Background: Various cell surface proteins of pathogenic mycobacteria have been implicated in virulence.
Results: A screen for secretion defects of specific cell surface proteins in Mycobacterium marinum identified predominantly lipooligosaccharide (LOS) biosynthesis mutants.
Conclusion: Defects in LOS biosynthesis alter the release of cell surface proteins.
Significance: Ten novel genes are described for LOS biosynthesis, and increased virulence is observed for a LOS-IV mutant.
Keywords: Bacterial Pathogenesis, Cell Wall, Gene Regulation, Lipid Synthesis, Mycobacterium, Protein Secretion, Zebrafish, Capsule, Lipooligosaccharides, Type VII Secretion
Abstract
The mycobacterial cell envelope is characterized by the presence of a highly impermeable second membrane, which is composed of mycolic acids intercalated with different unusual free lipids, such as lipooligosaccharides (LOS). Transport across this cell envelope requires a dedicated secretion system for extracellular proteins, such as PE_PGRS proteins, which are specific mycobacterial proteins with polymorphic GC-rich sequence (PGRS). In this study, we set out to identify novel components involved in the secretion of PE_PGRS proteins by screening Mycobacterium marinum transposon mutants for secretion defects. Interestingly, most mutants were not affected in secretion but in the release of PE_PGRS proteins from the cell surface. These mutants had insertions in a gene cluster associated with LOS biosynthesis. Lipid analysis of these mutants revealed a role at different stages of LOS biosynthesis for 10 novel genes. Furthermore, we show that regulatory protein WhiB4 is involved in LOS biosynthesis. The absence of the most extended LOS molecule, i.e. LOS-IV, and a concomitant accumulation of LOS-III was already sufficient to reduce the release of PE_PGRS proteins from the mycobacterial cell surface. A similar effect was observed for major surface protein EspE. These results show that the attachment of surface proteins is strongly influenced by the glycolipid composition of the mycobacterial cell envelope. Finally, we tested the virulence of a LOS-IV-deficient mutant in our zebrafish embryo infection model. This mutant showed a marked increase in virulence as compared with the wild-type strain, suggesting that LOS-IV plays a role in the modulation of mycobacterial virulence.
Introduction
Mycobacterium tuberculosis is the causative agent of tuberculosis, one of the world's major infectious diseases that is responsible for an estimated 1.4 million deaths annually (1). One of the major problems in controlling this disease is that the classical antibiotic treatment regimens for tuberculosis are lengthy and require four different drugs. These long treatments are needed due to the high intrinsic resistance of M. tuberculosis to antibiotics and the occurrence of highly resistant persister forms. The natural antibiotic resistance of M. tuberculosis is partially due to its exceptionally hydrophobic and complex cell envelope structure. Although mycobacteria are classified as high GC Gram-positives, recent cryo-transmission electron microscopy unequivocally showed that the mycobacterial cell envelope consists of two membranes: a cytoplasmic membrane and a unique outer membrane (2–4). The inner leaflet of this atypical outer membrane consists of long chain fatty acids called mycolic acids, which are covalently linked to a periplasmic arabinogalactan polymer. The outer leaflet of the outer membrane is formed by a range of different and mostly unique (glyco)lipids, which are extractable with organic solvents. This diverse group of extractable glycolipids includes lipoarabinomannan, phosphatidylinositol mannoside, trehalose dimycolate, phenolic glycolipid, and lipooligosaccharide (LOS)2 (5). Together with the mycolic acids, they form a thick permeability barrier.
To facilitate transport of virulence factors and other surface molecules over the mycobacterial cell envelope, dedicated secretion systems are required. Recently, a group of such secretion systems has been identified in mycobacteria, which was classified as type VII secretion systems (T7SS) (6). M. tuberculosis contains five different T7SS, each of which is encoded by a specific gene cluster, ESX-1 to ESX-5. Of these T7SS, the ESX-5 secretion system is predicted to be the most recently evolved system and is restricted to the slow growing mycobacteria, including all major pathogens (7). Several T7SS were found to mediate the secretion of small proteins belonging to the WXG100 family (8), such as EsxN, which is secreted by ESX-5. In addition, ESX-5 has been shown to be responsible for the secretion of different members of the PE and PPE protein families (9, 10). These enigmatic protein families are unique to mycobacteria and highly expanded among slow growing mycobacteria, especially Mycobacterium kansasii, Mycobacterium marinum, and species of the M. tuberculosis complex (11). Their names are derived from a conserved N-terminal motif of Pro-Glu (PE) and Pro-Pro-Glu (PPE) residues, respectively, although the homology regions are in fact considerably larger (12).
The PE and PPE families can be subdivided into different subfamilies, of which the PE_PGRS family is the largest, consisting of 67 members in M. tuberculosis and 148 members in M. marinum (12, 13). PE_PGRS proteins are characterized by multiple tandem repeats of glycine and alanine residues (14). Because of this highly specific amino acid composition, the encoding genes have a very high percentage of GC residues, which is reflected in their name (polymorphic GC-rich sequence). The large expansion of PE_PGRS genes on the genome of M. tuberculosis suggests a role in virulence or antigenic variation. However, although PE_PGRS proteins indeed seem to be located at the cell surface, their function remains largely unknown. Secretion and surface localization of PE_PGRS proteins have recently been shown to depend on an intact ESX-5 secretion system (10).
In the search for additional essential components of the ESX-5 system, we conducted a transposon mutant screen in M. marinum for impaired PE_PGRS secretion. Surprisingly, most PE_PGRS secretion mutants were located in the genomic region described to be involved in LOS biosynthesis (15). LOS is a trehalose-based glycolipid present in the mycobacterial outer membrane. This glycolipid has been described for several mycobacterial species, including M. marinum (16) and Mycobacterium canettii of the M. tuberculosis complex (17). Interestingly, other species of the M. tuberculosis complex have lost the ability to produce LOS. In contrast to several other mycobacterial glycolipids, the length and composition of LOS are highly variable between different species. M. marinum produces under laboratory conditions four different LOS structures of increasing size, in which different unusual sugar moieties are sequentially added to a core of acylated trehalose (15). The genes responsible for LOS biosynthesis of M. marinum have been proposed to include mmar_2302 through mmar_2344 (15, 18). So far, only six mutants affected in this genomic region have been characterized and were demonstrated to be involved in LOS biosynthesis (15, 16, 18–20). In this study, we have identified 10 novel genes, both expected and unexpected, with a role at different stages of LOS biosynthesis. We also show that transcriptional regulatory protein WhiB4 affects LOS biosynthesis gene expression. Interestingly, PE_PGRS proteins appear to be more firmly attached to the cell surface of these LOS mutants. This, together with the diversity and high number of the LOS biosynthesis mutants found, indicates an intriguing relation between PE_PGRS release and cell wall composition.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
Wild-type and mutant M. marinum strains MUSA (21) and E11 (22, 23) were routinely grown at 30 °C in Middlebrook 7H9 (Difco) liquid medium and on Middlebrook 7H10 plates (Difco) supplemented with 10% Middlebrook albumin/dextrose/catalase and 0.05% Tween 80 (BD Biosciences) or only oleic acid/albumin/dextrose/catalase, respectively. Escherichia coli strain DH5α was used for amplification and manipulation of plasmid DNA. Antibiotic concentrations used were 25 μg/ml kanamycin, 50 μg/ml hygromycin, and 30 μg/ml or 10 μg/ml chloramphenicol for M. marinum or E. coli, respectively.
Construction and Screening of M. marinum Transposon Library
Transposon mutagenesis was performed on M. marinum E11 or MUSA using the mycobacterium-specific phage ϕMycoMarT7 containing the mariner-like transposon Himar1 (24). The resulting transposon insertion mutant library was plated on nitrocellulose filters (Millipore HATF08250) that were placed on 7H10 agar plates for a double filter assay as described previously (9). After colonies appeared on the nitrocellulose filters, these filters were placed on top of a second nitrocellulose filter on a fresh plate and incubated overnight at 30 °C. Subsequently, the bottom filter containing the secreted proteins was stained for the presence of PE_PGRS proteins using a mouse monoclonal antibody that recognizes the PGRS domain of PE_PGRS proteins (mAb 7C4.1F7) (10). To obtain large amounts of purified antibody, clone mAb 7C4.1F7 was cultured in Iscove's modified Dulbecco's medium supplemented with 1% FCS. Subsequently, the antibodies were purified using a gradient of ammonium sulfate on thiophilic agarose (AFFI-T, KemEnTec) and dialyzed against PBS. After incubation with the PGRS antibody and a conjugate (goat anti-mouse horseradish peroxidase (HRP)), the presence of PE_PGRS proteins on the filter was visualized by staining with 4-chloronaphthol/3,3-diaminobenzidine. Colonies with reduced amounts of secreted PE_PGRS proteins were selected and re-checked for PE_PGRS secretion defects in a second double filter assay. In a similar assay, a selection of mutants was also tested with antiserum directed against the surface protein EspE (4, 25).
