Background: Presenilin is essential for neuronal survival in adult brains. Notch is a key mediator of presenilin function in developing brains.
Results: Deletion of Notch1/Notch2 in excitatory neurons of the adult cortex does not cause neurodegeneration or reduction of Notch expression.
Conclusion: Notch does not mediate presenilin-dependent survival of adult cortical neurons.
Significance: Notch expression is undetectable in excitatory neurons of the adult cortex.
Keywords: Alzheimer Disease, Neurons, Neuroscience, Presenilin, Secretases, γ-Secretase, Conditional Knockout, Neuronal Survival
Abstract
Activation of Notch signaling requires intramembranous cleavage by γ-secretase to release the intracellular domain. We previously demonstrated that presenilin and nicastrin, components of the γ-secretase complex, are required for neuronal survival in the adult cerebral cortex. Here we investigate whether Notch1 and/or Notch2 are functional targets of presenilin/γ-secretase in promoting survival of excitatory neurons in the adult cerebral cortex by generating Notch1, Notch2, and Notch1/Notch2 conditional knock-out (cKO) mice. Unexpectedly, we did not detect any neuronal degeneration in the adult cerebral cortex of these Notch cKO mice up to ∼2 years of age, whereas conditional inactivation of presenilin or nicastrin using the same αCaMKII-Cre transgenic mouse caused progressive, striking neuronal loss beginning at 4 months of age. More surprisingly, we failed to detect any reduction of Notch1 and Notch2 mRNAs and proteins in the cerebral cortex of Notch1 and Notch2 cKO mice, respectively, even though Cre-mediated genomic deletion of the floxed Notch1 and Notch2 exons clearly took place in the cerebral cortex of these cKO mice. Furthermore, introduction of Cre recombinase into primary cortical cultures prepared from postnatal floxed Notch1/Notch2 pups, where Notch1 and Notch2 are highly expressed, completely eliminated their expression, indicating that the floxed Notch1 and Notch2 alleles can be efficiently inactivated in the presence of Cre. Together, these results demonstrate that Notch1 and Notch2 are not involved in the age-related neurodegeneration caused by loss of presenilin or γ-secretase and suggest that there is no detectable expression of Notch1 and Notch2 in pyramidal neurons of the adult cerebral cortex.
Introduction
Notch receptors are type I transmembrane proteins and are involved in a variety of cell-fate decisions during development (1). Upon ligand binding, Notch undergoes a proteolytic cleavage at the extracellular juxtamembrane region (site 2) by tumor necrosis factor-α-converting enzyme, a member of a disintegrin and metalloprotease (ADAM) family (2–5). The resulting C-terminal fragment is further cleaved at transmembrane domain site 3 by γ-secretase (6–9), releasing the Notch intracellular domain, which translocates to the nucleus where it binds to CSL3 (CBF1/RBP-Jκ/Su(H)/Lag-1) and relieves the transcriptional suppression imposed by CSL. Presenilin (PS) encoded by the PSEN genes, two major genes linked with familial Alzheimer disease (AD), is an essential component of the γ-secretase complex and is required for the Notch intracellular domain production (7, 9). Our previous genetic analysis in mice showed that Notch is a key mediator of PS/γ-secretase function in the developing brain (10–13). In addition, genetic ablation of either Notch1 or Notch2 results in phenotypes that resemble PS1−/− or PS−/− mice (14–19). In contrast, Notch3−/− or Notch4−/− mice do not show embryonic lethality (20, 21), suggesting their unessential function in embryonic development.
In the central nervous system, the Notch signaling pathway is important for neural stem cell maintenance and proper neurogenesis in the embryonic brain (11–13, 22–25) as well as the adult brain (26–29). Interestingly, besides expression in the germinal zone where neural stem cells reside, Notch has been reported to express in terminally differentiated neurons in the adult cerebral cortex (30–32). Furthermore, altered expression of Notch receptors, its ligand Dll1, and effector Hes1 have been implicated in several brain disorders including AD, Down syndrome, and prion disease (33–35). Thus, despite its low expression in mature neurons, Notch may play an important role in the adult brain.
We previously reported that conditional inactivation of PS in excitatory neurons of the cerebral cortex causes progressive memory impairment and age-related neuronal degeneration (36, 37), raising the possibility that loss of PS function may underlie dementia and neurodegeneration in AD (38). Furthermore, similar inactivation of nicastrin, another key component of the γ-secretase complex, also leads to memory impairment and neurodegeneration (39), suggesting that γ-secretase-dependent activity of PS is important for neuronal survival. However, the molecular basis by which PS promotes neuronal survival in the adult brain remains to be elucidated. Although many γ-secretase substrates have been reported, most of them were identified using overexpression systems and cell lines (40, 41). Beyond the well established physiological substrates of γ-secretase, Notch and amyloid precursor protein (APP) (9, 42), the significance of γ-secretase-mediated cleavage of other substrates is often unclear. Based on prior genetic studies which demonstrated that Notch is an important downstream mediator of presenilin function in the developing brain (6, 7, 10, 12, 13, 43, 44), it was widely assumed that Notch is still a functional target of presenilin in the adult brain. In this study, we address this important question directly through the generation of Notch conditional knock-out (cKO) mice using the same αCaMKII-Cre transgenic mouse line that was used previously to inactivate presenilin and nicastrin selectively in mature excitatory neurons of the cerebral cortex beginning at the third to fourth postnatal week (36, 39, 45). Because Notch1 and Notch2 are reportedly the only two Notch receptors expressed in neurons of the adult rodent brain (46–48), we generated Notch1, Notch2, and Notch1/2 cKO mice to determine whether Notch1 and Notch2 are similarly required for neuronal survival as presenilin and nicastrin. Surprisingly, these three lines of mutant mice do not exhibit any sign of neurodegeneration. In addition, these mutant mice do not show any reduction in the levels of Notch mRNAs and proteins in their cerebral cortices, where Cre-mediated genomic deletion of the floxed Notch exons occurred correctly. In contrast to these results, there was no detectable Notch1 and Notch2 proteins in primary cortical cultures derived from homozygous floxed Notch1/2 neonatal pups infected with a Cre-expressing lentivirus, which we previously showed to infect all primary cultured neurons (49, 50), indicating that the floxed Notch1 and Notch2 alleles can be efficiently inactivated in the presence of Cre. Collectively, these results demonstrate that Notch1 and Notch2 are not involved in presenilin-dependent neuronal survival in the adult cerebral cortex and that there is no detectable Notch1 and Notch2 expression in excitatory pyramidal neurons of the hippocampus and the neocortex of the adult brain.
EXPERIMENTAL PROCEDURES
Generation of Notch cKO Mice
The generation of floxed Notch1 (fNotch1) and floxed Notch2 (fNotch2) mice (11, 51) and αCaMKII-Cre transgenic mice (45) was described previously. Exon1 and exon3 were floxed for the Notch1 and Notch2 genes, respectively. Forebrain-specific Notch1 cKO mice and littermate control were obtained by crossing fNotch1/fNotch1 mice with fNotch1/fNotch1; αCaMKII-Cre (Notch1 cKO) mice. Likewise, Notch2 cKO mice were obtained by crossing fNotch2/fNotch2 mice to fNotch2/fNotch2; αCaMKII-Cre (Notch2 cKO) mice. Notch1/Notch2 cDKO mice were obtained by crossing fNotch1/fNotch1;fNotch2/fNotch2 mice with fNotch1/fNotch1;fNotch2/fNotch2;αCaMKII-Cre (Notch1/Notch2 cDKO) mice. We used female mice carrying the αCaMKII-Cre transgene for breeding to reduce the number of offspring bearing germ line deletions. All experimental mice were in the C57BL6/J 129 hybrid background, and littermates were used as controls. All procedures relating to animal care and treatment conformed to the Institutional and NIH guidelines.
PCR Genotype
Genomic PCR was performed using the primers for the deleted, the floxed, and the wild-type Notch alleles. For the Notch1 gene, the following primers were used: 5′-ATTGAAAGCACATATGGAGAT-3′ (forward primer at ∼2600 nt upstream of exon1), 5′-GTATAAGCATGAAGTGGTCCA-3′ (reverse primer at ∼2200 nt upstream of exon1), and 5′-CTCAGTTCAAACACAAGATACGA-3′ (reverse primer at ∼1200 nt downstream of exon1). The first and the second primers amplify the wild-type and the floxed Notch1 alleles, giving rise to PCR products of 400 and 500 bp, respectively. The first and the last primers amplify the deleted Notch1 allele, yielding a 600-bp PCR product. For the Notch2 gene, the following primers were used: 5′-AGCACTCAGTTGTGAAGGAGC-3′ (forward primer at ∼500nt upstream of exon3), 5′-TGTTAGATACCAGCCTGGGAG-3′ (forward primer at ∼350 nt downstream of exon3), and 5′-TCCCTTCAAACTCTCCAAAGG-3′ (reverse primer at ∼700 nt downstream of exon3). The first and the last primers amplify the deleted Notch2 allele and give rise to a 489-bp PCR product, whereas the second and the last primers amplify the wild-type and the floxed Notch2 alleles, resulting in PCR products of 343 and 383 bp, respectively. PCR products were separated and analyzed in 2% agarose gel.
Primary Cortical Neuronal Cultures
Dissociated cortical neuronal cultures were prepared from newborn fNotch1/fNotch1;fNotch2/fNotch2 pups as described previously (52). Briefly, cerebral cortex was dissected from the brain of neonates. Neurons were dissociated by trypsin treatment (Sigma; 2.5 mg/ml for 10 min at 37 °C), triturated with a siliconized Pasteur pipette, and then plated at 1.5 × 105 cells/cm2 onto 12-well plates coated with Matrigel (BD Biosciences). Cultures were maintained in minimal essential medium, 5 g/liter glucose, 0.1 g/liter transferrin (Calbiochem), 0.25 g/liter insulin (Sigma), 0.3 g/liter glutamine, 5% fetal bovine serum (HyClone), 2% B-27 supplement (Invitrogen), and 2–4 μm cytosine arabinoside (Sigma) at 37 °C in a humidified incubator gassed with 95% air and 5% CO2 until 2–10 days in vitro (DIV) for biochemical analyses.
