Abstract
The endothelial NO synthase (eNOS) is regulated by diverse protein kinase pathways, yet eNOS activity ultimately depends on the ubiquitous calcium regulatory protein calmodulin (CaM). In these studies, we establish that CaM itself undergoes phosphorylation in endothelial cells and that CaM phosphorylation attenuates eNOS activation. Using [32P]orthophosphoric acid biosynthetic labeling, we found that CaM is a phosphoprotein in bovine aortic endothelial cells (BAEC) and that the kinase CK2 promotes CaM phosphorylation in BAEC. Phosphorylation of CaM by purified CK2 in vitro reduces the Vmax of immunopurified eNOS by a factor of 2 but has no effect on the KA for CaM or calcium. Additionally, [32P]orthophosphoric acid biosynthetic labeling of mutant CaM-transfected BAEC revealed that phosphorylation of Ser-81 to alanine mutant CaM (“phosphonull” S81A mutant) is dramatically reduced relative to WT, whereas phosphorylation of the “phosphomimetic” Ser-81 to aspartate (S81D) mutant is unchanged. Further studies using Escherichia coli-expressed and phenyl-Sepharose-purified CaM mutants revealed that the S81A mutation abrogates in vitro CK2-mediated phosphorylation of CaM, whereas phosphorylation of the S81D CaM mutant by CK2 is preserved. Additionally, we found that the phosphomimetic S101D CaM mutant is impaired in its ability to activate eNOS. Taken together, these results suggest that phosphorylation of CaM inhibits eNOS catalysis and proceeds in a hierarchical manner, initially requiring phosphorylation of the CaM Ser-81 residue. We conclude that CaM phosphorylation may represent a unique pathway in the regulation of eNOS signaling and thereby may play a role in modulating NO-dependent vascular responses.
The vascular system uses a network of cell signaling pathways to maintain a delicate homeostatic balance. The endothelial NO synthase (eNOS) is a calmodulin-dependent enzyme that plays a key role in many of these signaling cascades by catalyzing the conversion of l-arginine and oxygen to l-citrulline and the labile gas NO (1). Because NO has fundamental effects on many aspects of vascular physiology (2), it is not surprising that eNOS is tightly regulated by a number of mechanisms. Subcellular localization, acylation, phosphorylation, and direct interaction with other proteins, including calmodulin (CaM), caveolin, and heat shock protein 90, have been shown to modulate eNOS (3). Recently, the phosphorylation of eNOS in endothelial cells has been extensively characterized (4–9); however, the role of phosphorylation pathways in modulating the protein partners of eNOS, such as CaM, is much less well understood.
CaM is a ubiquitously expressed, highly conserved acidic protein that mediates a broad range of intracellular calcium-regulated enzymes (10, 11), including eNOS (12). CaM is comprised of 148 aa and has a “dumbbell” structure: a long flexible central helix joining two pairs of calcium-binding helix–loop–helix motifs, known as EF hands (13). Upon binding calcium, CaM exposes hydrophobic regions on its surface, which are important for mediating interactions with its targets.
CaM modulates a broad range of phosphorylation pathways, and many of CaM's target proteins are themselves phosphorylated. Furthermore, CaM itself has been shown to be a phosphoprotein (reviewed in ref. 14). CaM isolated from rat liver is phosphorylated on the central helix (Thr-79 and Ser-81, ref. 15) and the third EF hand (Ser-101, ref. 15). The ubiquitously expressed kinase CK2 (formerly known as casein kinase 2) phosphorylates CaM in vitro on these residues (15–17) and has been implicated in insulin-induced CaM phosphorylation in hepatocytes (18).
Although CaM has been identified as a phosphoprotein in a few cell types (14), phosphorylation of CaM in endothelial cells has not been previously described to our knowledge. The interactions between eNOS and CaM have been extensively characterized, and in endothelial cells, it appears that eNOS plays a significant role in modulating diverse CaM-dependent processes by binding up to ≈25% of the limited pool of total cellular CaM (19). Hence, the question of whether phosphorylation of CaM modifies CaM–eNOS interactions may have important ramifications on numerous fundamental processes in endothelium beyond NO synthesis. In these studies, we show that CaM is a phosphoprotein in endothelial cells and CK2 activity promotes CaM phosphorylation in a hierarchical manner. Furthermore, we demonstrate that phosphorylation of CaM attenuates its allosteric activation of eNOS.
Methods
CaM Plasmid Constructs. Site-specific mutations were generated by PCR-based site-directed mutagenesis; all PCR-generated constructs were validated by nucleotide sequence analysis. CaM cDNA as described (20) was subcloned into the pMEX8 cloning vector (MoBi-Tec, Goettingen, Germany) between the EcoR1 and PstI sites of the polylinker. This plasmid was the template for PCRs that generated fragments used for subcloning CaM WT and mutant cDNAs in the pET23(+) prokaryotic expression vector (Novagen). These pET23(+) vectors encoding CaM WT or mutant plasmids served as templates for generating the FLAG epitope-tagged CaM PCR products, which were subcloned into the eukaryotic expression vector pcDNA3 (Invitrogen) (for cloning details see Supporting Methods, which is published as supporting information on the PNAS web site).
Cell Culture and Transfection. Bovine aortic endothelial cells (BAEC) were obtained from Cell Systems (Kirkland, WA) and cultured as described (9). Transfections were performed with FuGENE6 (Roche Molecular Biochemicals) by using the manufacturer's protocol, and cells were studied 48 h after transfection.
