Background: In yeast, eIF4A and eIF4F are essential for efficient translation initiation, but our understanding of their mechanisms of action is rudimentary.
Results: A detailed biochemical analysis of yeast eIF4A, alone and within eIF4F, is described.
Conclusion: The eIF4G subunit confers specificity for unwinding duplexes with 5′-overhangs on yeast eIF4F.
Significance: The observed 5′-end specificity may play a role in ribosomal scanning along mRNA.
Keywords: Enzyme Kinetics, Protein Synthesis, Protein-Nucleic Acid Interaction, RNA, Translation Initiation Factors, Cap-binding Complex, RNA Helicase, eIF4A, eIF4F, Scanning
Abstract
During eukaryotic translation initiation, the 43 S ribosomal pre-initiation complex is recruited to the 5′-end of an mRNA through its interaction with the 7-methylguanosine cap, and it subsequently scans along the mRNA to locate the start codon. Both mRNA recruitment and scanning require the removal of secondary structure within the mRNA. Eukaryotic translation initiation factor 4A is an essential component of the translational machinery thought to participate in the clearing of secondary structural elements in the 5′-untranslated regions of mRNAs. eIF4A is part of the 5′-7-methylguanosine cap-binding complex, eIF4F, along with eIF4E, the cap-binding protein, and the scaffolding protein eIF4G. Here, we show that Saccharomyces cerevisiae eIF4F has a strong preference for unwinding an RNA duplex with a single-stranded 5′-overhang versus the same duplex with a 3′-overhang or without an overhang. In contrast, eIF4A on its own has little RNA substrate specificity. Using a series of deletion constructs of eIF4G, we demonstrate that its three previously elucidated RNA binding domains work together to provide eIF4F with its 5′-end specificity, both by promoting unwinding of substrates with 5′-overhangs and inhibiting unwinding of substrates with 3′-overhangs. Our data suggest that the RNA binding domains of eIF4G provide the S. cerevisiae eIF4F complex with a second mechanism, in addition to the eIF4E-cap interaction, for directing the binding of pre-initiation complexes to the 5′-ends of mRNAs and for biasing scanning in the 5′ to 3′ direction.
Introduction
Translation initiation in eukaryotes is a complex process involving more than a dozen initiation factors. It begins with the binding of a ternary complex composed of eukaryotic initiation factor 2, methionyl initiator tRNA (Met-tRNAi), and GTP to the 40 S ribosomal subunit to form the 43 S pre-initiation complex (PIC).2 This process is promoted by eIF1, -1A, and -3. The 43 S PIC is then thought to be recruited to the 5′-end of the mRNA with the help of eIF3, -4F, and -4B and poly(A)-binding protein (PABP) and to then move along the mRNA in a 5′ to 3′ direction to locate the start codon (1–4). To do this, ribosomes must often bind to and navigate through regions of the mRNA that contain secondary or tertiary structures and/or bound proteins. Eukaryotic initiation factors 4A and 4F have been implicated in playing key roles in resolving these barriers to mRNA recruitment and scanning.
eIF4A is an ∼50-kDa DEAD-box RNA helicase containing seven conserved sequence motifs that are important for its ATP binding and hydrolysis and RNA unwinding activities (5–8). The mammalian protein exists in three isoforms: eIF4AI, eIF4AII, and eIF4AIII (9). In Saccharomyces cerevisiae, two duplicate genes, TIF1 and TIF2, encode identical copies of eIF4A (10). eIF4F, also called the cap-binding complex, is a heterotrimer made up of eIF4E, the cap-binding protein; eIF4G, a large scaffolding protein that binds RNA and a number of initiation factors; and eIF4A.
The crystal structure of yeast eIF4A revealed that the protein is dumbbell shaped, with N- and C-terminal domains connected by a flexible 11-residue linker (11). eIF4A was shown to directly interact with eIF4G through the HEAT/4A binding domain of the latter (12, 13). These studies revealed that eIF4G contacts both the N- and C-terminal RecA domains of eIF4A, fixing them in space such that the residues from each domain involved in ATP binding/hydrolysis and RNA binding now face each other, poised for catalysis (12, 13). Consistent with these findings, the interaction between mammalian eIF4A and eIF4G results in a large enhancement of RNA-stimulated ATPase and RNA unwinding activities of eIF4A (14–18).
Simultaneous binding of eIF4E to the mRNA cap and eIF4G is thought to direct the 43 S complex to the 5′-end of the mRNA via interactions between eIF4G and components in the PIC. However, the mechanisms through which eIF4F facilitates binding of the PIC to the 5′-end of the mRNA remain obscure.
In this study, we have biochemically characterized yeast eIF4A and the eIF4F complex. Our results show that, unlike its human ortholog, yeast eIF4F has a distinct preference for unwinding RNA duplexes with 5′-overhangs. The RNA-binding sites of eIF4G confer this specificity for duplexes with 5′-overhangs both by enhancing their unwinding and by suppressing the unwinding of duplexes with 3′-overhangs. Our data indicate that this intrinsic bias toward interacting with the 5′-end of mRNA substrates is independent of the presence of the 5′-cap and may serve as an additional mechanism to ensure that 43 S PIC loading takes place at the 5′-end of mRNAs. In addition, this intrinsic directionality of unwinding by the eIF4F complex could play a role in ribosomal scanning and location of the initiation codon.
EXPERIMENTAL PROCEDURES
Proteins, Nucleic Acids, and Buffers
eIF4A, eIF4A-Rh (eIF4A labeled with TAMRA), eIF4E·eIF4G, and deletion mutants of eIF4G were purified as described previously (19–21). DNA oligonucleotides were synthesized by Integrated DNA Technologies (IDT) (Coralville, IA) and purified by PAGE in 8 m urea. The RNA oligonucleotides were either ordered from Thermo (formerly Dharmacon; Table 1) or transcribed in vitro using T7 RNA polymerase as described previously (19). The purity of the RNA was checked by end labeling with [γ-32P]ATP (PerkinElmer Life Sciences) using the enzyme T4-polynucleotide kinase (New England Biolabs), and running on a 20% polyacrylamide (19:1) 8 m urea gel. The concentrations of the oligonucleotides were determined from absorbance at 260 nm using their calculated extinction coefficients.
TABLE 1.
RNA substrates
N6 is the 3′-amino group with a 6-carbon linker (3′-C6-NH2).
| Sequence | Name |
|---|---|
| 5′-GGCUGUAACU-3′ | ds10 |
| 3′-N6CCGACAUUGA-5′ | |
| 5′-GGCUGUAACUAAAUGAGAGCCAAGUGGAGAAAGAAGAGAA-3′ | 3′-30-nt-ds10 |
| 3′-N6CCGACAUUGA-5′ | |
| 5′-GGAGACCACAUCGAUUCAAUCUUCCGAAAUCGGCUGUAACU-3′ | 5′-30-nt-ds10 |
| 3′-N6CCGACAUUGA-5′ | |
| 5′-GGCUGUAACUAAACUAGACGAAAGU-3′ | 3′-15-nt-ds10 |
| 3′-CCGACAUUGA-5′ | |
| 5′-GAACUAGACGAAAGUGGCUGUAACU-3′ | 5′-15-nt-ds10 |
| 3′-CCGACAUUGA-5′ | |
| 5′-GGCUGUAACUCUAAAGCCUUCUAACUUAGCUACACCAGAGG-3′ | 3′-5 seq-30-nt-ds10 |
| 3′-N6CCGACAUUGA-5′ | |
| 5′-GGCUGUAACUUUCAAUCGAAAUGAGAGCCAAGUGGAGAAAGAAGAGAA-3′ | 3′-30-nt-ss10–10 |
| 5′-AGTTACAGCCCN6–3′ | ss10 DNA |
| 5′-GGAAUCUCUCUCUCUCUCUAUGCUCUCUCUCUCUCUCUCUCUC-Fl-3′ | 75.1-Fl mRNA |
| 5′-GUUGCGAUUGAAUCN6–3′ | ss14-C6-NH2 RNA |
Unwinding Substrates
Three different unwinding substrates were used in this study (Table 1). The sequence of the 10-bp duplex region was designed to possess a GC content of ∼50% (Tm = 48 °C under the reaction conditions) to ensure stable base-pairing between the two strands. The sequence of the 3′- and the 5′-overhangs were derived from a naturally occurring mRNA for the ribosomal protein L41A (RPL41A). The duplexes were generated by heating the two strands to 95 °C for 5 min followed by slow cooling to 4 °C. The extent of base-pairing was checked by 5′-end-labeling one of the strands with [γ-32P]phosphate, annealing the two strands, and analyzing by native 22% (19:1) TBE-PAGE.
