Background: Sequence similarity between related proteins is often too low to allow inference of function.
Results: Applying phylogenetic networks to protein structural data improves resolution of deep evolutionary and functional relationships across the ferritin superfamily.
Conclusion: Structure-based phylogenetic networks facilitate inference of protein phylogeny and function beyond the “twilight zone” of sequence similarity.
Significance: Structural phylogenetics provides an important complement to sequence-based analyses.
Keywords: Evolution, Ferritin, Molecular Evolution, Phylogenetics, Protein Evolution, Protein Structure, Ribonucleotide Reductase, Functional Prediction
Abstract
In the postgenomic era, bioinformatic analysis of sequence similarity is an immensely powerful tool to gain insight into evolution and protein function. Over long evolutionary distances, however, sequence-based methods fail as the similarities become too low for phylogenetic analysis. Macromolecular structure generally appears better conserved than sequence, but clear models for how structure evolves over time are lacking. The exponential growth of three-dimensional structural information may allow novel structure-based methods to drastically extend the evolutionary time scales amenable to phylogenetics and functional classification of proteins. To this end, we analyzed 80 structures from the functionally diverse ferritin-like superfamily. Using evolutionary networks, we demonstrate that structural comparisons can delineate and discover groups of proteins beyond the “twilight zone” where sequence similarity does not allow evolutionary analysis, suggesting that considerable and useful evolutionary signal is preserved in three-dimensional structures.
Introduction
Ever since Carl Linnaeus humbly stated, Deus creavit, Linnaeus disposuit, biologists have sought to organize biological entities into natural groupings. Following Darwin's recognition that evolution occurs through modification with descent, the most robust classification schemes have been based on evolutionary principles (1), and a major pursuit in modern biology is the systematic classification of biological species using molecular phylogenies (2). With the emergence of high throughput methods of data collection in the molecular sciences, biology stands on the cusp of a flood of data that dwarfs previous attempts at cataloging the diversity of life. For instance, high throughput sequencing data generated by the Global Ocean Sampling program identified 61.2 million new proteins and 1700 new protein clusters with no sequence level similarities to known proteins (3). However, structural genomics efforts reveal that of the >40% of proteins with no known function in public databases, approximately two-thirds show structural similarity to known proteins, despite no significant sequence similarities (4). This suggests that, more than sequence, structure may provide the most productive way of categorizing the protein universe.
Several important databases exist that attempt to provide high level organization to the protein universe, including Pfam (5), which is sequence-based, and SCOP (6) and CATH (7), which both use protein structural information. These databases all use measures of either sequence or structural similarity as a means of organizing proteins into families or superfamilies. Although these databases are invaluable in charting broad structural relationships, in many cases they offer conflicting classifications, and known evolutionary relationships between individual superfamily constituents are not included. To gain a better appreciation of this issue, we examined the classification of ferritin-like proteins across these three databases and sought to establish whether the application of phylogenetic methods to protein structural data (8–10) can augment classification within a well sampled superfamily, rich in data on biological function of its members. We report that structural phylogenies of the ferritin-like superfamily recover informative relationships between superfamily members that reflect known evolutionary relationships and functional roles. We conclude that phylogenetic tools can provide an important complement to established structural classifications.
EXPERIMENTAL PROCEDURES
Structure Selection
Protein Data Bank codes for all solved structures in the ferritin-like superfamily in SCOP (6) release 1.75 (a.25.1) were downloaded, and one structure from each species in each protein group was selected. The half-ferritin SCOP family and the rubrerythrin-like protein from Sulfolobus tokodaii (Protein Data Bank code 1j30) were excluded because they form dimers where two helices from each monomer form the four-helix bundle and thus do not align with the other structures. In general, the highest resolution structure was chosen, unless it had mutations or other departures from the biologically active form, and a structure that was more similar to the biologically active form was present. In addition, we used the search capability of the SSM tool from EBI (11) to search for related proteins that were included in the analysis.
Three-dimensional Structural Alignment, Distance Measurement, and Construction of Phylogenetic Network
All of the structures were aligned against all other structures using the SSM tool from EBI (11). For each pair of sequences, two alignments were retrieved, using either of the two sequences as query and the other as target, and the alignment with the best Q-score was chosen. Q-scores, like most other quality indicators of structural alignments, are similarity scores (take values between 0 and 1), i.e., they increase with similarity, whereas distances used in phylogenetic methods need to decrease with similarity. We constructed distances by calculating 1 minus Q-scores. The NeighborNet network (12) was constructed using SplitsTree 4 (13).
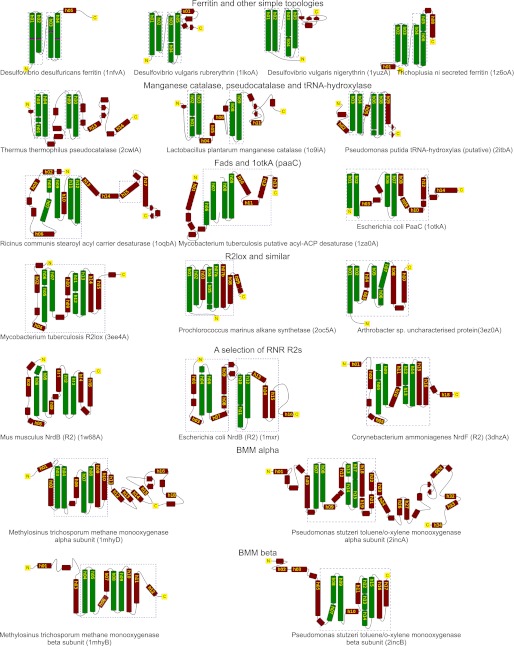
The topological diagrams were constructed manually using the pairwise alignments as models. Proteins were chosen to illustrate the majority of topologies. To illustrate that the evolutionarily conserved parts of the substrate-oxidizing ferritin-like proteins (ribonucleotide small subunit (RNR R2s),4 fatty acid desaturases (Fads), and bacterial multicomponent monooxygenases (BMMs, etc.)) extends beyond the four-helix bundle, we chose the mouse RNR R2 (1w68A) as a reference and identified with dashed blue boxes the parts of each structure that align well with 1w68A.
Our functional classification is based on the annotation of each protein in the Protein Data Bank entries of the individual proteins. In most cases the annotation in Protein Data Bank is clear, but especially in the Dps group, nomenclature is ambiguous with proteins known to be related to a Dps having another name. The classifications of Fads, RNR R2s, BMM α, and BMM β subunits are in general detailed in the Protein Data Bank. We did not assess their correctness independently.