Biosynthetic Labeling. BAEC were washed twice with DMEM without phosphate and then incubated with this medium containing 10% dialyzed FBS and 40–100 μCi/ml [32P]orthophosphoric acid ([32Pi]) (900 Ci/mmol) for 4 h. When indicated, BAEC were treated with a kinase inhibitor or vehicle, washed twice with phosphate-free DMEM, and harvested.
Preparation of Cellular Lysates and Immunoprecipitation of CaM. Cell lysates and immunoprecipitations were prepared in a manner similar to that described (18) with some modifications. After washing with phosphate-free DMEM, BAEC were scraped in 100 mM Tris·HCl (pH 7.4), 0.1% SDS, 100 mM NaF, 30 mM Na4P2O7, 2 mM Na3VO4, 5 mM EDTA, 1 mM PMSF, 10 μg/ml aprotinin, 2 μg/ml leupeptin, 2 μg/ml antipain, 2 μg/ml soybean trypsin inhibitor, 2 μg/ml lima bean trypsin inhibitor, and 50 ng/ml pepstatin and immediately frozen in an ethanol/solid CO2 bath for 5 min. Lysates were then thawed and diluted with immunoprecipitation buffer (50 mM Tris·HCl, pH 7.4/1.25% Triton X-100/180 mM NaCl/5 mM NaF/1 mM Na4P2O7/1 mM β–glycerophosphate/2mMNa3VO4/6 mM EDTA). The solution was precleared by incubating with IgG1 mAb and protein G at 4°C. The precleared supernatant was incubated with anti-CaM, anti-FLAG, or isotype control IgG1 mAb for 2 h and then with protein G for 1 h at 4°C. Immune complexes were then processed as described (18).
Assay of NOS Enzymatic Activity of eNOS Immune Complexes. eNOS was immunoprecipitated from BAEC as described (9), and the immune complexes were then washed three times with 20 mM Tris·HCl (pH 7.4), 300 mM NaCl, and 1 mM DTT. NOS activity of the immune complexes was assayed by measuring the conversion of [3H]l-arginine to [3H]l-citrulline (9) for 15 min at 30°C in the presence of various concentrations of CaM or EGTA. Free calcium concentrations were determined at fixed EGTA concentrations by using an established algorithm (www.stanford.edu/~cpatton/webmaxcS.htm).
Expression of CaM in Escherichia coli and CaM Purification. CaM was expressed in E. coli and purified to homogeneity by one-step phenyl-Sepharose chromatography as described by Persechini et al. (21). Purified CaM was then desalted on a Bio-Rad 10 DG column and concentrated under vacuum. Generally, 1.5 mg of pure CaM was obtained per liter of bacterial culture, and purity was confirmed by Coomassie staining of the gel obtained after SDS/PAGE of the purified preparation. Protein concentrations in the purified CaM preparations were determined by amino acid composition analysis, performed by the Biopolymer Laboratory at Harvard Medical School after acid hydrolysis (in 6 M HCl) of the sample; amino acids in the acid hydrolysates were resolved and quantitated by using reverse-phase HPLC. Protein concentrations were confirmed with the Bio-Rad protein assay kit, and the results of these two methods were within 5% of one another.
In Vitro Phosphorylation of CaM and Purification of Phospho-CaM. CaM was phosphorylated as described (16), using 0.5 units of CK2 in a total volume of 70 μl for 2 h at 30°C. Stoichiometry of CaM phosphorylation was determined, and phospho-CaM was purified as described (16).
Statistical Analysis. All experiments were performed at least three times in duplicate. Mean values for individual experiments were expressed as mean ± SE. Densitometric values were corrected for the amount of protein present by normalizing for the signal on the anti-FLAG immunoblots. Statistical differences were assessed by ANOVA followed by t test. P < 0.05 was considered statistically significant.
Results
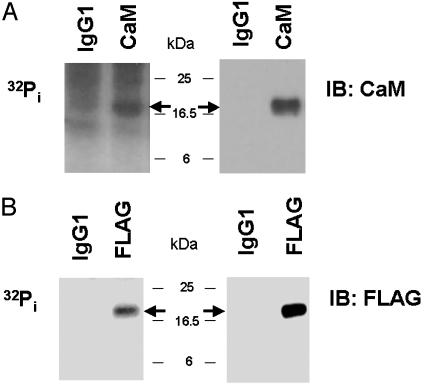
Phosphorylation of CaM in BAEC. We first used [32Pi] biosynthetic labeling experiments to determine whether CaM was phosphorylated in intact endothelial cells. CaM was immunoprecipitated from [32Pi] biosynthetically labeled BAEC with a specific anti-CaM mAb (18). As shown in Fig. 1A, CaM is a phosphoprotein in BAEC; however, the prominent background of the autoradiograph analyzed after immunoprecipitation with this anti-CaM antibody led us to search for an alternative strategy with a more favorable signal/noise ratio. We constructed a plasmid encoding CaM cDNA tagged on its carboxyl terminus with the FLAG epitope and transfected BAEC with this plasmid. Transfected BAEC were biosynthetically labeled with [32Pi], and the recombinant CaM was immunoprecipitated with an antibody directed against the FLAG epitope. As shown in Fig. 1B, the recombinant CaM is phosphorylated, and the background signal of the autoradiograph is minimal. Hence, further biosynthetic labeling studies evaluating CaM phosphorylation in intact endothelial cells used transfections of FLAG epitope-tagged CaM.