All experiments discussed in this study were carried out in “Recon Buffer” containing 30 mm HEPES-KOH, pH 7.4, 3 mm Mg(OAc)2, 2 mm dl-dithiothreitol (DTT), and 100 mm KOAc, pH 7.4, at 26 °C unless specified otherwise. In all assays requiring ATP (or its analogs), it was added with a stoichiometric amount of MgCl2 (i.e. as ATP·Mg2+).
Fluorescence Labeling of Oligonucleotides
The 14-mer RNA (or the 10-mer DNA) oligonucleotide containing a C6-NH2 at the 3′-end (Table 1) was mixed with TAMRA-SE (Invitrogen) in the ratio of 1:10 in 0.1 m sodium tetraborate buffer, pH 7.5. The reaction was carried out at room temperature overnight. The free dye was separated from the labeled oligonucleotide using Bio-Gel-P6 (Bio-Rad) spin columns. The efficiency of labeling and the concentration of the labeled oligonucleotides were determined spectrophotometrically using the extinction coefficients of the oligonucleotides and free dye.
Equilibrium RNA Binding Assays
Two different fluorescence-based techniques were used to measure the equilibrium binding constant (Kd) for RNA, depending on the protein in question. The Kd value for eIF4E·eIF4G binding to single-stranded (ss) RNA was measured by monitoring the change in fluorescence anisotropy of the labeled ssRNA upon protein binding. The Kd value for eIF4A binding to single-stranded (ss) and double-stranded (ds) RNA was measured by monitoring the change in fluorescence intensity upon protein binding.
Monitoring Fluorescence Anisotropy
In these experiments, 30 nm 75.1-Fl (Table 1) mRNA was titrated against increasing concentration of eIF4A or eIF4E·eIF4G. The fluorophore was excited at 490 nm, and the fluorescence emission was monitored at 515 nm. Protein binding to ssRNA resulted in an increase in fluorescence anisotropy, with little or no change in fluorescence intensity. The ratio of the change in anisotropy (r − r0) to the maximal change (rmax) (fraction bound) was plotted against the protein concentration to obtain the RNA binding curves for each protein. The data were then fit to a quadratic function given by Equation 1,
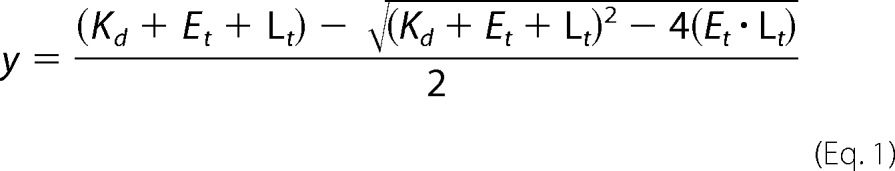 |
where Kd is the equilibrium binding constant; Et is the total protein (eIF4A) concentration, and Lt is the total ligand (RNA) concentration. Binding of eIF4A and eIF4E·eIF4G to the 75.1-Fl ssRNA was shown to have 1:1 stoichiometry (see below).
In the experiment measuring the stoichiometry of eIF4A binding to ssRNA, 5 μm 75.1-Fl (Table 1) mRNA was titrated against increasing concentration of eIF4A. Protein binding to ssRNA was monitored using fluorescence anisotropy, as described above. The fraction of ssRNA bound was plotted as a function of protein concentration, and the data were fit to Equation 1. The point of inflection of the binding curve indicates the stoichiometry of binding.
Monitoring Fluorescence Intensity
In these experiments, 5 nm ss14-C6-NH2 RNA or ds10 RNA labeled with 5-TAMRA (Table 1) was titrated against increasing concentrations of eIF4A. The fluorophore was excited at 550 nm, and the fluorescence emission was monitored at 580 nm. Protein binding to ssRNA resulted in an increase in fluorescence anisotropy, accompanied by a large change in fluorescence intensity. This change in fluorescence intensity (with unpolarized light) was used to monitor protein binding to RNA. The ratio of the change in intensity (F − F0) to the maximal change (Fmax) (fraction bound) was plotted against the protein concentration to obtain the RNA binding curves for each RNA substrate. The data were then fit with a hyperbola given by Equation 2,
 |
where A is the amplitude; E is the enzyme concentration, and Kd is the equilibrium binding constant for the protein-RNA interaction.
Equilibrium Protein Binding Assays
In these experiments, 30 nm eIF4A-Rh was titrated against increasing concentrations of eIF4E·eIF4G WT or ΔRNA mutant proteins. The Kd value of eIF4E binding to eIF4G (< 15 nm) is such that this complex is stable at all concentrations used in the titration. The fluorophore was excited at 550 nm, and the fluorescence emission was monitored at 580 nm. eIF4E·eIF4G binding to eIF4A resulted in an increase in fluorescence anisotropy, with little or no change in fluorescence intensity. The fraction bound was plotted against the protein concentration to obtain the eIF4A binding curves for each protein. The data were then fit to Equation 1.
Unwinding Assay
In these experiments, 100 nm eIF4A (with or without 300 nm eIF4E·eIF4G) was mixed with 10 nm dsRNA substrate (Table 1) and 3.0 μm ssDNA trap (Table 1). The reaction was initiated by mixing in 3 mm ATP. A reaction mix containing all components except ATP was considered t = 0. Aliquots were removed at various times (1–120 min) and mixed with native gel loading dye, containing 0.002% bromphenol blue and 40% sucrose. The time points were quenched by loading onto a running 16 or 22% (19:1) native polyacrylamide TBE gel. The resolved dsRNA and ssRNA were analyzed using a PhosphorImager (GE Healthcare). The fraction of unwound substrate was calculated and corrected for the presence of ssRNA at time 0 using Equation 3,
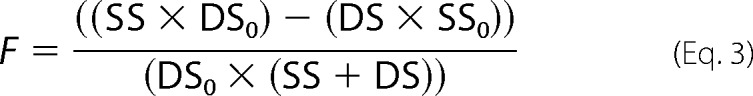 |
where F is the fraction of unwound substrate; DS and SS are radioactivities of duplex and unwound substrate bands at a given time, respectively; and DS0 and SS0 are the radioactivities of duplex and unwound substrates at time 0, respectively (22). Under our reaction conditions, <5% of the RNA was in the ssRNA form at the beginning of the reaction (t = 0).
Steady-state ATPase Assay
In these experiments, 200 nm eIF4A was mixed with 200 nm eIF4E·eIF4G to form the eIF4F complex. This enzyme complex was then pre-incubated with 5 μm ssRNA. The reaction was initiated by the addition of 5 mm ATP containing trace [γ-32P]ATP. The reaction was incubated at 26 °C, and aliquots were withdrawn at various times (15–180 min), and formic acid added to 1 n to stop the reaction. 0.5 μl of each time point was spotted onto a polyethyleneimine-cellulose TLC plate. Pi was separated from ATP using 0.4 m potassium phosphate buffer, pH 3.4, as the chromatographic solvent and quantified using a PhosphorImager.
In the experiments measuring the Km value for ATP, the RNA concentration was kept at saturating levels at 5 μm, and the concentration of ATP was varied from 20 μm to 5 mm. In these experiments, 200 nm eIF4F complex was used at low ATP concentrations (20 μm to 0.5 mm), and 500 nm eIF4F complex was used at higher ATP concentrations (1–5 mm ATP) to achieve an optimal signal to noise ratio. In the experiments measuring the Km value for RNA, the ATP concentration was kept at 5 mm (or 10 mm when measuring dsRNA-stimulated ATPase activity of eIF4F ΔRNA mutants), and the concentration of RNA was varied from 20 nm to 10 μm. The concentration of the eIF4F complex was kept at 500 nm. The active enzyme fraction was assumed to be 100%. Note that in the latter experiments, the measured Km values are not true Km values because [E] > Km. Thus, these values are reported as upper limits (Table 2).
TABLE 2.
