinity nucleosome-positioning sites, and not at random along the plasmid template nor on the enhancer and/or promoter (14). Taken together, the data suggest that saturated, defined nucleosomal arrays that contain specifically positioned nucleosomes are formed during assembly. Notably, nucleosome-saturated 601177×N arrays spontaneously form chromatin fibers in buffers similar in composition to the transcription buffer used in this study (42, 43).
Evaluation of the relative rates of enhancer-promoter communication entailed linearization of the chromatin templates followed by a single-round transcription assay (Fig. 6, B and C). We previously found that enhancer-promoter communication is the rate-limiting step in the entire transcription reaction and that the yield of transcripts is directly proportional to the level of communication on both DNA and chromatin templates (2, 15). Furthermore, in the case of histone-free DNA, the experimental results quantitatively mirrored the expected end-to-end properties of flexible DNA chains (12).
The assembly of saturated chromatin arrays of different lengths with intact core histones resulted in strong and comparable activation of transcription on all 601177×N templates (Fig. 6, C and D). Thus chromatin structure can support efficient enhancer-promoter communication over distances from 0.7 to at least 4.5 kb in vitro. As expected, the extent of activation by the assembly of intact chromatin became progressively lower as the length of the DNA spacer was increased, possibly due to the progressively increasing average spatial separation of the enhancer and promoter. The magnitude of activation between 0.7 and 4.5 kb, however, differed by only ∼1.5-fold, an indication that intact chromatin can maintain enhancer-promoter communication that is almost distance-independent in the constructs considered here. If the chromatin constructs were rigid fibers with the assumed nucleosomal repeat, the stimulatory effect of chromatin assembly on enhancer-dependent transcription would strongly and periodically depend on the number of nucleosomes on the intervening DNA (Fig. 6A). No strong periodic differences in the efficiency of activation were detected (Fig. 6D). The data do not support a rigid fiber model and suggest that the constructs are intrinsically dynamic structures that can support long-range communication in chromatin.
In striking contrast to enhancer-promoter communication in intact chromatin, the level of communication in tailless chromatin strongly decreases as enhancer-promoter spacing increases (Fig. 6D). Thus the rate of enhancer-promoter communication over 0.7 kb decreased only by ∼20% after removal of histone tails, but that over 4.5 kb dropped ∼4-fold. Taken together, the data suggest (in agreement with the predictions of our computations) that: (i) histone tails are essential for enhancer-promoter communication over large distances in saturated arrays of nucleosomes that form chromatin fibers and (ii) histone tails mediate long-range enhancer-promoter communication in chromatin.
DISCUSSION
Our composite findings show that internucleosomal interactions involving the histone tails can account for long-distance regulatory communication along chromatin. Our study of the bead-spring model, which represents chromatin at a length scale corresponding to many hundreds of nucleosomes, indicates that chromatin interactions induced by histone-tail contacts give rise to transient collapsed structures that nucleate long-range communication (Fig. 2). In addition, we have developed a mesoscale model of chromatin to decipher the effects of nucleosomal fine structure and linker DNA deformability on long-range polymeric interactions. Our Monte Carlo simulations of chains decorated with 4–50 evenly spaced nucleosomes reveal a natural compaction and curvature induced by the histone tails that enhance the likelihood of intrastrand loop formation and the concomitant association of bound, widely spaced proteins (Figs. 3–5). Our in vitro experimental system allows for analysis of enhancer-promoter communication over different distances on physiologically relevant, saturated arrays of precisely positioned nucleosomes that form higher-order chromatin structures (Fig. 1). Using this system, we have shown (Fig. 6) that: (i) efficient enhancer-promoter communication in chromatin occurs over distances from 0.7 to at least 4.5 kb and only weakly depends on the assumed rotational positioning of the communicating DNA elements within the assembled fiber; (ii) the assembled fiber is a dynamic structure that supports efficient communication over a variety of enhancer-promoter distances; and (iii) histone N-terminal tails are essential for long-range communication in chromatin.