Fig. 1.
Immunoprecipitation of 32P-CaM from biosynthetically labeled BAEC. Shown are an autoradiograph and immunoblot (IB) of CaM immunoprecipitated from [32Pi] biosynthetically labeled BAEC. BAEC (A) or BAEC transfected with FLAG-tagged CaM cDNA (B) were biosynthetically labeled and then immunoprecipitated with either an isotype control IgG1 antibody or antibodies directed against CaM (A) or the FLAG epitope tag (B). Immune complexes were analyzed by autoradiography and immunoblotting with an anti-CaM (A) or anti-FLAG (B) antibody. The arrows indicate the expected location of CaM.
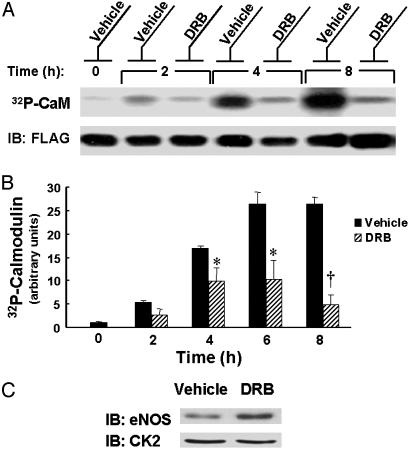
Effects of CK2 Inhibitor on CaM Phosphorylation. We next sought to identify which kinases are involved in modulating basal CaM phosphorylation in BAEC. BAEC were transfected with FLAG epitope-tagged CaM and biosynthetically labeled with [32Pi]. The labeled cells were then treated with various kinase inhibitors, and the recombinant CaM was immunoprecipitated with an anti-FLAG antibody and subjected to autoradiography. In preliminary experiments, we found that the phosphatidylinositol 3-kinase inhibitor wortmannin, the mitogen-activated protein kinase pathway inhibitor PD98059, and the protein kinase C inhibitor calphostin C do not have any effect on CaM phosphorylation (data not shown). However, treatment of BAEC with 50 μM 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), an inhibitor of the kinase CK2, robustly inhibits phosphorylation of CaM as shown in Fig. 2 A and B. In addition, DRB treatment of BAEC leads to a small, but reproducible, decrease in total cellular eNOS protein abundance without changing CK2 protein abundance (Fig. 2C). This observation is consistent with the work of Cieslik et al. (22), who reported that treatment of endothelial cells with DRB decreases eNOS promoter activity but does not change CK2 protein levels in nuclear extracts.
Fig. 2.
Effects of CK2 inhibitor on [32Pi] biosynthetic labeling of CaM. (A) Shown is an autoradiograph and immunoblot of CaM immunoprecipitated from [32Pi] biosynthetically labeled BAEC. BAEC were transfected with FLAG-tagged CaM cDNA, biosynthetically labeled with [32Pi], and then treated with vehicle or the CK2 inhibitor DRB (50 μM) for the indicated times at 37°C. CaM was immunoprecipitated with an antibody against the FLAG epitope. The immunoprecipitated proteins were analyzed by autoradiography and immunoblotting (IB) an anti-FLAG antibody. (B) The results of densitometric analysis of pooled data from the experiment shown in A and similar experiments, plotting the fold increase in the 32P-CaM signal, relative to the signal obtained for CaM labeling as analyzed in vehicle-treated cells at t = 0. * indicates P < 0.05, and † indicates P < 0.01 relative to vehicle-treated cells at the indicated time. (C) Immunoblots (IB) of CK2α and eNOS from lysates of DRB-treated BAEC. BAEC were treated with 50μM DRB or vehicle for 6 h at 37°C. Lysates were analyzed in immunoblots probed with an antibody directed against CK2α or eNOS.
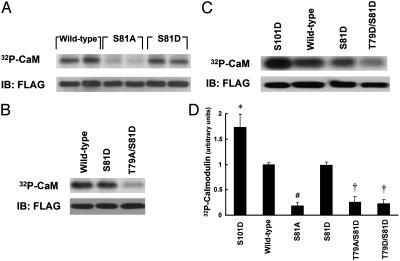
Phosphorylation of CaM Phosphorylation-Site Mutants. CaM isolated from rat liver has previously been shown to be phosphorylated on Thr-79, Ser-81, and Ser-101 (15). Using standard molecular cloning approaches, we generated a series of “phosphonull” and “phosphomimetic” CaM mutants by individually or collectively changing these phosphorylation sites to alanine (A) or aspartate (D), respectively. BAEC were transfected with FLAG epitopetagged WT or mutant CaM constructs and biosynthetically labeled with [32Pi]. Proteins were immunoprecipitated with the anti-FLAG antibody. As shown in Fig. 3 A and D, mutating the Ser-81 residue to alanine (S81A) markedly inhibits the incorporation of 32P into CaM. This dramatic reduction in [32Pi] labeling may result directly from elimination of Ser-81 phosphorylation; an alternative hypothesis is that the phosphorylation state of Ser-81 influences the kinetics of phosphorylation or dephosphorylation at another site(s). Therefore, we also evaluated the 32P incorporation of the phosphomimetic S81D mutant and the T79A/S81D mutant in biosynthetically labeled BAEC. Interestingly, the S81D mutant incorporates a comparable amount of 32P as WT CaM, whereas phosphorylation of the T79A/S81D mutant is attenuated (Fig. 3 A, B, and D). Additionally, as shown in Fig. 3 C and D, we found that phosphorylation of the T79D/S81D CaM mutant is reduced in comparison to WT, but the S101D mutant is hyperphosphorylated.