Lentivirus Production and Infection
Production of recombinant lentiviruses is achieved by transfecting HEK293T cells with three plasmids by FuGENE6 (Roche Applied Science). Vesicular Stomatitis Virus glycoprotein (VSVg) and Δ8.9 are plasmids encoding the envelope and the gag/pol/tat proteins of lentivirus, respectively. pFUGW-EGFP-NLS-Cre and pFUGW-EGFP-NLS were previously described (49, 53). Viruses were harvested 48 h after transfection by collecting the medium from transfected cells and filtrated. Titer of the lentivirus was estimated by measuring the GFP-positive cells with flow cytometry after the infection of diluted lentivirus to HEK293 cells. Cortical neurons at DIV1 were infected with each lentivirus at a 3–4 multiplicity of infection.
Western Blot
The cortices were dissected from brains and homogenized in 1 ml of radioimmune precipitation assay buffer (150 mm NaCl, 50 mm Tris-Cl (pH 7.4), 2 mm EDTA, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, protease inhibitors and phosphatase inhibitors mixture from Sigma). The protein concentration was measured by BCA assay (Pierce), and the same amount of protein per lane (30 μg for cortex lysate, 10 μg for cortical neuronal culture) was separated in NuPAGE Novex 3–8% Tris acetate gel (Invitrogen). Proteins were transferred to nitrocellulose or PDVF membrane, and the membranes were blocked in 5% nonfat milk, Tris-buffered saline (TBS) for 1 h. After that, the membranes were incubated at 4 °C overnight with the primary antibody against Notch1 rabbit polyclonal (used for primary neuronal cultures in Fig. 5 and mouse brains in supplemental Fig. S1; a kind gift of A. Israel), Notch1 rabbit monoclonal (used for mouse brains in Fig. 7; clone D1E11, Cell Signal Technology, #3608), Notch2 rat monoclonal (used for primary neuronal cultures in Fig. 5 and mouse brains in supplemental Fig. S1; clone C651.6DbHN, Developmental Studies Hybridoma Bank University of Iowa), Notch2 rabbit monoclonal (used for mouse brains in Fig. 7; clone D76A6, Cell Signal Technology #5732), Vasolin containing protein (VCP) rabbit polyclonal (Santa Cruz), and α-tubulin mouse monoclonal (Sigma). The membrane was then incubated with IRDye 800CW or IRDye 680-labeled secondary antibodies (LI-COR Bioscience) at room temperature for 1 h. Specific signals were developed by Odyssey Infrared Imaging System (LI-COR Biosciences).
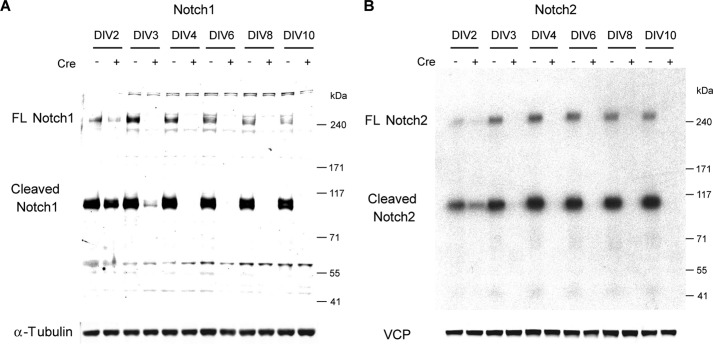
FIGURE 5.
Elimination of Notch1 and Notch2 proteins in primary cortical neuronal cultures after Cre transduction. Shown is the time course of Notch1 (A) or Notch2 (B) inactivation in fNotch1/fNotch1;fNotch2/fNotch2 cortical neuronal cultures after Cre transduction. The lentivirus carrying the cDNA encoding either a GFP/defective Cre (Cre−) or a GFP/functional Cre fusion protein (Cre+) was introduced to infect cortical neuronal cultures derived from fNotch1/fNotch1;fNotch2/fNotch2 neonate pups at DIV1. Total cell lysates were collected at the designated days (DIV2–10) and were analyzed (10 μg of lysates per lane) by Western blotting using specific antibodies for the Notch1 (rabbit polyclonal, from A. Israel) or the Notch2 (rat monoclonal, clone C651.6DbHN from the Hybridoma Bank, University of Iowa) C-terminal domain. α-Tubulin or VCP (Vasolin containing protein) was used as a loading control. Although most Notch proteins were subjected to S1 cleavage (Cleaved, ∼95 kDa), full-length (FL) Notch (∼270 kDa) could be easily detected in this culture system. In the presence of Cre, inactivation of Notch1 and Notch2 occurs rapidly, and loss of Notch1 and Notch2 proteins is complete by DIV4.
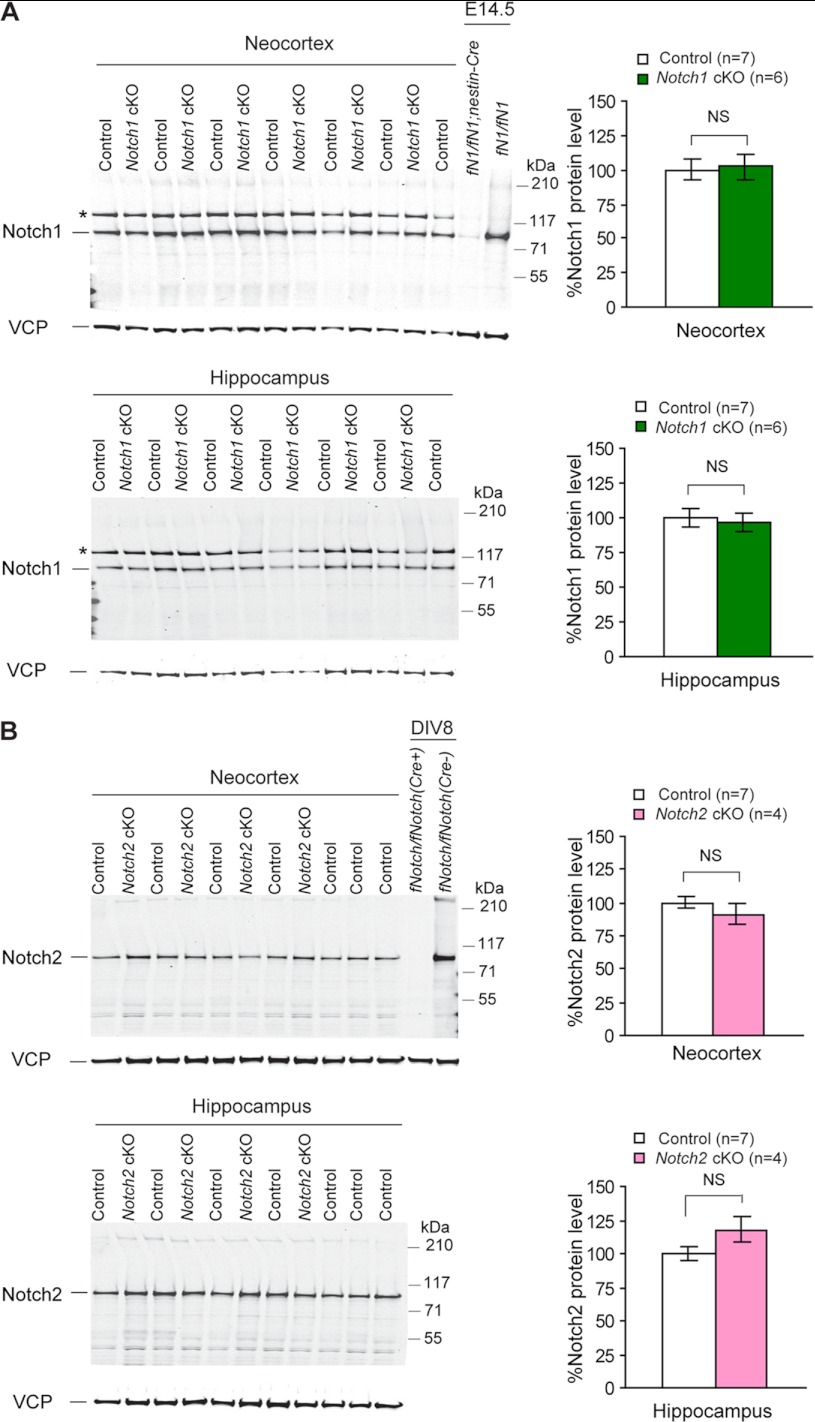
FIGURE 7.
Unchanged levels of Notch1 and Notch2 proteins in the cerebral cortex of Notch cKO mice. A, shown are Western analysis (left) and quantification (right) of Notch1 proteins in the neocortex and the hippocampus of adult Notch1 cKO (n = 6) and littermate control (n = 7) mice using a Notch1 rabbit monoclonal antibody (clone D1E11, Cell Signal Technology). B, shown are Western analysis (left) and quantification (right) of Notch2 proteins in the neocortex and hippocampus of adult Notch2 cKO (n = 4) and control (n = 7) mice using a Notch2 rabbit monoclonal antibody (clone D76A6, Cell Signal Technology). No significant difference of Notch1 or Notch2 proteins was detected between cKO and control samples, indicating that despite the deletion of the floxed exons at the genomic DNA level, protein expression is not altered in cKO cortical samples and suggesting that there is little Notch expression in these excitatory pyramidal neurons of the adult cerebral cortex where Cre is expressed under the control of the αCaMKII promoter. Total protein lysates of E14.5 embryonic brains of Nestin-Cre-driven Notch1 cKO (fNotch1/fNotch1;Nestin-Cre) mice (A) or neuronal cultures (DIV8) derived from fNotch1/fNotch1;fNotch2/fNotch2 (fNotch/fNotch) postnatal pups (B) are included as controls. All values are normalized to that of VCP (Vasolin containing protein) protein, which is used as loading control. The Notch1 antibody (rabbit monoclonal) used here recognizes a nonspecific signal that is only present in the adult brain (*). All data are expressed as the mean ± S.E. Statistical analysis was performed using two-tailed unpaired Student's t test. NS, not significant.