Identification of Transposon Insertion Site
To identify the location of the transposon, DNA was isolated, and ligation-mediated PCR was performed using primers bampt2, salgD, pSalg, and pMyco1 (supplemental Table S1) as described previously (9). The resulting DNA fragments were sequenced and compared with the M. marinum genome sequence. To determine the orientation of the transposon, genomic DNA of the mutants was used in a PCR with one of the primers that anneal to either end of the transposon, T7_1 or T7_2, together with a gene-specific primer (supplemental Table S1).
SDS-PAGE and Immunoblotting
Mycobacteria were grown routinely to mid-logarithmic phase. Subsequently, the bacteria were washed twice in Middlebrook 7H9 medium without supplement to remove all traces of bovine serum albumin (BSA) and grown overnight in Middlebrook 7H9 liquid medium supplemented with 0.2% dextrose and optionally 0.05% Tween 80. Secreted proteins present in cell-free supernatants were obtained by centrifugation and subsequent passage through a 0.45-μm filter and concentrated by precipitation with 10% trichloroacetic acid in 10% acetone. Bacterial pellets were disrupted with 425–600-μm glass beads (Sigma) in PBS for 3 min using a Mini-beadbeater (BioSpec Products). Subsequently, the protein concentration of these samples was measured using the BCA protein assay (Pierce) to ensure equal loading. Based on wild-type A600 measurements, an equivalent of 0,1 and 0,6 absorbance for pellet and supernatant, respectively, were loaded on 10 or 15% SDS-polyacrylamide gels (Bio-Rad).
Proteins were visualized by immunoblotting using mouse monoclonal antiserum against GroEL2 (CS44, Colorado State University), rabbit antiserum reactive to EsxN (rMtb9.9A (26)), the ESAT-6-specific mouse monoclonal antiserum Hyb76-8 (27), and the antibody against PE_PGRS that was described above. The presence of bound HRP-conjugated secondary antibodies was detected via chemiluminescence (Pierce) using a CCD camera (Bio-Rad).
For the extraction of surface proteins from plate-grown bacteria, a loop-full of bacteria grown on 7H10 plates was resuspended in PBS, spun down, and washed again in PBS. Subsequently, bacteria were incubated with 0.5% Genapol X-080 (Sigma) for 30 min at room temperature and centrifuged to separate bacterial cells from extracted fractions. Both fractions were analyzed by SDS-PAGE and immunoblotting using anti-PGRS, anti-GroEL, and anti-EspE as described above.
Molecular Cloning
M. marinum E11 chromosomal DNA was used as a template to amplify the papA3 gene using Pfu polymerase (Fermentas) with gene-specific primers (supplemental Table S1). The resulting PCR product was ligated into the cloning vector pJet1.2 (Fermentas), subsequently isolated by BglII restriction, and cloned in BamHI-digested shuttle vector pSMT3-eGFP (28) under control of the hsp60 promoter, resulting in pSMT3-papA3-eGFP. Both the whiB4 gene and the operon mmar_2319-2322 were amplified from M. marinum E11 chromosomal DNA using Phusion polymerase (Finnzymes) and specific primers containing anchoring restriction sites (supplemental Table S1). The resulting PCR products and integration vector pUC-Int-cat (10) were digested with XbaI and either EcoRV or StuI for the mmar_2319-2322 operon. Subsequent ligation resulted in plasmids pUC-Int-cat-whiB4 and pUC-Int-cat-mmar_2319-22. All resulting complementation constructs were introduced by electroporation in the M. marinum wecE (A3 and A8), papA3 (A5), and whiB4 (A9) transposon mutants as described previously (9).
RNA Extraction and Quantitative Real Time PCR
Bacterial cultures of 15 ml were pelleted at A600 of 0.8–2.0 and immediately dissolved in 1 ml of cold TRIzol (Invitrogen). Subsequently, cells were lysed with using a mini-beadbeater for 2 min, followed by 10 min of incubation at 60 °C. After centrifugation, the lysate was used to extract RNA after incubation with 200 μl of chloroform and centrifugation. Next, the RNA in the aqueous phase was precipitated with 2-propanol and, after resuspension, treated with DNase (Fermentas). cDNA was generated from ∼200 ng of RNA using the SuperScript VILO cDNA synthesis kit (Invitrogen). Quantitative real time PCR (RT-PCR) was performed using the SYBR GreenER qPCR kit (Invitrogen) and the Lightcycler 480 (Roche Applied Science) to amplify parts of the LOS biosynthesis region with specific primers (supplemental Table S1). Ct values were normalized to values obtained for the mycobacterial household gene sigA.
Lipid Extraction and Analysis
Polar lipids were extracted and analyzed as described in Burguiere et al. (16).
Zebrafish Embryo Infection
To visualize bacteria in zebrafish embryos, a vector containing a red fluorescent marker, pSMT3-mCherry (29), was introduced by electroporation into M. marinum E11 WT, wecE mutant A3, and A8 and A3 complemented with pUC-Int-cat-mmar_2322-19. For infections, a culture with an A600 of ∼1.0 was centrifuged at 3000 × g, resuspended in PBS with 0.3% Tween, and subsequently incubated for 10 min to obtain a single cell suspension. After another centrifugation step, the pellet was resuspended in 1 ml of PBS and four times diluted in phenol red for the final injection solution. Zebrafish embryos were obtained and infected as described by van der Sar et al. (30). The equivalent of one injection was plated on 7H10 plates to determine the inoculum. At 5 days post-infection, the embryos were anesthetized in 0.02% buffered 3-aminobenzoic acid methyl ester (MS222, Sigma), and the infection was monitored by visualizing red fluorescence using a Leica MZ 16FA stereomicroscope. The relative fluorescence compared with wild-type infection was quantified by a specifically designed dedicated analysis software program (31). In addition, a random selection of the infected zebrafish embryos was subsequently selected for whole embryo plating to determine bacterial counts (31).
RESULTS
Identification of PE_PGRS Secretion Mutants Indicate Link to LOS Biosynthesis
In a previous screen, three mutants deficient for PPE41 secretion were identified in M. marinum, all with a transposon insertion in the ESX-5 cluster, two in gene eccA5, and one in mmar_2676 (9, 10). Further analysis demonstrated that these mutants were unable to secrete various other PPE proteins and also PE_PGRS proteins (10). To identify additional proteins involved in ESX-5-dependent secretion, we decided to perform a different transposon mutant screen, this time screening for impaired PE_PGRS secretion using a monoclonal antiserum raised against the PGRS domain of PE_PGRS33 (Rv1818c). This antibody specifically interacts with the PGRS domain of multiple proteins (10) and could therefore potentially recognize all 148 putative PE_PGRS proteins of M. marinum.
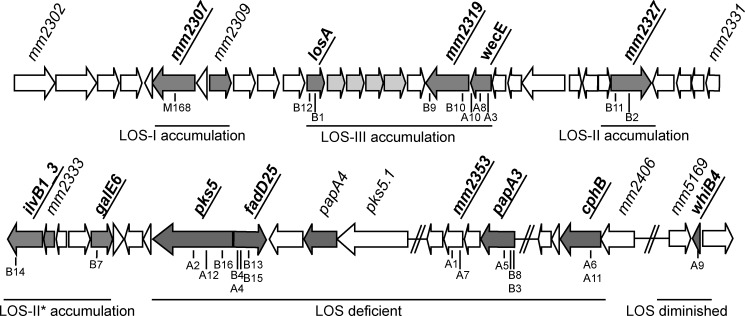
A mariner transposon library of ∼12,000 mutants of M. marinum strain E11 was plated on nitrocellulose filters and screened for PE_PGRS secretion in a double filter screening assay (see under “Experimental Procedures”). Mutants affected in PE_PGRS secretion were selected and retested in a second double filter assay. This resulted in 26 mutants that were consistently reduced or negative in PE_PGRS secretion. The mariner transposon insertion sites of all 26 mutants were determined by ligation-mediated PCR followed by sequencing (Table 1). Although the number of secretion-deficient mutants was much higher than expected for ESX-5 mutants (9, 10), it was still surprising that none of the PE_PGRS secretion mutants were found to be disrupted in the ESX-5 cluster. However, remarkably, for 23 of the 26 mutants, the transposon-insertion site was located in the same genomic region between genes mmar_2313 and mmar_2405, with several mutants containing a transposon at different positions in the same gene (Table 1 and Fig. 1). Part of this genomic region (mmar_2302 to mmar_2344) has been described previously to be involved in the biosynthesis of LOS (15, 18).
TABLE 1.