Fig. 3.
[32Pi] biosynthetic labeling of mutant CaM. (A–C) Autoradiographs and immunoblots (IB) of transfected CaM immunoprecipitated from [32Pi] biosynthetically labeled BAEC. BAEC were transfected with WT or mutant CaM cDNA constructs as indicated and biosynthetically labeled with [32Pi]. The lysates were immunoprecipitated with an antibody directed against the FLAG epitope. Immune complexes were analyzed by SDS/PAGE and autoradiography, and membranes were then probed with an anti-FLAG antibody. (D) The results of densitometric analysis of pooled data from the experiments shown in A–C and similar experiments, plotting the fold increase in the 32P-CaM signal, relative to the signal obtained for CaM labeling as analyzed in cells transfected with WT CaM. * indicates P < 0.05, and # indicates P < 0.001 relative to WT CaM-transfected cells; † indicates both P < 0.005 relative to WT CaM-transfected cells and P < 0.01 relative to S81D mutant CaM-transfected cells.
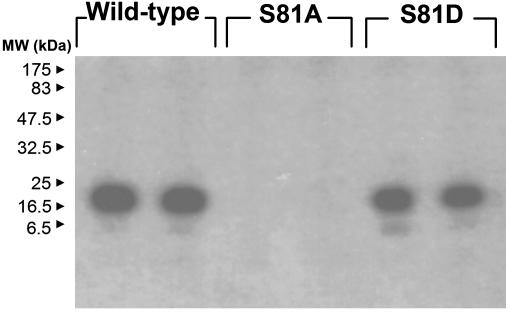
CK2 Treatment of CaM Ser-81 Phosphorylation-Site Mutants in Vitro. Having identified the prominent role of the Ser-81 residue in regulating CaM phosphorylation in BAEC, we next explored the role of this residue in CK2-induced CaM phosphorylation in vitro. WT, S81A, and S81D mutant CaM were expressed in E. coli, purified to homogeneity with phenyl-Sepharose chromatography, and then phosphorylated in vitro with CK2 and [γ–32P]ATP. Proteins were then analyzed by SDS/PAGE and autoradiography. As shown in Fig. 4, mutation of the Ser-81 residue to alanine abrogates the phosphorylation of CaM, whereas the S81D CaM mutant restores the phosphorylation phenotype.
Fig. 4.
Effects of Ser-81 mutation on in vitro CK2-induced CaM phosphorylation. Shown is an autoradiograph of WT and mutant CaM phosphorylated with [γ-32P]ATP in the presence of CK2. Purified WT and mutant CaM were phosphorylated with CK2 as described in Methods, and this phosphorylation reaction was analyzed by SDS/PAGE and autoradiography. Each lane represents an independent phosphorylation reaction.
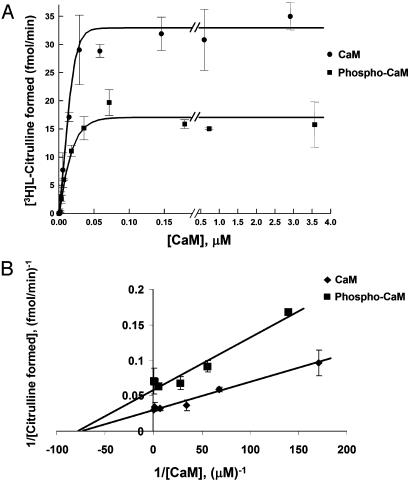
Effects of CaM Phosphorylation on CaM-Induced eNOS Activity. We next explored whether phosphorylation of CaM has functional consequences on the activation of eNOS, a physiologically important enzyme that is a major target of CaM in BAEC (19). In these studies, purified CaM was phosphorylated in vitro with CK2 to a stoichiometry ranging from 0.8 to 1.0 mol of phosphate/mol of CaM. Vehicle- and CK2-treated CaM preparations were once again purified to homogeneity over phenyl-Sepharose, and the protein concentrations were determined by amino acid analysis. CaM or CK2-phosphorylated CaM was added to eNOS immunopurified from BAEC, and the eNOS enzymatic activity was assayed by measuring the conversion of [3H]l-arginine to [3H]l-citrulline as described in Methods. As shown in Fig. 5, the maximal activity (Vmax) of eNOS in the presence of phosphorylated CaM is reduced by a factor of 2 in comparison to the Vmax in the presence of CaM (35.5 ± 5.0 fmol of [3H]l-citrulline formed per min vs. 17.6 ± 1.5 fmol/min, P < 0.05). In contrast, the affinity of phosphorylated CaM or CaM for eNOS is similar (KA of 13.1 vs. 16.2 nM, respectively, P = not significant). This KA of CaM is in good agreement with our previously determined value of 20 nM for E. coli-expressed eNOS (23).
Fig. 5.
Effects of CaM phosphorylation on eNOS enzymatic activity. (A) Enzymatic activity assay of immunopurified eNOS at various concentrations of CaM or phosphorylated CaM. eNOS was immunoprecipitated from BAEC with an anti-eNOS antibody. Purified preparations of vehicle- or CK2-treated CaM were added at various concentrations to the immune complexes, and the NOS activity was measured as described in Methods. The concentrations of CaM and phosphorylated CaM were determined by amino acid analysis. (B) A double reciprocal plot of the data in A.