Kinetic parameters for RNA stimulated ATPase activity of eIF4A and eIF4F
| kcat | km | |
|---|---|---|
| min−1 | μm | |
| ssRNA stimulated, increasing ATP concentration | ||
| WT eIF4Fa | 1.0 ± 0.7 | 75 ± 50 |
| WT eIF4F | 3.1 ± 1.3 | 160 ± 40 |
| eIF4F ΔRNA1 | 4.5 ± 1.2 | 620 ± 120 |
| eIF4F ΔRNA2 | 5.4 ± 0.5 | 1400 ± 390 |
| eIF4F ΔRNA3 | 4.6 ± 1.1 | 380 ± 45 |
| ssRNA-stimulated, increasing RNA concentration | ||
| WT eIF4Fa | 0.92 ± 0.5 | <0.065 |
| WT eIF4F | 3.1 ± 0.8 | <0.15 |
| eIF4F ΔRNA1 | 4.1 ± 0.5 | <0.15 |
| eIF4F ΔRNA2 | 4.3 ± 0.3 | <0.15 |
| eIF4F ΔRNA3 | 4.5 ± 0.5 | <0.15 |
| dsRNA-stimulated, increasing ATP concentration | ||
| eIF4A | 0.2 ± 0.05 | 225 ± 50 |
| WT eIF4F | NDb | ND |
| eIF4F ΔRNA1 | 5.9 ± 2.0c | 3270 ± 680c |
| eIF4F ΔRNA2 | 3.2 ± 0.4c | 2100 ± 150c |
| eIF4F ΔRNA3 | 4.4 ± 1.1c | 4100 ± 900c |
a Measured with capped RNA.
b ND means not determined because of low signal to noise with the slow rates observed.
c Values were projected from fits; all errors shown are mean deviations; n ≥ 2.
The molar ATP hydrolyzed per molar enzyme was plotted against time, and the slopes of the linear regions provided the initial velocities. These initial velocities were subtracted for any background ATPase observed in the absence of any nucleic acid. eIF4A showed negligible levels of background ATPase, whereas the background ATPase rate of eIF4F ranged between 0.05 and 0.075 min−1. Plots of the corrected initial velocities versus substrate concentration (with saturating concentration of the second substrate) were then fit with the Michaelis-Menten equation to obtain kcat and Km values.
Enzymatic Capping of RNA
RNA was capped using the vaccinia virus capping enzyme (purified as described in Ref. 20) in Capping Buffer (50 mm Tris-HCl, pH 7.8, 1.25 mm MgCl2, 6 mm KCl, 2.5 mm DTT). 0.5 mm GTP, 100 μm S-adenosylmethionine, 50 μm ssRNA, 125 nm capping enzyme, and 1 unit/μl RiboLock RNase Inhibitor (Fermentas) were added, and the reaction was incubated at 37 °C for 60 min. Additional enzyme was added to the reaction after 30 min for a final concentration of 250 nm capping enzyme. The reactions were quenched with EDTA, phenol/chloroform-extracted, and spin column-purified using Bio-Gel P6 columns (Bio-Rad). The capping efficiency was determined by adding trace [γ-32P]GTP to a small aliquot of the cold capping reaction, followed by analysis of the percentage incorporation of [γ-32P]GTP by electrophoresis. The capping efficiency ranged between 70 and 75%.
RESULTS
RNA Binding Specificity of Yeast eIF4A
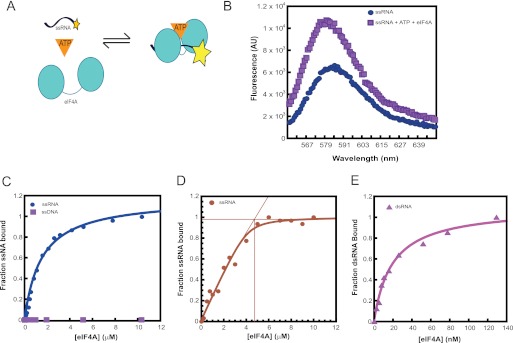
RNA helicases are typically characterized by their ability to bind RNA and couple this RNA binding activity to nucleotide hydrolysis. Thus, as a first step toward a thorough characterization of S. cerevisiae eIF4A, we examined the ability of the enzyme to bind RNA. We developed a fluorescence-based equilibrium binding assay for this activity. eIF4A binding to 3′-TAMRA-labeled ssRNA (ss14; Table 1) in the presence of ATP (or the nonhydrolyzable ATP analog ADP·[AlF4]−) results in an enhancement of the fluorescence intensity of the labeled RNA (Fig. 1, A and B). We used this as the signal to monitor the binding of eIF4A to ss- and dsRNA substrates. As was observed for mammalian eIF4A (23), binding of yeast eIF4A to ssRNA is strongly coupled to ATP binding; we could only detect binding to ssRNA in the presence of ATP (or ADP·[AlF4]−, data not shown). In the presence of saturating ATP, yeast eIF4A bound ssRNA with a Kd of ∼1.5 μm (Fig. 1C) similar to the affinity observed with the human factor (23).
FIGURE 1.
Equilibrium binding of eIF4A to RNA substrates. A, schematic representation of the fluorescence-based assay to monitor eIF4A binding to RNA. B, emission wavelength scan of 3′-TAMRA-labeled RNA alone (●) or eIF4A·RNA·ATP complex (■). The excitation wavelength was 550 nm. eIF4A binding to RNA results in an enhancement in the fluorescence intensity with a peak around 585 nm. C, equilibrium binding curves for eIF4A and ssRNA or ssDNA. Increasing concentrations of eIF4A were titrated against 5 nm ssRNA in the presence of 5 mm ATP. D, stoichiometry of eIF4A binding to ssRNA. Increasing concentrations of eIF4A were titrated against 5 μm ssRNA, in the presence of 5 mm ATP. E, equilibrium binding curve for eIF4A and dsRNA. Increasing concentrations of eIF4A were titrated against 5 nm dsRNA, in the presence of 5 mm ATP. C–E, the fraction bound was plotted as a function of increasing eIF4A concentrations to obtain the binding curves for the interaction between eIF4A and RNA. C and E, the data were fit with an equation for hyperbolic ligand binding to obtain the equilibrium binding constant for eIF4A and the different RNA substrates. D, data were fit with the quadratic form of the binding equation because the concentration of labeled ssRNA was greater than the Kd value. eIF4A binds to ssRNA with a Kd of ∼1.5 μm and a stoichiometry of 1:1. It binds dsRNA with a Kd of ∼20 nm. All binding experiments were carried out in “Recon buffer” containing 30 mm HEPES-KOH, pH 7.4, 3 mm Mg(OAc)2, 2 mm dl-dithiothreitol (DTT), and 100 mm KOAc, at 26 °C.
Monitoring binding of a different, pyrimidine-rich ssRNA labeled on its 3′-end with fluorescein (75.1-Fl; Table 1) using fluorescence anisotropy yielded a Kd of ∼1.9 μm (data not shown), indicating that the fluorescence enhancement assay is an accurate readout of the eIF4A-ssRNA interaction and suggesting ssRNA binding is not sequence-specific.
Unlike the mammalian protein, which has no appreciable dsRNA binding capability, yeast eIF4A bound dsRNA more tightly than it bound ssRNA, with a Kd of ∼20 nm (Fig. 1D). Unlike ssRNA binding, eIF4A binding to dsRNA was only moderately influenced by ATP binding (Kd for eIF4A·dsRNA in the absence of ATP ∼70 nm, data not shown). Similar experiments could not be carried out for the eIF4F complex due to the affinity of the interaction between yeast eIF4A and eIF4E·eIF4G (Kd ∼30 nm) (20), which precluded measurements at low complex concentrations.
Incorporation of Yeast eIF4A into the eIF4F Complex Strongly Enhances its ssRNA-dependent ATPase Activity
We next explored the abilities of S. cerevisiae eIF4A and eIF4F to hydrolyze ATP when stimulated by RNA substrates. In these assays, eIF4A or eIF4F (generated by adding saturating, equimolar concentrations of eIF4A to eIF4E·eIF4G) was incubated with saturating concentrations of RNA substrates (5 μm ssRNA or dsRNA, Table 1). The reaction was initiated by the addition of 5 mm ATP containing trace [γ-32P]ATP. The reactions were allowed to proceed for 0–180 min at 26 °C, quenched at different times with formic acid, and analyzed by polyethyleneimine-cellulose thin layer chromatography.
Our experiments showed that the ATPase activities of eIF4A and eIF4F are stimulated by both ssRNA and dsRNA, albeit to different extents. eIF4A exhibits no detectable ATP hydrolysis activity in the absence of RNA and has a 30-fold higher rate of ATP hydrolysis when stimulated by dsRNA than by ssRNA (Fig. 2). eIF4F, however, has robust ATPase activity when stimulated by ssRNA but 2 orders of magnitude lower activity with dsRNA (Fig. 2). Thus, the ATPase of yeast eIF4A on its own is activated better by dsRNA than by ssRNA, consistent with its ability to bind dsRNA more tightly than ssRNA, whereas eIF4A as part of the eIF4F complex has a strong preference for ssRNA.
FIGURE 2.