Multiple Alignment and Phylogeny of RNR R2
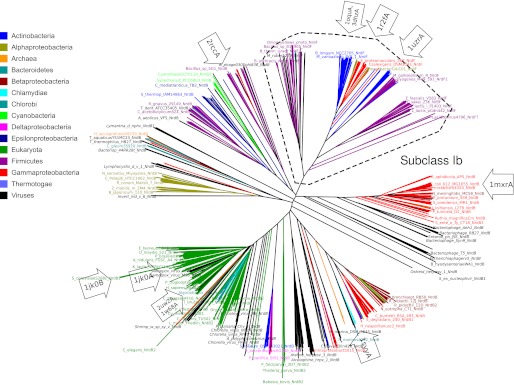
All unique NrdB and NrdF (subclass Ib) sequences were selected from the RNR database (14) and aligned using Muscle (15). A phylogeny was estimated from the full alignment using the BioNJ (16) algorithm as implemented in SplitsTree 4.6 (13). Fig. 5 was produced with the Dendroscope tree viewer (17).
FIGURE 5.
A BioNJ tree from a sequence alignment of RNR R2. The BioNJ tree was estimated from an alignment of all unique RNR R2 sequences in the RNRdb (17). The largest bacterial phyla, eukaryotes, and archaea are indicated by colored labels (see inset legend), viruses have labels in italics, and the subclass Ib is marked with a dashed box. Proteins with solved three-dimensional structures are indicated with labeled arrows. The figure was produced using Dendroscope (17).
Source File Deposition
Source files (Q-score matrix, SplitsTree network, pairwise three-dimensional alignment Jmol scripts, and Dendroscope RNR R2 tree) will be deposited in the data repository Dryad.
RESULTS
Current Classification of Ferritin-like Proteins across Major Databases
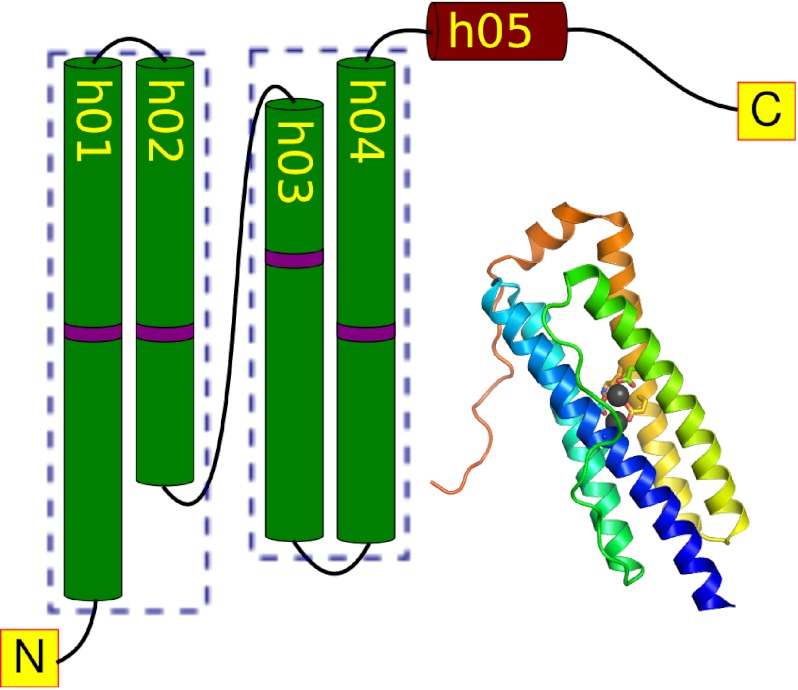
The Ferritin-like superfamily is characterized by the presence of a common four-helix bundle, characterized by a crossover section between helices 2 and 3, resulting in a distinctive up-down-down-up arrangement of helices (Fig. 1). Most ferritin-like proteins bind a pair of redox-active metal ions, at similar metal-binding sites coordinated by carboxylates and histidines in a shared fold (18). Characterized members of the ferritin-like protein superfamily are known to perform four broad roles, spanning iron storage, oxygen and radical detoxification, substrate oxidation, and radical generation.
FIGURE 1.
The ferritin fold. Proteins in the ferritin superfamily are characterized by a four-helix bundle coordinating a pair of metal ions. The helices are arranged in a characteristic up-down-down-up topology. The location of the metal coordinating residues is indicated in purple. The figure is based on the Desulfovibrio desulfuricans bacterioferritin structure (Protein Data Bank code 1nfv).
As summarized in Table 1, the functions associated with characterized members of this superfamily are diverse (see also Ref. 19 for a recent review of the functional spectrum of ferritin-like proteins). However, the full diversity of functions known among characterized members of this superfamily is not reflected in the broad scale classifications in the SCOP, CATH and Pfam databases. By contrast, the low sequence similarities across this superfamily make it practical to generate sequence-based phylogenies only across subsets of the superfamily. Consequently, superfamilies such as this are missed by efforts to marry structural information with sequence-based phylogenies for superfamilies in or beyond the twilight zone of sequence similarity (20–30% pairwise sequence identity) (20, 21). We summarize briefly how these databases and phylogenies inform current classification of the ferritin-like superfamily.
TABLE 1.
The largest and best characterized ferritin-like protein families
Ferritin-like proteins can be broadly characterized into four functional categories: iron storage, reactive oxygen species (ROS) protection (in which we include biotic oxidation of Fe(II)), substrate oxidation, and radical generation. Dashes are used where corresponding groups are not found in a classification scheme.
| Protein | Abbreviation | Functional category | Metals | References |
|---|---|---|---|---|
| Ferritins | Ferritins | Iron storage | Fe2 | Ref. 48 |
| Bacterioferritins | Bacterioferritins | Iron storage | Fe2 | Ref. 48 |
| Dps | ROS protection, iron storage | Fe2 | Ref. 49 | |
| Manganese catalases | Manganese catalases | ROS protection | Mn2 | |
| Fatty acid desaturases | Fads | Substrate oxidation (introducing a double bond) | Fe2 | Ref. 50 |
| Bacterial multicomponent monooxygenases | BMMs | Substrate oxidation (e.g., methane, phenol and other aromatic compounds) | Fe2 | Ref. 27 |
| Class I ribonucleotide reductase radical generating component | RNR R2s | Radical generation | Fe2, Mn2, or MnFe | Ref. 25 |
The SCOP database (6) describes structural similarities between proteins, classifying proteins hierarchically based on solved three-dimensional structures. SCOP superfamilies contain protein families that are assumed to be evolutionarily related based on sequence and structural similarity and functional commonalities. The “Ferritin-like proteins” superfamily in SCOP contains nine individual families (Table 2). The largest family, “ferritin,” contains ferritins, bacterioferritins, and Dps proteins, plus a number of less well characterized proteins. The next largest family, “ribonucleotide reductase-like,” contains the activating subunit of class I ribonucleotide reductases (RNR R2), BMMs, and Fads, plus a few poorly characterized proteins. The “manganese catalase family” is likewise a sizable family, whereas the other families are small and contain few solved structures, with few representatives of known function (Table 2).
TABLE 2.
