Background: Many bacterial pathogens reorganize the host actin cytoskeleton during infection.
Results: We show here that an enterohemorrhagic Escherichia coli (EHEC) protein, EspFU, targets the mammalian host protein TOCA1 to help drive actin “pedestal” assembly.
Conclusion: TOCA1 augments actin polymerization in these cell surface protrusions.
Significance: Our results suggest that EspFU acts as a scaffold to recruit multiple actin assembly factors to promote colonization.
Keywords: Actin, Bacterial Adhesion, Bacterial Pathogenesis, Cell Motility, Cell Signaling, Cytoskeleton, Membrane
Abstract
Many bacterial pathogens reorganize the host actin cytoskeleton during the course of infection, including enterohemorrhagic Escherichia coli (EHEC), which utilizes the effector protein EspFU to assemble actin filaments within plasma membrane protrusions called pedestals. EspFU activates N-WASP, a host actin nucleation-promoting factor that is normally auto-inhibited and found in a complex with the actin-binding protein WIP. Under native conditions, this N-WASP/WIP complex is activated by the small GTPase Cdc42 in concert with several different SH3 (Src-homology-3) domain-containing proteins. In the current study, we tested whether SH3 domains from the F-BAR (FCH-Bin-Amphiphysin-Rvs) subfamily of membrane-deforming proteins are involved in actin pedestal formation. We found that three F-BAR proteins: CIP4, FBP17, and TOCA1 (transducer of Cdc42-dependent actin assembly), play different roles during actin pedestal biogenesis. Whereas CIP4 and FBP17 inhibited actin pedestal assembly, TOCA1 stimulated this process. TOCA1 was recruited to pedestals by its SH3 domain, which bound directly to proline-rich sequences within EspFU. Moreover, EspFU and TOCA1 activated the N-WASP/WIP complex in an additive fashion in vitro, suggesting that TOCA1 can augment actin assembly within pedestals. These results reveal that EspFU acts as a scaffold to recruit multiple actin assembly factors whose functions are normally regulated by Cdc42.
Introduction
A variety of intracellular microbial pathogens trigger actin polymerization at their surface to drive directional motility within mammalian cells. For years, filamentous-actin (F-actin)2 “comet tail” formation by Listeria monocytogenes and Shigella flexneri have served as powerful model systems for studying the mechanisms by which cells control cytoplasmic actin assembly (1, 2). More recently, a group of extracellular pathogens that adhere to the cell surface and reorganize the underlying cytoskeleton into dynamic F-actin “pedestals” have been used to help decipher how cells regulate actin assembly during plasma membrane remodeling (3). These bacteria include enterohemorrhagic Escherichia coli (EHEC) (serotype O157), a major cause of bloody diarrhea and pediatric kidney failure (4, 5).
Actin pedestal biogenesis requires translocation of EHEC effector proteins into the host cell via a type III secretion system (6–8). Among the many effectors, only two are known to directly drive pedestal formation: Tir and EspFU (also known as TccP) (9). Tir acts as a plasma membrane receptor for intimin, an adhesin expressed on the bacterial surface, and intimin-Tir interactions result in clustering of the C-terminal cytoplasmic domain of Tir. A 12-residue peptide within this portion of Tir harbors its essential signaling function (10–12), and it binds to the I-BAR (inverse-BAR) domains of the membrane-deforming host cell proteins IRSp53 and IRTKS (13, 14). SH3 domains from IRSp53 and IRTKS, in turn, recruit EspFU (13, 14), which possesses multiple 47-residue proline-rich peptide repeats (EspFU(R1–6)) (15, 16). This repeat region is the most crucial bacterial component of the signaling machinery that triggers actin pedestal biogenesis, because a Tir-EspFU hybrid protein in which the C terminus of Tir is replaced with the EspFU repeats is fully capable of driving pedestal assembly (9).
One major function of the EspFU repeat region is to recruit WASP and N-WASP (15, 16), a pair of actin nucleation-promoting factors. WASP, which is expressed in hematopoietic cells, and N-WASP, which is expressed ubiquitously, utilize C-terminal WH2-connector-acidic (WCA) domains to stimulate the Arp2/3 complex, the major nucleator of branched actin filament networks in mammalian cells (17, 18). Normally, N-WASP adopts an auto-inhibited conformation in which this WCA region is prevented from activating Arp2/3 by an intramolecular interaction with a central GTPase-binding domain (GBD). The canonical mechanism of N-WASP activation involves binding of the small GTPase Cdc42 to a portion of the GBD that lies adjacent to the segment that contacts the WCA domain. This changes the GBD conformation, relieves auto-inhibition, and allows the WCA domain to stimulate the Arp2/3 complex (19).
Although Cdc42 is sufficient to activate recombinant N-WASP in vitro, several N-WASP-interacting proteins, including WIP, are known to influence N-WASP regulation in the cytoplasm. For example, activation of the native N-WASP/WIP complex by Cdc42 requires a protein named TOCA1 (20). TOCA1 harbors an N-terminal F-BAR domain that tubulates membranes, a central homology region-1 (HR1) domain that binds Cdc42, and a C-terminal SH3 domain that can interact with N-WASP. Two TOCA1 homologs, FBP17 and CIP4, also contain this F-BAR-HR1-SH3 domain organization.
Although TOCA1, FBP17, and CIP4 each participate in plasma membrane dynamics (21–23), their membrane tubulation activities differ significantly from one another (24–26), implying that they are regulated by divergent mechanisms or that they have distinct cellular roles. TOCA1 has been shown to also function with N-WASP during neuronal morphogenesis and in filopodial protrusions of the plasma membrane (26–28). Moreover, TOCA1 appears to recruit the N-WASP/WIP complex to drive actin assembly at the tips of reconstituted filopodia-like structures in vitro (29). TOCA1 additionally contributes to the formation of actin tails that drive endosome motility (20, 25), and can recruit N-WASP/WIP to liposomes in vitro, where actin assembly rates might be influenced by membrane curvature (30). Similarly, TOCA1 has been implicated in actin tail assembly by Shigella. In the Shigella system, TOCA1 increases actin tail length, although the mechanisms by which it is recruited have not been defined (31).
Like Cdc42, EspFU is able to activate WASP and N-WASP. However, in contrast to the aforementioned mechanism of WASP activation by Cdc42, EspFU directly competes with the WCA domain for binding to the auto-inhibitory segment of the GBD (32, 33). A short amphipathic helix near the N terminus of each 47-residue EspFU repeat binds to this region of the GBD with high affinity and therefore acts as a potent activator of WASP and N-WASP (32, 33), as well as the N-WASP/WIP complex (9). Importantly, actin assembly is further amplified by the multivalency of EspFU, because the repeats synergize in activating recombinant N-WASP derivatives and the N-WASP/WIP complex in vitro (9, 33, 34).
Interestingly, each N-WASP-binding helix within an EspFU repeat lies adjacent to a proline-rich sequence, suggesting that EspFU may interact with multiple SH3 domains that could potentially modulate N-WASP activity even further. In fact, these proline-rich motifs are involved in binding to the SH3 domain-containing I-BAR proteins IRSp53 and IRTKS (13, 14). However, the ability of these and other BAR protein relatives to regulate N-WASP during pedestal biogenesis has not been explored. Given that the SH3 domain of the F-BAR protein TOCA1 is known to cooperate with Cdc42 to activate N-WASP in uninfected cells, we sought to test whether the TOCA1 family of proteins plays a role in EspFU-mediated actin assembly during pedestal formation. We found that TOCA1 stimulates actin pedestal assembly, whereas FBP17 and CIP4 inhibit this process. TOCA1 is recruited to sites of pedestal biogenesis in a manner dependent on its SH3 domain, which recognizes EspFU, and these two proteins activate the N-WASP/WIP complex in an additive manner. These results highlight an important role for TOCA1 in pedestal biogenesis and define a key scaffolding function for EspFU.