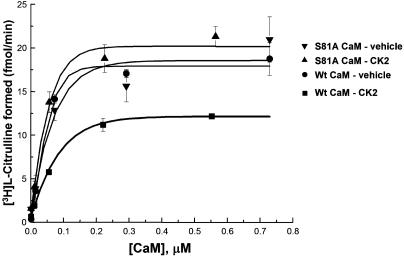
We then used the lack of CK2-induced S81A CaM phosphorylation in vitro (Fig. 4) as a means to confirm that the decreased eNOS activation of WT CaM treated with CK2 is indeed a result of CaM phosphorylation. WT and S81A mutant CaM were treated with CK2 or vehicle and then purified to homogeneity. These preparations were added to eNOS immunopurified from BAEC, and NOS activity was measured as described in Methods. The stoichiometry of phosphorylation of the CK2-treated WT CaM was 1.1 mol of phosphate/mol of CaM, whereas CK2 treatment of the S81A mutant led to negligible phosphate incorporation. As shown in Fig. 6, WT CaM treated with CK2 has a reduced ability to maximally activate eNOS compared to WT CaM treated with vehicle. In contrast, the Vmax of eNOS in the presence of purified S81A CaM treated with CK2 or vehicle is equivalent and essentially the same as that of vehicle-treated WT CaM.
Fig. 6.
Effects of phosphonull Ser-81 alanine CaM mutation on eNOS activation. Shown is an enzymatic activity assay of immunopurified eNOS at various concentrations of purified WT or S81A CaM treated with CK2 or vehicle. eNOS was immunoprecipitated from BAEC with an anti-eNOS antibody. Various concentrations of purified preparations of CK2- or vehicle-treated WT or S81A CaM were added to the immune complexes, and NOS activity was measured as described in Methods.
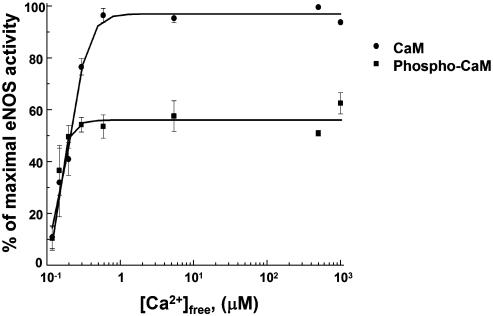
We also evaluated the effects of CK2-catalyzed CaM phosphorylation on the affinity of CaM for calcium. The enzymatic activity of immunopurified eNOS was measured in the presence of purified vehicle- or CK2-treated CaM (100 nM), 1 mM CaCl2, and various concentrations of EGTA (Fig. 7). The calcium concentration at which the half-maximal activation of eNOS is observed in the presence of purified CaM does not differ significantly from that in the presence of phospho-CaM (197 ± 30 μM vs. 160 ± 27 μM, P = not significant). This result is in agreement with our prior results that used other CaM-dependent enzymes to demonstrate that phosphorylation of CaM does not change the protein's affinity for calcium (24).
Fig. 7.
Calcium-dependent activation of phosphorylated CaM. Shown is an enzymatic activity assay of eNOS immunopurified from BAEC in the presence of CaM or phosphorylated CaM at increasing free calcium concentrations. eNOS was immunoprecipitated from BAEC, and the NOS activity of the immune complexes was measured in the presence of purified preparations of vehicle- or CK2-treated CaM, 1 mM CaCl2, and various concentrations of EGTA as described in Methods.
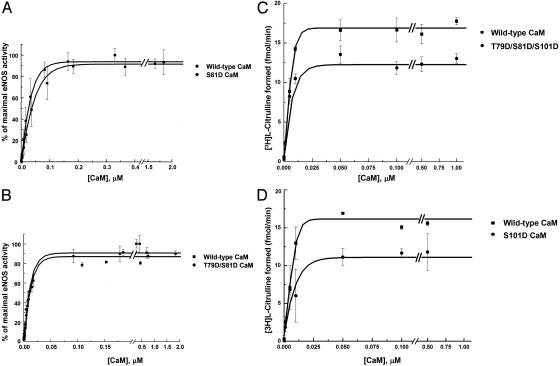
Allosteric Activation of eNOS by Site-Specific Phosphomimetic CaM Mutants. WT CaM and phosphomimetic CaM mutants, in which a specific phosphorylation site(s) was mutated to aspartate (D), were expressed in E. coli and purified to homogeneity. Varying concentrations of the purified CaM were then added to eNOS immunopurified from BAEC, and eNOS enzymatic activity of the immune complexes was determined as described above. As shown in Fig. 8 A and B, eNOS activation kinetics in the presence of S81D and T79D/S81D CaM mutants do not differ from those of WT CaM. However, in comparison to WT CaM-induced eNOS activation, the Vmax of eNOS is reduced in the presence of T79D/S81D/S101D CaM (by 29 ± 1%, P < 0.001) or S101D CaM (by 30 ± 2%, P < 0.05), whereas the KA does not change significantly (Fig. 8 C and D).
Fig. 8.