Our experiments with nucleosomal arrays of different lengths suggest that at least two types of deformation contribute to the overall mobility of our chromatin constructs. Precise alignment of the enhancer and promoter over a short distance from each other (4–7 nucleosomes) is impossible without changes in the local structure of the fiber. To account for efficient short-range communication, we propose that nucleosomes on neighboring gyres of the fiber can move relative to each other. Such nucleosome repositioning within the fiber could be accomplished by spontaneous partial uncoiling of nucleosomal DNA from the histone octamer (49). Communication over larger distances (10–25 nucleosomes) cannot occur only by this mechanism because extensive uncoiling of nucleosomal DNA is unlikely. Therefore communication over large distances must involve bending of the chromatin fiber as a whole (Fig. 5) and take advantage of the deformability of the DNA linkers and the electrostatic interactions of nucleosomes. Such features would also occur in shorter chains.
As noted above, our experimental design was based on chromatin fiber models with 6- and 12-nucleosome repeats deduced from histone cross-linking and low-resolution structural studies (42, 43). Neither model accounts for our findings. For example, if the six-nucleosome repeat applied, the construct with seven nucleosomes would have produced much higher levels of RNA than that with four nucleosomes. That is, enhancer and promoter sites separated by seven nucleosomes would lie on roughly the same face of such a fiber and thus come into more frequent contact than sites with four intervening nucleosomes, which would lie on nearly opposite faces of the fiber. On the other hand, a four-nucleosome spacer may not be sufficiently long to form an organized fiber. Similarly, if the fiber model with a 12-nucleosome repeat were correct, the construct with 13 nucleosomes would have produced much higher levels of RNA than a similarly organized structure with seven nucleosomes (Fig. 6A).
Determination of the nucleosome repeat of constructs like ours in solution requires the preparation and analysis of the long-range interactions on an extensive series of new templates. The monotonic decay of transcript levels found here (Figs. 3B and 6D) suggests that the enhancer and promoter lie on the same face of the examined constructs. Preliminary simulations based on the current mesoscale model reveal an oscillatory pattern of contact frequencies indicative of a structural repeat shorter than six nucleosomes. The constructs described here lie near the peaks in the predicted oscillations, and simulated arrays of intermediate lengths with the same nucleosomal spacing contribute to both peaks and valleys in the profiles. The differences in compaction between the predicted structures and the published images of similar nucleosome constructs (42, 43) may reflect the different conditions of our experiments and computations (low divalent salt) when compared with those needed to visualize compacted fibers (100 mm MgCl2 or the addition of linker histones).
Our simulations add a new perspective on the looping of chromatin not considered in previous research. The likelihood of looping chromatin into closed structures of the sizes detected in cells (∼100 nucleosomes) is often treated analytically (50, 51), and only recently has it been estimated in direct computations (52). The transient binary interactions introduced in our coarse-grained model allow for isolated long-range internucleosomal contacts not considered in previous work. Our mesoscale treatment of nucleosome-decorated DNA is consistent with previous simulations, which take explicit account of the positively charged histone proteins and the DNA polyelectrolyte backbone (20, 53). For example, the average number of internucleosomal contacts per core particle found here (Fig. 4A) is comparable with the values found in Langevin dynamics simulations of interacting charged beads (used to approximate nucleosomes) in the presence of low levels of Mg2+ (20), and the reduction in contact number found in the absence of the charged histone tails is similar to the loss of contacts determined upon reduction of bead charge. Our explicit treatment of nucleosomal and linker DNA, however, generates different distributions of nucleosome-nucleosome contacts from those reported in simulations that represent the linker by a few beads and confine the histone-bound DNA to an idealized pathway (53), i.e. a preponderance of contacts with nucleosomes at positions ±2 here (Fig. 4) versus roughly equivalent numbers of contacts at ±1 and ±2 in related simulations with short linkers (3–4 beads, each spanning ∼9 bp steps). Like many others (50, 54–56), we find that chromatin is a highly efficient communication device and a highly flexible polymer. Our computed estimates of enhancer-promoter contacts in chromatin versus free DNA, however, exceed the measured levels by roughly 4-fold (note the different scales in Fig. 3B versus Fig. 6D). As noted above, communication in chromatin could be limited by the probability of enhancer-promoter juxtaposition, such as found on relaxed and supercoiled DNA (12). This idea is consistent with data showing that the rates of communication on supercoiled DNA and chromatin are comparable (15).