EXPERIMENTAL PROCEDURES
Molecular Biology
For RT-PCR of primate TOCA1, FBP17, CIP4, and β-actin, cDNA prepared from COS7 cells with TRIzol reagent and the M-MLV reverse transcriptase (Invitrogen) was subsequently amplified using the following primer pairs: gtgggatcagttcgacagct and tcgtccttcttgcagatgca, ctgggatcagtttgacaact and gtgctttacggccatcgtga, gtgggatcagttcgaggtgc and aagtgcatcttcctctcctg, gctcgtcgtcgacaacggct and ggtcatcttctcgcggttgg. Each product corresponds to ∼350 nt of 5′ coding sequences. For cloning, human TOCA1 derivatives were PCR-amplified from plasmids (20) and inserted into the KpnI and XbaI sites of pBTM116 to create fusions to an N-terminal LexA DNA-binding domain (DBD) and to use with EspFU-Gal4 activation domain (GAD) fusions in yeast 2-hybrid assays (15). For expression of GFP-TOCA1 fusions in mammalian cells, TOCA1 derivatives were subcloned from the pBTM116 vector into the KpnI and XbaI sites of pKC425 (35). The GFP-FBP17 plasmid (Addgene 22229) has been described previously (21). For expression and purification of TOCA1 fusions containing an N-terminal His6 tag, TOCA1 derivatives were subcloned from the pKC425 vector into pKC-FastBac (36). For expression and purification of SH3 domains containing an N-terminal GST tag, DNA fragments encoding residues 481–546 of TOCA1, 551–617 of FBP17, 375–440 of IRSp53, and 337–403 of IRTKS were inserted into the BamHI and NotI sites of pGEX4T (GE Healthcare). The plasmid encoding GST-Nck-SH3(B) has been described previously (37). Plasmids for expression and purification of EspFU derivatives containing an N-terminal His10-tag and C-terminal 5myc-tag have been described (9). The HN-Tir-EspFU(R1–6) plasmid that encodes a hybrid effector in which the cytoplasmic C terminus of Tir has been replaced with the repeat region of EspFU was also described elsewhere (9), as were all other constructs (9, 12, 15, 35).
Protein Purification
His-EspFU-myc fusion proteins were previously expressed and purified from E. coli (9). Similar to procedures detailed elsewhere (36), His-tagged TOCA1 derivatives were expressed in Sf9 cells and purified using Ni-NTA-agarose beads (Qiagen), while GST-tagged SH3 domains were purified from E. coli using GST-bind resin (Novagen). The EspFU peptide (Biotin-AHA-HIPPAPNWPAPPPPVQ) was synthesized by the Tufts University Core Facility. His-tagged N-WASP/WIP and Arp2/3 complexes were also expressed and purified from Sf9 insect cells as described (9, 36). To maintain an auto-inhibited N-WASP/WIP complex, freeze-thaw cycles and periods of storage on ice were minimized.
Transfections and Infections
COS7 cells were cultured in DMEM with 10% FBS at 37 °C in 5% CO2, and all transfections were performed as described previously (9). TOCA1 siRNAs were acquired from Invitrogen, while FBP17 and CIP4 siRNAs were purchased from Ambion. Commercially-available antibodies that we used to test TOCA1 expression did not properly recognize native or recombinant TOCA1 on immunoblots, so we verified the potency of siRNAs by RT-PCR and by measuring their ability to prevent expression of GFP-TOCA1. To experimentally cluster Tir-EspFU fusions, cells were treated with antibodies that bind the Tir extracellular domain prior to adding formalin fixed Staphylococcus aureus particles, as described previously (9). Infections with non-pathogenic E. coli expressing intimin have also been described (9). This experimental system, in which Tir-EspFU fusions are expressed in mammalian cells, allows us to specifically examine the effect of dominant negative proteins or RNAi on EspFU-mediated signaling, rather than any proximal events. It bypasses complications that may arise from inactivation of host actin assembly factors that facilitate type III translocation (38), as well as any problems derived from the presence of other EHEC effectors or toxins.
Microscopy
Infected cells were fixed using 2.5% paraformaldehyde for 35 min and permeabilized with 0.1% Triton-X-100 in PBS. To visualize Tir-EspFU derivatives and Staphylococcus aureus particles, cells were treated with the anti-HA.11 primary antibody (Covance) and Alexa350 or Alexa488 anti-mouse secondary antibodies (Invitrogen). F-actin was detected using 4U/ml Alexa568-phalloidin (Invitrogen), and bacteria were identified with 1 μg/ml DAPI (Sigma). To assess the dominant negative effect of GFP-N-WASP or -TOCA1 fusion proteins on pedestal formation mediated by Tir-EspFU fusions, cells co-expressing high levels of these proteins were first identified by bright GFP and HA fluorescence. To measure pedestal formation efficiencies, the percentage of cells harboring at least 15 adherent particles and 10 actin pedestals was measured. At least 100 cells were examined per sample per experiment. To measure pedestal intensities, individual phalloidin-stained pedestals were chosen randomly and outlined using ImageJ. The mean pixel intensity of this region was then multiplied by its 2-dimensional area. This value was compared with a non-pedestal containing area in the same cell to yield a normalized value. Pedestals from control samples treated with a nonspecific siRNA or with a GFP plasmid were set to a value of 1, and intensities for other experimental conditions were adjusted proportionally. Measurements were acquired from at least 10 pedestals on at least 6 cells per condition. Statistical significance was determined using nonparametric Mann-Whitney or parametric ANOVA tests.
EspFU Interaction Assays
For pulldown assays, cobalt-conjugated magnetic particles (Invitrogen) were coated with His-EspFU-myc derivatives for 1h. After removal of excess unbound EspFU, beads were incubated with cell extract for 1 h in 50 mm NaPO4 pH 7.4, 150 mm NaCl, and 0.015% Triton X-100 as described previously (9). Bound proteins were eluted by boiling in SDS-PAGE sample buffer. For Far-Western assays, 1 μg of TOCA1 derivatives were subjected to SDS-PAGE, transferred to nitrocellulose membranes, and probed with soluble His-EspFU-myc derivatives, similar to experiments described previously (15). Bound EspFU was detected using anti-myc antibodies (Sigma). Yeast two-hybrid assays and quantification of β-gal activity and 3-AT resistance were described elsewhere (15). For EspFU peptide-binding assays, 96-well MaxiSorp ELISA plates (Nunc), were coated with different concentrations (10–100 μm) of GST fusion proteins in 100 mm sodium carbonate buffer (pH 9.6). Wells were washed using PBS with 0.1% Tween-20, blocked using PBS with 3.5% BSA, and probed with the EspFU peptide (10 μg/ml) in PBS plus 0.2% BSA. Bound peptide was then fixed using PBS with 3% paraformaldehyde. Following washes using PBS with 0.2% BSA and 5% milk, bound peptide was detected using streptavidin-horseradish peroxidase (HRP; BIOSOURCE) and the colorimetric reagent o-phenylenediamine dihydrochloride (Sigma).
Immunoblotting
To prepare extracts, transfected cells were collected in PBS with 2 mm EDTA and lysed in 50 mm Tris-HCl pH 7.6, 50 mm NaCl, 1% Triton X-100, 1 mm PMSF, and 10 μg/ml of aprotinin, leupeptin, pepstatin, and chymostatin (EMD Bioscience), prior to mixing with SDS-PAGE sample buffer. Protein samples were boiled and analyzed by 10% SDS-PAGE before staining with Coomassie Blue or transferring to nitrocellulose membranes and staining with Ponceau S. Membranes were blocked in PBS with 5% milk before probing with anti-GFP or anti-Myc antibodies, as described previously (36). Following washes, membranes were treated with HRP-conjugated antibodies and detected using enhanced chemiluminescence (GE Healthcare).
Actin Assembly Assays
Pyrene-actin assays typically contained 2.0 μm actin (5% pyrene-labeled), 20 nm recombinant N-WASP/WIP complex, and 10–20 nm Arp2/3 complex. Polymerization and barbed end concentrations were measured and calculated as described previously (36).