A comparison between the SCOP structure database and the Pfam sequence-based protein family database
Families in the SCOP superfamily “ferritin-like proteins” are compared with families in the Pfam clan “Ferritin.” For Pfam, only families containing solved structures are included. Pfam (version 24.0) in general contains a more complete list of structures than SCOP (version 1.75).
| SCOP family | Pfam family | Structuresa | Description |
|---|---|---|---|
| Ferritin | Ferritin | 138 | Ferritin, bacterioferritin, and Dps |
| Ribonucleotide reductase-like | Ribonuc_red_sm | 48 | RNR R2 |
| Phenol_Hydrox | 39 | BMM | |
| FA_desaturase | 7 | Fad | |
| AMB4284-like and Ferritinsb | Rubrerythrin | 20 | Rubrerythrin |
| Manganese catalase (T-catalase) | Mn_catalase | 6 | Manganese catalase |
| YciF-like | DUF892 | 2 | Unknown function |
| MiaE-like | MiaE | 1 | tRNA- ([2])-hydroxylase |
| Half-ferritin | — | 1 | Unknown function |
| PMT1231-like | — | 1 | Unknown function |
| Rv2844-like | — | 1 | Unknown function |
| — | DUF2383 | 1 | Unknown function |
| — | Coat_F | No | Contribute to Bacillales spore coat |
| — | COQ7 | No | Ubiquinone biosynthesis |
| — | DUF2202 | No | Unknown function |
a Number of solved structures referred to in Pfam, except in the SCOP families half-ferritin and PMT1231-like, which have no corresponding Pfam families.
b Two proteins in the Pfam rubrerythrin family are classified in the SCOP Ferritins family, whereas the other two make up the SCOP AMB4284-like family.
The CATH database (7) also aims to classify proteins based on their three-dimensional structures. In CATH, ferritin-like proteins are classified into two distinct “topologies”: “ferritin” and “ribonucleotide reductase, subunit A” (the CATH choice of “subunit A” as part of the name is unfortunate, because the letter A is more commonly used for the larger subunit of RNRs), each containing a single “homologous superfamily.” CATH thus yields less structured information about relationships between individual proteins than SCOP.
Pfam is a protein domain classification database built on sequence similarity. There are 12 Pfam families containing ferritin-like proteins, with a number of sequences in each family. The families are collected in the “ferritin” clan, where a clan incorporates two or more families that appear homologous (5). By and large, the classification in Pfam corresponds well with the SCOP classification (Table 1). The most notable difference is that Pfam carries separate RNR R2 (“Ribonuc_red_sm”), BMM (“Phenol_Hydrox”), and Fad (“FA_desaturase_2”) families, providing a somewhat clearer picture of functional divisions, at least of the substrate oxidizing group of ferritin-like proteins (Table 2).
At the highest level (superfamily inclusion), the classification of the ferritin-like superfamily appears broadly consistent across these databases but does differ in the amount of information provided regarding the relationships and functions of superfamily constituents (Table 2). Significantly, relationships within the superfamily do not capture either expected relationships based on function (Table 1) or on sequence-based phylogenetic trees (22–24). For instance, none identify whether ferritins, bacterioferritins, and Dps proteins represent evolutionarily distinct proteins, and although Pfam separates the ribonucleotide reductase R2 family from BMMs (albeit with the somewhat misleading name “Phenol_Hydrox”), the division of BMM components into the paralogous α and β subunits (22) is not captured in Pfam.
Although the classification in all three databases is hierarchical, and broadly accurate, they do not encompass all levels of functional and evolutionary information, so in this respect, they provide only partial classifications. This extends to protein structure also; BMMs and RNR R2s are structurally more complex proteins than ferritins, but only the CATH classification recognizes these different topologies (SCOP places them in the same family), and although Pfam places them in different families, the Pfam classification does not provide any indication as to whether BMMs and RNR R2s are more closely related to one another than to other ferritin-like proteins.
Indeed, a potential drawback with existing database classification schemes is that all make use of predefined ranks (superfamilies and families in SCOP, clans and families in PFAM, and topology and homologous superfamily in CATH). In that biological entities do not evolve following a predefined scheme, biological relationships within protein superfamilies may be better described using a hierarchy without predefined ranks similar to what have been proposed for organismal taxonomy and nomenclature (see, for example, the PhyloCode website). To examine whether evolutionary relationships between proteins can be accurately described using structural data, we generated phylogenies using three-dimensional structure alignments to examine whether both broad superfamily relationships and more fine scale functional groupings could be recovered for a protein superfamily with good protein structural coverage.
Structural Phylogenetic Network of Ferritin-like Proteins
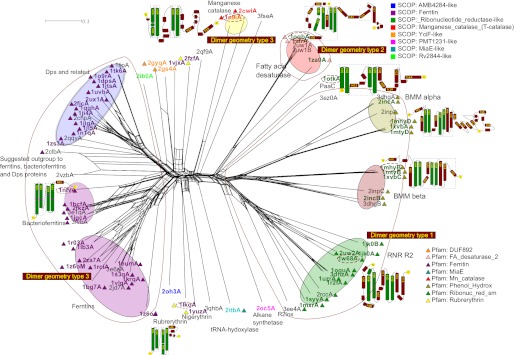
Sequence-based phylogeny has been central in establishing evolutionary relationships within and between protein families. However, in the case of the ferritin-like superfamily, the low degree of sequence similarity precludes the use of sequence-based analyses. We therefore sought to examine the phylogeny of the ferritin-like superfamily using a combination of structure-based alignment methods and traditional, sequence alignment-based phylogeny. Our data set consisted of 62 structures representing all structurally solved ferritin-like proteins in SCOP (version 1.75) and Pfam (version 24.0). We also added 18 ferritin-like proteins identified by the search function of SSM (11). Each pair of three-dimensional structures was aligned using SSM. We chose Q-scores as distances between structure-aligned pairs, because this takes both alignment length and alignment quality (root mean square deviation) into account (11). From the Q-score-based pairwise distances, we constructed a phylogenetic network using the distance-based network method NeighborNet (12) (Fig. 2) instead of a standard distance-based tree building method, such as neighbor joining. If there is conflict in the distance matrix regarding which structures are most similar, some of that conflict will be visible in a network, but not in a tree. A network thus provides a more informative view of possible relationships.
FIGURE 2.
Structure alignment based phylogenetic network of the ferritin-like superfamily. Structures were pairwise aligned using the SSM tool from EBI (14), and quality scores (Q-scores) were transformed to distance measurements by subtraction of the Q-score from one to form the basis for this NeighborNet network (17). SCOP families are indicated with colored labels (see inset legend), Pfam families are indicated with colored triangles near labels (see inset legend), related proteins are identified via the SSM tool from EBI (11) with black labels, and our functional classification are indicated with colored ellipses. Dimerization geometries for the three major groups are demarcated; proteins outside these groups either do not form dimers or form dimers with singleton geometries. Topology cartoons for a selection of important folds have been inset with the corresponding labels marked by dashed ellipses. The dashed boxes in the topologies indicate how much of the protein aligns well with mouse RNR R2 structure (1w68A). See Fig. 3 for a description of dimerization types.