Finally, our studies suggest that efficient communication over a large distance (more than 1.5 kb) critically and selectively depends on the presence of intact tails on the core histones. This observation implies that communication over a distance could constitute a separate regulated step during execution of various intranuclear DNA-targeted processes. Specifically, histone tails are subject to intensive modifications (57); many of these modifications (e.g. acetylation or phosphorylation) change the charges of histone tails, destabilize higher-order chromatin structure (e.g. histone H4 acetylation at position 16 (H4K16Ac) (58, 59)), and most likely affect chromatin dynamics. We propose that in addition to their other important functions, some histone tail modifications could affect the rate of enhancer-promoter communication, perhaps in a regulated way. In particular, H4K16Ac is associated with active chromatin (60) and could facilitate distant communication in chromatin.
Acknowledgment
We thank Tim Richmond for the plasmid containing 601177×12 nucleosome-positioning sequences.
This work was supported, in whole or in part, by National Institutes of Health Grants GM34809 (to W. K. O.) and HG03470 (to A. M. S.) and National Institutes of Health Instrumentation Award RR022375 (to A. M. S. and W. K. O.). This work was also supported by National Science Foundation Grant 7046342 and a Government of the Russian Federation (Order 220) research grant (to V. M. S.).

This article contains supplemental methods and references, Figs. S1–S6, and supplemental Table S1.
- NtrC
- nitrogen regulatory protein C
- NPS
- nucleosome-positioning sequence.
REFERENCES
- 1. Bellomy G. R., Record M. T., Jr. (1990) Stable DNA loops in vivo and in vitro: roles in gene regulation at a distance and in biophysical characterization of DNA. Prog. Nucleic Acid Res. Mol. Biol. 39, 81–128 [DOI] [PubMed] [Google Scholar]
- 2. Liu Y., Bondarenko V., Ninfa A., Studitsky V. M. (2001) DNA supercoiling allows enhancer action over a large distance. Proc. Natl. Acad. Sci. U.S.A. 98, 14883–14888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shore D., Baldwin R. L. (1983) Energetics of DNA twisting. I. Relation between twist and cyclization probability. J. Mol. Biol. 170, 957–981 [DOI] [PubMed] [Google Scholar]
- 4. Jackson V. (1990) In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry 29, 719–731 [DOI] [PubMed] [Google Scholar]
- 5. Taylor W. H., Hagerman P. J. (1990) Application of the method of phage T4 DNA ligase-catalyzed ring closure to the study of DNA structure. II. NaCl dependence of DNA flexibility and helical repeat. J. Mol. Biol. 212, 363–376 [DOI] [PubMed] [Google Scholar]
- 6. Bondarenko V. A., Liu Y. V., Jiang Y. I., Studitsky V. M. (2003) Communication over a large distance: enhancers and insulators. Biochem. Cell Biol. 81, 241–251 [DOI] [PubMed] [Google Scholar]
- 7. de Laat W., Klous P., Kooren J., Noordermeer D., Palstra R. J., Simonis M., Splinter E., Grosveld F. (2008) Three-dimensional organization of gene expression in erythroid cells. Curr. Top. Dev. Biol. 82, 117–139 [DOI] [PubMed] [Google Scholar]
- 8. Schoenfelder S., Clay I., Fraser P. (2010) The transcriptional interactome: gene expression in 3D. Curr. Opin. Genet. Dev. 20, 127–133 [DOI] [PubMed] [Google Scholar]
- 9. Vologodskii A., Cozzarelli N. R. (1996) Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys. J. 70, 2548–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang J., Schlick T., Vologodskii A. (2001) Dynamics of site juxtaposition in supercoiled DNA. Proc. Natl. Acad. Sci. U.S.A. 98, 968–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Czapla L., Swigon D., Olson W. K. (2006) Sequence-dependent effects in the cyclization of short DNA. J. Chem. Theory Comput. 2, 685–695 [DOI] [PubMed] [Google Scholar]