RESULTS
Multiple F-BAR-HR1-SH3 Proteins Are Expressed in Host Cells
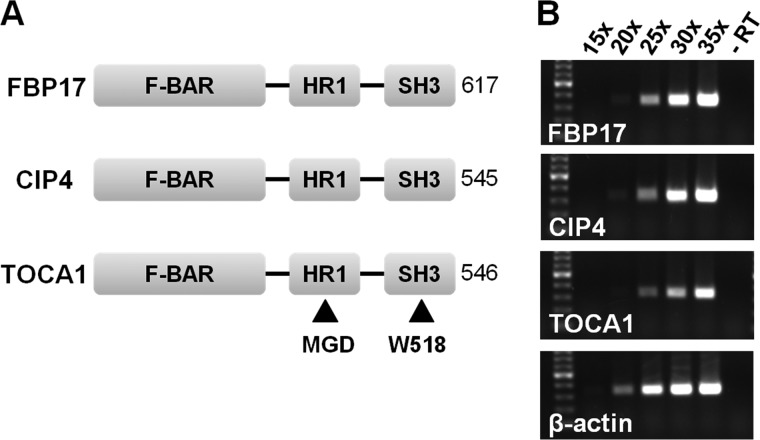
The observation that EspFU targets N-WASP to drive actin assembly prompted us to explore whether TOCA1, which normally cooperates with Cdc42 to activate the native N-WASP/WIP complex, might promote pedestal formation. Mammalian cells encode TOCA1 plus two closely-related homologs, FBP17 and CIP4, each of which contains an F-BAR-HR1-SH3 domain organization (Fig. 1A). These 3 proteins are all thought to regulate N-WASP activity along plasma membrane rearrangements such as those that occur during endocytosis (21–23). Therefore, we first sought to determine which of the F-BAR-HR1-SH3 proteins were expressed in COS7 cells, a commonly-employed host cell line for examining actin pedestal formation. RT-PCR using primer pairs to amplify portions of the 5′ coding sequences of each of these transcripts yielded products of the predicted size, indicating that TOCA1, FBP17, and CIP4 are all expressed in this cell line (Fig. 1B).
FIGURE 1.
COS7 cells express all three members of the F-BAR-HR1-SH3 family. A, the domain organization of FBP17, CIP4, and TOCA1 is shown (20). B, RNA isolated from COS7 cells was reverse transcribed and subjected to 15–35 cycles of PCR using primers for human FBP17, CIP4, TOCA1, and a β-actin control. The resulting ∼350 bp products and a 100 bp ladder (highlighted band, 500 bp) were visualized on ethidium bromide-stained 1% agarose gels. No PCR products were detected when the reverse transcription step was omitted (-RT).
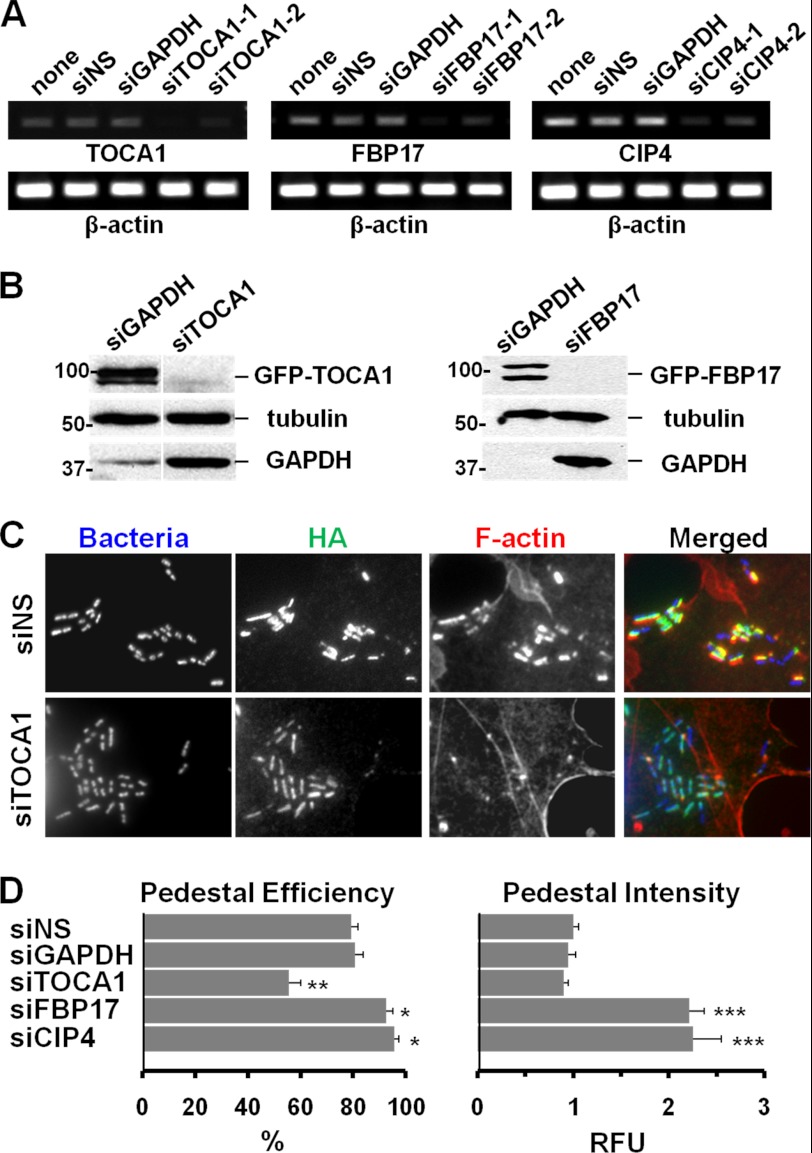
To explore a functional role for each of these F-BAR proteins in actin pedestal formation, we next reduced their expression using RNAi. For measuring the efficacy of different siRNAs, we performed RT-PCR on RNA isolated from COS7 cells treated with independent siRNAs against TOCA1, FBP17, or CIP4, and from control cells treated with an siRNA against GAPDH, with a nonspecific siRNA, or without any siRNA treatment. RNAs targeting TOCA1, FBP17, or CIP4 triggered a reduction in their corresponding transcripts while quantities of the control β-actin transcript were unchanged (Fig. 2A). Thus, these siRNAs silence their respective mRNA targets.
FIGURE 2.
TOCA1 depletion inhibits pedestal assembly whereas FBP17 or CIP4 depletion enhances it. A, COS7 cells treated with no siRNAs, a nonspecific (NS) siRNA, or independent siRNAs to GAPDH, TOCA1, FBP17, or CIP4 were subjected to RT-PCR for TOCA1, FBP17, CIP4, and β-actin (25 cycles) and agarose gel electrophoresis. B, cells treated with an siRNA to GAPDH or with pairs of siRNAs to TOCA1 or FBP17 were transfected with plasmids encoding GFP-TOCA1 or GFP-FBP17 and analyzed by immunoblotting for GFP, β-tubulin, or GAPDH. C, cells treated with siRNAs and transfected with HA-Tir-EspFU(R1–6) were infected with E. coli expressing intimin, fixed, and stained with HA antibodies to identify transfected cells, with phalloidin to detect F-actin, and with DAPI to visualize bacteria. D, pedestal formation efficiencies were determined by calculating the % of infected cells harboring 10 or more actin pedestals. Data represent the mean ± S.E. from 4–7 experiments. Pedestal intensities in relative fluorescent units (RFU) were determined by measuring the total F-actin pixel intensity in the pedestal. Data represent the mean ± S.E. from ∼70 pedestals per sample across 3–4 experiments. The significant differences in pedestal efficiency and intensity that were observed using siRNA pairs were verified using individual siRNAs to TOCA1 and FBP17 (data not shown). For comparisons to siNS: *, p < 0.05; **, p < 0.01; ***, p < .0001.