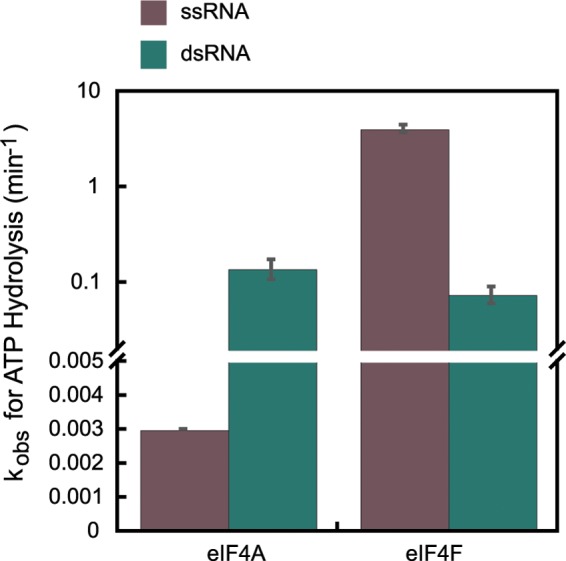
Steady-state ATPase activities of eIF4A and eIF4F. ATPase activities of WT eIF4A and eIF4F were measured as described under “Experimental Procedures.” Briefly, ATP (5 mm) was added to the reaction mixture containing eIF4A or eIF4F (200 nm) with saturating single-stranded or double-stranded nucleic acids substrates (5 μm). The reaction was allowed to proceed for 0–180 min at 26 °C in Recon buffer. The reaction was quenched with formic acid, and the products were resolved by polyethyleneimine-cellulose TLC and quantified using ImageQuant. The molar ATP hydrolyzed per molar enzyme was plotted as a function of time. The data were fit with an equation for a straight line. The slope of this line gave the initial rate of ATP hydrolysis. The ATPase rates reported here have been corrected for the low level of background ATPase observed in the absence of any nucleic acid (see “Experimental Procedures”).
Incorporation of Yeast eIF4A into the eIF4F Complex Induces a Marked Preference for Unwinding RNA Substrates with 5′-Overhangs
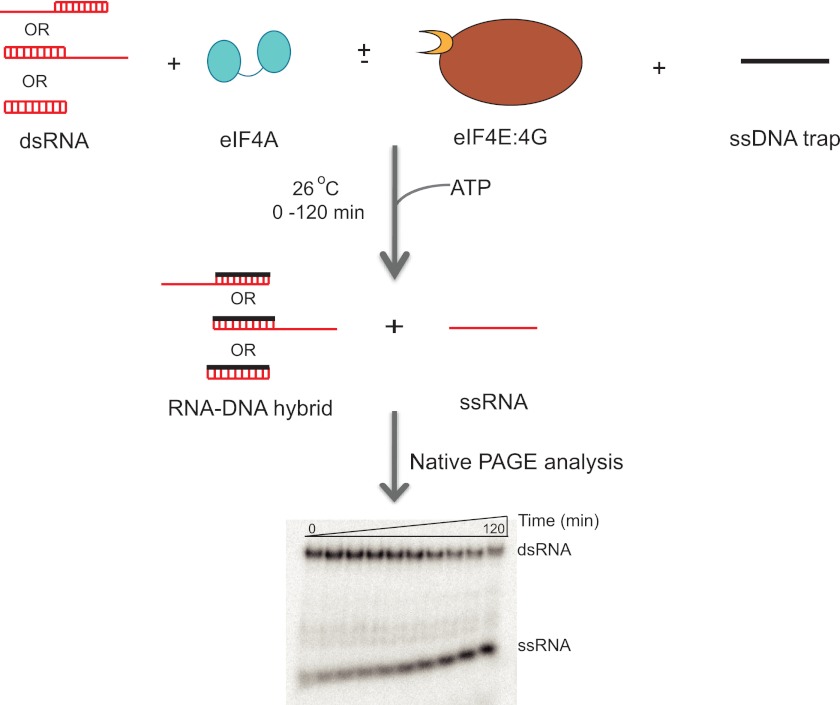
We next examined the RNA unwinding activity of yeast eIF4A and the eIF4F complex. Briefly, 100 nm eIF4A (with 300 nm eIF4E·eIF4G, when working with eIF4F) was incubated with 10 nm dsRNA (Table 1). The reaction was initiated by the addition of ATP and a DNA trap oligonucleotide to a final concentration of 3 mm and 3 μm, respectively. The reactions were allowed to proceed for 0–120 min and quenched by loading on a running 16% (19:1) native PAGE TBE gel (Fig. 3). The DNA trap used in the experiment had the same sequence as the radiolabeled strand of the duplex substrate. The trap served to base pair with the unlabeled ssRNA generated during the unwinding reaction, thereby preventing re-annealing of the labeled RNA strand and making its dissociation irreversible. Experiments performed with varying ratios of eIF4A to eIF4E·eIF4G showed that the 3-fold excess of eIF4E·eIF4G did not affect the rate of ATPase or RNA unwinding of the eIF4F complex (data not shown). In addition, control experiments with the trap DNA oligonucleotide showed that it did not bind to eIF4A (Fig. 1C) or eIF4E·eIF4G (Fig. 6B) at the concentrations used in the experiments. Moreover, ssDNA did not stimulate the ATPase activity of the eIF4F complex (data not shown). The trap by itself did not displace the RNA strand from the duplex on the time scale of the assay (data not shown). However, the trap was essential in the presence of eIF4A or eIF4F for observing RNA unwinding, consistent with it serving to capture the unlabeled RNA strand and prevent reannealing with the labeled strand. The rate of unwinding with eIF4A alone was linearly proportional to the concentration of the enzyme between 0.05 and 1.0 μm, indicating that it was not saturating for binding to the RNA. In contrast, the rate of unwinding by eIF4F did not change between 0.075 and 0.3 μm, indicating the concentration used in subsequent unwinding experiments (0.1 μm) was saturating.
FIGURE 3.
Schematic representation of the RNA unwinding assay. A flow chart of the RNA unwinding assay. Briefly, the dsRNA substrate (10 nm) was incubated with eIF4A (100 nm, with or without 300 nm eIF4E·eIF4G) and DNA trap (3 μm). The DNA trap has the same sequence as the labeled RNA oligonucleotide (top strand of duplexes in red) and serves to prevent reannealing of the two RNA strands after unwinding. The unwinding reaction was initiated by the addition of ATP (3 mm). The time course (0–120 min) was monitored at 26 °C and analyzed by native PAGE cooled with a circulating water bath. A representative unwinding gel is shown here. The gel was scanned and quantified using a PhosphorImager. As with the RNA binding and ATPase, the reaction was carried out in Recon buffer.
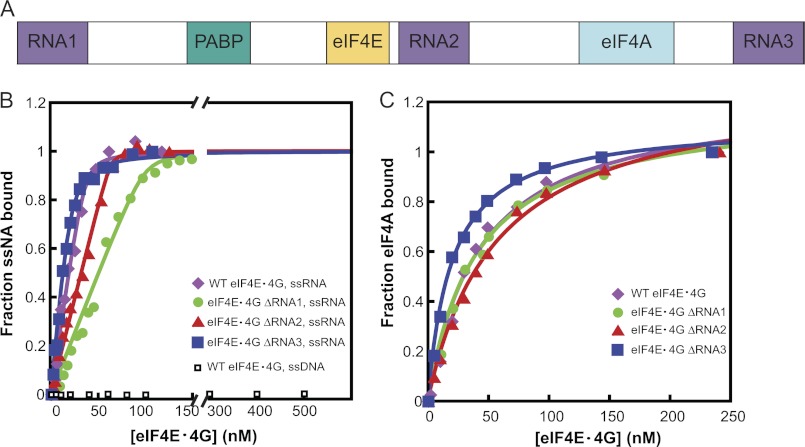
FIGURE 6.
ssRNA and eIF4A binding to eIF4G RNA domain deletion mutants. A, schematic representation of yeast eIF4G, showing the locations of the RNA binding domains (RNA1–RNA3), the canonical PABP-binding domain (PABP), the eIF4E-binding domain (eIF4E), and the eIF4A-binding domain (eIF4A). B, equilibrium binding constants (Kd) for ssRNA and the different ΔRNA domain mutants of eIF4G were measured using fluorescence anisotropy as described under “Experimental Procedures.” Briefly, the change in fluorescence anisotropy of 75.1-Fl RNA (Table 1) was monitored as a function of increasing concentrations of WT eIF4E·eIF4G or ΔRNA mutants. The curves were then fit with a quadratic binding equation to obtain the Kd value for the interaction between eIF4E·eIF4G and ssRNA. All the mutants bound to ssRNA very tightly, with Kd values ≤30 nm. Because of this tight binding, only upper limits for Kd values could be obtained. The inflection point of the curve indicates 1:1 binding between the WT eIF4E·eIF4G complex and the ssRNA. WT eIF4E·eIF4G did not bind detectably to ssDNA. C, Kd value for binding of eIF4A to the different ΔRNA mutants of eIF4E·eIF4G was measured using fluorescence anisotropy as described under “Experimental Procedures.” Briefly, the change in fluorescence anisotropy of TAMRA-labeled eIF4A was monitored as a function of increasing concentrations of eIF4E·eIF4G ΔRNA mutants. The titration curves were then fit with a hyperbolic ligand binding equation to obtain the Kd value for eIF4E·eIF4G and eIF4A. All mutants bound eIF4A with Kd ≤30 nm. B and C, ●, eIF4E·eIF4G ΔRNA1; ▴, eIF4E·eIF4G ΔRNA2; ■, eIF4E·eIF4G ΔRNA3; and ♦, WT eIF4E·eIF4G. B, □, WT eIF4E·eIF4G binding to ssDNA. The experiments were performed at 26 °C in Recon buffer. The concentrations of labeled ssRNA and eIF4A were each 30 nm in B and C, respectively.