- 12. Polikanov Y. S., Bondarenko V. A., Tchernaenko V., Jiang Y. I., Lutter L. C., Vologodskii A., Studitsky V. M. (2007) Probability of the site juxtaposition determines the rate of protein-mediated DNA looping. Biophys. J. 93, 2726–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Polikanov Y. S., Rubtsov M. A., Studitsky V. M. (2007) Biochemical analysis of enhancer-promoter communication in chromatin. Methods 41, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Polikanov Y. S., Studitsky V. M. (2009) Analysis of distant communication on defined chromatin templates in vitro. Methods Mol. Biol. 543, 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rubtsov M. A., Polikanov Y. S., Bondarenko V. A., Wang Y. H., Studitsky V. M. (2006) Chromatin structure can strongly facilitate enhancer action over a distance. Proc. Natl. Acad. Sci. U.S.A. 103, 17690–17695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katritch V., Bustamante C., Olson W. K. (2000) Pulling chromatin fibers: computer simulations of direct physical micromanipulations. J. Mol. Biol. 295, 29–40 [DOI] [PubMed] [Google Scholar]
- 17. Langowski J. (2006) Polymer chain models of DNA and chromatin. Eur. Phys. J. E. Soft Matter 19, 241–249 [DOI] [PubMed] [Google Scholar]
- 18. Arya G., Schlick T. (2006) Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. U.S.A. 103, 16236–16241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arya G., Schlick T. (2009) A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A 113, 4045–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang Y., Lyubartsev A. P., Korolev N., Nordenskiöld L. (2009) Computer modeling reveals that modifications of the histone tail charges define salt-dependent interaction of the nucleosome core particles. Biophys. J. 96, 2082–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Angelov D., Vitolo J. M., Mutskov V., Dimitrov S., Hayes J. J. (2001) Preferential interaction of the core histone tail domains with linker DNA. Proc. Natl. Acad. Sci. U.S.A. 98, 6599–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng C., Hayes J. J. (2003) Intra- and internucleosomal protein-DNA interactions of the core histone tail domains in a model system. J. Biol. Chem. 278, 24217–24224 [DOI] [PubMed] [Google Scholar]
- 23. Mergell B., Everaers R., Schiessel H. (2004) Nucleosome interactions in chromatin: fiber stiffening and hairpin formation. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 70, 011915 [DOI] [PubMed] [Google Scholar]
- 24. Chodaparambil J. V., Barbera A. J., Lu X., Kaye K. M., Hansen J. C., Luger K. (2007) A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat. Struct. Mol. Biol. 14, 1105–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou J., Fan J. Y., Rangasamy D., Tremethick D. J. (2007) The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 14, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 26. Zheng G., Czapla L., Srinivasan A. R., Olson W. K. (2010) How stiff is DNA? Phys. Chem. Chem. Phys. 12, 1399–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Czapla L., Swigon D., Olson W. K. (2008) Effects of the nucleoid protein HU on the structure, flexibility, and ring closure properties of DNA deduced from Monte Carlo simulations. J. Mol. Biol. 382, 353–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee S. Y., De La Torre A., Yan D., Kustu S., Nixon B. T., Wemmer D. E. (2003) Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 17, 2552–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murakami K. S., Darst S. A. (2003) Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 13, 31–39 [DOI] [PubMed] [Google Scholar]
- 30. Ramakrishnan V. (1997) Histone structure and the organization of the nucleosome. Annu. Rev. Biophys. Biomol. Struct. 26, 83–112 [DOI] [PubMed] [Google Scholar]
- 31. Widom J. (2001) Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 34, 269–324 [DOI] [PubMed] [Google Scholar]
- 32. Poirier M. G., Oh E., Tims H. S., Widom J. (2009) Dynamics and function of compact nucleosome arrays. Nat. Struct. Mol. Biol. 16, 938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Banères J. L., Martin A., Parello J. (1997) The N-tails of histones H3 and H4 adopt a highly structured conformation in the nucleosome. J. Mol. Biol. 273, 503–508 [DOI] [PubMed] [Google Scholar]
- 34. Lee K. M., Hayes J. J. (1997) The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proc. Natl. Acad. Sci. U.S.A. 94, 8959–8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jacobson H., Stockmayer W. H. (1950) Intramolecular reaction in polycondensations. I. The theory of linear systems. J. Chem. Phys. 18, 1600–1606 [Google Scholar]
- 36. Walter W., Kashlev M., Studitsky V. M. (2004) Transcription through the nucleosome by mRNA-producing RNA polymerases. Methods Enzymol. 377, 445–460 [DOI] [PubMed] [Google Scholar]