To confirm that RNAi was capable of reducing expression at the protein level, we also tested whether siRNAs could deplete GFP-tagged versions of TOCA1 or FBP17 in transfected cells. Indeed, siRNA pairs targeting TOCA1 and FBP17 almost completely blocked expression of GFP-TOCA1 and GFP-FBP17, respectively (Fig. 2B). These results provide proof-in-principle that our RNAi-based approaches are effective for examining the cellular roles of F-BAR family members.
TOCA1 Enhances Actin Pedestal Biogenesis, whereas FBP17 and CIP4 Inhibit This Process
To test whether TOCA1, FBP17, or CIP4 is important for actin pedestal assembly, we next combined siRNA-mediated gene silencing with functional assays for actin pedestal formation. To examine EspFU-mediated actin assembly while bypassing any requirements for the actin polymerization machinery during type III effector translocation from EHEC (38), we expressed a Tir-EspFU fusion protein in the plasma membrane of siRNA-treated host cells. To trigger pedestal assembly, we then clustered this hybrid effector by treating cells with a non-pathogenic strain of E. coli that expresses intimin or with anti-Tir coated S. aureus particles (9). Just as biomimetic approaches involving ActA-coated beads and IcsA-expressing E. coli are used to examine the actin-based motility mechanisms of Listeria and Shigella (39–41), this experimental system allows us to specifically study the EspFU-mediated actin assembly mechanisms that are used by EHEC.
Quantification of the fraction of infected cells that contained visibly detectable F-actin pedestals revealed that cells treated with nonspecific or GAPDH control siRNAs formed pedestals at high efficiencies (∼80%). Interestingly, the percentage of cells with pedestals was roughly one-third lower (∼55%) when treated with a pair of siRNAs to TOCA1 (Fig. 2, C and D). In contrast, treatment with siRNAs to FBP17 or CIP4 caused a significant increase (to ∼95%) in the efficiency of pedestal formation (Fig. 2D). Taken together, these results suggest that TOCA1 normally plays a positive role in actin pedestal assembly, while FBP17 and CIP4 have negative roles in this process.
As a second measure of actin pedestal formation, we quantified the intensity of F-actin staining within pedestals. Surprisingly, pedestals formed in cells treated with siRNAs to FBP17 or CIP4 were more than twice as bright as pedestals in control cells (Fig. 2D), implying that EspFU can drive the assembly of more densely-packed actin networks when either of these two proteins are absent. TOCA1 depletion did not have any apparent effects on pedestal intensity (Fig. 2D), consistent with the possibility that this protein plays a role in initiating actin assembly into pedestals but is not required for increasing the amount of actin filaments found in pedestals at steady-state.
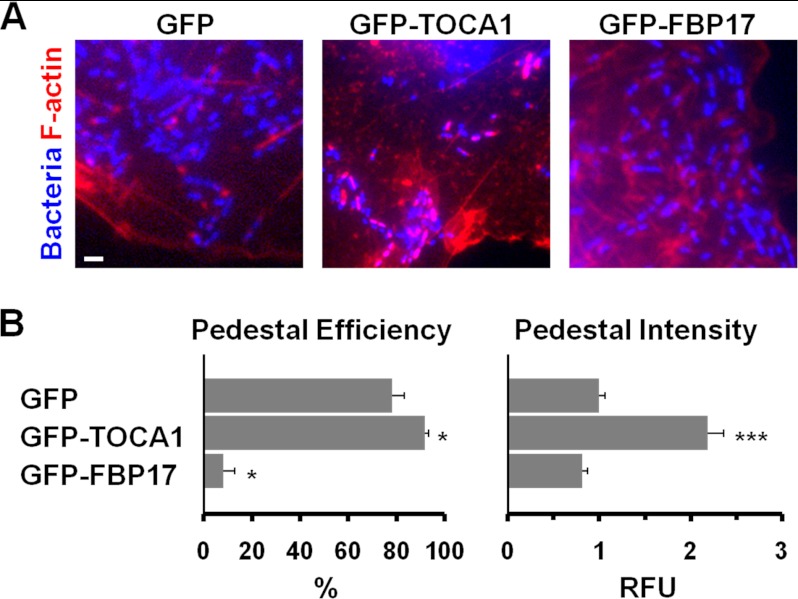
To further explore the role of F-BAR proteins in pedestal formation, we also assessed actin pedestal efficiency and intensity when GFP-tagged versions of TOCA1 or FBP17 were overexpressed in host cells. When expressed at low levels by themselves, each fusion protein was found throughout COS7 cells and at the plasma membrane. However, only GFP-FBP17 caused a dramatic induction of membrane tubulation (data not shown), similar to previous observations (21). In accordance with depletion experiments suggesting that TOCA1 protein levels positively correlate with pedestal assembly (Fig. 2, C and D), GFP-TOCA1 overexpression caused a significant increase in the efficiency of pedestal formation and more than doubled the intensity of F-actin staining within pedestals (Fig. 3, A and B). In addition, consistent with a negative correlation between FBP17 protein levels and pedestal assembly, GFP-FBP17 overexpression caused a dramatic (∼8-fold) decrease in the efficiency of pedestal formation (Fig. 3, A and B). Overall, the combination of siRNA-mediated loss-of-function and overexpression analyses indicates that TOCA1 contributes positively to EspFU-mediated actin pedestal biogenesis, whereas FBP17 (and probably CIP4) play inhibitory roles in pedestal assembly.
FIGURE 3.
TOCA1 overexpression enhances pedestal assembly while FBP17 overexpression inhibits it. A, COS7 cells co-expressing HA-Tir-EspFU(R1–6) and GFP fusion proteins were infected with E. coli expressing intimin, fixed, and stained with DAPI to identify bacteria and phalloidin to label F-actin. The scale bar is 1 μm in length. B, pedestal formation efficiencies were determined by calculating the % of infected GFP-expressing cells harboring 10 or more actin pedestals. Data represent the mean ± S.E. from 3–4 experiments. Pedestal intensities in relative fluorescent units (RFU) were determined by measuring the total F-actin pixel intensity in the pedestal. Data represent the mean ± S.E. from ∼70 pedestals per sample across 3–4 experiments. For comparisons to GFP: *, p < 0.05; ***, p < .0001.
TOCA1 Localizes to Actin Pedestals in an SH3 Domain-dependent Manner
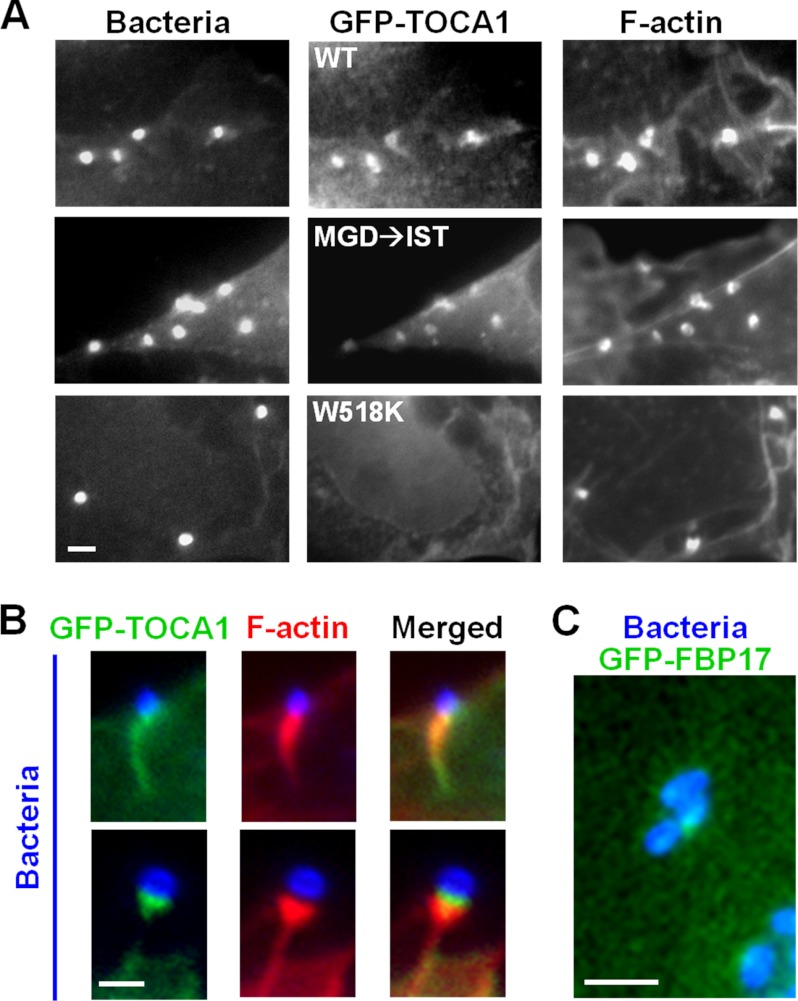
Since TOCA1 had a positive function in actin pedestal formation, we next examined the localization of GFP-TOCA1 during EspFU-mediated actin assembly. Interestingly, GFP-TOCA1 showed a significant enrichment in pedestals, compared with the rest of the cell (Fig. 4, A and B). GFP-FBP17, even though it blocked pedestal formation, could also be recruited to sites of EspFU clustering. However, localization of the TOCA1 fusion was much more prominent than that of FBP17 (Fig. 4, B and C). GFP-TOCA1 usually localized throughout the entire pedestal, and it sometimes appeared enriched near the pedestal tip adjacent to the bacterium (Fig. 4B, lower panels).