It is immediately clear from the network in Fig. 2 that the broad outline of classification in SCOP, CATH, and Pfam is recovered through our structure-based phylogeny. The two large SCOP families, ferritins (Fig. 2, violet labels) and ribonucleotide reductase-like (Fig. 2, green labels), form two separate groups in the network with smaller groups scattered mostly in the ferritin part of the network.
The Pfam family Ferritins (Fig. 2, violet triangles) is slightly better defined than the SCOP family (Fig. 2, violet labels), which also includes four structures in the Pfam Rubrerythrin family (Fig. 2, yellow triangles: 1lkoA, 1yuzA, 2fzfA and 1vjxA). None of the existing classifications subdivide ferritins into the three functional classes ferritins, bacterioferritins, and Dps (Fig. 2, pink and violet ellipses). In contrast, our structure-based network recovers them as likely clans (a possible monophyletic group in an unrooted phylogeny (26)).
The SCOP family Ribonucleotide reductase-like (Fig. 2, green labels) is very broad, and our network suggests clear subdivisions that correspond well with established functional groups such as Fads, RNR R2s, BMM α, and BMM β (marked by separate ellipses in Fig. 2). In general, these functional groups are better classified in Pfam, where they form three families: FA_desaturases_2 (pink triangles), Ribonuc_red_sm (green triangles) and Phenol_Hydrox (brown triangles), than in SCOP, except that the Pfam family Phenol_Hydrox contains both the catalytic α subunits, as well as the non-metal-binding β subunits of BMMs.
Topology of Proteins Supports Network
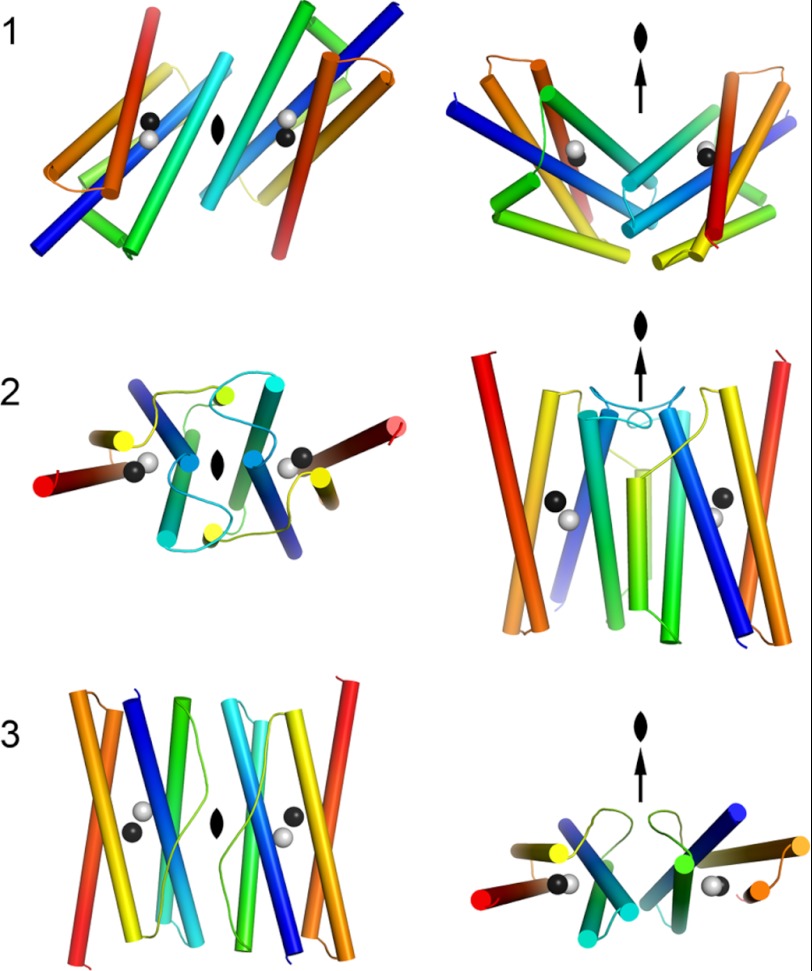
To test the robustness of our phylogenetic network, we surveyed the dimerization geometries and analyzed the general topology of the proteins. Most ferritin-like proteins form dimers, although sometimes as parts of larger quaternary structures such as the 12-meric Dps proteins and 24-meric ferritins and bacterioferritins. Despite the almost infinite possible number of dimer interaction geometries, we find that the majority of dimeric ferritin-like proteins fall into three large groups (Fig. 3 shows an explanation of the three geometries). Dimerization geometry is remarkably conserved and confirms some of the major clans in our network, dimer geometry type 1 for RNR R2s and BMMs and type 2 for Fads (Fig. 2). Only dimer geometry type 3 is paraphyletic, being a feature of two functional categories: ferritins, bacterioferritins, and Dps proteins on the one hand and manganese catalases on the other (Fig. 2).
FIGURE 3.
Schematic drawings of the dimerization architectures of ferritin-like proteins. The metal-binding four-helix bundle and crossover connection is displayed and colored from the N terminus (blue) to the C terminus (red) to illustrate the three different dimer architectures indicated in Fig. 2. The left column is viewed along the 2-fold axis, and the right column is viewed perpendicular to the 2-fold axis.
We next inspected all pairwise three-dimensional alignments manually to establish whether any regions outside the four-helix bundle show similarities to structures from distant parts of the network. Seven topological classes are shown in Fig. 2. To qualitatively visualize the spatial extent of structural similarity in the more complex proteins, the RNR R2 from mouse (1w68A) was used as a reference structure (see “Experimental Procedures”) and the portions that align well structurally to this are marked with dashed blue boxes (a larger selection of topological diagrams can be found in Fig. 4). The qualitative assessment lends support to the subdivision of the SCOP ribonucleotide reductase-like family into separate Fad, RNR R2, BMM α, and BMM β clans in our network (Fig. 2). The differences between three-dimensional alignments of the smaller ferritin proteins (Ferritins, Bacterioferritins, and Dps) are too subtle to convey with topological diagrams.
FIGURE 4.
A selection of representative topologies. Manually created topological diagrams with an emphasis on the region around the metal coordinating four long helices. These four helices align well between all pairs in our data set. To qualitatively visualize the spatial extent of structural similarity in the more complex proteins, a reference structure was chosen (mouse RNR R2 (1w68A)), and the region that aligned well to this structure is marked with dashed boxes.