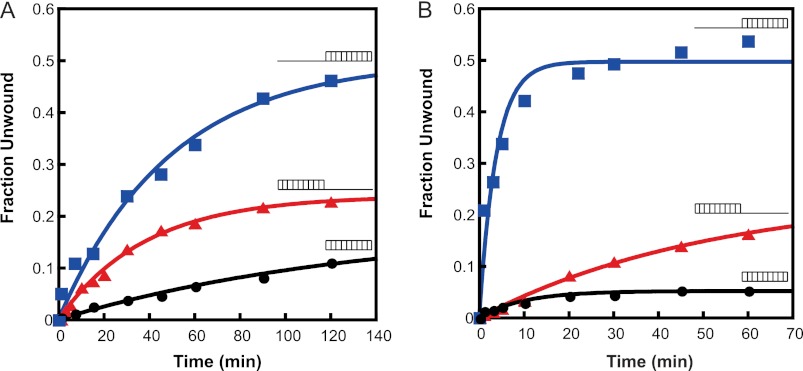
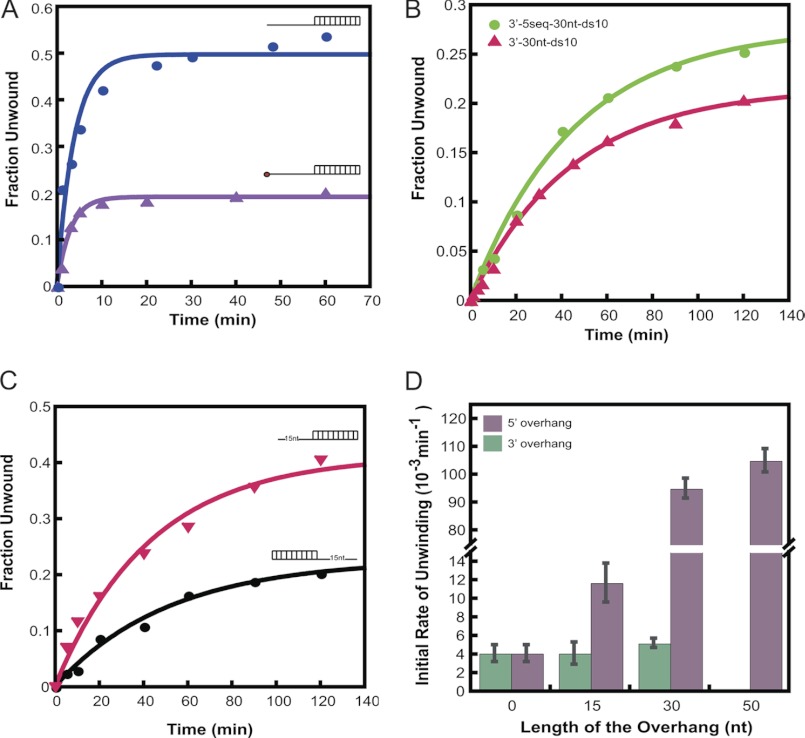
Three different duplexes (Table 1) were used to study the unwinding activity of eIF4A and eIF4F as follows: (i) a 10-bp duplex with a 30-nucleotide 3′-overhang; (ii) a 10-bp duplex with a 30-nucleotide 5′-overhang; and (iii) a 10-bp duplex with no overhangs (in all cases the 10-bp duplex was the same). Our results showed that both eIF4A and eIF4F were capable of unwinding all three duplexes (Fig. 4) and that the unwinding activities of both were ATP-dependent (data not shown). eIF4A unwound duplexes with 5′- and 3′-overhangs at similar rates, with a slight (1.5-fold) preference for unwinding duplexes with a 5′-overhang (Fig. 4A and Table 3). However, eIF4F unwound the duplex with a 5′-overhang 20-fold faster than the duplexes with a 3′-overhang or no overhang (Fig. 4B and Table 3). The rate of unwinding of the duplex with the 5′-overhang was 10-fold higher with eIF4F than eIF4A alone, whereas the rate was the same for eIF4A and eIF4F with the duplex with a 3′-overhang. To rule out any sequence effects on the unwinding rate of eIF4F, an RNA substrate was made that had the same sequence within the duplex region but now had the same sequence in its 3′-overhang as that of the 5′-overhang substrate (3′-5 seq-30 nt-ds 10; Table 1). eIF4F unwinds this substrate with similar kinetics as the original 3′-overhang RNA (Fig. 5B; Table 3), demonstrating that the preference for a 5′-overhang is not due to the sequence of the overhangs. In all of the three duplexes used above, one of the strands carried a C6NH2 linker at its 3′-end to facilitate chemical labeling with a fluorophore (Table 1). Control experiments using an RNA oligonucleotide lacking this linker indicated that its presence did not affect the results (data not shown).
FIGURE 4.
Substrate specificity of RNA unwinding by eIF4A and eIF4F. The RNA unwinding activities of eIF4A and eIF4F were measured with three different unwinding substrates (Table 1). Conditions were the same as in Fig. 3. A, representative plot of the unwinding kinetics of eIF4A with the different RNA duplexes. B, representative plot for the unwinding kinetics of WT eIF4F with the different RNA duplexes. The substrates used (Table 1) are schematically represented next to each time course. The initial rates of unwinding (Table 3) were obtained using the slope of the linear portion of the exponential plots.
TABLE 3.
Initial rates of RNA unwinding
| 5′-30-nt ds10 |
3′-30-nt ds10 |
ds10 |
||||
|---|---|---|---|---|---|---|
| Unwinding ratea | Amplitude | Unwinding ratea | Amplitude | Unwinding ratea | Amplitude | |
| eIF4A | 9.5 ± 0. 7 | 0.46 ± 0.04 | 6.5 ± 0.81 | 0.22 ± 0.02 | 1.2 ± 0.22 | 0.16 ± 0.02 |
| WT eIF4F | 113 ± 17.5 | 0.49 ± 0.05 | 4.9 ± 0.3 | 0.21 ± 0.04 | 4.7 ± 0.61 | 0.05 ± 0.02 |
| (4.7 ± 1.4)b | 0.23 ± 0.05 | |||||
| WT eIF4Fc | 67 ± 3.4 | 0.20 ± 0.01 | NDd | ND | ||
| WT eIF4Fe | 10.6 ± 1.1 | 0.40 ± 0.02 | 4.3 ± 1.2 | 0.18 ± 0.02 | ND | |
| eIF4F ΔRNA1 | 40 ± 5.5 | 0.55 ± 0.03 | 18 ± 2.4 | 0.61 ± 0.01 | 3.4 ± 0.3 | 0.15 ± 0.02 |
| eIF4F ΔRNA2 | 9.9 ± 0.65 | 0.81 ± 0.05 | 13 ± 2.7 | 0.84 ± 0.05 | 7.3 ± 0.4 | 0.23 ± 0.02 |
| eIF4F ΔRNA3 | 6.3 ± 0.23 | 0.83 ± 0.4 | 11.4 ± 0.9 | 0.88 ± 0.06 | 1.8 ± 0.2 | 0.06 ± 0.01 |
FIGURE 5.