FIGURE 4.
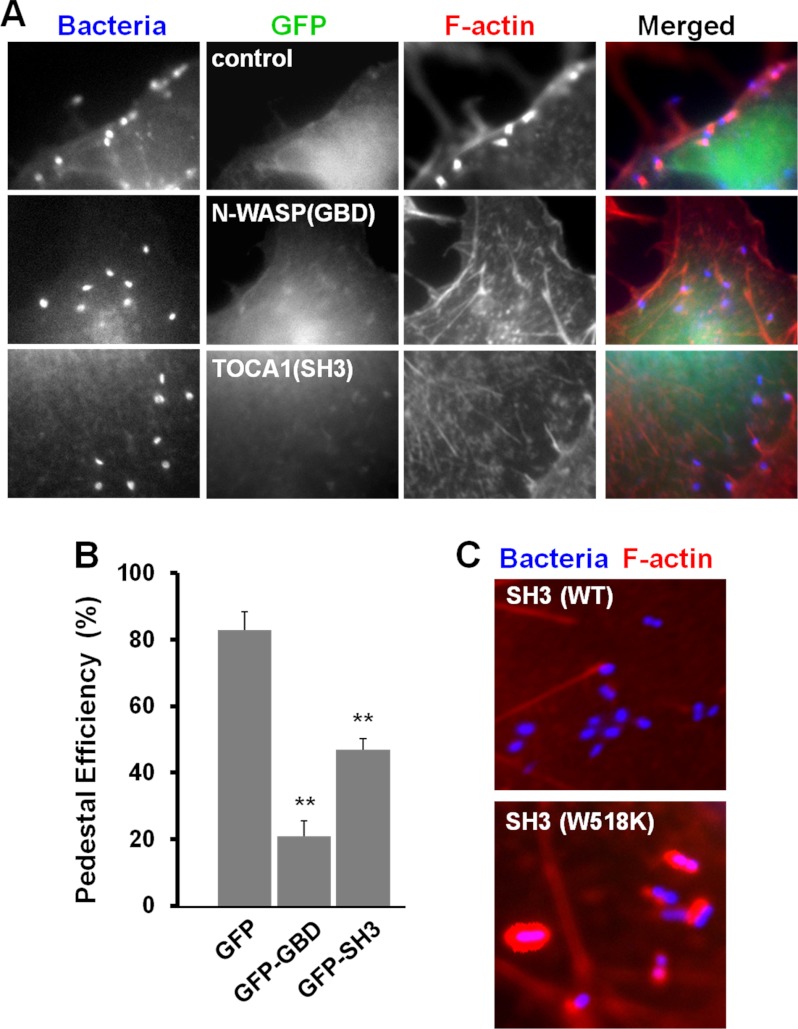
TOCA1 localizes to EspFU-derived actin pedestals in an SH3-dependent manner. A, COS7 cells were co-transfected with plasmids encoding HN-Tir-EspFU(R1–6) (9) and either wild type GFP-TOCA1 or mutants with inactivated HR1 (MGD → IST) or SH3 (W518K) domains. The Tir-EspFU fusion along with S. aureus were visualized by anti-HA and anti-mouse antibody staining. F-actin was detected with phalloidin. All scale bars are 1 μm in length. B, magnified examples of GFP-TOCA1 localization to actin pedestals are shown. C, magnified example of GFP-FBP17 recruitment to intimin-expressing bacteria is shown.
TOCA1 contains an HR1 domain that binds Cdc42 and an SH3 domain that interacts with proline-rich peptides, so we next tested whether its recruitment to pedestals could still occur when such interactions were disrupted by the previously-characterized MGD → IST mutations in the HR1 domain or a W518K mutation in the SH3 domain (20). Importantly, whereas the TOCA1 MGD → IST mutant localized to pedestals, the W518K mutant did not (Fig. 4A), suggesting that SH3 domain interactions are important for TOCA1 recruitment to pedestals.
Given this requirement for the TOCA1 SH3 domain in recruitment to pedestals, we predicted that overexpression of the SH3 domain by itself might act as a dominant negative inhibitor of actin pedestal formation. To test this possibility, we measured the capacity of the Tir-EspFU fusion protein to assemble actin pedestals in cells expressing high levels of GFP-TOCA1(SH3). Consistent with the decrease in pedestal formation observed in TOCA1 siRNA-treated cells, expression of the TOCA1 SH3 domain caused about a 2-fold reduction in the efficiency of pedestal formation (Fig. 5, A and B), indicating that the SH3 domain of TOCA1 is indeed capable of disrupting EspFU-mediated actin assembly. A similar (but somewhat greater) inhibition of pedestal formation was observed when we overexpressed the best known target of EspFU, the N-WASP GBD (Fig. 5, A and B). Thus, these dominant negative studies further support a role for TOCA1 in actin pedestal assembly.
FIGURE 5.
The isolated SH3 domain of TOCA1 inhibits EspFU-mediated actin pedestal assembly. A, cells co-expressing HA-Tir-EspFU(R1–6) and high levels of GFP fusion proteins carrying the N-WASP GBD or TOCA1 SH3 domain were treated with Tir antibodies and S.aureus, fixed, and stained with HA antibodies to identify both transfected cells and S.aureus, and with phalloidin to detect F-actin. Cells expressing low levels of GFP-N-WASP(GBD) and GFP-TOCA-1(SH3) showed strong recruitment of the GFP fusions to sites of S.aureus adherence (data not shown). B, pedestal formation efficiencies were determined by calculating the % of co-transfected cells harboring 10 or more actin pedestals. Data represent the mean ± S.E. from three experiments. For comparisons to GFP: **, p < 0.01. C, cells co-expressing HA-Tir-EspFU(R1–6) and GFP-TOCA1 fusion proteins carrying a wild type or mutant SH3 domain were infected with E. coli expressing intimin, fixed, and stained with DAPI to identify bacteria and phalloidin to label F-actin.
To investigate the basis of the dominant negative effect of the GFP-TOCA1(SH3) fusion, we introduced a W518K mutation into this construct. In contrast to cells overexpressing the wild type SH3 domain, cells overexpressing this mutant generated actin pedestals quite strongly (Fig. 5C), indicating that binding to proline-rich sequences is required for the inhibitory activity of the SH3 domain. In addition, the dominant negative effect of the TOCA1 SH3 domain is likely specific to signaling from EspFU, and not due to ectopic N-WASP activation or sequestration, because overexpression of an SH3 domain from the adaptor protein Nck did not affect actin pedestal formation (13). Presumably the TOCA1 SH3 domain acts in a dominant negative fashion because it inhibits EspFU from recruiting endogenous TOCA1.