Effects of phosphomimetic CaM mutants on eNOS enzymatic activity. Shown are the results of enzymatic activity assays of immunoprecipitated eNOS at various concentrations of purified WT or mutant CaM in which putative phosphorylation sites were changed to aspartate. eNOS was immunopurified from BAEC with an antibody directed against eNOS. Various concentrations of purified WT or mutant CaM were added to the immune complexes, and NOS activity was measured as described in Methods.
Discussion
These studies provide evidence that the ubiquitous calcium regulatory protein CaM is a phosphoprotein in endothelial cells and that phosphorylation of CaM attenuates its ability to activate eNOS. The importance of delineating signaling pathways that modulate the CaM–eNOS interaction is emphasized by the fact that eNOS activity absolutely depends on CaM. Phosphorylation of eNOS itself has been shown to occur on several residues and modulate enzymatic activity in a complex fashion. The role of phosphorylation of CaM in the regulation of eNOS has been evaluated here.
We explored CaM phosphorylation by studying the incorporation of 32P into CaM in cultured endothelial cells biosynthetically labeled with [32Pi] and documented that CaM is a phosphoprotein in endothelial cells (Fig. 1). In analyzing a series of kinase inhibitors, we found that the activity of CK2 potentates CaM phosphorylation in BAEC (Fig. 2). Although the CK2 inhibitor DRB has been used to evaluate the role of CK2 in other endothelial signaling pathways (22), to our knowledge no CK2 inhibitor has previously been used to evaluate pathways controlling CaM phosphorylation in any cell type. Indeed, the only existing evidence supporting a role for CK2 in mediating CaM phosphorylation in vivo is indirect: phosphopeptides generated from CaM isolated from insulin-treated rat hepatocytes overlap significantly with those isolated from in vitro CK2-phosphorylated CaM (18).
Our results establish that CaM phosphorylation has functional consequences on the activation of eNOS, a key target enzyme in endothelial cells. Within the limitations of performing kinetic analyses of a partially purified enzyme preparation, we have established that the phosphorylation of CaM markedly reduces the maximal activation of eNOS, but does not change the affinity for eNOS (Fig. 5). Intracellular free Ca2+-CaM is in limited supply as the concentration of CaM-binding proteins in intact cells outnumbers the concentration of CaM by 2-fold (25), and competition for this limited free Ca2+-CaM pool facilitates cross-talk between multiple CaM-dependent signaling pathways (26). Recently, it has been proposed that eNOS plays a key role in regulating a broad range of CaM-dependent signaling cascades in endothelial cells by modulating CaM availability (19). The eNOS–CaM interaction is high affinity and dynamic, and based on stoichiometry, eNOS has the potential to bind up to ≈25% of CaM in BAEC (the total CaM and eNOS concentrations in homogenates are 25.9 ± 1.5 and 5.6 ± 0.6 pmol/mg total protein, respectively; ref. 19). Thus, we speculate that phosphorylation of CaM may represent a means of decreasing cellular eNOS activity without changing the availability of CaM for other signaling pathways.
Interestingly, the inhibitory effects of CK2-catalyzed CaM phosphorylation on eNOS activation kinetics described above are distinct from the reported effects on neuronal NOS (nNOS) kinetics: Quadroni et al. (27) found that in vitro CK2 treatment of CaM increases the nNOS maximal activity. Although eNOS and nNOS have similar features, their amino acid sequences are only 60% identical. Important differences exist in terms of the biological function and catalysis of these NOS isoforms and in the nature of their interaction with CaM (28–33). Given these disparities, it is perhaps not surprising that the phosphorylation of CaM affects the kinetics of eNOS and nNOS differently.
CaM has previously been reported to be phosphorylated on Thr-79, Ser-81, and Ser-101 in rat liver (15), but the individual roles of these sites in phosphorylation of CaM in vivo have not been characterized. To begin to explore this issue, we constructed a series of epitope-tagged phosphonull and phosphomimetic mutants in which the candidate phosphorylation sites were individually or collectively mutated to alanine or aspartate, respectively. These mutants were transfected into BAEC, and the cells were biosynthetically labeled with [32Pi]. We found that the conversion of Ser-81 to alanine dramatically attenuates CaM phosphorylation, whereas phosphorylation of the S81D mutant is indistinguishable from that of WT (Fig. 3). Furthermore, phosphorylation of the T79A/S81D and T79D/S81D mutants are significantly inhibited. These results are in good agreement with our in vitro CaM phosphorylation studies: CK2-catalyzed phosphorylation of purified CaM is abrogated by the S81A mutation, whereas phosphorylation of the S81D mutant is preserved (Fig. 4). Taken together, these results suggest that under basal conditions in BAEC, CaM is constitutively phosphorylated on Ser-81 and that the kinetics of phosphorylation and dephosphorylation of this residue are slow. The phosphorylation of Ser-81 appears to be required for phosphorylation of another residue(s), most likely Thr-79 (as shown in Fig. 3, the phosphorylation of Thr-79 does not appear to be required for subsequent CaM phosphorylation). The similarity of the in vitro and in vivo results provide further evidence that CK2 may phosphorylate CaM in BAEC and suggest that CaM phosphorylation may occur in a sequential manner. In combination with other kinases, CK2 has previously been shown to phosphorylate some proteins in a hierarchical fashion (34). Our results suggest that CK2 itself may phosphorylate proteins in a hierarchical manner. Although the regulation of phosphorylation of specific sites of CaM is complex and inter-related, this complexity is not uncommon among multiply phosphorylated proteins (35), such as eNOS (9, 36).