Transposon insertion sites of mutants affected in PE_PGRS secretion
| Gene | Annotation | (Putative) function | Mutant | PE_PGRS secretion on filtera | Colony morphologyb | Insertion in gene (bp 5′) | Orientation transposonc |
|---|---|---|---|---|---|---|---|
| mmar_1008 | Conserved hypothetical membrane protein | B5 | +/− | S | 437 | NT | |
| mmar_2307 | Hypothetical transmembrane protein | M168 | − | R | 1000 | NT | |
| mmar_2313 | losA | Involved in assembly of the LOS-IV glycolipid | B12 | +/− | S/W | 88 | NT |
| B1 | +/− | S/W | 349 | NT | |||
| mmar_2319 | Conserved hypothetical transmembrane protein | B9 | +/− | S/W | 1548 | + | |
| B10 | +/− | S/W | 223 | − | |||
| mmar_2320 | wecE | Pyridoxal phosphate-dependent enzyme | A10 | +/− | S/W | 839 | − |
| A8 | +/− | S/W | 755 | − | |||
| A3 | +/− | S/W | 215 | + | |||
| mmar_2327 | – | Conserved hypothetical transmembrane protein | B11 | +/− | S | 145 | NT |
| B2 | +/− | S | 773 | NT | |||
| mmar_2332 | ilvB1_3 | Acetolactate synthase | B14 | +/− | S/W | 1683 | + |
| mmar_2336 | galE6 | UDP-glucose 4-epimerase | B7 | +/− | S | 283 | NT |
| mmar_2340 | pks5 | Polyketide synthase | B16 | − | R | 1051 | − |
| A12 | − | R | 2141 | − | |||
| A2 | − | R | 3265 | − | |||
| mmar_2341 | fadD25 | Fatty acyl AMP ligase | B4 | − | R | 330 | − |
| A4 | − | R | 373 | + | |||
| B13, B15d | − | R | 798 | − | |||
| mmar_2353 | UDP-glycosyltransferase | A1 | − | R | 751 | NT | |
| A7 | − | R | 103 | − | |||
| mmar_2355 | papA3 | Conserved polyketide synthase-associated protein | A5 | − | R | 691 | + |
| B8 | − | R | 139 | − | |||
| B3 | − | R | 120 | + | |||
| mmar_2405 | cphB | Cyanophycinase | A6, A11d | − | R | 496 | + |
| mmar_4419 | Conserved hypothetical protein | B17 | +/− | S | 373 | NT | |
| mmar_5170 | whiB4 | Transcriptional regulatory protein Whib-like | A9 | − | R | 38 | − |
a +/− indicates reduced, and − indicates negative.
b S indicates smooth; R indicates rough; and S/W indicates smooth-wrinkled.
c Orientation of the transposon is as follows: +, promoter activity in the same direction as the tested gene; −, promoter activity against the direction of the gene; NT, not tested.
d Mutants were detected with a transposon insertion at the exact same location in the gene.
FIGURE 1.
Genetic locus involved in LOS biosynthesis. Genes involved in LOS biosynthesis obtained from our screen are depicted in bold and underlined, with the location of individual mutants indicated below the dark gray arrows. LOS biosynthesis defects were shown previously for gene disruption of mm2309, losA, mm2332, mm2333, and papA4 (15, 16, 18, 20) and deletion of region mm2314 to mm2317 (19) (depicted in light gray as a single responsible gene was not assigned). Similar LOS biosynthesis defects are observed in disruption of adjacent genes or genes in close proximity to each other, suggesting genomic clustering. The defects are explained below the gray arrows.
After identification of the transposon insertion sites, the orientation of the inserted transposons was determined. This analysis showed that the transposons inserted in the same gene were commonly found in both orientations (Table 1), ruling out transposon-induced alterations of gene expression. Finally, to investigate strain dependence, we have also performed a small screen for PE_PGRS secretion defects in M. marinum strain MUSA. The identified PE_PGRS secretion mutant M168 was also located in the LOS biosynthesis cluster (Table 1), which suggests that the link between LOS biosynthesis and PE_PGRS secretion is not strain-specific. In conclusion, the diversity and high number of detected mutants in the LOS biosynthesis region indicate a strong link between LOS production and PE_PGRS secretion.
All Mutants within the LOS Gene Cluster Show Specific Defects in LOS Biosynthesis
M. marinum produces four different LOS structures designated LOS-I to LOS-IV. The biosynthesis of these LOS variants is proposed to be sequential. The structure of LOS-I is 3-O-Me-Rhap-(1–3)-Glcp-(1–3)-Glcp-(1–4)-Glcp-(1–1)-Glcp (16). The addition of xylose together with one or two molecules of the highly unusual sugar caryophyllose (19) produces LOS-II and LOS-III, respectively. The molecule added to produce LOS-IV has been characterized as a heterogenic group of mainly one acidic form of an N-acylated 4-amino-4,6-dideoxy-Galp residue (resulting in LOS-IVc) with a minor population of two neutral forms (LOS-IVa and -b) (32). The biosynthetic pathway for LOS and the genes involved is largely unknown. Only two of the genes identified in this screen have been previously described to be involved in LOS production, i.e. losA (mmar_2313) was shown to have a role in LOS-IV production, whereas an ilvB1_3 (mmar_2332) mutant produces only LOS-I and a LOS-II intermediate, lacking caryophyllose (LOS-II*) (15, 16). To determine whether the genes disrupted in the newly identified mutants have a role in LOS biosynthesis as well, their polar lipid profiles were examined by two-dimensional thin layer chromatography (Fig. 2, A and B, and Table 2).
FIGURE 2.
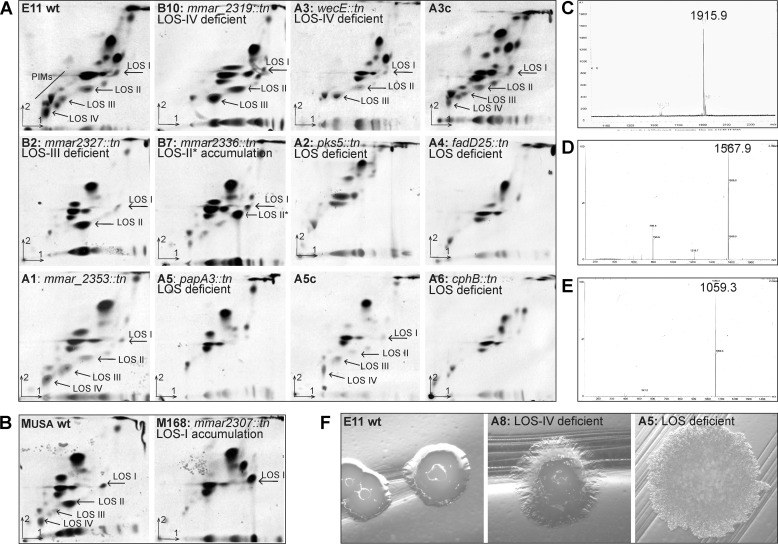
Secretion mutants show specific defects in LOS production. A, 14C-labeled polar lipids were extracted from M. marinum E11 wild-type, mutants B10, A3, and A3c (complemented with pUC-Int-cat-mmar_2319-22), B2, B7, A2, A4, A1, A5, and A5c (complemented with pSMT3-papA3-eGFP), A6; and B, MUSA wild-type and mutant M168. Subsequently, they were separated on two-dimensional TLC using the solvent system chloroform/methanol/water (60:30:6, v/v/v) in the first direction and chloroform/acetic acid/methanol/water (40:25:3:6, v/v/v) in the second direction and visualized by autoradiography. The LOS structures produced by the mutants are marked by arrows and the major biosynthesis defect is described for each mutant. C–E, per-O-methylated LOS-III, LOS-II, and LOS-I isolated from mutant A8, B2, and M168, respectively, were analyzed by MALDI-MS. C, signal was obtained at 1915.9 m/z (M + Na), which corresponds to the mass of the tetraglucose core, methylated rhamnose, xylose with two caryophyllose residues (i.e. LOS-III). D, signal of 1567.9 m/z corresponds to LOS-II; E, LOS-I is represented by a signal of 1059.3 m/z. F, LOS deficiency results in a rough-dry colony morphology, as shown for mutant A5. Mutants such as A8, which have a defect in higher order LOS biosynthesis, have an intermediate smooth-wrinkled phenotype.
TABLE 2.
LOS biosynthesis defects of selected mutants
| Tested mutant | LOS biosynthesis |
|---|---|
| M168 (mmar_2307) | No LOS-II, -III, -IV; accumulation LOS-I |
| B10 (mmar_2319) | No LOS-IV; accumulation LOS-II, -III |
| A3/A8 (wecE) | No LOS-IV; accumulation LOS-III |
| B2 (mmar_2327) | No LOS-III, -IV; accumulation LOS-II |
| B7 (galE6) | No LOS-II, -III, -IV; accumulation LOS-II* |
| A2 (pks5) | No LOS |
| A4 (fadD25) | No LOS |
| A1/A7 (mmar_2353) | Wild type |
| A5 (papA3) | No LOS |
| A6 (cphB) | No LOS |
| A9 (whiB4) | Diminished LOS |
Both mutants A3 and A8, disrupted in a different location of mmar_2320 with opposite directions of the transposon, showed a distinctive two-dimensional TLC pattern with a specific defect in the biosynthesis of LOS-IV and a concomitant accumulation of LOS-III (shown for A3 in Fig. 2A). Analysis of this product of mutant A8 by MALDI-MS (Fig. 2C) and NMR (data not shown) confirmed that the accumulating intermediate was indeed LOS-III. mmar_2320 codes for WecE. A WecE homologue of E. coli has been shown to be a sugar aminotransferase (33). Therefore, it probably has a function in the biosynthesis of the unusual sugar residue of LOS-IV. Complementation of the A3 mutant with integrative vector pUC-int-cat containing the intact mmar_2319-2322 operon (A3c) restored LOS-IV biosynthesis (Fig. 2A). Similar to the wecE mutants A3 and A8, disruption of downstream gene mmar_2319 (mutant B10), encoding a hypothetical transmembrane protein, resulted in LOS-IV deficiency (Fig. 2A). To exclude the possibility that the LOS biosynthesis defect observed for the wecE mutant A3 is caused by a polar effect on mmar_2319, the expression levels of this gene in mutant A3 were compared with wild-type levels by quantitative real time PCR (qRT-PCR). This showed that mmar_2319 expression was not decreased but was in fact elevated in the A3 mutant, possibly due to the promoter activity encoded by the transposon. Increased mmar_2319 expression levels were equally high for the complemented mutant A3c with restored LOS biosynthesis (results not shown), suggesting that wecE disruption is indeed responsible for the observed LOS-IV deficiency of the A3 mutant.