Effect of a 5′-cap and ssRNA sequence on RNA unwinding by eIF4F. The RNA unwinding activity of wild-type eIF4F was measured with different dsRNA substrates (Table 1). A, eIF4F-catalyzed unwinding of 5′-30-nt-ds10 substrate (Table 1) with (▴) or without (●) a 5′-7mG cap. B, comparison of eIF4F-catalyzed unwinding of a 10-bp duplex with two different 30-nt 3′-overhangs. The kinetics of unwinding are the same with the standard 3′-overhang (3′-30-nt-ds10; Table 1 (▴)) and one that is identical to the 30-nt single-stranded region of the 5′-overhang dsRNA substrate (3′-5 seq-30 nt-ds 10; Table 1 (●)). C, eIF4F-catalyzed unwinding of a 10-bp duplex substrate with a 15-nt 5′-overhang (▾) and a 15-nt 3′-overhang (●) (Table 1). D, dependence of eIF4F unwinding rate upon lengths of 3′- or 5′-overhangs. The unwinding rate of a 10-bp duplex with a 50-nt 3′-overhang was not measured. Conditions were the same as in Fig. 3.
We next determined the effect of the length of the overhangs on the rate of unwinding of the duplex by eIF4F. Keeping the sequence of the duplex region the same, we generated new RNA substrates with 15 nucleotide 5′- or 3′-overhangs. eIF4F preferentially unwound the duplex with the 15-nt 5′-overhang relative to the substrate with the 15-nt 3′-overhang (Fig. 5C), although with a smaller preference than that observed with the substrates with 30-nt overhangs (2.5- and 20-fold preference with 15- and 30-nt overhangs, respectively). This reduced preference was because the rate of unwinding of the 5′-15-nt overhang substrate was ∼10-fold slower than the rate with the substrate with a 5′-30-nt overhang, and decreasing the 3′-overhang from 30 to 15 nucleotides did not affect the rate of unwinding (Table 3). No further increase in unwinding rate above that with the 30-nt overhang was observed with an identical substrate with a 50-nt 5′-overhang (Fig. 5D). Unwinding of the substrate with a 50-nt 3′-overhang was not carried out.
All the experiments discussed above were carried out on substrates lacking a 5′-m7G cap. Because eIF4F contains the cap-binding protein eIF4E, which specifically binds the m7G-cap, we wanted to test the effect of a 5′-cap on the unwinding of the RNA substrates. We therefore enzymatically capped the 5′-end of the RNA of the duplex carrying the 30-nt 5′-overhang using the vaccinia virus capping enzyme (see under “Experimental Procedures”), and we tested if this changed the kinetics of unwinding of the RNA duplex by eIF4F. Capping resulted in only a small reduction in the unwinding rate and about a 2-fold decrease in amplitude (Fig. 5A and Table 3) indicating that the cap does not enhance the ability of eIF4F to unwind duplexes with a 5′-overhang.
RNA Binding Domains in eIF4G Direct the 5′-End Specificity for Unwinding by Yeast eIF4F
Yeast eIF4G, in addition to having specific binding sites for interaction with eIF4A, eIF4E, and PABP also possesses three RNA binding regions, defined as RNA1 (residues 1–82), RNA2 (residues 492–539), and RNA3 (residues 883–952) (24–26) (Fig. 6A). A recent study by Park et al. (21) showed that RNA1 interacts with RNA and PABP to promote assembly of the eIF4F-mRNA-PABP messenger ribonucleoprotein, and also identified critical roles for RNA2 and RNA3 downstream of the assembly of this messenger ribonucleoprotein, presumably in PIC attachment or scanning.
To assess the roles played by these three RNA-binding sites in RNA unwinding by eIF4F, we used three deletion constructs of eIF4G, each lacking one of the RNA binding domains, in RNA unwinding assays. These same constructs were previously studied in vivo by Park et al. (21). The proteins were co-expressed with eIF4E and purified as described previously (see under “Experimental Procedures”) (20). Equilibrium RNA binding measurements using fluorescence anisotropy (Fig. 6B) revealed that all three deletion constructs of eIF4G bound ssRNA tightly, with Kd values <30 nm. With two of the deletion mutants, eIF4G ΔRNA1 and eIF4G ΔRNA2, the titration point shifted toward higher protein concentrations, consistent with a lower concentration of functional RNA binding sites in solution with these mutants (Fig. 6B). Thus, in all subsequent assays care was taken to use these mutants in 3–5-fold molar excess of eIF4A to ensure optimal RNA binding.
We also measured the ability of each eIF4G deletion construct to bind eIF4A. Equilibrium binding of eIF4G ΔRNA constructs to eIF4A-Rh was monitored using fluorescence anisotropy. All three deletion constructs bound eIF4A-Rh with a Kd of <30 nm (Fig. 6C), similar to the value previously reported for the interaction with wild-type eIF4G·eIF4E (20).
The mutant proteins were then bound to eIF4A to generate the respective eIF4F complexes. The mutant eIF4F complexes were tested for their ATPase and RNA unwinding activities. At saturating ATP and ssRNA, all the eIF4F mutants had similar rate constants for ATP hydrolysis (kcat values in Table 2; 4–5 min−1), close to the value for the WT complex (3 min−1), and much faster than that observed for isolated eIF4A (<0.01 min−1). These higher rates provide additional evidence that eIF4A is bound to the eIF4E·eIF4G complex in all cases and that this binding enhances the ATPase activity of the factor. Although the maximal rates of ATP hydrolysis were similar to WT eIF4F, the mutant complexes had varying levels of defect in their apparent interaction with ATP at saturating concentrations of ssRNA (Km values in Table 2). Individually, deletion of RNA1, RNA2, and RNA3 increased the Km values for ATP by 4-, 12-, and 2-fold, respectively, relative to the WT eIF4F complex (Table 2). These data suggest that removal of the RNA-binding sites in eIF4G alters the ground state conformation of eIF4A within the eIF4F complex. This alteration in its conformation is further reflected by the fact that when stimulated by 5 μm dsRNA, in the presence of 5 mm ATP, the ΔRNA mutants of eIF4F show significant levels of dsRNA stimulated ATPase activity (kobs of 4.5, 2.0, and 2.1 min−1 for eIF4F ΔRNA1, ΔRNA2, and ΔRNA3, respectively; Table 2), whereas the activity for the WT complex is very low (0.14 min−1; Fig. 2).
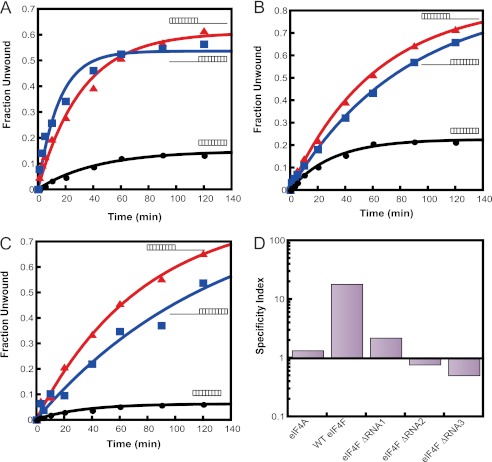
We next analyzed the RNA unwinding activities of the mutant eIF4F complexes. Deletion of any of the RNA binding regions reduced the observed rates of unwinding of the RNA substrate with the 30-nt 5′-overhang at saturating ATP concentration: 2-fold for eIF4F ΔRNA1 and 10–15-fold for ΔRNA2 and ΔRNA3 (Table 3; Fig. 7, A–C, respectively). Strikingly, all the deletion mutants actually increased the rates of unwinding of the RNA substrate with the 30-nt 3′-overhang (2–4-fold increases) as compared with the wild-type enzyme (Table 3; Fig. 7). The net effect of each of the deletions is to greatly reduce (ΔRNA1) or eliminate (ΔRNA2 and ΔRNA3) the preference of eIF4F for unwinding substrates with a 5′-overhang (Fig. 7D). These data indicate that the RNA binding sites together stimulate unwinding of duplexes with a 5′-overhang and suppress unwinding of duplexes with a 3′-overhang.
FIGURE 7.
Substrate specificity of mutant eIF4F complexes. The RNA unwinding activities of the eIF4F ΔRNA mutants were measured with three different substrates (Table 1). Representative time courses of unwinding are shown with the different RNA duplexes catalyzed by eIF4F ΔRNA1 (A), eIF4F ΔRNA2 (B), and eIF4F ΔRNA3 (C). The substrates are schematically represented next to each time course. The unwinding rate constants (Table 3) were obtained by fitting each plot to a first-order exponential equation. D, specificity index, defined as the ratio of the unwinding rate constant for the duplex with the 30-nt 5′-overhang to that of the duplex with the 30-nt 3′-overhang, for each eIF4F complex. A specificity index >1 indicates a preference for unwinding the duplex with the 5′-overhang and a value <1 indicates a preference for the duplex with the 3′-overhang. Conditions were as in Fig. 3, except in experiments with eIF4F ΔRNA1 500 nm eIF4E·eIF4G was used.