Northern Blot
Total RNAs were isolated with TRI reagent (Sigma) according to manufacture's instruction. For Northern blot, ∼20 μg of total RNA were separated in formaldehyde agarose gels and transferred into nylon membrane (Amersham Biosciences). Hybridization was performed using [α-32P]dCTP-labeled probes specific for Notch1 (408-bp coding sequence from exons 6–8), Notch2 (260-bp coding sequence from exon3), and GAPDH (452-bp coding sequence from exons 5–7). Specific signals were detected by autoradiography with Hyperfilm (Amersham Biosciences). Signal intensities were quantified by ImageJ software (NIH). Statistic significance was calculated using Student's t test.
Nissl Staining and Stereology
Mice were anesthetized with CO2 and perfused with phosphate-buffered saline including heparin and procaine. We took out the brain, and hemibrains were further immersed in 4% paraformaldehyde at 4 °C for 3 h and then processed for paraffin embedding. Serial sagittal sections were collected by microtome at 10 μm in thickness. Sections from every 40 slides were deparaffinized, dehydrated, and stained with 0.5% Cresyl Violet (Sigma). Brain volumes were measured by BioQuant image analysis software.
Immunohistochemistry
Paraffin-embedded brain sections were deparaffinized, alcohol-dehydrated, and blocked in 5% normal horse serum, TBS for 1 h. Then sections were reacted with primary antibodies against NeuN (1:400, Chemicon) or GFAP (1:500, Sigma) at 4 °C overnight. These slides were then incubated with biotinylated secondary antibody (Vector Laboratories Inc.) at room temperature for 1 h. Specific signals were developed by Vectastain Elite ABC kit and DAB peroxidase substrate (Vector Laboratories, Inc.) and analyzed by BX50 microscope system (Olympus).
Counting Number of Cortical Neurons
The NeuN-stained sections (total number is 6 sections per animal, which spaced 0.4 mm apart each other) were analyzed by an unbiased neuronal counting by the fractionators and optical dissector method and showed the live image on the BioQuant image analysis software, which connected to the Leica DMRB microscope with a CCD camera. Forty optical dissectors were used to count the entire cortex area. Each optical dissector was a 50 × 50-μm sampling box. Then the number of neurons can be counted with an indicator of NeuN-positive cells through the 100× oil-immersion lens. The total number of neurons should be sum of the bilateral cortex neurons from all the picked slides. The genotype of each section was also blind to the experimenter. The coefficient of error from the counting technique was <0.10. Finally, the average number of neurons was calculated per genotype (n = 4 per genotype). Values are reported as the means ±S.E.
RESULTS
Notch1, Notch2, and Notch1/2 cKO Mice Do Not Exhibit Age-related Neurodegeneration
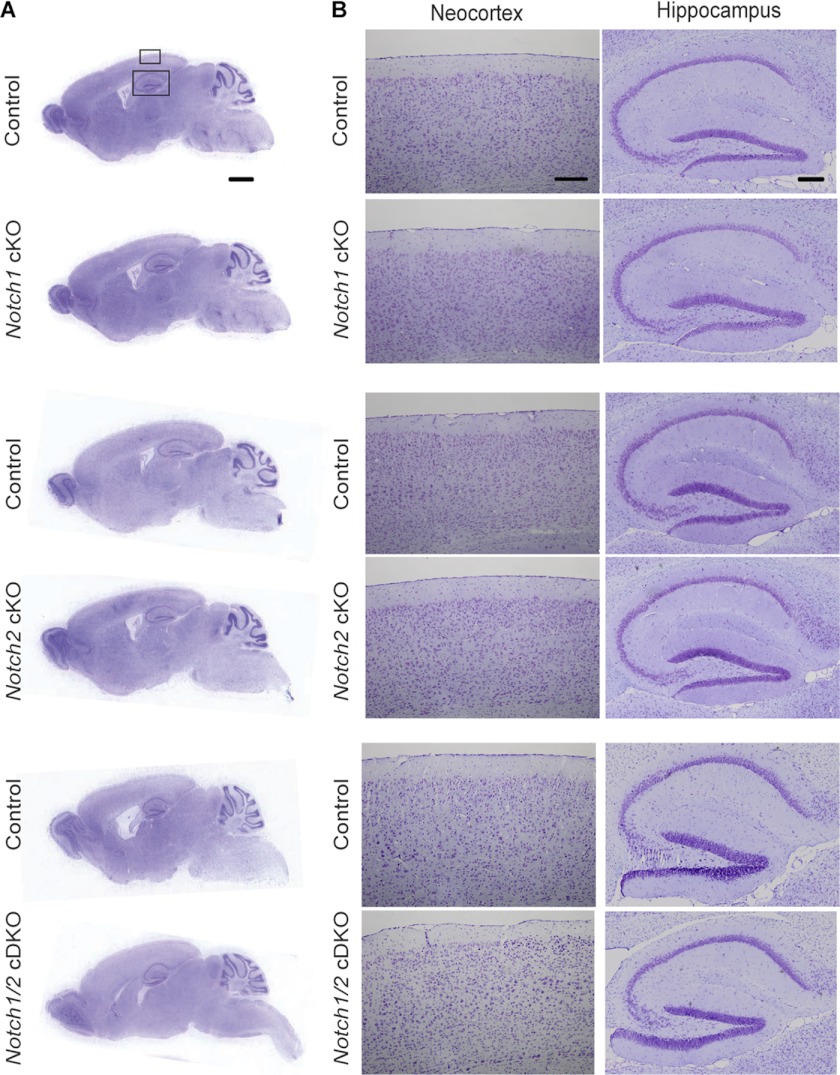
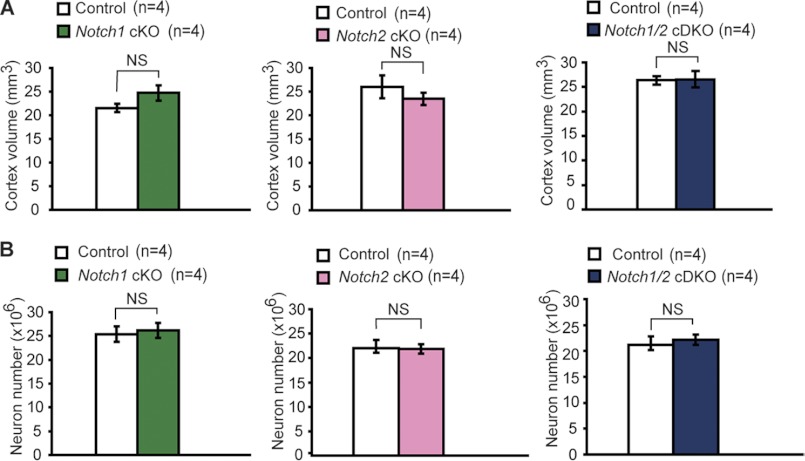
Prior genetic studies have shown that activation of Notch receptors are presenilin- or γ-secretase-dependent and that Notch receptors are key mediators of presenilin function in cortical development (10–12, 14–19, 54–56). It was, therefore, widely assumed that Notch may still be a target of presenilin in mediating adult brain function and that γ-secretase inhibitors may have unwanted side effects due to its inhibition of Notch function. To address this question, we generated Notch1, Notch2, and Notch1/2 cKO mice using the αCaMKII-Cre transgenic mouse, which we used previously to generate presenilin and nicastrin cKO mice (39, 45). In the presence of the αCaMKII-Cre transgene, Cre recombinase is expressed under the control of the α-calcium calmodulin-dependent kinase II promoter in excitatory neurons of the cerebral cortex beginning at approximately postnatal day 18 (45). Using the same Cre line, we will be able to compare directly the consequence of conditional deletion of Notch1/2 with the phenotypes of presenilin and nicastrin cKO mice to determine whether presenilin and γ-secretase promote neuronal survival through the Notch signaling pathway. Presenilin and nicastrin cKO mice develop age-related neurodegeneration, and by 6–9 months the cerebral cortex shows severe atrophy and dramatic loss of cortical volume and neurons (36, 37, 39). Our histological analysis of Notch1, Notch2, and Notch1/2 cKO mice, however, did not yield any cortical atrophy (Fig. 1). Nissl staining of paraffin-embedded series sagittal sections showed no gross histological change in brain morphology in aged Notch1 cKO (24 months), Notch2 cKO (24 months), and Notch1/2 cKO (12 months) mice (Fig. 1, A and B). We then measured cortical volume using stereological methods. Unlike PS cDKO mice, which showed progressive loss of cortical volume and neuron number (35% loss of cortical volume and 18% lost of cortical neurons at 6 months of age), the volume of the cerebral cortex was not significantly different (n = 4 per genotype) in each of the three Notch cKO mice relative to their respective littermate controls (Fig. 2A). Furthermore, we performed immunohistochemical analysis using an antibody specific for NeuN, which specifically stains the nucleus of neurons, followed by quantification of the number of NeuN-positive cells using unbiased stereological methods. Again we found similar numbers of neurons in the neocortex in each of the three Notch cKO mice compared with their respective littermate control mice (Fig. 2B). These results indicate that conditional deletion of Notch1 and/or Notch2 in excitatory pyramidal neurons of the cerebral cortex does not cause loss of cortical neurons or volume during the life span of mice.