The interaction of phosphorylation sites in CaM does not appear to be limited to Thr-79 and Ser-81, as the S101D phosphomimetic mutant displays increased phosphorylation compared to WT CaM (Fig. 3). This result suggests that phosphorylation of Ser-101 may itself facilitate phosphorylation at other sites. In addition, our in vitro eNOS activity assays with the purified S101D CaM mutant suggest that phosphorylation of Ser-101 is likely to play a role in mediating the decreased activation of eNOS upon CaM phosphorylation (Fig. 8). The Ser-101 residue resides in the loop of the third EF hand, a region of CaM not implicated in CaM–eNOS contact by a recent crystallographic analysis (29). These crystallographic data are consistent with our present work, suggesting that phosphorylation of Ser-101 of CaM influences eNOS catalysis but not CaM–eNOS binding.
These studies have shown that CaM phosphorylation attenuates eNOS activity and implicate CK2 in signaling pathways that modulate CaM phosphorylation in endothelial cells. Regulation of CK2 itself, however, remains an enigmatic and controversial area (37, 38). CK2 has been found in specialized plasmalemma domains termed caveolae (39), which are enriched in a broad range of signaling molecules (40, 41), including eNOS and CaM (42). Moreover, CK2 has been shown to catalyze in vitro phosphorylation of the scaffolding protein caveolin, the principal protein in caveolae (39). We have previously shown that eNOS undergoes a regulatory cycle of enzymatic activation and inhibition based on its reversible association with Ca2+–CaM and caveolin, respectively (23, 43). It is possible that CK2 plays a role in regulating this cycle through catalyzing phosphorylation of CaM or caveolin and thereby altering allosteric regulation of eNOS. Taken together, our results suggest that CaM phosphorylation may influence eNOS-dependent signaling pathways in endothelial cells and thereby modulate vascular homeostasis.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health grants: National Heart, Lung, and Blood Institute Grant HL46457 and National Institute of General Medical Sciences Grant GM36259 (to T.M.). D.M.G. was supported by a Physician Postdoctoral Research Fellowship from the Howard Hughes Medical Institute. D.B.S is supported by grants from the National Institutes of Health.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NOS, NO synthase; eNOS, endothelial NOS; CaM, calmodulin; BAEC, bovine aortic endothelial cells; [32Pi], [32P]orthophosphoric acid; DRB, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole.
References
- 1.Stuehr, D. J. (1999) Biochim. Biophys. Acta 1411, 217–230. [DOI] [PubMed] [Google Scholar]
- 2.Loscalzo, J. & Welch, G. (1995) Prog. Cardiovasc. Dis. 38, 87–104. [DOI] [PubMed] [Google Scholar]
- 3.Fleming, I. & Busse, R. (1999) Cardiovasc. Res. 43, 532–541. [DOI] [PubMed] [Google Scholar]
- 4.Dimmeler, S., Fleming, I., Fisslthaler, B., Hermann, C., Busse, R. & Zeiher, A. M. (1999) Nature 399, 601–605. [DOI] [PubMed] [Google Scholar]
- 5.Fulton, D., Gratton, J. P., McCabe, T. J., Fontana, J., Fujio, Y., Walsh, K., Franke, T. F., Papapetropoulos, A. & Sessa, W. C. (1999) Nature 399, 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris, M. B., Ju, H., Venema, V. J., Liang, H., Zou, R., Michell, B. J., Chen, Z. P., Kemp, B. E. & Venema, R. C. (2001) J. Biol. Chem. 276, 16587–16591. [DOI] [PubMed] [Google Scholar]
- 7.Kou, R., Greif, D. & Michel, T. (2002) J. Biol. Chem. 277, 29669–29673. [DOI] [PubMed] [Google Scholar]
- 8.Michell, B. J., Harris, M. B., Chen, Z. P., Ju, H., Venema, V. J., Blackstone, M. A., Huang, W., Venema, R. C. & Kemp, B. E. (2002) J. Biol. Chem. 277, 42344–42351. [DOI] [PubMed] [Google Scholar]
- 9.Greif, D. M., Kou, R. & Michel, T. (2002) Biochemistry 41, 15845–15853. [DOI] [PubMed] [Google Scholar]
- 10.Stull, J. T. (2001) J. Biol. Chem. 276, 2311–2312. [DOI] [PubMed] [Google Scholar]
- 11.Chin, D. & Means, A. R. (2000) Trends Cell Biol. 10, 322–328. [DOI] [PubMed] [Google Scholar]
- 12.Forstermann, U., Pollock, J. S., Schmidt, H. H., Heller, M. & Murad, F. (1991) Proc. Natl. Acad. Sci. USA 88, 1788–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babu, Y. S., Sack, J. S., Greenhough, T. J., Bugg, C. E., Means, A. R. & Cook, W. J. (1985) Nature 315, 37–40. [DOI] [PubMed] [Google Scholar]