DISCUSSION
In this study we have characterized the biochemical properties of S. cerevisiae eIF4A, a DEAD-box RNA helicase essential for translation initiation. We studied the RNA binding of the enzyme, RNA stimulated ATPase and RNA unwinding activities and compared and contrasted these functions between eIF4A in its free form and within the eIF4F cap-binding complex. We also assessed the roles of the three RNA-binding sites within eIF4G in modulating these activities.
RNA Helicase Activity of eIF4A and eIF4F
Yeast eIF4A, in its free form, is an extremely poor ATPase having only weak RNA-stimulated ATP hydrolysis activity. Interestingly, it is stimulated more by dsRNA than by ssRNA, although in both cases this activity is quite modest (Fig. 2). This result is consistent with the fact that yeast eIF4A binds dsRNA with ∼100-fold higher affinity (Kd ∼20 nm; Fig. 1C) than it binds ssRNA (Kd ∼1.5 μm; Fig. 1D). This behavior is in stark contrast with that of the mammalian ortholog, which binds ssRNA with a similarly low affinity (Kd ∼3 μm) but exhibits more robust ssRNA-stimulated ATPase activity and no detectable dsRNA binding (23). As seen with the mammalian protein, ssRNA binding by yeast eIF4A is strongly coupled to ATP binding (23). This observation may be explained by the fact that eIF4A in its free form adopts an extended “dumbbell” structure, with two compact RecA domains connected by a flexible 18-Å long linker (11). In the absence of ssRNA or ATP, the domains are not strictly oriented relative to one another, and thus the binding sites are not organized. When the two RecA domains are brought together by the binding of either ATP or ssRNA, the full substrate binding and active sites are formed, explaining the cooperative interactions between the two substrates (11). In contrast, the structural basis behind the differential dsRNA binding activities of the yeast and mammalian proteins is unclear. Further experiments will be required to elucidate the nature and significance of dsRNA binding to yeast eIF4A.
When incorporated into the eIF4F complex, the rate constant for ssRNA-stimulated ATP hydrolysis by eIF4A increases ≥250-fold as compared with the factor by itself. It was previously demonstrated that the physical interaction between eIF4A and eIF4G enhances the RNA helicase activity of the mammalian protein (14–16). NMR and x-ray crystallographic studies of the complex between eIF4A and the HEAT domain of eIF4G showed that eIF4G interacts with both RecA domains of eIF4A bringing the two lobes closer together (12, 13). Thus, in the eIF4F complex it is proposed that the inter-domain cleft in eIF4A, which contains all the conserved residues for the RNA helicase activity, is poised to bind and hydrolyze ATP.
Consistent with its low dsRNA-stimulated ATPase activity, eIF4F shows <5% unwinding of blunt duplexes under our assay conditions (Fig. 4B). Introduction of a single-stranded region to the duplex improves the unwinding activity of eIF4F although to different extents depending on the orientation of the overhang. The greatest enhancement in unwinding rate and amplitude is achieved in the presence of a 30-nt 5′-overhang (Fig. 4B, Fig. 5D, and Table 3). eIF4F unwinds this duplex at an ∼20-fold higher rate as compared with the blunt duplex (Fig. 5D and Table 3). This enhancement provided by a single-stranded region in the unwinding substrate is consistent with the high ssRNA-stimulated ATPase activity of eIF4F (Fig. 2). Decreasing the 5′-overhang from 30- to 15-nt decreases the unwinding rate by ∼7-fold, whereas increasing the length of the overhang from 30- to 50-nt does not increase it further, indicating that eIF4F has an optimal 5′-overhang length of ∼20–30 nucleotides.
The preference of the enzyme for unwinding duplexes with a 5′-overhang is intriguing because the mammalian factor unwinds RNA duplexes with either a 3′- or a 5′-overhang with similar efficiencies (27, 28). It implies that S. cerevisiae eIF4F has an inherent polarity to its unwinding activity, which enables it to distinguish between 5′- and 3′-ends in an mRNA substrate, even without the 5′-m7G cap-eIF4E interaction. It is possible that this function of eIF4F, which we have shown is mediated by the eIF4G subunit, was taken up by another protein in mammals, for example, by one or more of the eIF3 subunits that are present in higher eukaryotes but not in S. cerevisiae (3).
Although eIF4A on its own has a preference for unwinding substrates with ssRNA overhangs, it does not strongly discriminate between 5′- and 3′-overhangs (Fig. 4A and Table 3). This behavior seems to be conserved between the mammalian and the yeast proteins (27). However, unlike the mammalian and plant factors (17, 29–31), RNA unwinding by S. cerevisiae eIF4A and eIF4F is not stimulated by S. cerevisiae eIF4B (data not shown), consistent with previous reports (32). However, we cannot rule out the possibility that eIF4B stimulates the ATPase and/or unwinding activity of eIF4F within the context of the pre-initiation complex.
RNA Helicase Activity of eIF4F on Capped mRNA Substrates
The 5′-m7G cap on mRNA substrates had only small effects on the biochemical properties of eIF4F in the in vitro assays we employed in this work. As expected, eIF4F appears to bind the capped substrate more tightly than the uncapped substrate, as judged by the Km values with respect to ssRNA (Km ≤65 and 160 nm, with and without a cap, respectively; Table 2). The relatively modest apparent effect of the cap on RNA binding is consistent with recent results with mammalian eIF4F (33). The kcat for ATP hydrolysis is 3-fold lower with capped ssRNA than uncapped ssRNA (1 versus 3 min−1 for capped and uncapped mRNA, respectively; Table 2). It is possible that the capped RNA is more constrained in its mode of binding to the eIF4F complex, resulting in a lower extent of ATPase activation.
RNA-binding Sites of eIF4G Play Critical Roles in Setting the Directionality of RNA Unwinding by the eIF4F Complex
To explore the roles of the RNA-binding sites in eIF4G in the activities of eIF4F, we constructed eIF4F complexes using derivatives of eIF4G lacking one of the three sites. All three versions of eIF4G bound eIF4A with the same affinity as the WT protein (Fig. 6C). The resulting eIF4F complexes all had kcat values for ssRNA-stimulated ATP hydrolysis similar to that of the wild-type complex (Table 2). These data indicate that all the mutants are bound to eIF4A at the concentrations used and that they couple the RNA binding to ATP hydrolysis to similar extents. However, all the mutants had ∼10-fold higher kcat values for dsRNA-stimulated ATP hydrolysis activity as compared with wild-type eIF4F indicating that deleting any of the RNA binding domains in eIF4G resulted in a partial loss of the ssRNA specificity seen with wild-type eIF4F. It also resulted in an increased Km value for ATP with all the mutants (Table 2), with eIF4F ΔRNA2 having the largest defect (∼10-fold) and eIF4F ΔRNA1 and eIF4F ΔRNA3 having more moderate defects (3.5- and 2-fold, respectively; Table 2). These increases in the Km values for ATP suggest that deletion of the RNA binding domains in eIF4G influences the conformation of eIF4A within the eIF4F complex.
Analysis of the RNA unwinding activities of the mutant complexes showed that in all three cases, the observed rate of unwinding the duplex with the 5′-overhang was reduced (2-, 10-, and 15-fold, for ΔRNA1, ΔRNA2, and ΔRNA3, respectively) relative to the WT complex, whereas the rate of unwinding of the duplex with the 3′-overhang was increased 2–4-fold. To quantitatively describe the unwinding specificity of eIF4A and eIF4F complexes, we computed a specificity index by obtaining the ratio of the unwinding rates of the duplex with the 5′-overhang to that of the duplex with the 3′-overhang. Although eIF4F exhibits a high specificity index of ∼20, indicating a strong preference for unwinding duplexes with 5′-overhangs, eIF4A and each of the ΔRNA mutants have a low specificity index, near or below 1 (Fig. 7D). These data indicate that the RNA-binding sites in eIF4G cooperate to impart a directionality to RNA unwinding by yeast eIF4F, with RNA2 and RNA3 playing important roles in enhancing the use of substrates with 5′-overhangs and all three sites inhibiting use of substrates with 3′-overhangs.
Overall, removal of RNA1 from eIF4G is the least disruptive to eIF4F function in RNA unwinding of the three deletions. This result is in agreement with recent in vivo data showing that deletion of RNA1 only slightly exacerbated the cell growth defect conferred by a point mutation in the eIF4E binding domain, whereas deletion of RNA2 or RNA3 was lethal in combination with the same point mutation (21). It is also consistent with the observation that when mammalian eIF4GI is truncated in such a way that its RNA binding domain (residues 642–681, analogous to RNA2 of S. cerevisiae eIF4G) is removed, it results in the loss of ribosomal scanning function in rabbit reticulocyte lysates (34).