There is a clear similarity between the tertiary structures of all subgroups of the SCOP family ribonucleotide reductase-like. All but three proteins (1otkA, 2oc5A, and 3ez0A) have N-terminal secondary structure elements that align at least partially to the mouse RNR R2 (1w68A), and parts of the structure directly following the four-helix bundle also align well (Figs. 2 and 4). In contrast, the comparably topologically complex manganese catalases (1o9iA and 2cwlA) do not align with mouse RNR R2 (our reference structure; see “Experimental Procedures”) outside the four-helix bundle. We can thus conclude that there is an evolutionarily conserved core in the SCOP ribonucleotide reductase-like family that is larger than the four-helix bundle, implying that the substrate oxidizing proteins have evolved from a more recent common ancestor than the common ancestor of all ferritin-like proteins.
Known Functional Groups Are Recovered in Network and Can Be Further Subdivided
The functional groups we have marked with ellipses (Fig. 2) are all possible monophyletic groups. Using sequence-based phylogenies (22, 27), BMMs have been suggested to have evolved by duplication and divergence, leading to distinct di-iron binding catalytic (α) and non-metal binding (β) subunits. Although substrate specificity is generally considered low in BMMs (27), we can identify one distinct subgroup both in the α and β clans containing only proteins annotated as soluble methane monooxygenases (1mhyD B, 1mtyD B, and 1xvbA C for α and β components, respectively), and another subgroup consisting of toluene monooxygenases (2incA B and 3dhgA B). This indicates that our structural analysis is able to recover relatively recent evolutionary relationships in addition to the more distant examples described above. Indeed, within both clans, we see that the phenol hydroxylase subunits (2inpC A) also appear distinct within this part of the network.
In the Fad group, there is a distinct cluster of plant Fads (2uw1, 1afrA, and 1oqbA), whereas the Mycobacterium tuberculosis protein (1za0A) appears more distantly related. Unfortunately, this is the only solved structure of a bacterial Fad. It is also one of a paralogous pair and not the one considered functional. The structure of the functional Fad has not yet been possible to solve (28). With such a skewed data set, it is difficult to judge how well our structure-based network identifies evolutionary relationships within the Fad group.
In the better surveyed ribonucleotide reductases, we find good agreement between structure- and sequence-based analyses. Several members of the subclass Ib of RNR R2s have recently been shown to contain a di-manganese center that requires a flavodoxin-like protein (NrdI) to be oxidized (29). In support of this subclass, solved structures from subclass Ib (3dhzA, 1r2fA, 1uzrA, and 1oquA) form a distinct clan in our network (Fig. 2), and we independently recover sequence-based phylogenetic support for the existence of a subclass Ib cluster (Fig. 5), consistent with monophyly despite including sequences from diverse bacterial phyla.
Interestingly, Pfam classifies the R2lox (R2-like ligand-binding oxidase) from M. tuberculosis (3ee4A) as a Ribonuc_red_sm, possibly because of its sequence similarity to RNR R2 proteins (30). In our network this structure clearly occupies an outgroup position relative to the RNR R2 structures. This is functionally consistent with its ligand-binding pocket, which indicates that it is a substrate oxidizing enzyme, and its lack of competence as an RNR R2 (30, 31). Alignments of R2 and R2lox proteins carry too few informative sites for reliable phylogeny. We have therefore not been able to independently verify whether R2lox has evolved from within the R2 clade (which might be expected given the much broader evolutionary distribution of R2 sequences; aerobic RNR sequences are widespread in bacteria and eukaryotes and have been transferred into Archaea (23)) or whether it shares a common ancestor with R2, as suggested by our structure-based analyses (Fig. 2). This highlights a potentially important caveat in going from structural phylogeny to evolutionary interpretation; structural divergence could in some cases result in an artifact analogous to the well documented long branch attraction artifact observed in sequence-based phylogenetics (32), so the same caution should be exercised in interpreting phylogeny based on structure.
DISCUSSION
We show that structure-based phylogenetic analysis can be a robust method to delineate and even discover functionally important protein groups. We recover well known groups of ferritin-like proteins such as RNR R2s, BMMs, Fads, and the three subtypes of ferritins. The exploration of such methods as a complement to high level classification (5–7) is necessitated by the exponential growth of available structural information with a large fraction of structures of unknown function (33) and deserves now to be applied more systematically to larger structural genomics data sets.
Using a phylogenetic network (Fig. 2) enables us to assess the degree to which the signal in protein structure fits an evolutionary tree. Whereas there is some noise (this is essentially true of all phylogenies based on biological data), the data are clearly tree-like. Furthermore, no evolutionary processes—such as protein fusion events—in this data suggest reticulate evolution (hybridization or other non-tree-like evolution). Consequently, we can represent the main features of the network as a bifurcating tree (Fig. 6) to aid interpretation.
FIGURE 6.
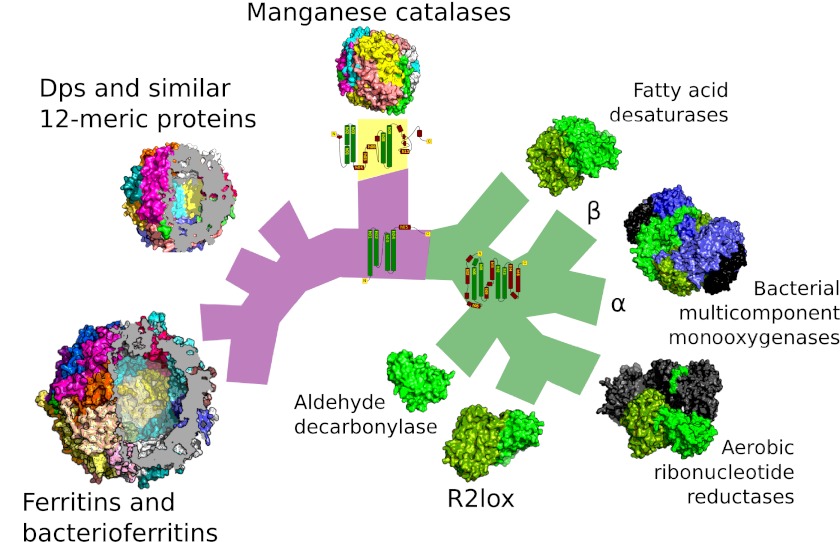
Schematic view of the relationships between the major ferritin-like protein families. A schematic tree based on our network analysis of ferritin-like proteins. Common branch color indicates shared monomer topology, illustrated with an example topological diagram per group. Violet, the simplest topology consisting only of the four-helix bundle; yellow, the manganese catalase topology; green, the core of the topology shared by substrate oxidizing enzymes and ribonucleotide reductase radical generating subunits. Tree tips are populated with quaternary structures of representatives of each family, approximately to scale. Proteins that are not part of the ferritin-like superfamily but that form part of a holoenzyme are colored dark gray. Proteins with similar quaternary structures are represented by a single example (ferritins and bacterioferritins, and Dps and similar 12-meric proteins). The iron-storing proteins have been sectioned to underscore the fact they are hollow. Bacterial multicomponent monooxygenases contain two paralogous ferritin-like subunits: α and β (blue and green, respectively, in the diagram). A diverse group of topologically simple proteins such as rubrerythrins are not shown so as to aid readability (see Fig. 2 for the full set of relationships).