- 37. Polach K. J., Lowary P. T., Widom J. (2000) Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J. Mol. Biol. 298, 211–223 [DOI] [PubMed] [Google Scholar]
- 38. Woodcock C. L., Grigoryev S. A., Horowitz R. A., Whitaker N. (1993) A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc. Natl. Acad. Sci. U.S.A. 90, 9021–9025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ben-Haïm E., Lesne A., Victor J. M. (2001) Chromatin: a tunable spring at work inside chromosomes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 64, 051921 [DOI] [PubMed] [Google Scholar]
- 40. Schiessel H., Gelbart W. M., Bruinsma R. (2001) DNA folding: structural and mechanical properties of the two-angle model for chromatin. Biophys. J. 80, 1940–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wedemann G., Langowski J. (2002) Computer simulation of the 30-nanometer chromatin fiber. Biophys. J. 82, 2847–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R. R., Richmond T. J. (2004) Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306, 1571–1573 [DOI] [PubMed] [Google Scholar]
- 43. Routh A., Sandin S., Rhodes D. (2008) Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. U.S.A. 105, 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davey C. A., Sargent D. F., Luger K., Maeder A. W., Richmond T. J. (2002) Solvent-mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319, 1097–1113 [DOI] [PubMed] [Google Scholar]
- 45. Lowary P. T., Widom J. (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 46. Schalch T., Duda S., Sargent D. F., Richmond T. J. (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fiber. Nature 436, 138–141 [DOI] [PubMed] [Google Scholar]
- 47. Robinson P. J., Fairall L., Huynh V. A., Rhodes D. (2006) EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl. Acad. Sci. U.S.A. 103, 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bondarenko V. A., Jiang Y. I., Studitsky V. M. (2003) Rationally designed insulator-like elements can block enhancer action in vitro. EMBO J. 22, 4728–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Staynov D. Z., Proykova Y. G. (2008) Topological constraints on the possible structures of the 30-nm chromatin fiber. Chromosoma 117, 67–76 [DOI] [PubMed] [Google Scholar]
- 50. Ringrose L., Chabanis S., Angrand P. O., Woodroofe C., Stewart A. F. (1999) Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J. 18, 6630–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rosa A., Becker N. B., Everaers R. (2010) Looping probabilities in model interphase chromosomes. Biophys. J. 98, 2410–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Diesinger P. M., Kunkel S., Langowski J., Heermann D. W. (2010) Histone depletion facilitates chromatin loops on the kilobase-pair scale. Biophys. J. 99, 2995–3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Perišić O., Collepardo-Guevara R., Schlick T. (2010) Modeling studies of chromatin fiber structure as a function of DNA linker length. J. Mol. Biol. 403, 777–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Laybourn P. J., Kadonaga J. T. (1992) Threshold phenomena and long-distance activation of transcription by RNA polymerase II. Science 257, 1682–1685 [DOI] [PubMed] [Google Scholar]
- 55. Stein A., Dalal Y., Fleury T. J. (2002) Circle ligation of in vitro assembled chromatin indicates a highly flexible structure. Nucleic Acids Res. 30, 5103–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Poirier M. G., Bussiek M., Langowski J., Widom J. (2008) Spontaneous access to DNA target sites in folded chromatin fibers. J. Mol. Biol. 379, 772–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bannister A. J., Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 59. Robinson P. J., An W., Routh A., Martino F., Chapman L., Roeder R. G., Rhodes D. (2008) 30-nm chromatin fiber decompaction requires both H4-K16 acetylation and linker histone eviction. J. Mol. Biol. 381, 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bell O., Schwaiger M., Oakeley E. J., Lienert F., Beisel C., Stadler M. B., Schübeler D. (2010) Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation, and replication timing. Nat. Struct. Mol. Biol. 17, 894–900 [DOI] [PubMed] [Google Scholar]
- 61. Bondarenko V., Liu Y. V., Ninfa A. J., Studitsky V. M. (2003) Assay of prokaryotic enhancer activity over a distance in vitro. Methods Enzymol. 370, 324–337 [DOI] [PubMed] [Google Scholar]