FIGURE 1.
Normal gross morphology of the cerebral cortex of Notch cKO mice. A and B, shown is Nissl staining of comparable sagittal brain sections (10 μm, paraffin) of aged Notch cKO and littermate control mice (24 months for Notch1 cKO, 24 months for Notch2 cKO, 12 months for Notch1/2 cDKO). Representative images of sagittal brain sections at low magnification (A) and the boxed areas of the neocortex and hippocampus at a higher magnification (B) are shown. Scale bar, 1 mm (A) and 200 μm (B).
FIGURE 2.
No cerebral atrophy and neuronal loss in the neocortex of aged Notch cKO mice. A, shown is stereological measurement of the cortical volume from Notch cKO and littermate control brains (16–24 months for Notch1 cKO, 22–26 months for Notch2 cKO, 6–12 months for Notch1/2 cDKO mice). Values are presented per hemisphere. There is no significant (NS) difference in the neocortical volume between Notch cKO mice and their respective littermate controls (n = 4 per genotype, p > 0.05). B, shown is stereological quantification of neuronal number in the neocortex using NeuN staining. Similar numbers of cortical neuron are present between Notch cKO mice and the littermate control mice (n = 4 per genotype, p > 0.05). NS, not significant.
Because astrogliosis is often associated with ongoing neurodegeneration (36, 39, 57–64), we further looked for the presence of astrogliosis in these Notch cKO mice. We performed immunohistochemical analysis on glial fibrillary acidic protein (GFAP), a marker of astrogliosis, using brain sections of aged Notch cKO mice and littermate controls. We did not detect GFAP-immunoreactive astrogliosis in the hippocampus (Fig. 3A) and the neocortex (data not shown) in each of the three Notch cKO mice even at ∼24 months of age, whereas we could easily see significant increases of GFAP immunoreactivity in the hippocampus of nicastrin cKO mice at 9 months of age (Fig. 3A), when prominent neurodegeneration had taken place (39). Western analysis also showed no up-regulation of GFAP in the cerebral cortex of Notch1 and Notch2 cKO mice compared with littermate controls at 14–23 months of age (Fig. 3B), whereas 10-fold up-regulation of GFAP was detected in the cortex of PS cDKO mice at 6 months of age (58). Together, these results demonstrate the lack of neurodegeneration in Notch1 and Notch2 single and double cKO mice.
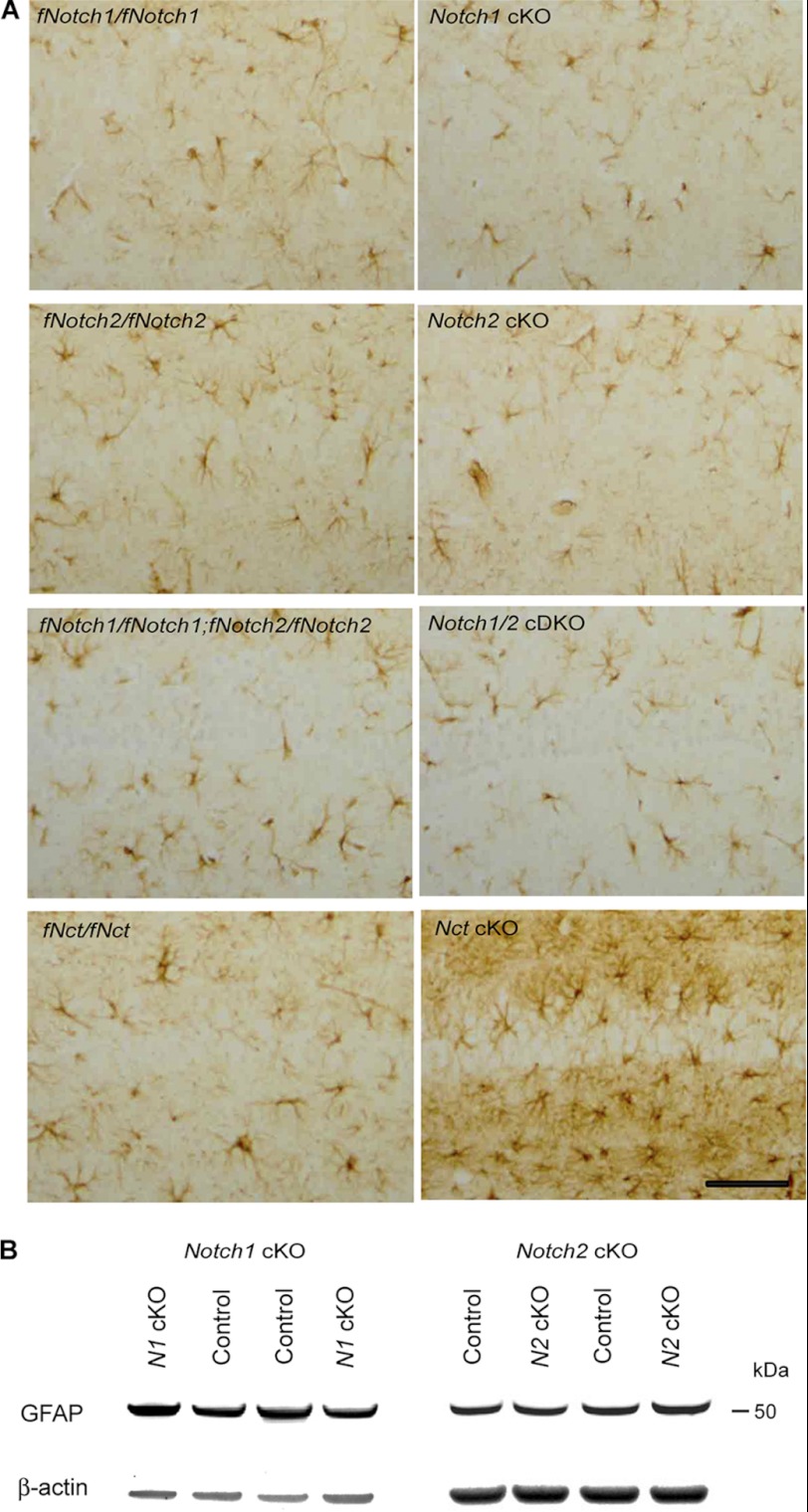
FIGURE 3.
No astrogliosis or up-regulation of GFAP in the cerebral cortex of Notch cKO mice. A, shown is GFAP immunohistochemical analysis of Notch1 cKO, Notch2 cKO, and Notch1/2 cDKO mice using paraffin sections of Notch cKO and littermate control mice at 12–26 months old of age. Representative views of hippocampal area CA1 at comparable levels are shown. There is no increase in GFAP immunoreactivity in any of the Notch cKO mice compared with the respective controls (fNotch1/fNotch1 for Notch1 cKO, fNotch2/fNotch2 for Notch2 cKO, fNotch1/fNotch1;fNotch2/fNotch2 for Notch1/2 cDKO). GFAP immunostaining of Nicastrin (Nct) cKO and littermate control (fNct/fNct) brains at 9 months of age is also included as the positive control for reactive astrogliosis. Scale bar, 100 μm. B, shown is Western analysis of GFAP in the cerebral cortex of Notch1 or Notch2 cKO mice. The level of GFAP expression (50 kDa) is similar in cortical lysates of Notch1 cKO and littermate control mice at 23 months of age or Notch2 cKO and littermate control mice at 14 months of age. β-Actin is used as loading control.
Floxed Notch1 and Notch2 Alleles Are Excised Correctly in Cerebral Cortex of Notch1 and Notch2 cKO Mice
The lack of neurodegeneration and other phenotypes in Notch1 and Notch2 cKO mice prompted us to examine whether the floxed Notch1 and Notch2 alleles (exon1 for Notch1, exon3 for Notch2) are appropriately excised in the cerebral cortex of these cKO mice. We, therefore, performed PCR using genomic DNA samples purified from the neocortex, the hippocampus, and the tail of Notch cKO mice and littermate controls at 2–3 months of age. PCR was performed using 3 primers that can amplify and distinguish all three alleles (floxed, deleted, and wild-type) for Notch1 and Notch2. In the neocortex and the hippocampus, PCR products derived from both the correctly deleted allele (600 bp for Notch1 and 489 bp for Notch2) and the floxed allele (500 bp for Notch1 and 383 bp for Notch2) are present in Notch1 and Notch2 cKO mice, respectively, whereas only the floxed allele-derived single product was amplified in their littermate controls (Fig. 4, A and B). In contrast to genomic DNAs obtained from the cortical samples, single PCR products corresponding to the appropriate floxed allele were detected in the tail DNA of Notch1 and Notch2 cKO as well as their respective littermate controls (Fig. 4, A and B). The presence of the PCR products corresponding to the floxed Notch1 and Notch2 alleles in the hippocampus and the neocortex of the Notch1 and Notch2 cKO mice, respectively, is due to the selective expression of αCaMKII-Cre in excitatory pyramidal neurons (45, 50, 65). Thus, the floxed Notch alleles remain intact in cells other than excitatory neurons, such as interneurons, glia, and neural stem cells in the cerebral cortex where genomic DNA was isolated. Indeed we observed a similar proportion of PCR products derived from the deleted and the floxed alleles in the cerebral cortex of PS cDKO mice (data not shown), in which we used the same αCaMKII-Cre transgenic mice. These results confirm that excision of the floxed Notch1 and Notch2 exons occurred correctly in the cerebral cortex of Notch1 and Notch2 cKO mice, respectively.