A tree naturally inspires an evolutionary interpretation, but to what extent can a three-dimensional alignment distance tree be interpreted as an evolutionary tree? A matrix of pairwise distances will yield a tree or network using distance-based phylogenetic methods, but this does not imply that there is an evolutionary signal in all distance matrices. Much, however, points to this being the case. Trees have been constructed from structural similarity scores previously (8–10), and structure is generally better conserved than sequence (34). In addition, given that our network identifies relationships equivalent to those based on functionally delineated groups and sequence-based phylogenies, we conclude that it does likely carry significant evolutionary signal. However, it is important to recognize that this type of analysis may be as prone to artifact as any other analysis used to infer evolutionary history; in light of the inescapable specter of phylogenetic error (35), what follows below is a cautious evolutionary interpretation of our results.
Ferritins, Bacterioferritins, and Dps Likely Evolved from Common Ancestor
Our results (Figs. 2 and 6) strongly support a common ancestry of the 24-meric ferritins and bacterioferritins and the 12-meric Dps proteins. Although these are small and topologically similar, there is no evidence of misclassification in any of these subgroups (indicated with ellipses in Fig. 2); all three subgroups are recovered, and other ferritin-like proteins of similarly simple architecture (e.g., the Pfam Rubrerythrin and DUF892 families) are distant from these groups in our network. This indicates that our approach is robust even for small, topologically similar proteins.
Consistent with a common origin for ferritins, bacterioferritins, and Dps proteins, dimerization geometry is common across this group. Although dimer geometry type 3, which all exhibit (Fig. 2), does not define a clan in the network, function is broadly consistent with common ancestry for this group. Although the primary role of Dps proteins in many organisms seems to be DNA protection, like ferritins and bacterioferritins they are also involved in iron storage (36), suggesting this as a possible ancestral function for the three protein groups.
Bacterioferritins and Dps proteins each have a defining feature that strengthens the differentiation within the network. Bacterioferritins differ from standard ferritins in that the former bind heme groups in the interface between subunits (37). The function of the heme groups is not entirely understood but seems connected to the reduction of the ferric precipitate and release of ferrous iron (38). We therefore suggest that the bacterioferritins diverged from the ferritins following acquisition of heme group binding.
Dps proteins do not have a di-iron-binding site in the standard position inside the four-helix bundle but instead bind metals in the dimer interface. The intersubunit di-iron site appears to be particularly well suited to use hydrogen peroxide as the metal oxidant and confers peroxide tolerance to the organism (39, 40). Protection against reactive oxygen species is a recurrent functional theme among ferritin-like proteins and a likely ancestral function to the superfamily.
Several structures provide possible clues as to the deeper evolutionary history of the ferritin-bacterioferritin-Dps group. Of particular interest, 2clbA from Sulfolobus solfataricus and 2vzbA from Bacteroides fragilis are possible outgroups of Dps (and possibly of the entire group). Both possess the same type 3 dimerization geometry found across the group, and similar to Dps, both are annotated as 12-mers. Their metal-binding sites are housed within the four-helix bundle, in contrast to Dps proteins (41, 42). It has been suggested that the S. solfataricus protein (2clbA) may resemble the common ancestor of the ferritin, bacterioferritin, Dps group (24), something that our network is broadly consistent with in that 2clbA is distinct from all three clans and falls plausibly near the base of this wider clan. There is considerable conflict in this part of the tree, so although an outgroup position is plausible, it is by no means certain. Similarity to an hypothesized ancestral state is appealing. It is noteworthy that this protein is present in both bacteria and archaea; there is of course no way to directly relate extant structures to a now extinct ancestor on available data.
Substrate Oxidation Evolved by Evolutionary Capture of Reactive Di-iron-oxygen Species
The substrate oxidizing proteins (e.g., Fad, RNR R2, and BMM) form a large clan in our network (Figs. 2 and 6). Individual members align well with each other both on the N-terminal and the C-terminal side of the four-helix bundle, defining a larger common structural core in the substrate oxidizing proteins (Fig. 6). Notably, outside the four-helix bundle, none of these protein groups align with the manganese catalases, which are the only proteins outside the substrate-oxidizing group with comparable topological complexity (Fig. 6).
The existence of a clan supports evolution from a common ancestor. Dimer geometry within the group has diverged however: RNR R2s and BMM α and β components share a common geometry (type 1), but the Fad group is distinct (type 2). Despite this difference, the overall similarities in topology and function across the clan led us to conclude that Fads share a common ancestor with RNR R2s and BMMs. We suggest that this group likely evolved from non-substrate-oxidizing ferritin-like proteins by evolutionary capture of a reactive di-iron-oxygen species. Mutations allowing entry of a substrate near the di-metal site, and ensuing selection for the catalyzed reaction provides a simple and chemically plausible evolutionary model for the emergence of this enzymatic capacity.
Evolution of RNR R2 Proteins and R2lox
The well studied RNR R2 proteins in our network are cofactors required for generation in the large component of RNRs of a transient thiyl radical necessary for reduction of ribonucleotides to deoxyribonucleotides. RNR R2s are structurally closely related and share a common function, indicating evolution from a common ancestor (Figs. 2 and 6). Subclass Ia is a di-iron/tyrosyl cofactor, whereas subclass Ib is a dimanganese/tyrosyl cofactor. Subclass Ib forms a clan in the network (3dhzA, 1r2fA, 1uzrA, and 1oquA; Fig. 2), a result independently obtained in our sequence-based phylogenetic analyses (Fig. 5). The agreement between structure- and sequence-based trees is also consistent with the functional division (43, 44), which is not recognized in existing protein family classifications. Together, these provide strong independent lines of evidence for the presence of fine scale evolutionary signal in protein structure and the distinct evolutionary histories of subclasses Ia and Ib (23, 43).
Again, deeper evolutionary relationships are identifiable from our analyses, as exemplified by the relationship between RNR R2 proteins, R2lox (3ee4A) and alkane synthetase (2oc5A). We clearly identify a relationship between R2lox and R2 in our network (Fig. 2), consistent with previous observations of significant sequence similarity between them (45). The R2lox structure differs from R2s in that a fatty acid is bound near the di-iron center (30). The 2oc5 protein was recently characterized as an aldehyde decarbonylase (46), and the reaction mechanism was suggested to be an ω-carbon oxidation of a fatty acid aldehyde bound to coenzyme A (47). Although sequence similarity between 2oc5 and R2lox/RNR R2 is low (best blast alignment between R2lox and 2oc5 spans only 34 amino acids, 26.5% identity; no significant alignment is detected between mouse RNR R2 (1w68) and 2oc5), our phylogenetic network indicates an evolutionary relationship that provides context for the functional bridge spanning the aldehyde decarbonylase 2oc5, R2lox, and the RNR R2 cofactors.