Also other mutants in the LOS region showed various defects in LOS biosynthesis. Mutant B2, disrupted in mmar_2327 encoding a multiple transmembrane protein, lost the ability to produce both LOS-III and LOS-IV and accumulated large amounts of LOS-II (Fig. 2, A and D). The mmar_2336 mutant B7, a gene encoding a putative UDP-glucose 4-epimerase, appeared to accumulate LOS-II* (Fig. 2A), a phenotype similar to the previously described mutant in ilvB1_3 (also identified in our screen) (15). The MUSA mutant M168 in mmar_2307 appeared to accumulate LOS-I (Fig. 2, B and E), similar to that observed for a mmar_2309 gene disruption (15).
The two-dimensional TLC patterns for polar lipids of the mmar_2340 and mmar_2341 mutants A2 and A4 revealed that they were both completely deficient in LOS production, as none of the four M. marinum LOS structures were visible (Fig. 2A). This suggests that these genes have a role in the biosynthesis of the core-acylated trehalose structure of LOS. Gene mmar_2340 encodes a putative polyketide synthase, Pks5, which was already suggested to be involved in the synthesis of the acyl chains of LOS, based on lipid analysis of a Mycobacterium smegmatis mutant disrupted in the orthologue of pks5 (34). The downstream gene mmar_2341 is annotated as a fatty acyl AMP ligase, FadD25, which has a proposed role in loading an adenylated metabolite onto the Pks5 multienzyme for extension. Therefore, it is not surprising that disruption of this gene resulted in a phenotype similar to the pks5 mutant.
The presence of LOS in mycobacteria has been associated with smooth colony morphology. For instance, M. kansasii strains devoid of LOS have a rough colony morphology, whereas all tested strains with LOS were smooth (35). In M. marinum defects in higher order LOS biosynthesis have also been correlated with altered colony morphology, described as a smooth-wrinkled phenotype (15, 16). Our mutants with a shorter or modified LOS production also showed a smooth-wrinkled phenotype, whereas all isolates devoid of LOS exhibited a characteristic rough and dry colony morphology (Fig. 2F and Table 1) similar to the M. kansasii strains.
Identification of Unexpected Genes Involved in LOS Biosynthesis
Interestingly, the rough-dry phenotype was also observed for some mutants located downstream of the proposed LOS biosynthesis region, i.e. in genes mmar_2353, mmar_2355, and mmar_2405, and for one mutant located far from the LOS biosynthesis region, in gene mmar_5170 (Table 1). The mutants disrupted in genes mmar_1008 and mmar_4419 exhibited wild-type colony morphology and were therefore not analyzed further. To examine whether these four aggregating mutants have a role in LOS production, their polar lipid content was analyzed.
The two-dimensional TLC analysis of mutant A5 (mmar_2355) confirmed that this mutant did not produce LOS (Fig. 2A). This gene encodes the conserved polyketide synthase-associated protein PapA3. PapA3 might function as an acyltransferase associated with Pks5, explaining its role in the biosynthesis of the LOS core structure of acylated trehalose. LOS production of mutant A5 could be restored with a shuttle vector containing an intact copy of the papA3 gene (Fig. 2A). Mutants A7 and A1 with a disruption in the gene mmar_2353, located downstream of papA3 in the same operon, were both not visibly affected in LOS production as shown by two-dimensional TLC analysis, although the total amount of LOS may be reduced (shown for A1 in Fig. 2A), which would be in line with the observed colony phenotype and genomic location of this mutant.
Disruption of mmar_2405 (mutant A6) also resulted in an LOS-deficient phenotype (Fig. 2A). Surprisingly, this gene is annotated as a homologue of a cyanophycinase and, among the genus Mycobacterium, is unique to M. marinum. Cyanophycinase is a serine protease specializing in the degradation of cyanophycin, a large polymer of arginine and aspartic acid used as nitrogen storage by cyanobacteria (36). The M. marinum cyanophycinase is actually a fusion of two copies of cyanophycinase, positioned head-to-tail linked by a short stretch of amino acids, with one copy mutated in its active site residues (37). Apparently, this putative protease is necessary for biosynthesis or transport of the LOS core structure. Together, these results show that the LOS biosynthesis region is even more extended than proposed by Ren et al. (15) (Fig. 1).
WhiB4 Regulates Expression of Several LOS Biosynthesis Genes
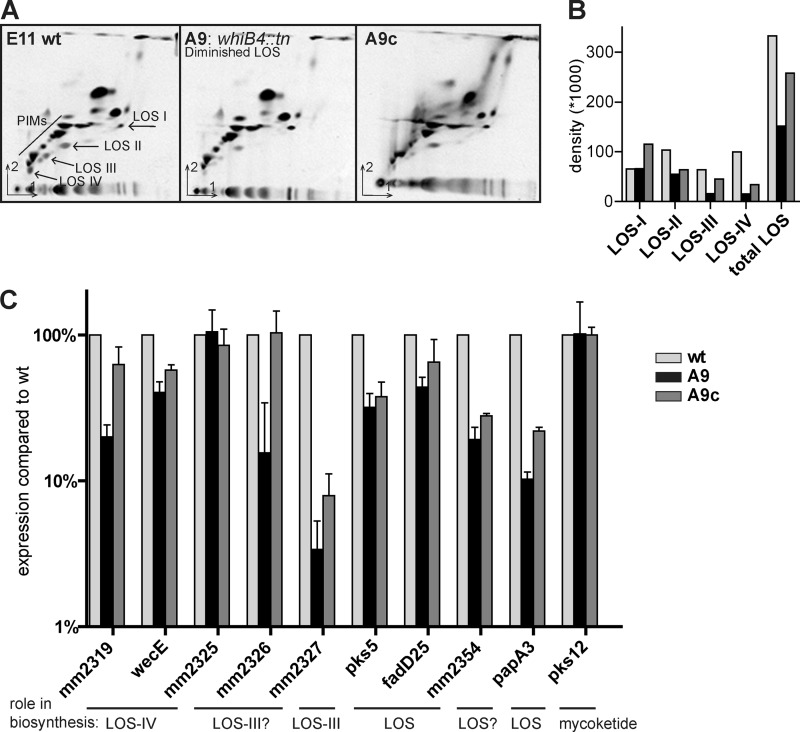
Mutant A9 is disrupted in a gene located outside the LOS biosynthesis region, i.e. mmar_5170, coding for WhiB4. WhiB4 belongs to the family of WhiB-like proteins, putative transcriptional regulatory proteins characterized by an iron-sulfur cluster, of which six are annotated in M. marinum. In accordance with the colony morphology, two-dimensional TLC analysis of polar lipids of the whiB4 mutant showed that LOS production was affected in this mutant. LOS biosynthesis is highly diminished, although some traces of LOS, especially LOS-I, seem to be present (Fig. 3A). Reintroduction of an intact copy of the whiB4 gene restored LOS biosynthesis to levels almost comparable with wild type (Fig. 3A). Quantification of the different LOS structures produced by wild type, A9, and A9c shows that total LOS biosynthesis of mutant A9 is roughly 50% percent diminished and is most strongly observed for LOS-III and LOS-IV biosynthesis (Fig. 3B).
FIGURE 3.
WhiB4 has a role in LOS biosynthesis by regulation LOS gene expression. A, two-dimensional TLC of 14C-labeled polar lipids extracted from M. marinum E11 wild-type, whiB4 mutant A9, and A9c (complemented with pUC-Int-cat-whiB4) as described for Fig. 2A/B. B, quantification of LOS structures. Equal counts of polar lipids (20,000 cpm) were loaded for two-dimensional TLC, and after development the density of each visible LOS spot was measured. C, relative expression levels of mmar_2319, wecE, mmar_2325 to mmar_2327, pks5, fadD25,mmar_2354, papA3, and pks12, shown for A9 and A9c compared with wild-type expression levels. Total RNA was isolated from M. marinum cultures (A600: 0.8–2.0) from three independent replicates, and subsequently qRT-PCR was performed. Ct values were normalized to expression of household gene sigA. Expression of mmar_2326-27 and papA3 is clearly diminished in the whiB4 mutant A9, although other genes involved in LOS biosynthesis are only slightly affected. Expression levels of mmar_2325 and negative control pks12 are comparable with wild type.