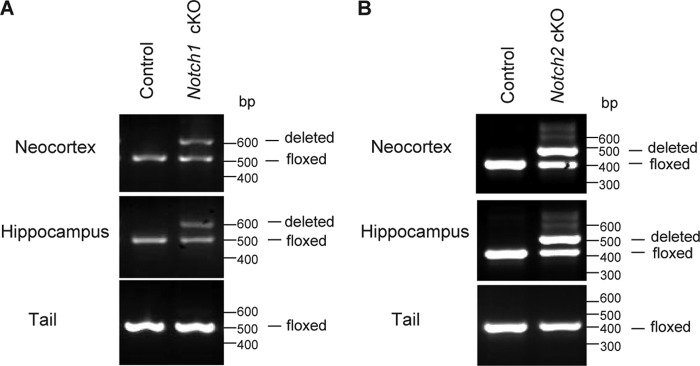
FIGURE 4.
Deletion of the floxed Notch sequences in the cerebral cortex of Notch1 and Notch2 cKO mice. A and B, PCR using genomic DNA isolated from the neocortex and the hippocampus as well as the tail of Notch1 cKO (A) and Notch2 cKO (B) mice is shown. PCR was performed using three primers that can amplify and distinguish the floxed, deleted, and wild-type alleles. Only one PCR product (500 bp for the floxed Notch1 allele, 383 bp for the floxed Notch2 allele) is produced in the DNA samples from the neocortex, the hippocampus, and the tail of control (fNotch1/fNotch1 for Notch1 cKO, fNotch2/fNotch2 for Notch2 cKO) mice. In cKO mice, one PCR product (500 bp for the floxed Notch1 allele, 383 bp for the floxed Notch2 allele) is produced in the tail DNA sample, whereas two PCR products representing the floxed and the deleted (600 bp for Notch1, 489 bp for Notch2) alleles are produced in DNA samples from the neocortex and hippocampus, suggesting mosaic cell populations (some carrying the floxed allele, some carrying the deleted allele) in the cortex.
Introduction of Cre Recombinase Efficiently Eliminated Notch1 and Notch2 Expression in Cultured Cortical Neurons Carrying Floxed Notch1 and Notch2 Alleles
Notch proteins are expressed highly in primary neuronal cultures compared with adult brains (30, 31). To determine whether introduction of Cre recombinase can effectively eliminate expression of Notch1 and Notch2 proteins in cultured cortical neurons bearing the floxed Notch alleles, we used a Cre-expressing lentivirus, which can efficiently infect all cultured neurons based on our earlier studies (49, 50). We, therefore, developed primary cortical neuronal cultures from neonates carrying homozygous floxed Notch1 and Notch2 alleles (fNotch1/fNotch1;fNotch2/fNotch2) in which both Notch1 and Notch2 can be inactivated upon introduction of the Cre recombinase. After one day in vitro (DIV1), we introduced a lentivirus carrying the cDNA encoding a functional Cre recombinase into the neuronal cultures and continued to culture primary neurons until the designated days. Western analysis was performed on the collected total cell lysates using antibodies specific for the C-terminal region of Notch1 or Notch2. Levels of both full-length Notch (∼270 kDa) and furin (S1)-cleaved products (∼95 kDa) were unchanged over time in cultures infected with the control lentivirus, which carries the cDNA encoding a defective Cre recombinase (Cre−, Fig. 5, A and B). In contrast, Notch1 and Notch2 proteins are decreased in cortical cultures infected with a functional Cre lentivirus beginning at DIV2 and are completely absent by DIV4 (Cre+, Fig. 5, A and B). The reduction of Notch1 and Notch2 proteins after Cre transduction is more rapid than that of PS1 protein in cortical cultures derived from floxed PS1 neonates, in which PS1 protein is completely eliminated by DIV7 (49). This difference could be due to the easier accessibility of Cre recombinase to the Notch loci (66) or shorter half-life of Notch mRNAs and proteins (67–70).
Levels of Notch1 and Notch2 mRNAs and Proteins Are Unchanged in Cerebral Cortex of Notch1 and Notch2 cKO Mice
The genomic PCR and primary neuronal culture experiments confirmed that the floxed Notch alleles are deleted in the cerebral cortex of cKO mice and that inactivation of Notch is complete in cultured cortical neurons where Cre recombinase is efficiently introduced by lentivirus. We examined further whether Notch mRNA and protein expression are reduced in the cerebral cortex of Notch cKO mice similarly as in PS1 cKO and nicastrin cKO mice carrying the same αCaMKII-Cre mice (39, 45). Although excitatory neurons are only a subset of cell types in the cerebral cortex, our previously generated presenilin, nicastrin and CBP cKO mice using this αCaMKII-Cre mouse all showed ∼50% reduction of mRNAs and proteins in the cerebral cortex (39, 45, 71). Surprisingly, Northern analysis using total RNA isolated from the hippocampus and the neocortex of Notch cKO mice and littermate controls at 2 months or later ages showed similar levels of full-length Notch1 mRNAs (∼9.5 kb) in the neocortex and the hippocampus between Notch1 cKO mice and littermate controls (Fig. 6A). Likewise, levels of Notch2 mRNAs (∼10.5 kb) were unchanged between Notch2 cKO mice and controls (Fig. 6B). Western analysis using antibodies specific for Notch1 showed that levels of Notch1 proteins are much lower in hippocampal and neocortical lysates from 2–3-month-old mice compared with E14.5 embryonic brains when the same amount of total proteins (30 μg/lane) was analyzed using rabbit monoclonal antibody specific for C-terminal region of Notch1 (Fig. 7A). Because Notch proteins are subjected to a furin-mediated cleavage at the S1 site, releasing a C-terminal fragment (∼95 kDa), which is further cleaved by ADAM10 upon ligand binding (2–5, 72), we quantified the band of ∼95 kDa in the hippocampal and neocortical samples from Notch1 and Notch2 cKO mice as well as their respective littermate controls. Consistent with the unchanged Notch1 mRNA levels in cKO mice, levels of Notch1 proteins are also unchanged in the neocortex and hippocampus between Notch1 cKO mice and littermate controls, whereas expression of Notch1 was almost completely inactivated in embryonic brains of Notch1 cKO mice driven by Nestin-Cre transgenic mice (Fig. 7A), as shown previously (11). The higher molecular weight band (*) appears to be nonspecific, as this band is not present in samples from the embryonic brain or cultured neurons (Figs. 5A and 7A). In addition, the predicted molecular mass of the S1-cleaved Notch weight of the C-terminal fragment is ∼95 kDa (based on 877 amino acid residues in mouse Notch1). Western analysis using rabbit monoclonal antibodies specific for the C-terminal region of Notch2 also showed that levels of Notch2 proteins are lower in the adult brain (30 μg/lane) compared with cortical neuronal cultures (10 μg per lane) (Fig. 7B). Similar to Notch1, the predominant species detected by Western is the furin-cleaved C-terminal fragment (∼95 kDa). Consistent with the unchanged Notch2 mRNA levels in cKO mice, levels of Notch2 proteins are also unaltered in the neocortex and the hippocampus of Notch2 cKO mice relative to littermate controls, whereas expression of Notch2 was completely absent in fNotch1/fNotch1;fNotch2/fNotch2 cortical cultures 7 days after Cre transduction (Figs. 5B and 7B). We also used another set of antibodies (a rabbit polyclonal for Notch1, a rat monoclonal for Notch2) to confirm further our results. Again, similar levels of Notch1 or Notch2 proteins were found in Notch cKO mice and littermate controls (supplemental Fig. S1). Thus, postnatal deletion of the Notch alleles in excitatory neurons of the cerebral cortex does not affect the amount of Notch proteins detected in the cortex, in contrast to our earlier studies using the same αCaMKII-Cre transgenic mouse to generate PS1, nicastrin, CBP cKO mice, all of which exhibit ∼50% reduction of mRNAs and proteins in the cerebral cortex of cKO mice (39, 45, 71). Together, these results indicate that Notch1 and Notch2 are either normally not expressed or expressed at undetectable levels in the excitatory neurons of the hippocampus and the neocortex.
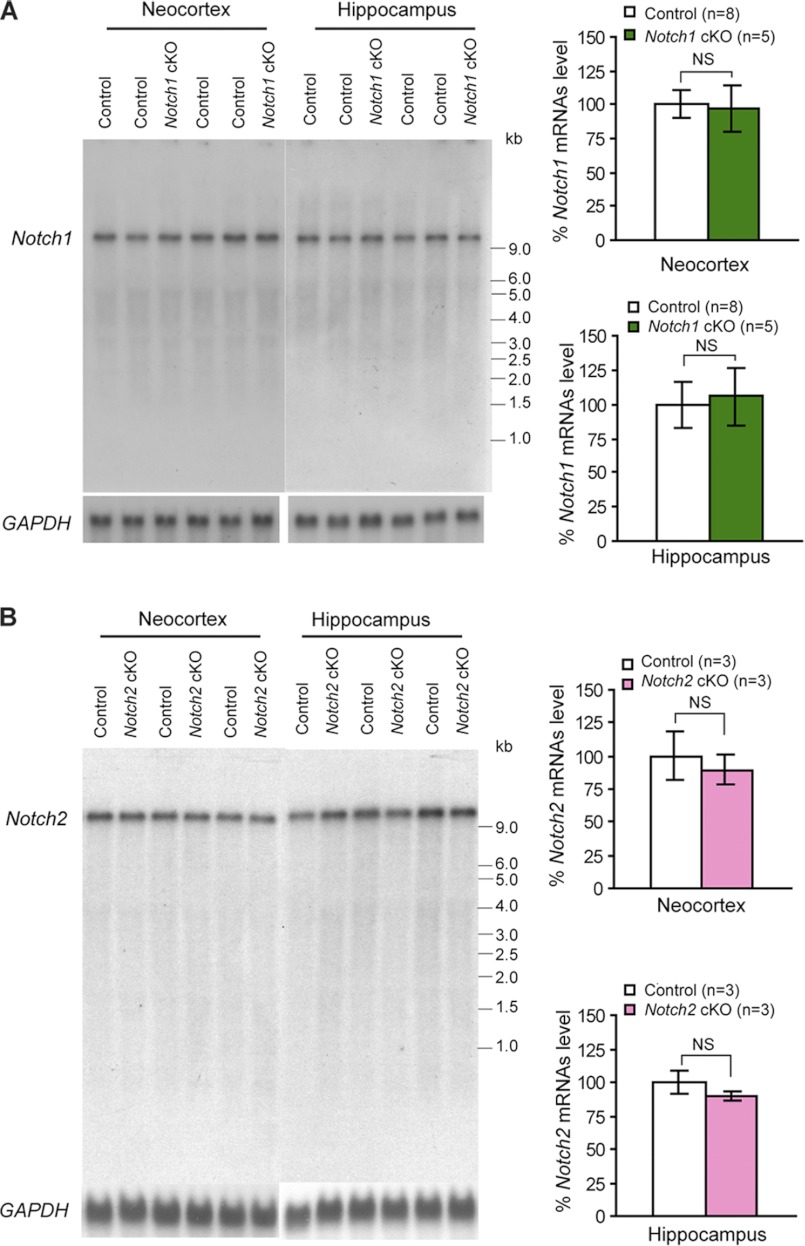
FIGURE 6.