To test whether TOCA1 SH3 overexpression affects N-WASP recruitment, we stained transfected cells with N-WASP antibodies. N-WASP localized to sites of clustered EspFU in the absence or presence of the functional SH3 fragment (data not shown), suggesting that the inhibitory effect of this TOCA1 derivative is not due to any obvious defect in N-WASP recruitment. Since it does not appear to be essential for N-WASP localization, a function for TOCA1 in actin pedestal formation may reside in its ability to facilitate N-WASP activation.
The TOCA1 SH3 Domain Binds to the Proline-rich Repeat Region of EspFU
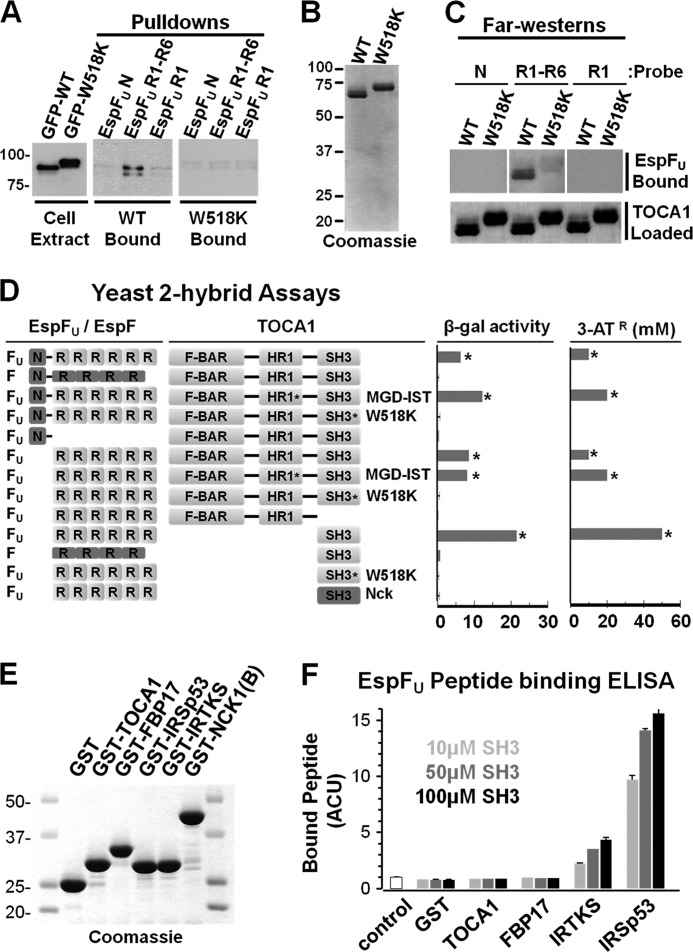
The observations that TOCA1 requires its SH3 domain for recruitment to actin pedestals and that SH3 overexpression can inhibit pedestal formation raised the possibility that this region of TOCA1 might associate with the proline-rich sequences of EspFU. To test whether EspFU and TOCA1 interact, we coated magnetic beads with purified recombinant His-tagged EspFU derivatives (9) and mixed these beads with extracts generated from cells expressing either GFP-TOCA1 wild type or the W518K mutant. Binding of TOCA1 to the beads was then assessed by immunoblotting for GFP. Whereas beads coated with EspFU(N), the control N-terminal EspFU translocation domain, did not pulldown either TOCA1 protein, EspFU(R1–6), the derivative containing the entire EspFU C-terminal repeat region (9), pulled down wild type GFP-TOCA1 (Fig. 6A), indicating that EspFU and TOCA1 interact in cell extracts. In contrast, EspFU(R1–6) did not pulldown the W518K mutant (Fig. 6A), implying that the TOCA1 SH3 domain is necessary for interacting with EspFU. Interestingly, EspFU(R1), a derivative containing only a single repeat (9), did not pull down substantial amounts of GFP-TOCA1 when compared with EspFU(R1–6) (Fig. 6A), suggesting that multiple repeats contribute to the interaction between EspFU and TOCA1.
FIGURE 6.
TOCA1 binds to the proline-rich repeat region of EspFU. A, COS7 cells were transfected with GFP-TOCA1 plasmids, lysed, and either analyzed by SDS-PAGE directly (Cell Extract) or mixed with beads coated with His-EspFU-myc derivatives containing either the N-terminal translocation domain (N), 6 proline-rich peptide repeats (R1–6), or a single repeat (R1). After precipitation, bead-associated TOCA1 derivatives (Pulldowns) were detected by blotting with GFP antibodies. The W518K mutant migrates through SDS-PAGE gels slower than the wild type protein (20). B, full-length N-terminally His-tagged TOCA1 derivatives were purified and analyzed by SDS-PAGE and Coomassie staining. C, His-TOCA1 wild type or W518K were transferred to nitrocellulose membranes following SDS-PAGE and probed with the His-EspFU-myc derivatives N, R1–6, or R1. Bound EspFU was detected by anti-myc immunoblotting. TOCA1 was detected with Ponceau S. Densitometry indicated that binding of R1-R6 to TOCA1 was reduced 4.1-fold by the W518K mutation (data not shown). D, plasmids encoding EspFU (or EspF) derivatives fused to the Gal4 transcriptional activation domain and TOCA1 (or Nck) fragments fused to the LexA DNA-binding domain were co-transformed into a yeast two-hybrid reporter strain. The MGD and W518K mutants were utilized where indicated. Data represent the mean ± S.D. of β-galactosidase activity for three co-transformants for each pairwise combination. Activation of the HIS3 reporter was determined by growth on media containing 3-AT. Similar results were obtained in multiple transformations. E, N-terminally GST-tagged SH3 domains were purified and analyzed by SDS-PAGE and Coomassie staining. F, ELISA plates were coated with 10, 50, or 100 μm GST-SH3 fusion proteins and probed with an N-terminally biotinylated EspFU peptide. Bound peptide was detected using a streptavidin-HRP conjugate, binding values were normalized to BSA-coated (control) wells and then plotted in adjusted colorimetric units (ACU). Data represent the mean ± S.E. of four samples.
To determine if the association between TOCA1 and EspFU could be direct, we purified recombinant His-tagged versions of wild type TOCA1 and the W518K mutant (Fig. 6B). These proteins were then subjected to SDS-PAGE, immobilized on nitrocellulose membranes, and probed with purified EspFU derivatives harboring zero, one, or six repeats. Binding of EspFU to TOCA1 in these Far-Western assays was then detected by immunoblotting for the C-terminal Myc tags on the EspFU proteins (9). Consistent with results from pulldown experiments, TOCA1 did not bind either EspFU(N) or EspFU(R1) in this assay (Fig. 6C). Moreover, wild type TOCA1 bound EspFU(R1–6), and a W518K mutation in TOCA1 reduced the amount of binding by more than 75% (Fig. 6C). These results indicate that the multivalent proline-rich region of EspFU can directly bind to TOCA1, and that a functional TOCA1 SH3 domain is required for this interaction.