Structural Evidence That BMM Components Evolved by Gene Duplication
Sequence-based phylogenetic analysis (22) has previously shown that the α and β components of BMM are related via gene duplication. Our network is clearly consistent with this scenario (Fig. 2). Moreover, there is a short, but distinct, common stem that divides BMMs from the RNR R2 branch, suggesting the BMM components duplicated following divergence from their common ancestor with RNR R2s. We likewise recover signal consistent with the existence of functionally meaningful subclasses within each BMM component clade, suggesting parallel functional diversification following the initial duplication and divergence event generating the α and β subunits. Specifically, the α (1xvbA, 1mhyD, and 1mtyD) and β (1xvbC, 1mhyB, and 1mtyB) subunits of the soluble methane monooxygenases are clearly distinct from the other members of each BMM subunit clade, as are the phenol hydroxylase subunits from Pseudomonas stutzeri (2inpA C). We also observe the same pattern for toluene monooxygenases (2incA B and 3dhgA B from P. stutzeri and P. mendocina). Our results clearly demonstrate that the soluble methane monooxygenases, phenol hydroxylases, and toluene monooxygenases evolved first by duplication, generating the α and β subunits, then diversification, generating the observed modern enzymatic diversity based on this heterodimeric architecture.
Conclusions
We have explored the potential utility of structure-based phylogeny to bridge the not insignificant gap between sequence-based phylogenies and high level structural classification databases, using the ferritin-like superfamily as a test case. Our results show the existence of two main groups within the ferritin-like protein superfamily, and we find good overall agreement between our broad structural survey, known protein functions and relationships revealed using sequence-based phylogenies. This approach provides sufficient resolution that we can both resolve distant relationships between structurally related proteins, yet still identify more recent evolutionary events identifiable using sequence data. We believe that this approach, based on phylogenetic analysis of three-dimensional structural alignments, promises to be useful both for functional and evolutionary categorization beyond the twilight zone of sequence similarity. An achievable goal is the up-scaling of this approach to the much larger data sets being generated through genome sequencing and structural genomics, which may help assign uncharacterized sequences and structures into specific families and generate functional leads for downstream experimental analysis. Three-dimensional alignment of a newly solved structure from an uncharacterized protein against all members of the best fitting superfamily followed by phylogenetic analysis would supply a hypothetical family membership, provided a sufficient number of diverse structures from the superfamily were previously solved. The method would also suggest new families or pinpoint areas of potential structural interest when no near neighbors to the new structure are found.
Acknowledgment
We thank D. Liberles for helpful comments on the manuscript.
This work was supported by grants from the Swedish Research Council (to A. M. P. and B. M. S.), and grants from the Swedish Research Council and the Swedish Foundation for Strategic Research (to M. H.).
- RNR R2
- ribonucleotide reductase small subunit
- Fad
- fatty acid desaturase
- BMM
- bacterial multicomponent monooxygenase
- Dps
- DNA-binding proteins from starved cells.
REFERENCES
- 1. Ridley M. (1986) Evolution and Classification: The Reformation of Cladism, Longman, London [Google Scholar]
- 2. Hillis D. M. (2007) Constraints in naming parts of the Tree of Life. Mol. Phylogenet. Evol. 42, 331–338 [DOI] [PubMed] [Google Scholar]
- 3. Rusch D. B., Halpern A. L., Sutton G., Heidelberg K. B., Williamson S., Yooseph S., Wu D., Eisen J. A., Hoffman J. M., Remington K., Beeson K., Tran B., Smith H., Baden-Tillson H., Stewart C., Thorpe J., Freeman J., Andrews-Pfannkoch C., Venter J. E., Li K., Kravitz S., Heidelberg J. F., Utterback T., Rogers Y. H., Falcón L. I., Souza V., Bonilla-Rosso G., Eguiarte L. E., Karl D. M., Sathyendranath S., Platt T., Bermingham E., Gallardo V., Tamayo-Castillo G., Ferrari M. R., Strausberg R. L., Nealson K., Friedman R., Frazier M., Venter J. C. (2007) The Sorcerer II Global Ocean Sampling expedition. Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jaroszewski L., Li Z., Krishna S. S., Bakolitsa C., Wooley J., Deacon A. M., Wilson I. A., Godzik A. (2009) Exploration of uncharted regions of the protein universe. PLoS Biol. 7, e1000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murzin A. G., Brenner S. E., Hubbard T., Chothia C. (1995) SCOP. A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540 [DOI] [PubMed] [Google Scholar]
- 7. Greene L. H., Lewis T. E., Addou S., Cuff A., Dallman T., Dibley M., Redfern O., Pearl F., Nambudiry R., Reid A., Sillitoe I., Yeats C., Thornton J. M., Orengo C. A. (2007) The CATH domain structure database. New protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 35, D291–D297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bujnicki J. M. (2000) Phylogeny of the restriction endonuclease-like superfamily inferred from comparison of protein structures. J. Mol. Evol. 50, 39–44 [DOI] [PubMed] [Google Scholar]
- 9. Breitling R., Laubner D., Adamski J. (2001) Structure-based phylogenetic analysis of short-chain alcohol dehydrogenases and reclassification of the 17β-hydroxysteroid dehydrogenase family. Mol. Biol. Evol. 18, 2154–2161 [DOI] [PubMed] [Google Scholar]
- 10. Garau G., Di Guilmi A. M., Hall B. G. (2005) Structure-based phylogeny of the metallo-β-lactamases. Antimicrob. Agents Chemother. 49, 2778–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 12. Bryant D., Moulton V. (2004) Neighbor-net. An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21, 255–265 [DOI] [PubMed] [Google Scholar]
- 13. Huson D. H., Bryant D. (2006) Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 [DOI] [PubMed] [Google Scholar]
- 14. Lundin D., Torrents E., Poole A. M., Sjöberg B. M. (2009) RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics 10, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Edgar R. C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gascuel O. (1997) BIONJ. An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14, 685–695 [DOI] [PubMed] [Google Scholar]
- 17. Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. (2007) Dendroscope. An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nordlund P., Eklund H. (1995) Di-iron-carboxylate proteins. Curr. Opin. Struct. Biol. 5, 758–766 [DOI] [PubMed] [Google Scholar]