Unchanged levels of Notch1 and Notch2 mRNAs in the cerebral cortex of Notch cKO mice. A, Northern blots (left) and quantification (right) of Notch1 mRNAs in Notch1 cKO (n = 5) and littermate control (n = 8) mice show similar levels of Notch1 mRNAs in total RNA samples isolated from the cortex of cKO and littermate controls. B, Northern blots (left) and quantification (right) of Notch2 mRNAs in Notch2 cKO (n = 3) and littermate control (n = 3) mice show similar levels of Notch2 mRNAs in control and Notch2 cKO mice. The level of Notch mRNAs is normalized to GAPDH mRNAs, and the value of littermate controls is set as 100%. All data are expressed as the mean ± S.E. Statistical analysis was performed using two-tailed unpaired Student's t test. NS, not significant.
DISCUSSION
In this study we generated three lines of conditional knock-out mice in which the Notch1 and/or Notch2 genes are selectively deleted in excitatory neurons of the postnatal forebrain to investigate whether Notch receptors are crucial for neuronal survival in the adult brain. Unexpectedly, we did not detect any neuronal degeneration in the adult cerebral cortex of Notch1,Notch2 cKO and Notch1/Notch2 cDKO mice up to ∼2 years of age (Figs. 1–3), whereas conditional inactivation of presenilin or nicastrin, each of which is essential for γ-secretase-mediated Notch activation, using the same αCaMKII-Cre transgenic mouse caused progressive neuronal loss beginning at 4 months of age (36, 37, 39). More surprisingly, we failed to detect any reduction of Notch1 and Notch2 mRNAs (Fig. 6) and proteins (Fig. 7 and supplemental Fig. S1) in the cerebral cortex of Notch1 and Notch2 cKO mice, respectively, even though Cre-mediated genomic deletion of the floxed Notch1 and Notch2 exons clearly took place in the cerebral cortex of these cKO mice (Fig. 4). Furthermore, introduction of Cre recombinase into primary cortical cultures prepared from the floxed Notch1/Notch2 mice, where Notch1 and Notch2 are highly expressed, completely eliminated their expression (Fig. 5), indicating that Cre can efficiently delete the floxed Notch1 and Notch2 alleles. Based on these findings, we conclude that in the adult cerebral cortex, Notch is not a key mediator of presenilin-dependent neuronal survival and that there is no detectable expression of Notch1 and Notch2 in excitatory neurons of the adult cerebral cortex, where presenilin is highly expressed. Our current study argues against possible functional association between presenilins and Notch in mature pyramidal neurons of the adult cerebral cortex, although before our current study it was widely assumed that Notch1/2 are expressed in these excitatory neurons and could mediate presenilin function during aging.
The failure to identify neurodegenerative phenotypes in our Notch cKO models was somewhat unexpected given the striking neurodegenerative phenotypes observed in presenilin and nicastrin cKO mice (36, 37, 39), the essential requirement of presenilin/γ-secretase in Notch activation (6–9), and the similarity of developmental phenotypes shared between presenilin and Notch germ line mutant mice (14–19). These findings highlight the difference in molecular targets regulated by presenilin in the mediation of its function in the developing and the adult brain. Although Notch is an important mediator of presenilin function in the developing brain, where they control the size of neural progenitor and neuronal population through the regulation of cell fate decision and apoptotic cell death (11–13, 22–25), the molecular target through which presenilin promotes neuronal survival in the adult cerebral cortex is entirely unknown. Although many γ-secretase substrates have been reported (40), most of them were identified in overexpression systems and cell lines, so it remains to be determined how many of them are physiological substrates of γ-secretase and which one(s) might be involved in mediating presenilin-dependent neuronal survival. Furthermore, the fact that presenilin and nicastrin cKO mice share similar phenotypes in memory impairment and neurodegeneration supports a γ-secretase-dependent involvement, but the reduction of presenilin levels in nicastrin cKO mice suggested that a γ-secretase-independent mechanism may still be at play (36, 37, 39).
We previously proposed that CBP (CREB-binding protein encoded by the Crebbp gene) is a putative downstream target of presenilin-mediated neuronal survival in the adult brain via Notch signaling, based on the reduced mRNA expression of the Crebbp gene and the CREB target genes and on the identification of the putative consensus sequences for CSL binding in the Crebbp promoter, which suggests that Notch activation should enhance CBP expression (36). However, conditional inactivation of CBP in excitatory neurons of the postnatal forebrain did not cause neuronal loss in the cerebral cortex of CBP cKO mice although these mutant mice did exhibit cognitive deficits (71). In addition, using CRE-luc reporter assays, we found that CRE-luc activity is not reduced in primary PS-null cortical cultures, arguing against CRE-dependent gene expression being a direct target of PS-mediated Notch signaling pathway (49). Therefore, the modest reduction of CREB-CBP activity may be the consequence of other molecular changes caused by loss of presenilin, as suggested by a recent report (73). Collectively, Notch is unlikely an essential target of presenilin in mediating neuronal survival in the adult cerebral cortex. Consistent with this interpretation, RBP-Jκ cKO mice, which were generated using independent αCaMKII-Cre lines, also failed to exhibit any phenotypes in excitatory neurons of the adult cerebral cortex,4,5 although adult neurogenesis and glial cell fate specification in the subventricular zone was affected (74). This is consistent with data from our in situ hybridization analysis that showed Notch2 signals in the subventricular zone, the rostral migratory stream, and the olfactory bulb of the adult brain.6 Although we cannot exclude the possibility of compensatory effects provided by other Notch family members, Notch3 or Notch4, in our Notch cKO mice, their reported expression patterns, Notch3 being expressed exclusively in endothelial cells of the adult brain (75) and Notch4 expression being undetectable in the adult brain (76), make this possibility unlikely.
The most surprising finding of our current study is perhaps the absence of the reduced expression of Notch1 and Notch2 mRNAs and proteins, which we anticipated to find in the cerebral cortex of cKO mice. Using the same αCaMKII-Cre transgenic line, which expresses Cre recombinase in most if not all excitatory pyramidal neurons in the cerebral cortex (45, 50), we previously generated PS, nicastrin, and CBP cKO mice, all of which showed ∼50% reduction of protein expression in the cerebral cortex (39, 45, 71). The remaining ∼50% protein is likely due to expression of these genes in other cell types, such as glia and interneurons, where Cre expression is not targeted. In Notch1 and Notch2 cKO mice, we were able to confirm that deletion of the floxed DNA sequences indeed took place selectively in the cerebral cortex (Fig. 4). Furthermore, in primary cortical cultures where Notch1 and Notch2 are relatively highly expressed compared with the adult brain, introduction of Cre recombinase efficiently eliminated all of the Notch proteins (Fig. 5). Thus, the only reasonable interpretation of these results is that there is no Notch1 and Notch2 expression in excitatory neurons of the cerebral cortex. Consistent with this interpretation, the deletion of the floxed exons in excitatory neurons of the cerebral cortex in Notch1 and Notch2 cKO mice has no detectable effect on Notch mRNA and protein levels. These results appear to be at odds with earlier studies showing high levels of Notch immunoreactivity in the nucleus of the pyramidal neurons in the adult cerebral cortex (30–32). However, our Northern blot, which showed a single clean band of the correct size on the entire blot, does not rely upon specificity of available Notch antibodies. We were able to find two Notch2 antibodies that are rather specific and recognize Notch2-specific bands on Western blots using lysates from adult cortical samples as well as primary cortical cultures (Figs. 5 and 7 and supplemental Fig. S1). Notch1 antibodies, however, recognize nonspecific bands on Western blots, especially when adult cortical lysates were used (Fig. 7 and supplemental Fig. S1), making them less reliable indicators of Notch expression in the adult brain.
In summary, despite of the prevailing concerns of unwanted side effects on Notch while using γ-secretase inhibitors for treatment of Alzheimer disease, our current genetic study shows that Notch is not required for age-dependent survival of cortical pyramidal neurons, which are particularly vulnerable in AD. Thus, Notch is not a target of presenilin in promoting survival of excitatory neurons in the adult cerebral cortex. Although presenilin is highly expressed in pyramidal neurons in the cortex, Notch expression is undetectable in the same neurons, making it even less likely that presenilin and Notch families, which are tightly linked in the same signaling pathway during development, functionally act together in the adult brain. Future investigation is needed to identify molecular targets, through which presenilin exerts its neuronal protection in the aging cerebral cortex. The identification of such targets may provide novel targets to combat neurodegeneration in AD.