Implications for Translation Initiation
Recruitment of the eIF4F complex to the 5′-end of the mRNA is thought to be mediated by the interaction of eIF4E with the 5′-m7G cap structure. The data presented above indicate that yeast eIF4F has a strong 5′-end preference for RNA unwinding, even in the absence of the m7G-cap. This inherent polarity of unwinding by the complex is conferred by the RNA-binding sites in eIF4G, particularly RNA2 and RNA3. These results help explain how uncapped messages can be accurately translated (20, 35–40) and suggest that this activity evolved as a second mechanism to ensure that 43 S PICs are only recruited to the 5′-ends of normal cellular messages.
The preference of the eIF4F complex for 5′-overhangs is also consistent with the need for the 43 S PIC to scan in a 5′ to 3′ direction on the mRNA during the search for the start codon (41, 42). The intrinsic 5′-directionality of RNA unwinding by the eIF4F complex could play a critical role in biasing the direction of scanning along the mRNA. For example, eIF4F might unwind RNA in a 5′ to 3′ direction on the leading edge of the ribosome, followed by diffusion of the PIC over the unwound region. Backsliding could be prevented by reformation of the mRNA structures behind the ribosome, or by binding of RNA binding proteins to this region, as proposed recently by Spirin (43).
Acknowledgments
We thank the members of our laboratories for their comments and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM62128 (to J. R. L.) and by the Intramural Research Program (to A. G. H.).
- PIC
- pre-initiation complex
- nt
- nucleotide
- PABP
- poly(A)-binding protein
- ss
- single-stranded
- TAMRA
- 5-carboxytetramethylrhodamine.
REFERENCES
- 1. Lorsch J. R., Dever T. E. (2010) Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J. Biol. Chem. 285, 21203–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hinnebusch A. G. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W. C., Sonenberg N. (2011) mRNA helicases. The tacticians of translational control. Nat. Rev. Mol. Cell Biol. 12, 235–245 [DOI] [PubMed] [Google Scholar]
- 5. Gorbalenya A. E., Koonin E. V. (1993) Helicases. Amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3, 419–429 [Google Scholar]
- 6. Schmid S. R., Linder P. (1992) DEAD protein family of putative RNA helicases. Mol. Microbiol. 6, 283–291 [DOI] [PubMed] [Google Scholar]
- 7. Pause A., Méthot N., Sonenberg N. (1993) The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol. Cell. Biol. 13, 6789–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blum S., Schmid S. R., Pause A., Buser P., Linder P., Sonenberg N., Trachsel H. (1992) ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 89, 7664–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nielsen P. J., Trachsel H. (1988) The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 7, 2097–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nielsen P. J., McMaster G. K., Trachsel H. (1985) Cloning of eukaryotic protein synthesis initiation factor genes. Isolation and characterization of cDNA clones encoding factor eIF-4A. Nucleic Acids Res. 13, 6867–6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caruthers J. M., Johnson E. R., McKay D. B. (2000) Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 97, 13080–13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oberer M., Marintchev A., Wagner G. (2005) Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 19, 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schütz P., Bumann M., Oberholzer A. E., Bieniossek C., Trachsel H., Altmann M., Baumann U. (2008) Crystal structure of the yeast eIF4A-eIF4G complex. An RNA helicase controlled by protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 105, 9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hinton T. M., Coldwell M. J., Carpenter G. A., Morley S. J., Pain V. M. (2007) Functional analysis of individual binding activities of the scaffold protein eIF4G. J. Biol. Chem. 282, 1695–1708 [DOI] [PubMed] [Google Scholar]
- 15. Pause A., Méthot N., Svitkin Y., Merrick W. C., Sonenberg N. (1994) Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13, 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korneeva N. L., First E. A., Benoit C. A., Rhoads R. E. (2005) Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J. Biol. Chem. 280, 1872–1881 [DOI] [PubMed] [Google Scholar]
- 17. Rogers G. W., Jr., Richter N. J., Lima W. F., Merrick W. C. (2001) Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276, 30914–30922 [DOI] [PubMed] [Google Scholar]
- 18. Rogers G. W., Jr., Richter N. J., Merrick W. C. (1999) Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 274, 12236–12244 [DOI] [PubMed] [Google Scholar]
- 19. Acker M. G., Kolitz S. E., Mitchell S. F., Nanda J. S., Lorsch J. R. (2007) Reconstitution of yeast translation initiation. Methods Enzymol. 430, 111–145 [DOI] [PubMed] [Google Scholar]
- 20. Mitchell S. F., Walker S. E., Algire M. A., Park E. H., Hinnebusch A. G., Lorsch J. R. (2010) The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol. Cell 39, 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park E. H., Walker S. E., Lee J. M., Rothenburg S., Lorsch J. R., Hinnebusch A. G. (2011) Multiple elements in the eIF4G1 N terminus promote assembly of eIF4G1·PABP mRNPs in vivo. EMBO J. 30, 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levin M. K., Wang Y. H., Patel S. S. (2004) The functional interaction of the hepatitis C virus helicase molecules is responsible for unwinding processivity. J. Biol. Chem. 279, 26005–26012 [DOI] [PubMed] [Google Scholar]
- 23. Lorsch J. R., Herschlag D. (1998) The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry 37, 2180–2193 [DOI] [PubMed] [Google Scholar]
- 24. Berset C., Zurbriggen A., Djafarzadeh S., Altmann M., Trachsel H. (2003) RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA 9, 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lamphear B. J., Kirchweger R., Skern T., Rhoads R. E. (1995) Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270, 21975–21983 [DOI] [PubMed] [Google Scholar]
- 26. Tarun S. Z., Jr., Sachs A. B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15, 7168–7177 [PMC free article] [PubMed] [Google Scholar]
- 27. Rogers G. W., Jr., Lima W. F., Merrick W. C. (2001) Further characterization of the helicase activity of eIF4A. Substrate specificity. J. Biol. Chem. 276, 12598–12608 [DOI] [PubMed] [Google Scholar]
- 28. Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. (1990) Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10, 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marintchev A., Edmonds K. A., Marintcheva B., Hendrickson E., Oberer M., Suzuki C., Herdy B., Sonenberg N., Wagner G. (2009) Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136, 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bi X., Goss D. J. (2000) Wheat germ poly(A)-binding protein increases the ATPase and the RNA helicase activity of translation initiation factors eIF4A, eIF4B, and eIF-iso4F. J. Biol. Chem. 275, 17740–17746 [DOI] [PubMed] [Google Scholar]
- 31. Bi X., Ren J., Goss D. J. (2000) Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 39, 5758–5765 [DOI] [PubMed] [Google Scholar]
- 32. Altmann M., Wittmer B., Méthot N., Sonenberg N., Trachsel H. (1995) The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J. 14, 3820–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaye N. M., Emmett K. J., Merrick W. C., Jankowsky E. (2009) Intrinsic RNA binding by the eukaryotic initiation factor 4F depends on a minimal RNA length but not on the m7G cap. J. Biol. Chem. 284, 17742–17750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prévôt D., Décimo D., Herbreteau C. H., Roux F., Garin J., Darlix J. L., Ohlmann T. (2003) Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 22, 1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Treder K., Kneller E. L., Allen E. M., Wang Z., Browning K. S., Miller W. A. (2008) The 3′-cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 14, 134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gallie D. R. (2001) Cap-independent translation conferred by the 5′-leader of tobacco etch virus is eukaryotic initiation factor 4G-dependent. J. Virol. 75, 12141–12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Gregorio E., Preiss T., Hentze M. W. (1998) Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′-end-dependent. RNA 4, 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tarun S. Z., Jr., Sachs A. B. (1997) Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol. Cell. Biol. 17, 6876–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Borman A. M., Kirchweger R., Ziegler E., Rhoads R. E., Skern T., Kean K. M. (1997) eIF4G and its proteolytic cleavage products. Effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA 3, 186–196 [PMC free article] [PubMed] [Google Scholar]
- 40. Ohlmann T., Rau M., Pain V. M., Morley S. J. (1996) The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 15, 1371–1382 [PMC free article] [PubMed] [Google Scholar]
- 41. Vassilenko K. S., Alekhina O. M., Dmitriev S. E., Shatsky I. N., Spirin A. S. (2011) Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 39, 5555–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E. (2004) Dynamics and processivity of 40 S ribosome scanning on mRNA in yeast. Mol. Microbiol. 51, 987–1001 [DOI] [PubMed] [Google Scholar]
- 43. Spirin A. S. (2009) How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian Ratchet model. Biochemistry 48, 10688–1069219835415 [Google Scholar]