Recently, WhiB3, a WhiB4 paralogue, was shown to specifically control lipid biosynthesis by regulating the expression of pks2 and pks3 (38). Therefore, we hypothesized that WhiB4 regulates the production of LOS by controlling expression of genes in the LOS biosynthesis cluster. To examine this, we compared three biological replicates of wild type, whiB4 mutant A9, and the complemented mutant A9c for their expression levels of different LOS biosynthesis genes in qRT-PCR (Fig. 3C). In our analysis, we included genes necessary for the LOS core structure, pks5, fadD25, and papA3, but also genes involved in specific steps of higher order LOS biosynthesis, mmar_2319, wecE (LOS-IV), and mmar_2327 (LOS-III). As a negative control, pks12 was included, a gene that is involved in the biosynthesis of phosphoglycolipid mycoketide (39). Interestingly, the whiB4 mutant showed diminished expression for many of the tested LOS genes but most apparent for mmar_2327 and papA3, which showed 4 and 10%, respectively, of wild-type expression levels (Fig. 3C). Other genes of these putative operons, namely mmar_2326 and mmar_2354, are likewise affected. The strong regulation effect on mmar_2327 might explain why the LOS biosynthesis defects are most evident for LOS-III and LOS-IV. These qRT-PCR data also show that complementation is only partial for many of the tested genes (Fig. 3C), which correlates with the two-dimensional TLC data (Fig. 3, A and B). In conclusion, WhiB4 has a role in LOS biosynthesis, which is likely by regulating expression of multiple genes of the LOS cluster.
LOS Deficiency Decreases PE_PGRS Release from the Cell Surface
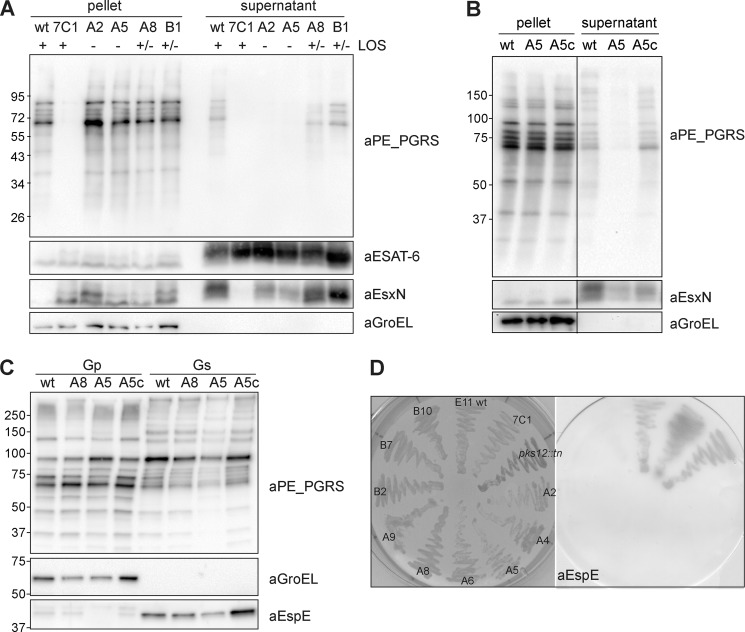
The ESX-5 mutant 7C1 is highly affected in both secretion and expression of PE_PGRS proteins (shown in Fig. 4A and Ref. 10). To determine whether the identified LOS mutants are similarly affected in expression and/or secretion of specific PE_PGRS proteins, a selection of these mutants was grown in liquid cultures, and secretion was analyzed by immunoblotting. Surprisingly, this analysis showed that production of PE_PGRS proteins was not affected, and the extracellular levels of PE_PGRS were comparable with wild-type levels for all tested LOS mutants (data not shown). This suggests that PE_PGRS production and secretion were normal under these conditions. In these experiments, we used culture medium supplemented with the detergent Tween 80, which is not present in 7H10 agar. Mycobacterial culture medium is routinely supplemented with this detergent to avoid bacterial aggregation (40). Because Tween 80 is known to (partially) remove mycobacterial surface structures, e.g. the capsule (4), we decided to re-test the mutants for PE_PGRS secretion in liquid cultures devoid of detergent. As PE_PGRS proteins are partially extracted by Tween 80 (4), their presence in the supernatant of wild-type M. marinum was reduced in culture conditions without detergent, but the overall pattern of secreted PE_PGRS was similar. The omission of Tween 80, however, almost completely abolished the extracellular accumulation of PE_PGRS for the LOS-deficient mutants, shown in Fig. 4A for a selection of the mutants. This release effect could be restored by complementation (shown for the A5 mutant Fig. 4B). No such PE_PGRS release defects could be observed for mutants deficient in higher order LOS biosynthesis (Fig. 4A). To investigate the extent of the release or secretion deficiency of the LOS mutants, their culture filtrates were also analyzed for the presence of ESX-5 substrate EsxN and ESX-1 substrate ESAT-6. Interestingly, the LOS-deficient mutants also showed a reproducible reduction in EsxN secretion, whereas the secretion of ESAT-6 was unaffected in all conditions tested (Fig. 4A). Next, we also tested the extraction of surface proteins with the mild detergent Genapol X-080. This analysis further demonstrated that PE_PGRS proteins expressed by the mutants were normally surface-localized (Fig. 4C), indicating that the lack of PE_PGRS secretion observed in the double filter assay was not caused by a transport defect. Taken together, these experiments demonstrate that LOS biosynthesis mutants are not disturbed in PE_PGRS translocation but show, in the absence of detergent, a stronger attachment of PE_PGRS proteins to the cell surface.
FIGURE 4.
LOS biosynthesis mutants show reduced PE_PGRS and EspE release. A, whole cells pellets and supernatant from M. marinum E11 wild-type, ESX-5 mutant 7C1, and PE_PGRS mutants A2, A5, A8, and B1 grown in 7H9 broth without Tween 80 were analyzed by immunoblot for expression and secretion of PE_PGRS, ESAT-6, and EsxN. GroEL2 was used as a negative control for lysis. LOS-deficient mutants A2 and A5 show a clear effect on the secretion of PE_PGRS and EsxN, although LOS-IV-deficient mutants A8 and B1 are unaffected. Results are representative of three independent experiments. B, immunoblot of whole cells pellets and supernatant from M. marinum WT and mutants A5 and A5c shows that the secretion defect of mutant A5 can be complemented. C, genapol X-080-treated whole cell pellets (Gp) and cell surface extracts (Gs) of bacteria grown on solid agar. No reproducible differences between E11 wild type and the tested mutants were observed for both PE_PGRS and EspE extractability. D, selected LOS mutants A2, A4, A5, A6, A8, A9, B2, B7, and B10 were plated on a nitrocellulose filter together with E11 wild-type, ESX-5 mutant 7C1, and a pks12 mutant. All LOS mutants are reproducibly negative for EspE secretion on double filter assay, whereas all the controls are positive for secretion.
This detergent-dependent increase in PE_PGRS cell surface attachment observed for LOS mutants indicates that in these mutants the capsule layer, which is also detergent-labile and has been shown to contain PE_PGRS proteins (4), may be affected. To further explore this possibility, we next studied the secretion of EspE, a major capsular protein (4). Using a double filter assay, we found that release of EspE was clearly affected in all tested LOS biosynthesis mutants (Fig. 4D). This effect was not seen for the negative control transposon mutant in pks12, which was obtained in a different screen.3 Similar to PE_PGRS proteins, the level of EspE extraction with the mild detergent Genapol X-080 did not differ from that of wild-type bacteria (Fig. 4C), showing that EspE was still surface-localized in the LOS mutants. Together, these results show that LOS biosynthesis defects result in a tighter surface attachment of capsular proteins, which presents itself as a secretion defect of these proteins in the double filter assay.