Acknowledgments
We thank Xiaoyan Zou for breeding and genotyping the mice. We also thank Dr. Alain Israel for providing anti-Notch1 rabbit polyclonal antibody, and Dr. Guiquan Chen for discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 NS042818 (to J. S.) and RC2 AG036614.

This article contains supplemental Fig. 1.
K. Tanigaki, Research Institute, Shiga Medical Center, personal communication.
R. Kopan, unpublished results.
M. Wines-Samuelson and J. Shen, unpublished data.
- CSL
- CBF1/RBP-Jκ/Su(H)/Lag-1
- PS
- presenilin
- AD
- Alzheimer disease
- cKO
- conditional knockout
- αCaMKII
- α-calcium calmodulin-dependent kinase II
- DIV
- days in vitro
- GFAP
- glial fibrillary acidic protein
- CBP
- CREB binding protein
- CREB
- cAMP response element-binding protein
- nt
- nucleotide(s)
- APP
- amyloid precursor protein
- VCP
- vasolin containing protein.
REFERENCES
- 1. Kopan R., Ilagan M. X. (2009) The canonical Notch signaling pathway. Unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Tetering G., van Diest P., Verlaan I., van der Wall E., Kopan R., Vooijs M. (2009) Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem. 284, 31018–31027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000) A novel proteolytic cleavage involved in Notch signaling. The role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 [DOI] [PubMed] [Google Scholar]
- 4. Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000) A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 [DOI] [PubMed] [Google Scholar]
- 5. Bozkulak E. C., Weinmaster G. (2009) Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol. Cell. Biol. 29, 5679–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ye Y., Lukinova N., Fortini M. E. (1999) Neurogenic phenotypes and altered Notch processing in Drosophila presenilin mutants. Nature 398, 525–529 [DOI] [PubMed] [Google Scholar]
- 7. Struhl G., Greenwald I. (1999) Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398, 522–525 [DOI] [PubMed] [Google Scholar]
- 8. Schroeter E. H., Kisslinger J. A., Kopan R. (1998) Notch-1 signaling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 [DOI] [PubMed] [Google Scholar]
- 9. De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J. S., Schroeter E. H., Schrijvers V., Wolfe M. S., Ray W. J., Goate A., Kopan R. (1999) A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 [DOI] [PubMed] [Google Scholar]
- 10. Wines-Samuelson M., Handler M., Shen J. (2005) Role of presenilin-1 in cortical lamination and survival of Cajal-Retzius neurons. Dev. Biol. 277, 332–346 [DOI] [PubMed] [Google Scholar]
- 11. Yang X., Klein R., Tian X., Cheng H. T., Kopan R., Shen J. (2004) Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 269, 81–94 [DOI] [PubMed] [Google Scholar]
- 12. Handler M., Yang X., Shen J. (2000) Presenilin-1 regulates neuronal differentiation during neurogenesis. Development 127, 2593–2606 [DOI] [PubMed] [Google Scholar]
- 13. Kim W. Y., Shen J. (2008) Presenilins are required for maintenance of neural stem cells in the developing brain. Mol Neurodegener 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donoviel D. B., Hadjantonakis A. K., Ikeda M., Zheng H., Hyslop P. S., Bernstein A. (1999) Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13, 2801–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Swiatek P. J., Lindsell C. E., del Amo F. F., Weinmaster G., Gridley T. (1994) Notch1 is essential for post-implantation development in mice. Genes Dev. 8, 707–719 [DOI] [PubMed] [Google Scholar]
- 16. Conlon R. A., Reaume A. G., Rossant J. (1995) Notch1 is required for the coordinate segmentation of somites. Development 121, 1533–1545 [DOI] [PubMed] [Google Scholar]
- 17. Hamada Y., Kadokawa Y., Okabe M., Ikawa M., Coleman J. R., Tsujimoto Y. (1999) Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 126, 3415–3424 [DOI] [PubMed] [Google Scholar]
- 18. Wong P. C., Zheng H., Chen H., Becher M. W., Sirinathsinghji D. J., Trumbauer M. E., Chen H. Y., Price D. L., Van der Ploeg L. H., Sisodia S. S. (1997) Presenilin 1 is required for Notch1 and DII1 expression in the paraxial mesoderm. Nature 387, 288–292 [DOI] [PubMed] [Google Scholar]
- 19. Shen J., Bronson R. T., Chen D. F., Xia W., Selkoe D. J., Tonegawa S. (1997) Skeletal and CNS defects in Presenilin-1-deficient mice. Cell 89, 629–639 [DOI] [PubMed] [Google Scholar]
- 20. Krebs L. T., Xue Y., Norton C. R., Sundberg J. P., Beatus P., Lendahl U., Joutel A., Gridley T. (2003) Characterization of Notch3-deficient mice. Normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis 37, 139–143 [DOI] [PubMed] [Google Scholar]
- 21. Krebs L. T., Xue Y., Norton C. R., Shutter J. R., Maguire M., Sundberg J. P., Gallahan D., Closson V., Kitajewski J., Callahan R., Smith G. H., Stark K. L., Gridley T. (2000) Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14, 1343–1352 [PMC free article] [PubMed] [Google Scholar]
- 22. Imayoshi I., Sakamoto M., Yamaguchi M., Mori K., Kageyama R. (2010) Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J. Neurosci. 30, 3489–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hitoshi S., Alexson T., Tropepe V., Donoviel D., Elia A. J., Nye J. S., Conlon R. A., Mak T. W., Bernstein A., van der Kooy D. (2002) Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 16, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mizutani K., Yoon K., Dang L., Tokunaga A., Gaiano N. (2007) Differential Notch signaling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351–355 [DOI] [PubMed] [Google Scholar]
- 25. Androutsellis-Theotokis A., Leker R. R., Soldner F., Hoeppner D. J., Ravin R., Poser S. W., Rueger M. A., Bae S. K., Kittappa R., McKay R. D. (2006) Notch signaling regulates stem cell numbers in vitro and in vivo. Nature 442, 823–826 [DOI] [PubMed] [Google Scholar]
- 26. Breunig J. J., Silbereis J., Vaccarino F. M., Sestan N., Rakic P. (2007) Notch regulates cell fate and dendrite morphology of newborn neurons in the postnatal dentate gyrus. Proc. Natl. Acad. Sci. U.S.A. 104, 20558–20563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ables J. L., Decarolis N. A., Johnson M. A., Rivera P. D., Gao Z., Cooper D. C., Radtke F., Hsieh J., Eisch A. J. (2010) Notch1 is required for maintenance of the reservoir of adult hippocampal stem cells. J. Neurosci. 30, 10484–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aguirre A., Rubio M. E., Gallo V. (2010) Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature 467, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andreu-Agulló C., Morante-Redolat J. M., Delgado A. C., Fariñas I. (2009) Vascular niche factor PEDF modulates Notch-dependent stemness in the adult subependymal zone. Nat. Neurosci. 12, 1514–1523 [DOI] [PubMed] [Google Scholar]
- 30. Redmond L., Oh S. R., Hicks C., Weinmaster G., Ghosh A. (2000) Nuclear Notch1 signaling and the regulation of dendritic development. Nat. Neurosci. 3, 30–40 [DOI] [PubMed] [Google Scholar]
- 31. Sestan N., Artavanis-Tsakonas S., Rakic P. (1999) Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science 286, 741–746 [DOI] [PubMed] [Google Scholar]
- 32. Alberi L., Liu S., Wang Y., Badie R., Smith-Hicks C., Wu J., Pierfelice T. J., Abazyan B., Mattson M. P., Kuhl D., Pletnikov M., Worley P. F., Gaiano N. (2011) Activity-induced Notch signaling in neurons requires Arc/Arg3.1 and is essential for synaptic plasticity in hippocampal networks. Neuron 69, 437–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fischer D. F., van Dijk R., Sluijs J. A., Nair S. M., Racchi M., Levelt C. N., van Leeuwen F. W., Hol E. M. (2005) Activation of the Notch pathway in Down syndrome. Cross-talk of Notch and APP. FASEB J. 19, 1451–1458 [DOI] [PubMed] [Google Scholar]
- 34. Ishikura N., Clever J. L., Bouzamondo-Bernstein E., Samayoa E., Prusiner S. B., Huang E. J., DeArmond S. J. (2005) Notch-1 activation and dendritic atrophy in prion disease. Proc. Natl. Acad. Sci. U.S.A. 102, 886–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nagarsheth M. H., Viehman A., Lippa S. M., Lippa C. F. (2006) Notch-1 immunoexpression is increased in Alzheimer and Pick disease. J. Neurol. Sci. 244, 111–116 [DOI] [PubMed] [Google Scholar]
- 36. Saura C. A., Choi S. Y., Beglopoulos V., Malkani S., Zhang D., Shankaranarayana Rao B. S., Chattarji S., Kelleher R. J., 3rd, Kandel E. R., Duff K., Kirkwood A., Shen J. (2004) Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron 42, 23–36 [DOI] [PubMed] [Google Scholar]
- 37. Wines-Samuelson M., Schulte E. C., Smith M. J., Aoki C., Liu X., Kelleher R. J., 3rd, Shen J. (2010) Characterization of age-dependent and progressive cortical neuronal degeneration in presenilin conditional mutant mice. PLoS One 5, e10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen J., Kelleher R. J., 3rd (2007) The presenilin hypothesis of Alzheimer disease. Evidence for a loss-of-function pathogenic mechanism. Proc. Natl. Acad. Sci. U.S.A. 104, 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tabuchi K., Chen G., Südhof T. C., Shen J. (2009) Conditional forebrain inactivation of nicastrin causes progressive memory impairment and age-related neurodegeneration. J. Neurosci. 29, 7290–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kopan R., Ilagan M. X. (2004) γ-Secretase. Proteasome of the membrane? Nat. Rev. Mol. Cell Biol. 5, 499–504 [DOI] [PubMed] [Google Scholar]