- 19. Krebs C., Bollinger J. M., Jr., Booker S. J. (2011) Cyanobacterial alkane biosynthesis further expands the catalytic repertoire of the ferritin-like “di-iron-carboxylate” proteins. Curr. Opin. Chem. Biol. 15, 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rost B. (1999) Twilight zone of protein sequence alignments. Protein Eng. 12, 85–94 [DOI] [PubMed] [Google Scholar]
- 21. Furnham N., Sillitoe I., Holliday G. L., Cuff A. L., Laskowski R. A., Orengo C. A., Thornton J. M. (2012) Exploring the evolution of novel enzyme functions within structurally defined protein superfamilies. PLoS Comput. Biol. 8, e1002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leahy J. G., Batchelor P. J., Morcomb S. M. (2003) Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 27, 449–479 [DOI] [PubMed] [Google Scholar]
- 23. Lundin D., Gribaldo S., Torrents E., Sjöberg B. M., Poole A. M. (2010) Ribonucleotide reduction. Horizontal transfer of a required function spans all three domains. BMC Evol. Biol. 10, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andrews S. C. (2010) The ferritin-like superfamily. Evolution of the biological iron storeman from a rubrerythrin-like ancestor. Biochim. Biophys. Acta 1800, 691–705 [DOI] [PubMed] [Google Scholar]
- 25. Nordlund P., Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 [DOI] [PubMed] [Google Scholar]
- 26. Wilkinson M., McInerney J. O., Hirt R. P., Foster P. G., Embley T. M. (2007) Of clades and clans. Terms for phylogenetic relationships in unrooted trees. Trends Ecol. Evol. (Amst.) 22,114–115 [DOI] [PubMed] [Google Scholar]
- 27. Merkx M., Kopp D. A., Sazinsky M. H., Blazyk J. L., Müller J., Lippard S. J. (2001) Dioxygen activation and methane hydroxylation by soluble methane monooxygenase. A tale of two irons and three proteins. Angew. Chem. Int. Ed. Engl. 40, 2782–2807 [DOI] [PubMed] [Google Scholar]
- 28. Dyer D. H., Lyle K. S., Rayment I., Fox B. G. (2005) X-ray structure of putative acyl-ACP desaturase DesA2 from Mycobacterium tuberculosis H37Rv. Protein Sci. 14, 1508–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cotruvo J. A., Jr., Stubbe J. (2010) An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry 49, 1297–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andersson C. S., Högbom M. (2009) A Mycobacterium tuberculosis ligand-binding Mn/Fe protein reveals a new cofactor in a remodeled R2-protein scaffold. Proc. Natl. Acad. Sci. 106, 5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Högbom M. (2011) Metal use in ribonucleotide reductase R2, di-iron, di-manganese and heterodinuclear-an intricate bioinorganic workaround to use different metals for the same reaction. Metallomics 3, 110–120 [DOI] [PubMed] [Google Scholar]
- 32. Bergsten J. (2005) A review of long-branch attraction. Cladistics 21, 163–193 [DOI] [PubMed] [Google Scholar]
- 33. Terwilliger T. C., Stuart D., Yokoyama S. (2009) Lessons from structural genomics. Annu. Rev. Biophys. 38, 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Illergård K., Ardell D. H., Elofsson A. (2009) Structure is three to ten times more conserved than sequence. A study of structural response in protein cores. Proteins 77, 499–508 [DOI] [PubMed] [Google Scholar]
- 35. Penny D., McComish B. J., Charleston M. A., Hendy M. D. (2001) Mathematical elegance with biochemical realism. The covarion model of molecular evolution. J. Mol. Evol. 53, 711–723 [DOI] [PubMed] [Google Scholar]
- 36. Bozzi M., Mignogna G., Stefanini S., Barra D., Longhi C., Valenti P., Chiancone E. (1997) A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J. Biol. Chem. 272, 3259–3265 [DOI] [PubMed] [Google Scholar]
- 37. Frolow F., Kalb A. J., Yariv J. (1993) Location of haem in bacterioferritin of E. coli. Acta Crystallogr. D Biol. Crystallogr. 49, 597–600 [DOI] [PubMed] [Google Scholar]
- 38. Weeratunga S. K., Gee C. E., Lovell S., Zeng Y., Woodin C. L., Rivera M. (2009) Binding of Pseudomonas aeruginosa apobacterioferritin-associated ferredoxin to bacterioferritin B promotes heme mediation of electron delivery and mobilization of core mineral iron. Biochemistry 48, 7420–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao G., Ceci P., Ilari A., Giangiacomo L., Laue T. M., Chiancone E., Chasteen N. D. (2002) Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277, 27689–27696 [DOI] [PubMed] [Google Scholar]
- 40. Ilari A., Latella M. C., Ceci P., Ribacchi F., Su M., Giangiacomo L., Stefanini S., Chasteen N. D., Chiancone E. (2005) The unusual intersubunit ferroxidase center of Listeria innocua Dps is required for hydrogen peroxide detoxification but not for iron uptake. A study with site-specific mutants. Biochemistry 44, 5579–5587 [DOI] [PubMed] [Google Scholar]
- 41. Gauss G. H., Benas P., Wiedenheft B., Young M., Douglas T., Lawrence C. M. (2006) Structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like dimetal binding site within a DPS-like dodecameric assembly. Biochemistry 45, 10815–10827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gauss G. H., Reott M. A., Rocha E. R., Young M. J., Douglas T., Smith C. J., Lawrence C. M. (2012) Characterization of the Bacteroides fragilis bfr gene product identifies a bacterial DPS-like protein and suggests evolutionary links in the ferritin superfamily. J. Bacteriol. 194, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sjöberg B. M. (2010) A never-ending story. Science 329, 1475–1476 [DOI] [PubMed] [Google Scholar]
- 44. Boal A. K., Cotruvo J. A., Jr., Stubbe J., Rosenzweig A. C. (2010) Structural basis for activation of class Ib ribonucleotide reductase. Science 329, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Högbom M. (2010) The manganese/iron-carboxylate proteins. What is what, where are they, and what can the sequences tell us? J. Biol. Inorg. Chem. 15, 339–349 [DOI] [PubMed] [Google Scholar]
- 46. Schirmer A., Rude M. A., Li X., Popova E., del Cardayre S. B. (2010) Microbial biosynthesis of alkanes. Science 329, 559–562 [DOI] [PubMed] [Google Scholar]
- 47. Warui D. M., Li N., Nørgaard H., Krebs C., Bollinger J. M., Jr., Booker S. J. (2011) Detection of formate, rather than carbon monoxide, as the stoichiometric coproduct in conversion of Fatty aldehydes to alkanes by a cyanobacterial aldehyde decarbonylase. J. Am. Chem. Soc. 133, 3316–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andrews S. C. (1998) Iron storage in bacteria. Adv. Microb. Physiol. 40, 281–351 [DOI] [PubMed] [Google Scholar]
- 49. Chiancone E., Ceci P. (2010) The multifaceted capacity of Dps proteins to combat bacterial stress conditions. Detoxification of iron and hydrogen peroxide and DNA binding. Biochim. Biophys. Acta 1800, 798–805 [DOI] [PubMed] [Google Scholar]
- 50. Shanklin J., Guy J. E., Mishra G., Lindqvist Y. (2009) Desaturases. Emerging models for understanding functional diversification of diiron-containing enzymes. J. Biol. Chem. 284, 18559–18563 [DOI] [PMC free article] [PubMed] [Google Scholar]