- 14.Benaim, G. & Villalobo, A. (2002) Eur. J. Biochem. 269, 3619–3631. [DOI] [PubMed] [Google Scholar]
- 15.Quadroni, M., James, P. & Carafoli, E. (1994) J. Biol. Chem. 269, 16116–16122. [PubMed] [Google Scholar]
- 16.Sacks, D. B., Davis, H. W., Crimmins, D. L. & McDonald, J. M. (1992) Biochem. J. 286, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meggio, F., Brunati, A. M. & Pinna, L. A. (1987) FEBS Lett. 215, 241–246. [DOI] [PubMed] [Google Scholar]
- 18.Joyal, J. L., Crimmins, D. L., Thoma, R. S. & Sacks, D. B. (1996) Biochemistry 35, 6267–6275. [DOI] [PubMed] [Google Scholar]
- 19.Tran, Q. K., Black, D. & Persechini, A. (2003) J. Biol. Chem. 278, 24247–24250. [DOI] [PubMed] [Google Scholar]
- 20.Sacks, D. B., Davis, H. W., Crimmins, D. L., Persechini, A. & McDonald, J. M. (1992) Biochem. Biophys. Res. Commun. 188, 754–759. [DOI] [PubMed] [Google Scholar]
- 21.Persechini, A., Blumenthal, D. K., Jarrett, H. W., Klee, C. B., Hardy, D. O. & Kretsinger, R. H. (1989) J. Biol. Chem. 264, 8052–8058. [PubMed] [Google Scholar]
- 22.Cieslik, K., Lee, C. M., Tang, J. L. & Wu, K. K. (1999) J. Biol. Chem. 274, 34669–34675. [DOI] [PubMed] [Google Scholar]
- 23.Michel, J. B., Feron, O., Sase, K., Prabhakar, P. & Michel, T. (1997) J. Biol. Chem. 272, 25907–25912. [DOI] [PubMed] [Google Scholar]
- 24.Sacks, D. B., Davis, H. W., Williams, J. P., Sheehan, E. L., Garcia, J. G. & McDonald, J. M. (1992) Biochem. J. 283, 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persechini, A. & Cronk, B. (1999) J. Biol. Chem. 274, 6827–6830. [DOI] [PubMed] [Google Scholar]
- 26.Persechini, A. & Stemmer, P. M. (2002) Trends Cardiovasc. Med. 12, 32–37. [DOI] [PubMed] [Google Scholar]
- 27.Quadroni, M., L'Hostis, E. L., Corti, C., Myagkikh, I., Durussel, I., Cox, J., James, P. & Carafoli, E. (1998) Biochemistry 37, 6523–6532. [DOI] [PubMed] [Google Scholar]
- 28.Nishida, C. R. & Ortiz de Montellano, P. R. (1999) J. Biol. Chem. 274, 14692–14698. [DOI] [PubMed] [Google Scholar]
- 29.Aoyagi, M., Arvai, A. S., Tainer, J. A. & Getzoff, E. D. (2003) EMBO J. 22, 766–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, M. & Vogel, H. J. (1994) J. Biol. Chem. 269, 981–985. [PubMed] [Google Scholar]
- 31.Salerno, J. C., Harris, D. E., Irizarry, K., Patel, B., Morales, A. J., Smith, S. M., Martasek, P., Roman, L. J., Masters, B. S., Jones, C. L., et al. (1997) J. Biol. Chem. 272, 29769–29777. [DOI] [PubMed] [Google Scholar]
- 32.Roman, L. J., Martasek, P., Miller, R. T., Harris, D. E., de La Garza, M. A., Shea, T. M., Kim, J. J. & Masters, B. S. (2000) J. Biol. Chem. 275, 29225–29232. [DOI] [PubMed] [Google Scholar]
- 33.Lane, P. & Gross, S. S. (2002) J. Biol. Chem. 277, 19087–19094. [DOI] [PubMed] [Google Scholar]
- 34.Picton, C., Woodgett, J., Hemmings, B. & Cohen, P. (1982) FEBS Lett 150, 191–196. [DOI] [PubMed] [Google Scholar]
- 35.Cohen, P. (2000) Trends Biochem. Sci. 25, 596–601. [DOI] [PubMed] [Google Scholar]
- 36.Bauer, P. M., Fulton, D., Boo, Y. C., Sorescu, G. P., Kemp, B. E., Jo, H. & Sessa, W. C. (2003) J. Biol. Chem. 278, 14841–14849. [DOI] [PubMed] [Google Scholar]
- 37.Litchfield, D. W., Dobrowolska, G. & Krebs, E. G. (1994) Cell. Mol. Biol. Res. 40, 373–381. [PubMed] [Google Scholar]
- 38.Pinna, L. A. (1990) Biochim. Biophys. Acta 1054, 267–284. [DOI] [PubMed] [Google Scholar]
- 39.Sargiacomo, M., Scherer, P. E., Tang, Z. L., Casanova, J. E. & Lisanti, M. P. (1994) Oncogene 9, 2589–2595. [PubMed] [Google Scholar]
- 40.Shaul, P. W. & Anderson, R. G. (1998) Am. J. Physiol. 275, L843–L851. [DOI] [PubMed] [Google Scholar]
- 41.Smart, E. J., Graf, G. A., McNiven, M. A., Sessa, W. C., Engelman, J. A., Scherer, P. E., Okamoto, T. & Lisanti, M. P. (1999) Mol. Cell. Biol. 19, 7289–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaul, P. W., Smart, E. J., Robinson, L. J., German, Z., Yuhanna, I. S., Ying, Y., Anderson, R. G. & Michel, T. (1996) J. Biol. Chem. 271, 6518–6522. [DOI] [PubMed] [Google Scholar]
- 43.Michel, J. B., Feron, O., Sacks, D. & Michel, T. (1997) J. Biol. Chem. 272, 15583–15586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.