To further define the interaction between TOCA1 and EspFU, TOCA1 was fused to the LexA-DBD for examination with a GAD-EspFU fusion in yeast two-hybrid assays (Fig. 6D). As expected, full-length EspFU associated with full-length TOCA1 in these studies, as determined by activation of LacZ and His3 reporter genes. The related proline-rich EHEC effector protein EspF, which is related to EspFU and also capable of activating N-WASP (42), did not activate reporter gene expression in the presence of TOCA1, indicating that this interaction was specific to EspFU. EspFU(R1–6), but not EspFU(N), also triggered reporter activation, confirming that the ability of EspFU to interact with TOCA1 is found in its repeat region. The central HR1 domain of TOCA1 that binds Cdc42 does not appear to mediate binding to EspFU, because an inactivating MGD mutation in the HR1 domain had no effect on the interaction with EspFU. However, as predicted from the pull-down and Far-Western assays, a W518K mutation in the SH3 domain abrogated the interaction of TOCA1 with full-length EspFU and with its isolated repeat region. Importantly, the SH3 domain alone was also sufficient to interact with the EspFU repeats. The specificity of the TOCA1 SH3-EspFU association was further reflected by the observations that the proline-rich repeats of EspF did not bind to the TOCA1 SH3 domain, and that the EspFU repeats did not interact with an SH3 domain from the adaptor protein Nck (Fig. 6D).
Aside from TOCA1, EspFU has been shown to bind other SH3 domain-containing proteins. The best characterized of these interactions are with the I-BAR proteins IRSp53 (14) and IRTKS (13). In fact, a single 47-residue EspFU repeat binds to the SH3 domain of IRTKS with a much higher affinity (∼500 nm) than typical proline-rich sequences (43). Therefore, to compare the ability of SH3 domains from different BAR family proteins to bind to EspFU, we generated GST-tagged versions of the SH3 domains from the F-BAR proteins TOCA1 and FBP17 and from the I-BAR proteins IRTKS and IRSp53 (Fig. 6E). Using a modified ELISA approach, we then coated microtiter wells with multiple concentrations of these SH3 domains and measured their ability to bind a biotinylated 16-residue EspFU peptide that encompasses its entire proline-rich sequence. Neither the SH3 domain from TOCA1 nor the SH3 domain from FBP17 bound to the EspFU peptide in these experiments (Fig. 6F), consistent with previous assays indicating that multivalency within EspFU is required for binding to TOCA1. In contrast, the SH3 domain from IRTKS and, to a greater extent, the SH3 domain from IRSp53 bound to the 16-residue proline-rich EspFU peptide (Fig. 6F), demonstrating that, unlike the SH3 domains from F-BAR proteins, the SH3 domains from these I-BAR proteins are capable of binding to the monovalent EspFU sequence with relatively high affinity.
Overall, this series of protein-protein interaction studies defines the specific association of the multivalent repeat region of EspFU with the SH3 domain of TOCA1, indicating that the binding sites on TOCA1 for EspFU and Cdc42 are distinct. Thus, just as EspFU is able to activate N-WASP in actin pedestals by a mechanism that is distinct from the way in which Cdc42 normally activates N-WASP (32, 33), EspFU can apparently promote TOCA1 recruitment in a manner distinct from the way in which Cdc42 normally recruits TOCA1.
TOCA1 Stimulates N-WASP/WIP-mediated Actin Assembly in Vitro
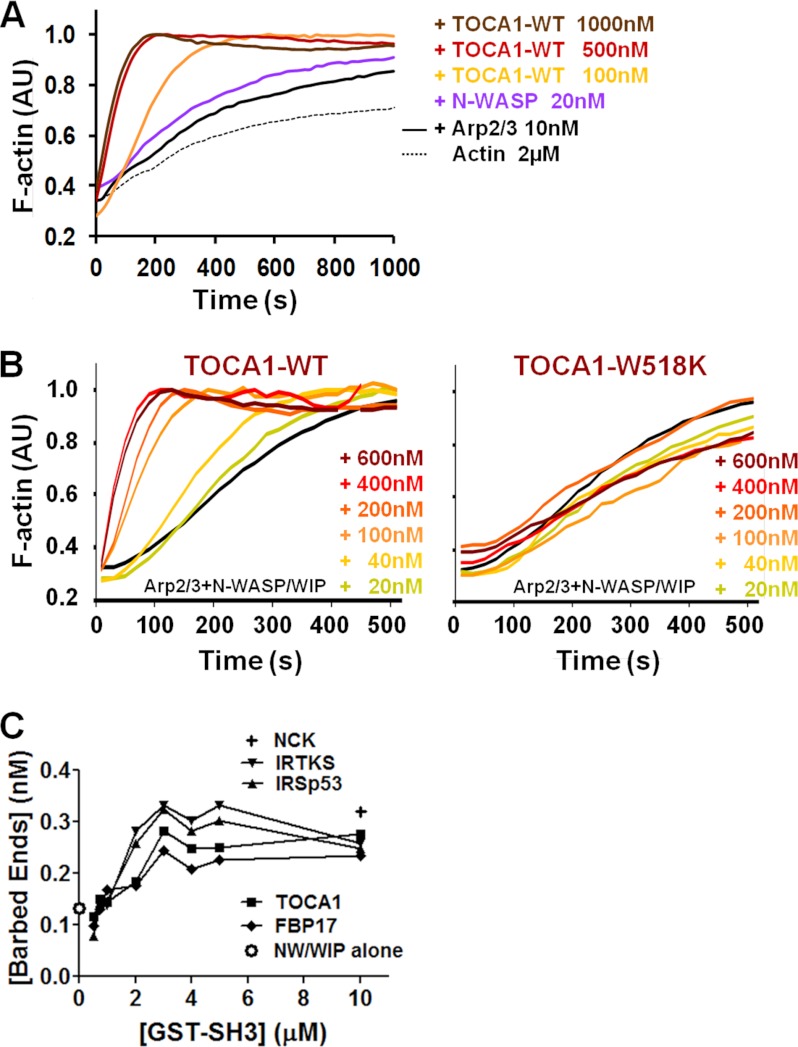
TOCA1 was initially identified based on its ability to act in concert with Cdc42 to activate the native N-WASP/WIP complex (20), so we next explored whether TOCA1 might contribute to pedestal formation by augmenting N-WASP activation. To test this possibility, we first measured the activity of recombinant TOCA1 toward N-WASP and the Arp2/3 complex in pyrene-actin polymerization assays in vitro. Interestingly, we found that TOCA1 by itself could promote actin assembly with N-WASP and the Arp2/3 complex (Fig. 7A). This activation occurred in a concentration-dependent manner, with maximal stimulation occurring at ∼1 μm TOCA1 (Fig. 7A).
FIGURE 7.
TOCA1 stimulates N-WASP/WIP-mediated actin assembly in vitro. A, actin (2 μm) was polymerized using 10 nm Arp2/3 complex, 20 nm N-WASP, and the indicated concentrations of TOCA1 wild type. Pyrene-actin fluorescence was measured in arbitrary units (AU) over time. B, actin (2 μm) was polymerized using 20 nm Arp2/3 complex, 20 nm N-WASP/WIP complex, and the indicated concentrations of TOCA1 wild type or the W518K mutant as in part A. C, actin (2 μm) was polymerized using 20 nm Arp2/3 complex, 20 nm N-WASP/WIP complex, and the indicated concentrations of GST-SH3 fusion proteins. Barbed end concentrations were calculated when F-actin reached its half-maximal value.
To test whether TOCA1 could also activate N-WASP in the presence of WIP, we measured the effect of TOCA1 on actin polymerization using the N-WASP/WIP and Arp2/3 complexes. TOCA1 also activated N-WASP/WIP, with maximal stimulation occurring at 400 nm TOCA1 (Fig. 7B). Moreover, the recombinant W518K mutant of TOCA1 did not accelerate N-WASP/WIP-mediated actin assembly at any of the protein concentrations we tested (Fig. 7B). Collectively, these results suggest that TOCA1 can act as an N-WASP/WIP activator by itself and that this activity requires a functional SH3 domain.