wecE Mutant Shows Increased Virulence in Zebrafish Embryo Infection Model
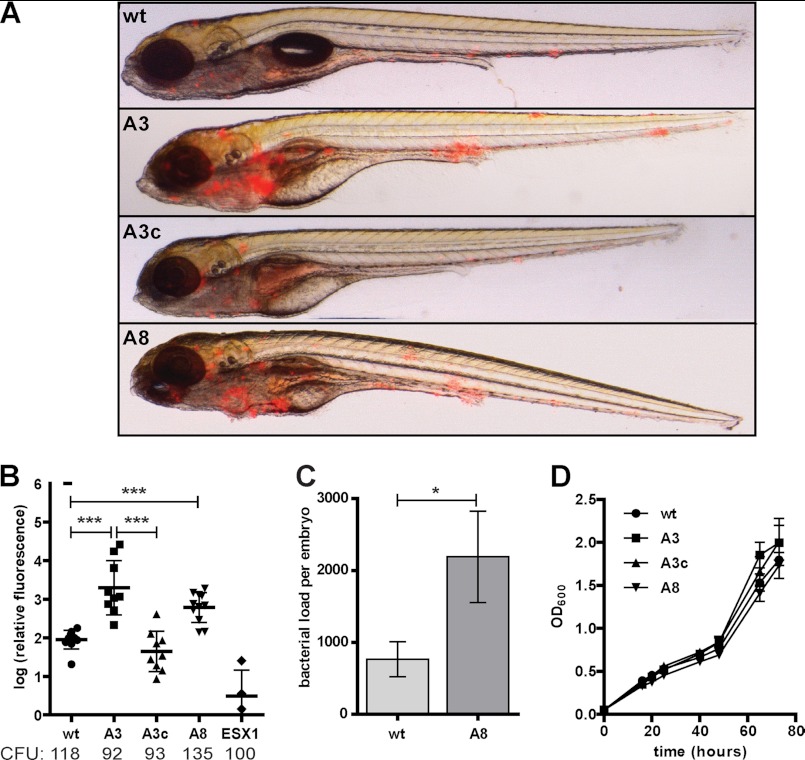
LOS has been shown to be antigenic (41), although its exact role in virulence has not yet been elucidated. Previous in vitro studies on the role of higher order LOS in M. marinum have suggested a role in macrophage cell entry and tumor necrosis factor α (TNF-α) response (15, 19). To study the effects of different levels of LOS deficiency in vivo, we decided to use the zebrafish embryo infection model. Infection of zebrafish embryos with (fluorescent) M. marinum leads to the formation of granuloma-like aggregates of (infected) macrophages, which can be monitored with fluorescence microscopy (42, 43). Attempts to study the virulence of the LOS-deficient mutants in this infection model were unsuccessful due to the excessive aggregation of these mutants upon growth in both solid and liquid medium. Therefore, we limited our analysis to the wecE mutants A3 and A8 that do not show this clumping effect.
Zebrafish embryos were infected with an inoculum of roughly 100 colony-forming units (cfu) of red fluorescent M. marinum E11, the LOS-IV deficient wecE mutants A3 and A8, and the complemented strain (A3c). The ESX-1 mutant eccCb1::tn was used as a negative control (31). Five days post-infection, the degree of infection and formation of early granulomas were monitored by measuring the relative red fluorescence. Remarkably, the embryos infected with the wecE mutants developed more and larger granulomatous structures than observed in wild-type infection (Fig. 5A). Close inspection of these aggregates showed that the localization of bacteria was similar to wild-type infection, i.e. all bacteria appeared intracellular (results not shown). This enhanced granuloma formation was quantified in three separate experiments as a 10-fold average increase for both mutants (Fig. 5B). Complementation of the wecE mutant A3, which was shown to restore LOS-IV biosynthesis (Fig. 2A), likewise restored infection to wild-type levels (Fig. 5B). In accordance with the fluorescence quantification, significantly more bacteria (3-fold) were recovered from embryos infected with the wecE mutant A8, as compared with a wild-type infection (Fig. 5C). The observed increased virulence in vivo is probably not due to a growth advantage, because the wecE mutants show no differences in growth rate when grown in culture medium (Fig. 5D). Our data demonstrate that disruption of wecE in M. marinum, which results in LOS-IV deficiency, causes hypervirulence in zebrafish embryos, which suggests that LOS-IV directly or indirectly plays a role in modulating innate immune response.
FIGURE 5.
LOS-IV biosynthesis mutant shows increased virulence in zebrafish embryo infection model. Infection of zebrafish embryos with red fluorescent wild-type M. marinum E11 (WT) and mutants A3 and A8, A3 complemented with pUC-Int-cat-mmar_2319-22 (A3c) and ESX-1 mutant eccCb1::tn (ESX1), were monitored at 5 days post-infection. A, representative overlay pictures of embryos infected with WT, A3, A3c, and A8 with ∼80 colony-forming units (CFU). B, automated quantification of infection based on red pixel intensity (31). Represented are relative infection levels compared with wild-type infection as a log of the percentage. Results of three replicate experiments, with the average injected cfu below each group, are shown with standard deviation (***, p < 0.001 by unpaired Student's t test). C, mean bacterial loads, measured as cfu, for zebrafish embryos infected for 5 days with WT and mutant A8 bacteria. Error bars represent mean ± S.E. D, growth curve of bacterial cultures in 7H9 + 0.05% Tween 80; mean A600 measurements of three independent cultures with standard deviation.
DISCUSSION
In M. marinum, PE_PGRS proteins are substrates for the ESX-5 secretion system (10). Therefore, by screening transposon mutants for defects in the secretion of PE_PGRS proteins, we expected to find mutants in the ESX-5 secretion system. However, our screen of ∼12,000 clones surprisingly did not result in the identification of novel ESX-5 mutants. A possible explanation for this unanticipated result is that different transposon mutagenesis studies in both M. marinum and M. tuberculosis have shown that mutants in most ESX-5 genes cannot be isolated (10, 24),4 indicating that these mutations might be lethal. Perhaps, a more saturating transposon mutant screen for PE_PGRS secretion in M. marinum might lead to the isolation of ESX-5 mutants similar to the ones identified previously (9, 10), for which indeed higher number of colonies were screened.
The high number of identified secretion mutants with a disruption in the LOS biosynthesis region and the large variety of biosynthesis defects found, ranging from LOS-IV to complete LOS deficiency, show a clear link between PE_PGRS secretion and LOS biosynthesis in the double filter assay. Surprisingly, all LOS mutants still produce wild-type levels of PE_PGRS proteins, and these PE_PGRS proteins are still (partially) surface-exposed. PE_PGRS proteins seem to be more tightly attached to the surface of LOS mutants, resulting in an apparent secretion defect in a double filter blot assay. Although this effect is stronger for the LOS negative mutants, already the absence of LOS-IV resulted in an altered surface attachment of PE_PGRS proteins, indicating that the terminal sugar moiety plays a role in this process. For the mutants deficient in higher order LOS, this tighter attachment of PE_PGRS proteins can already be neutralized by growing them in liquid culture with agitation. The LOS negative mutants need the supplemental addition of Tween 80 for the release of PE_PGRS proteins. It was previously shown that the presence of Tween 80 in culture medium (partially) removes the mycobacterial capsule layer, including some PE_PGRS proteins (4), which suggests that in LOS-deficient mutants capsular proteins in general are more tightly attached to the cell surface. The observation that the surface attachment of the major capsular protein EspE is also affected in all LOS mutants confirms this change in capsular properties. One possibility is that LOS functions as a detergent for these capsular proteins. However, co-culturing of an LOS mutant mixed with an ESX-5 mutant, which expresses all LOS structures, did not show cross-complementation in the double filter assay (results not shown). This suggests that the LOS of the ESX-5 mutant fails to function as a detergent for surface-localized PE_PGRS proteins of the LOS mutant. Interestingly, for both Mycobacterium bovis BCG and M. tuberculosis, which do not produce LOS, PE_PGRS release in the culture supernatant seems much lower than in M. marinum, although levels of secreted ESAT-6 are similar.4 The absence of LOS might explain these PE_PGRS secretion/release differences. Further studies of the capsule of the LOS mutants in M. marinum might give us more insight in the interaction of PE_PGRS proteins and the mycobacterial cell wall.
All M. marinum LOS-deficient mutants show a rough-dry colony morphology that resembles M. tuberculosis colony morphology, indicating a change in cell envelope structure. This rough-dry phenotype is also strongly associated with LOS deficiency in M. kansasii (35) but less clearly in Mycobacterium gastri (44) and M. canettii (45). In M. marinum, two mutants with rough-dry colony morphology, A1/A7 (mmar_2353) and A9 (whiB4), were still producing LOS. However, for the whiB4 mutant, LOS production was clearly diminished, suggesting that a decrease in total LOS content could be responsible for the rough-dry colony morphology. Although the mmar_2353 mutant did not clearly show diminished LOS production, this gene is located within the extended LOS biosynthesis cluster and could therefore play a role in LOS production.
Apart from the observed link between PE_PGRS surface attachment and LOS production, this study also led to the identification of 10 novel LOS biosynthesis mutants. We showed that the genes coding for Pks5 and FadD25 are both necessary for the synthesis of the core LOS structure, which supports their previously proposed role in the synthesis and activation of the fatty acids linked to the trehalose core (18). The gene disruption of papA3 also resulted in an LOS-deficient phenotype. Although this gene is located downstream of the putative LOS region, the role of an acyltransferase in the synthesis of the LOS core structure is not surprising. Interestingly, a papA4 mutant in M. marinum was also shown to be deficient in LOS biosynthesis, showing two different acyltransferases are necessary for the synthesis of the core LOS structure (18). A more unexpected finding is the involvement of the putative peptide protease CphB in LOS biosynthesis. Among the sequenced strains of the genus Mycobacterium, this protease is unique to M. marinum (37), and its function is unclear. It is hard to imagine a role for this peptidase in LOS biosynthesis, as LOS does not contain peptides. Perhaps, CphB also plays a role in gene regulation. Further research is necessary to clarify the role of this gene in LOS biosynthesis.