- 41. Haapasalo A., Kovacs D. M. (2011) The many substrates of presenilin/ϵγ-secretase. J. Alzheimers Dis. 25, 3–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Strooper B., Saftig P., Craessaerts K., Vanderstichele H., Guhde G., Annaert W., Von Figura K., Van Leuven F. (1998) Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391, 387–390 [DOI] [PubMed] [Google Scholar]
- 43. Levitan D., Greenwald I. (1995) Facilitation of lin-12-mediated signaling by sel-12, a Caenorhabditis elegans S182 Alzheimer disease gene. Nature 377, 351–354 [DOI] [PubMed] [Google Scholar]
- 44. Levitan D., Greenwald I. (1998) Effects of SEL-12 presenilin on LIN-12 localization and function in Caenorhabditis elegans. Development 125, 3599–3606 [DOI] [PubMed] [Google Scholar]
- 45. Yu H., Saura C. A., Choi S. Y., Sun L. D., Yang X., Handler M., Kawarabayashi T., Younkin L., Fedeles B., Wilson M. A., Younkin S., Kandel E. R., Kirkwood A., Shen J. (2001) APP processing and synaptic plasticity in presenilin-1 conditional knockout mice. Neuron 31, 713–726 [DOI] [PubMed] [Google Scholar]
- 46. Stump G., Durrer A., Klein A. L., Lütolf S., Suter U., Taylor V. (2002) Notch1 and its ligands Delta-like and Jagged are expressed and active in distinct cell populations in the postnatal mouse brain. Mech. Dev. 114, 153–159 [DOI] [PubMed] [Google Scholar]
- 47. Irvin D. K., Zurcher S. D., Nguyen T., Weinmaster G., Kornblum H. I. (2001) Expression patterns of Notch1, Notch2, and Notch3 suggest multiple functional roles for the Notch-DSL signaling system during brain development. J. Comp. Neurol. 436, 167–181 [PubMed] [Google Scholar]
- 48. Higuchi M., Kiyama H., Hayakawa T., Hamada Y., Tsujimoto Y. (1995) Brain Res. Mol. Brain Res. 29, 263–272 [DOI] [PubMed] [Google Scholar]
- 49. Watanabe H., Smith M. J., Heilig E., Beglopoulos V., Kelleher R. J., 3rd, Shen J. (2009) Indirect regulation of presenilins in CREB-mediated transcription. J. Biol. Chem. 284, 13705–13713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang C., Wu B., Beglopoulos V., Wines-Samuelson M., Zhang D., Dragatsis I., Südhof T. C., Shen J. (2009) Presenilins are essential for regulating neurotransmitter release. Nature 460, 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McCright B., Lozier J., Gridley T. (2006) Generation of new Notch2 mutant alleles. Genesis 44, 29–33 [DOI] [PubMed] [Google Scholar]
- 52. Kavalali E. T., Klingauf J., Tsien R. W. (1999) Activity-dependent regulation of synaptic clustering in a hippocampal culture system. Proc. Natl. Acad. Sci. U.S.A. 96, 12893–12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ho A., Morishita W., Atasoy D., Liu X., Tabuchi K., Hammer R. E., Malenka R. C., Südhof T. C. (2006) Genetic analysis of Mint/X11 proteins. Essential presynaptic functions of a neuronal adaptor protein family. J. Neurosci. 26, 13089–13101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gaiano N., Nye J. S., Fishell G. (2000) Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron 26, 395–404 [DOI] [PubMed] [Google Scholar]
- 55. Patten B. A., Peyrin J. M., Weinmaster G., Corfas G. (2003) Sequential signaling through Notch1 and erbB receptors mediates radial glia differentiation. J. Neurosci. 23, 6132–6140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schmid R. S., McGrath B., Berechid B. E., Boyles B., Marchionni M., Sestan N., Anton E. S. (2003) Neuregulin 1-erbB2 signaling is required for the establishment of radial glia and their transformation into astrocytes in cerebral cortex. Proc. Natl. Acad. Sci. U.S.A. 100, 4251–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ho A., Shen J. (2011) Presenilins in synaptic function and disease. Trends Mol. Med. 17, 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beglopoulos V., Sun X., Saura C. A., Lemere C. A., Kim R. D., Shen J. (2004) Reduced β-amyloid production and increased inflammatory responses in presenilin conditional knock-out mice. J. Biol. Chem. 279, 46907–46914 [DOI] [PubMed] [Google Scholar]
- 59. Casas C., Sergeant N., Itier J. M., Blanchard V., Wirths O., van der Kolk N., Vingtdeux V., van de Steeg E., Ret G., Canton T., Drobecq H., Clark A., Bonici B., Delacourte A., Benavides J., Schmitz C., Tremp G., Bayer T. A., Benoit P., Pradier L. (2004) Massive CA1/2 neuronal loss with intraneuronal and N-terminal-truncated Aβ42 accumulation in a novel Alzheimer transgenic model. Am. J. Pathol. 165, 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. LaFerla F. M., Tinkle B. T., Bieberich C. J., Haudenschild C. C., Jay G. (1995) The Alzheimer Aβ peptide induces neurodegeneration and apoptotic cell death in transgenic mice. Nat. Genet. 9, 21–30 [DOI] [PubMed] [Google Scholar]
- 61. Lewis J., McGowan E., Rockwood J., Melrose H., Nacharaju P., Van Slegtenhorst M., Gwinn-Hardy K., Paul Murphy M., Baker M., Yu X., Duff K., Hardy J., Corral A., Lin W. L., Yen S. H., Dickson D. W., Davies P., Hutton M. (2000) Neurofibrillary tangles, amyotrophy, and progressive motor disturbance in mice expressing mutant (P301L) Tau protein. Nat. Genet. 25, 402–405 [DOI] [PubMed] [Google Scholar]
- 62. Lucas J. J., Hernández F., Gómez-Ramos P., Morán M. A., Hen R., Avila J. (2001) Decreased nuclear β-catenin, Tau hyperphosphorylation, and neurodegeneration in GSK-3β conditional transgenic mice. EMBO J. 20, 27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sriram K., Benkovic S. A., Hebert M. A., Miller D. B., O'Callaghan J. P. (2004) Induction of gp130-related cytokines and activation of JAK2/STAT3 pathway in astrocytes precedes up-regulation of glial fibrillary acidic protein in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model of neurodegeneration. Key signaling pathway for astrogliosis in vivo? J. Biol. Chem. 279, 19936–19947 [DOI] [PubMed] [Google Scholar]
- 64. Yoshiyama Y., Higuchi M., Zhang B., Huang S. M., Iwata N., Saido T. C., Maeda J., Suhara T., Trojanowski J. Q., Lee V. M. (2007) Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351 [DOI] [PubMed] [Google Scholar]
- 65. Mayford M., Bach M. E., Huang Y. Y., Wang L., Hawkins R. D., Kandel E. R. (1996) Control of memory formation through regulated expression of a CaMKII transgene. Science 274, 1678–1683 [DOI] [PubMed] [Google Scholar]
- 66. Vooijs M., Jonkers J., Berns A. (2001) A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2, 292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shaye D. D., Greenwald I. (2002) Endocytosis-mediated down-regulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature 420, 686–690 [DOI] [PubMed] [Google Scholar]
- 68. Gonsalves F. C., Weisblat D. A. (2007) MAPK regulation of maternal and zygotic Notch transcript stability in early development. Proc. Natl. Acad. Sci. U.S.A. 104, 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cornell M., Evans D. A., Mann R., Fostier M., Flasza M., Monthatong M., Artavanis-Tsakonas S., Baron M. (1999) The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 152, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Qiu L., Joazeiro C., Fang N., Wang H. Y., Elly C., Altman Y., Fang D., Hunter T., Liu Y. C. (2000) Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 275, 35734–35737 [DOI] [PubMed] [Google Scholar]
- 71. Chen G., Zou X., Watanabe H., van Deursen J. M., Shen J. (2010) CREB binding protein is required for both short-term and long-term memory formation. J. Neurosci. 30, 13066–13077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Blaumueller C. M., Qi H., Zagouras P., Artavanis-Tsakonas S. (1997) Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90, 281–291 [DOI] [PubMed] [Google Scholar]
- 73. Boyles R. S., Lantz K. M., Poertner S., Georges S. J., Andres A. J. (2010) Presenilin controls CBP levels in the adult Drosophila central nervous system. PLoS One 5, e14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fujimoto M., Takagi Y., Muraki K., Nozaki K., Yamamoto N., Tsuji M., Hashimoto N., Honjo T., Tanigaki K. (2009) RBP-J promotes neuronal differentiation and inhibits oligodendroglial development in adult neurogenesis. Dev. Biol. 332, 339–350 [DOI] [PubMed] [Google Scholar]
- 75. Prakash N., Hansson E., Betsholtz C., Mitsiadis T., Lendahl U. (2002) Mouse Notch 3 expression in the pre- and postnatal brain. Relationship to the stroke and dementia syndrome CADASIL. Exp. Cell Res. 278, 31–44 [DOI] [PubMed] [Google Scholar]
- 76. Uyttendaele H., Marazzi G., Wu G., Yan Q., Sassoon D., Kitajewski J. (1996) Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development 122, 2251–2259 [DOI] [PubMed] [Google Scholar]