The other five LOS mutants detected in this screen show a specific defect in higher order LOS production and a concomitant accumulation of the lower order LOS structures. The genes affected in these mutants also show a specific spatial genomic clustering, according to their role in LOS biosynthesis. Mutants that show LOS-I accumulation, indicating a deficiency to synthesize, attach, or transport the xylose unit to the LOS structure, are found at the beginning of the LOS biosynthesis cluster, in genes mmar_2307 and mmar_2309. The region responsible for the synthesis and attachment of caryophyllose to produce LOS-II and LOS-III seems to be localized roughly between genes mmar_2327 and mmar_2336. Also a rather large part of the genome cluster, genes mmar_2313 to mmar_2320, seems to be reserved for the synthesis and transfer of the amino sugar necessary to produce LOS-IV. Together, these results show that the genomic locus involved in LOS biosynthesis is more extended than previously suggested.
The specific decrease in LOS production of the whiB4 mutant together with its predicted function in gene regulation suggests that this protein has a role in regulating LOS biosynthesis. In line with this hypothesis, we have shown that the expression of a number of LOS biosynthesis genes is strongly decreased in this mutant. Full complementation was only achieved for some genes, although we tested multiple complementing constructs (data not shown). The effect of WhiB4 on LOS gene regulation could either be direct or indirect, through a general effect on metabolism. Because the whiB4 mutation had no effect on the pks12 gene involved in mycoketide biogenesis, we favor the first option. Correspondingly, WhiB3 has been shown to regulate biosynthesis of various polyketides (38), which could indicate that WhiB proteins are important regulators of lipid biosynthesis in mycobacteria.
A number of microarray and qRT-PCR assays have shown that WhiB4 itself in M. tuberculosis is regulated in a variety of stress conditions such as macrophage infection and nutrient starvation (46–48), indicating it has an important role in infection. Biochemical and biophysical studies of recombinant M. tuberculosis WhiB4 showed that it has disulfide reductase activity upon removal of the iron-sulfur cluster (49), but it is unclear how this observation relates to transcriptional regulatory activity. M. tuberculosis does not produce LOS but does contain a large part of the LOS biosynthesis genome cluster, including a homologue of mmar_2327 (15), which might be involved in biosynthesis of a different glycolipid.
A mutation in the wecE gene of M. marinum, resulting in LOS-IV deficiency and concomitant LOS-III accumulation, leads to significantly increased bacterial growth and early granuloma formation in zebrafish embryos. Increased virulence as a result of a single mutation is rarely observed. For instance, in an extensive screen for mycobacterial virulence of 1,000 transposon mutants of M. marinum in zebrafish embryos, only a single hypervirulence mutant was found (31).5 These in vivo data highlight the importance of WecE as a factor to suppress virulence in early mycobacterial infection. Whether the observed hypervirulence is actually caused by the loss of LOS-IV, the accumulation of LOS-III, or the altered release of surface proteins, such as PE_PGRS, is not clear at this point. In contrast to these in vivo experiments, recent in vitro infections of murine macrophage cell lines with different M. marinum LOS mutants demonstrated impaired cell-entry efficiency (15). Also, the addition of purified LOS-IV to macrophages resulted in an altered pro-inflammatory response (19, 32). However, from our studies it seems that LOS-IV deficiency actually results in a more pronounced infection, suggesting a different mechanism in the context of zebrafish embryos. Increased virulence was also observed in mouse infection studies with different M. kansasii strains. These studies showed that smooth strains producing LOS were rapidly cleared from the animal organs, whereas the rough LOS-deficient strains showed greater persistence (35, 50). It has been proposed that the presence of LOS might act as a mask for other surface-associated factors, such as lipoarabinomannan and phenolic glycolipids (35); enhanced exposure of these glycolipids might result in a more virulent infection. The fact that LOS production is absent in most species of the M. tuberculosis complex, except for the ancient M. canettii, also fits with this hypothesis. Interestingly, in a screen for M. tuberculosis mutants with decreased ability to arrest phagosome maturation in human macrophages, a mutant was found in the homologue of wecE. M. tuberculosis does not produce LOS, and disruption of this gene was shown to be responsible for the increased biosynthesis of a sulfoglycolipid, which, in purified form, actually promotes phagosome acidification and therefore presumably mycobacterial killing (51). Further research is required to study the mechanism of the observed hypervirulence associated with LOS-IV deficiency, which could tell us more about the interaction of mycobacteria with its host.
Acknowledgments
We thank Ben Appelmelk for helpful discussions and advice. We also thank Ida Rosenkrands and Eric J. Brown for kindly providing the antiserum against ESAT-6 and EspE, respectively. We are grateful to Michael J. Brennan for providing the PGRS hybridoma clone and to Janneke Maaskant for efforts in selection and subculture of the PGRS hybridoma clone. We also thank Gunny van den Brink-van Stempvoort and Esther Stoop for their help and suggestions in the zebrafish embryo infections, and Roy Ummels, Eveline Weerdenburg, Iris Braat, and Wina Verlaat for their technical assistance.
This work was supported by an ECHO project grant from the Netherlands Organization for Scientific Research (to A. D. v. d. W.) and a Netherlands Organization for Scientific Research-VENI grant (to E. N. G. H.).

This article contains supplemental Table S1.
E. Stoop and A. M. van der Sar, manuscript in preparation.
R. Ummels, W. Bitter, and E. N. G. Houben, unpublished results.
E. Stoop and A. M. van der Sar, personal communication.
- LOS
- lipooligosaccharide
- PE_PGRS
- group of proteins from PE family with polymorphic GC-rich sequence
- T7SS
- type VII secretion system
- qRT-PCR
- quantitative real time PCR.
REFERENCES
- 1. World Health Organization (2011) 2011/2012 Tuberculosis Global Facts, WHO Press, Geneva, Switzerland [Google Scholar]
- 2. Hoffmann C., Leis A., Niederweis M., Plitzko J. M., Engelhardt H. (2008) Disclosure of the mycobacterial outer membrane. Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U.S.A. 105, 3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zuber B., Chami M., Houssin C., Dubochet J., Griffiths G., Daffé M. (2008) Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190, 5672–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sani M., Houben E. N., Geurtsen J., Pierson J., de Punder K., van Zon M., Wever B., Piersma S. R., Jiménez C. R., Daffé M., Appelmelk B. J., Bitter W., van der Wel N., Peters P. J. (2010) Direct visualization by cryo-EM of the mycobacterial capsular layer. A labile structure containing ESX-1-secreted proteins. PLoS Pathog. 6, e1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brennan P. J., Nikaido H. (1995) The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 6. Abdallah A. M., Gey van Pittius N. C., Champion P. A., Cox J., Luirink J., Vandenbroucke-Grauls C. M., Appelmelk B. J., Bitter W. (2007) Type VII secretion. Mycobacteria show the way. Nat. Rev. Microbiol. 5, 883–891 [DOI] [PubMed] [Google Scholar]
- 7. Gey Van Pittius N. C., Gamieldien J., Hide W., Brown G. D., Siezen R. J., Beyers A. D. (2001) The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2, RESEARCH0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pallen M. J. (2002) The ESAT-6/WXG100 superfamily and a new Gram-positive secretion system? Trends Microbiol. 10, 209–212 [DOI] [PubMed] [Google Scholar]
- 9. Abdallah A. M., Verboom T., Hannes F., Safi M., Strong M., Eisenberg D., Musters R. J., Vandenbroucke-Grauls C. M., Appelmelk B. J., Luirink J., Bitter W. (2006) A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol. 62, 667–679 [DOI] [PubMed] [Google Scholar]
- 10. Abdallah A. M., Verboom T., Weerdenburg E. M., Gey van Pittius N. C., Mahasha P. W., Jiménez C., Parra M., Cadieux N., Brennan M. J., Appelmelk B. J., Bitter W. (2009) PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol. Microbiol. 73, 329–340 [DOI] [PubMed] [Google Scholar]
- 11. Gey van Pittius N. C., Sampson S. L., Lee H., Kim Y., van Helden P. D., Warren R. M. (2006) Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 6, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 13. Stinear T. P., Seemann T., Harrison P. F., Jenkin G. A., Davies J. K., Johnson P. D., Abdellah Z., Arrowsmith C., Chillingworth T., Churcher C., Clarke K., Cronin A., Davis P., Goodhead I., Holroyd N., Jagels K., Lord A., Moule S., Mungall K., Norbertczak H., Quail M. A., Rabbinowitsch E., Walker D., White B., Whitehead S., Small P. L., Brosch R., Ramakrishnan L., Fischbach M. A., Parkhill J., Cole S. T. (2008) Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poulet S., Cole S. T. (1995) Characterization of the highly abundant polymorphic GC-rich-repetitive sequence (PGRS) present in Mycobacterium tuberculosis. Arch. Microbiol. 163, 87–95 [DOI] [PubMed] [Google Scholar]
- 15. Ren H., Dover L. G., Islam S. T., Alexander D. C., Chen J. M., Besra G. S., Liu J. (2007) Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol. Microbiol. 63, 1345–1359 [DOI] [PubMed] [Google Scholar]
- 16. Burguière A., Hitchen P. G., Dover L. G., Kremer L., Ridell M., Alexander D. C., Liu J., Morris H. R., Minnikin D. E., Dell A., Besra G. S. (2005) LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J. Biol. Chem. 280, 42124–42133 [DOI] [PubMed] [Google Scholar]
- 17. Daffe M., McNeil M., Brennan P. J. (1991) Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry 30, 378–388 [DOI] [PubMed] [Google Scholar]
- 18. Rombouts Y., Alibaud L., Carrère-Kremer S., Maes E., Tokarski C., Elass E., Kremer L., Guérardel Y. (2011) Fatty acyl chains of Mycobacterium marinum lipooligosaccharides. Structure, localization, and acylation by PapA4 (MMAR_2343) protein. J. Biol. Chem. 286, 33678–33688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rombouts Y., Burguière A., Maes E., Coddeville B., Elass E., Guérardel Y., Kremer L. (2009) Mycobacterium marinum lipooligosaccharides are unique caryophyllose-containing cell wall glycolipids that inhibit tumor necrosis factor-α secretion in macrophages. J. Biol. Chem. 284, 20975–20988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sarkar D., Sidhu M., Singh A., Chen J., Lammas D. A., van der Sar A. M., Besra G. S., Bhatt A. (2011) Identification of a glycosyltransferase from Mycobacterium marinum involved in addition of a caryophyllose moiety in lipooligosaccharides. J. Bacteriol. 193, 2336–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Talaat A. M., Reimschuessel R., Wasserman S. S., Trucksis M. (1998) Goldfish, Carassius auratus, a novel animal model for the study of Mycobacterium marinum pathogenesis. Infect. Immun. 66, 2938–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Puttinaowarat S., Thompson K. D., Adams A. (2000) Mycobacteriosis. Detection and identification of aquatic Mycobacterium species. Fish Vet. J. 5, 6–21 [Google Scholar]
- 23. van der Sar A. M., Abdallah A. M., Sparrius M., Reinders E., Vandenbroucke-Grauls C. M., Bitter W. (2004) Mycobacterium marinum strains can be divided into two distinct types based on genetic diversity and virulence. Infect. Immun. 72, 6306–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 25. Carlsson F., Joshi S. A., Rangell L., Brown E. J. (2009) Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog. 5, e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alderson M. R., Bement T., Day C. H., Zhu L., Molesh D., Skeiky Y. A., Coler R., Lewinsohn D. M., Reed S. G., Dillon D. C. (2000) Expression cloning of an immunodominant family of Mycobacterium tuberculosis antigens using human CD4+ T cells. J. Exp. Med. 191, 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harboe M., Malin A. S., Dockrell H. S., Wiker H. G., Ulvund G., Holm A., Jørgensen M. C., Andersen P. (1998) B-cell epitopes and quantification of the ESAT-6 protein of Mycobacterium tuberculosis. Infect. Immun. 66, 717–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hayward C. M., O'Gaora P., Young D. B., Griffin G. E., Thole J., Hirst T. R., Castello-Branco L. R., Lewis D. J. (1999) Construction and murine immunogenicity of recombinant Bacille Calmette Guérin vaccines expressing the B subunit of Escherichia coli heat labile enterotoxin. Vaccine 17, 1272–1281 [DOI] [PubMed] [Google Scholar]
- 29. Meijer A. H., van der Sar A. M., Cunha C., Lamers G. E., Laplante M. A., Kikuta H., Bitter W., Becker T. S., Spaink H. P. (2008) Identification and real time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev. Comp. Immunol. 32, 36–49 [DOI] [PubMed] [Google Scholar]
- 30. van der Sar A. M., Musters R. J., van Eeden F. J., Appelmelk B. J., Vandenbroucke-Grauls C. M., Bitter W. (2003) Zebrafish embryos as a model host for the real time analysis of Salmonella typhimurium infections. Cell. Microbiol. 5, 601–611 [DOI] [PubMed] [Google Scholar]
- 31. Stoop E. J., Schipper T., Huber S. K., Nezhinsky A. E., Verbeek F. J., Gurcha S. S., Besra G. S., Vandenbroucke-Grauls C. M., Bitter W., van der Sar A. M. (2011) Zebrafish embryo screen for mycobacterial genes involved in the initiation of granuloma formation reveals a newly identified ESX-1 component. Dis. Model. Mech. 4, 526–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rombouts Y., Elass E., Biot C., Maes E., Coddeville B., Burguière A., Tokarski C., Buisine E., Trivelli X., Kremer L., Guérardel Y. (2010) Structural analysis of an unusual bioactive N-acylated lipo-oligosaccharide LOS-IV in Mycobacterium marinum. J. Am. Chem. Soc. 132, 16073–16084 [DOI] [PubMed] [Google Scholar]
- 33. Hwang B. Y., Lee H. J., Yang Y. H., Joo H. S., Kim B. G. (2004) Characterization and investigation of substrate specificity of the sugar aminotransferase WecE from E. coli K12. Chem. Biol. 11, 915–925 [DOI] [PubMed] [Google Scholar]
- 34. Etienne G., Malaga W., Laval F., Lemassu A., Guilhot C., Daffé M. (2009) Identification of the polyketide synthase involved in the biosynthesis of the surface-exposed lipooligosaccharides in mycobacteria. J. Bacteriol. 191, 2613–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Belisle J. T., Brennan P. J. (1989) Chemical basis of rough and smooth variation in mycobacteria. J. Bacteriol. 171, 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Allen M. M. (1984) Cyanobacterial cell inclusions. Annu. Rev. Microbiol. 38, 1–25 [DOI] [PubMed] [Google Scholar]
- 37. Füser G., Steinbüchel A. (2007) Analysis of genome sequences for genes of cyanophycin metabolism: identifying putative cyanophycin metabolizing prokaryotes. Macromol. Biosci. 7, 278–296 [DOI] [PubMed] [Google Scholar]
- 38. Singh A., Crossman D. K., Mai D., Guidry L., Voskuil M. I., Renfrow M. B., Steyn A. J. (2009) Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsunaga I., Bhatt A., Young D. C., Cheng T. Y., Eyles S. J., Besra G. S., Briken V., Porcelli S. A., Costello C. E., Jacobs W. R., Jr., Moody D. B. (2004) Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J. Exp. Med. 200, 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parish T., Stoker N. G. (1998) Mycobacteria. Bugs and bugbears. Methods Mol. Biol. 101, 1–13 [DOI] [PubMed] [Google Scholar]
- 41. Hunter S. W., Murphy R. C., Clay K., Goren M. B., Brennan P. J. (1983) Trehalose-containing lipooligosaccharides. A new class of species-specific antigens from Mycobacterium. J. Biol. Chem. 258, 10481–10487 [PubMed] [Google Scholar]
- 42. Davis J. M., Clay H., Lewis J. L., Ghori N., Herbomel P., Ramakrishnan L. (2002) Real time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17, 693–702 [DOI] [PubMed] [Google Scholar]
- 43. Lesley R., Ramakrishnan L. (2008) Insights into early mycobacterial pathogenesis from the zebrafish. Curr. Opin. Microbiol. 11, 277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gilleron M., Vercauteren J., Puzo G. (1993) Lipooligosaccharidic antigen containing a novel C4-branched 3,6-dideoxy-α-hexopyranose typifies Mycobacterium gastri. J. Biol. Chem. 268, 3168–3179 [PubMed] [Google Scholar]
- 45. Lemassu A., Lévy-Frébault V. V., Lanéelle M. A., Daffé M. (1992) Lack of correlation between colony morphology and lipooligosaccharide content in the Mycobacterium tuberculosis complex. J. Gen. Microbiol. 138, 1535–1541 [DOI] [PubMed] [Google Scholar]
- 46. Rachman H., Strong M., Schaible U., Schuchhardt J., Hagens K., Mollenkopf H., Eisenberg D., Kaufmann S. H. (2006) Mycobacterium tuberculosis gene expression profiling within the context of protein networks. Microbes Infect. 8, 747–757 [DOI] [PubMed] [Google Scholar]
- 47. Geiman D. E., Raghunand T. R., Agarwal N., Bishai W. R. (2006) Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50, 2836–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Betts J. C., Lukey P. T., Robb L. C., McAdam R. A., Duncan K. (2002) Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731 [DOI] [PubMed] [Google Scholar]
- 49. Alam M. S., Garg S. K., Agrawal P. (2007) Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv. A [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol. Microbiol. 63, 1414–1431 [DOI] [PubMed] [Google Scholar]
- 50. Collins F. M., Cunningham D. S. (1981) Systemic Mycobacterium kansasii infection and regulation of the alloantigenic response. Infect. Immun. 32, 614–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brodin P., Poquet Y., Levillain F., Peguillet I., Larrouy-Maumus G., Gilleron M., Ewann F., Christophe T., Fenistein D., Jang J., Jang M. S., Park S. J., Rauzier J., Carralot J. P., Shrimpton R., Genovesio A., Gonzalo-Asensio J. A., Puzo G., Martin C., Brosch R., Stewart G. R., Gicquel B., Neyrolles O. (2010) High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog. 6, e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]