SH3 domains from many proteins are known to promote N-WASP activation, especially the adaptor proteins Grb2 and Nck1/Nck2 (37, 44). Given that TOCA1 and FBP17 from the F-BAR family (Fig. 4), along with IRSp53 and IRTKS from the I-BAR family (13, 14) are recruited to EspFU-driven actin pedestals, we next sought to compare the abilities of the SH3 domains from these proteins to activate the N-WASP/WIP complex. Therefore, we measured the effects of the GST-SH3 domains on actin polymerization in vitro. The efficiency of polymerization was quantified by measuring the concentration of F-actin barbed ends (a function of the rate of actin assembly at half of the maximal filament concentration) over a range of SH3 concentrations. Consistent with previous results using individual SH3 domains from other proteins in conjunction with N-WASP (37, 44), the SH3 domains from TOCA1, FBP17, IRSp53, and IRTKS all activated N-WASP/WIP, albeit inefficiently (Fig. 7C). As a point of reference, the maximal barbed end value for each reaction was comparable to that of a GST-tagged version of an SH3 domain from Nck1 (Fig. 7C) (37). The SH3 domains from the I-BAR proteins were slightly better N-WASP/WIP activators than the SH3 domains from the F-BAR proteins at most concentrations, while within the F-BAR family member TOCA1 was marginally more potent than FBP17 when used at concentrations higher than 2 μm (Fig. 7C). Thus, based on the ability of their SH3 domains to stimulate the actin nucleation machinery, there does not appear to be any dramatic advantage to recruiting any one specific BAR family member during pedestal formation. However, it is important to note that full-length TOCA1 was roughly 4-fold more potent than the minimal SH3 domain from TOCA1 (compare Fig. 8A to Fig. 7C), so TOCA1 sequences outside of the SH3 domain might also contribute to actin assembly during pedestal formation.
FIGURE 8.
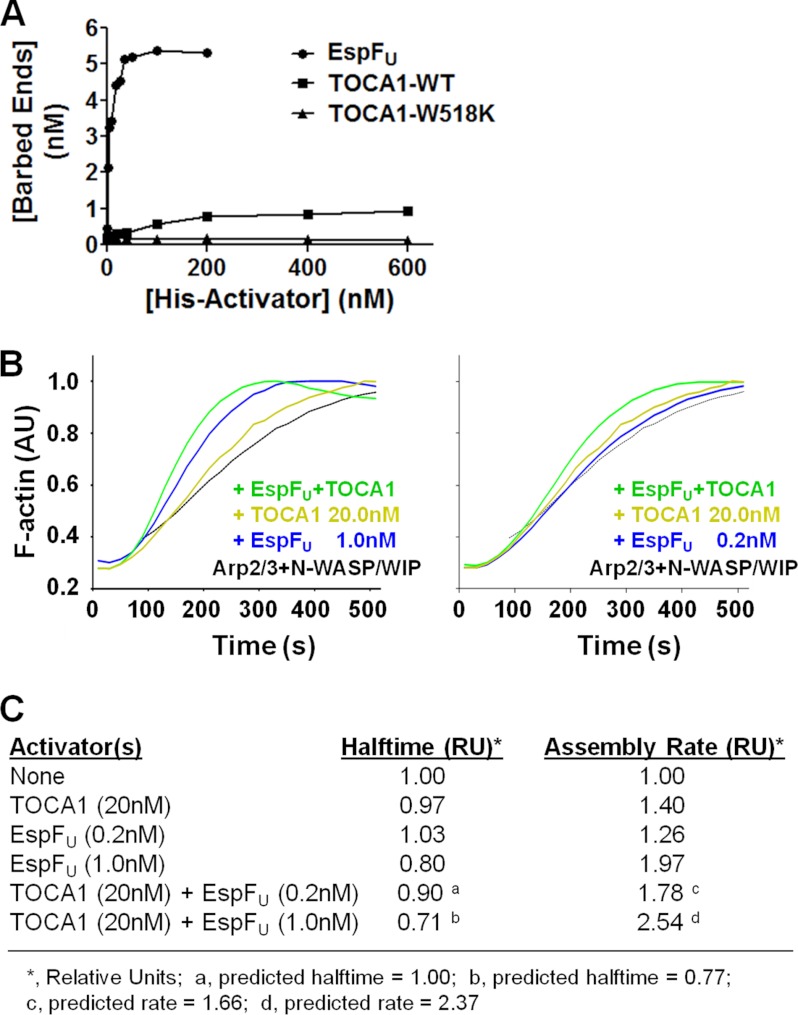
TOCA1 and EspFU activate N-WASP/WIP additively in vitro. A, actin (2 μm) was polymerized using 20 nm Arp2/3 complex, 20 nm N-WASP/WIP complex, and the indicated concentrations of His-tagged EspFU(R1–6), TOCA1 wild type, or the W518K mutant. Barbed end concentrations were calculated when F-actin reached its half-maximal value. B, actin (2 μm) was polymerized using 20 nm Arp2/3 complex, 20 nm N-WASP/WIP complex, and the indicated concentrations of TOCA1 and EspFU(R1–6). Pyrene-actin fluorescence was measured in arbitrary units (AU) over time. C, actin (2 μm) was polymerized as in part B, and the time to reach half of the maximal F-actin concentration (Halftime) and slope of assembly curves at 60% polymer (Assembly Rate) was measured and normalized to controls containing only actin, Arp2/3 complex, and N-WASP/WIP complex. The predicted Halftimes and Assembly Rates were calculated based on the premise that the effects of TOCA1 and EspFU on actin polymerization would be additive.
TOCA1 and EspFU Stimulate N-WASP/WIP-mediated Actin Assembly Additively in Vitro
The EspFU repeat region activates N-WASP with a potency that might be 100-fold greater than that of Cdc42 in vitro (32, 33). EspFU is also an extremely strong activator of the N-WASP/WIP complex (9), and comparisons of barbed end quantities in the presence of multiple concentrations of EspFU(R1–6) or full-length TOCA1 indicate that EspFU is also dramatically better than TOCA1 at activating N-WASP/WIP-mediated actin assembly (Fig. 8A). However, since maximal N-WASP activity normally occurs in response to multiple signaling inputs (20, 37, 44), we next sought to determine if EspFU and TOCA1 are capable of acting together to drive actin polymerization.
Because of the efficiency with which EspFU and, to a lesser extent, TOCA1, activates N-WASP/WIP, we had to use relatively low concentrations of each of these activators in pyrene-actin assembly assays. In the presence of 20 nm N-WASP/WIP and Arp2/3, 1 nm EspFU modestly increased the rate of actin assembly (Fig. 8B). 20 nm TOCA1 also resulted in a marginal acceleration of polymerization kinetics over N-WASP/WIP and Arp2/3 alone. Interestingly, when EspFU and TOCA1 were added to the N-WASP/WIP and Arp2/3 complexes together, actin polymerization occurred at rates faster than with either EspFU or TOCA1 individually (Fig. 8B). We also utilized EspFU at an even lower concentration, 0.2 nm, which by itself did not accelerate the time for the polymerization reaction to reach half of the maximal F-actin concentration (halftime; Fig. 8C). Nevertheless, this concentration of EspFU, in combination with 20 nm TOCA1, resulted in a decrease in the halftime and a measurable increase in the maximal polymerization rate (Fig. 8, B and C). A comparison of the halftimes and the maximum actin assembly rates triggered by the EspFU and TOCA1 proteins revealed that the stimulatory effects of the EspFU and TOCA1 mixtures were consistent with (but slightly higher than) those predicted from the simple addition of their individual activities (Fig. 8C). Thus, the actin nucleation-promoting activity of the N-WASP/WIP complex can be increased by the combined actions of EspFU and TOCA1.
DISCUSSION
One of the best-characterized ways in which actin polymerization is regulated involves activation of the nucleation-promoting factor N-WASP by the small GTPase Cdc42 (19). More recently, a bacterial molecule, the EHEC O157 effector protein EspFU, has been estimated to activate N-WASP with a potency that is orders of magnitude higher than Cdc42 and other individual N-WASP-binding proteins (32, 33). EspFU is a relatively simple protein: a type III translocation sequence followed by a 47-residue peptide repeated between 2 and 8 times (45). Included within each repeat is an N-terminal amphipathic helix that binds to N-WASP (32, 33) and a C-terminal proline-rich sequence that is recognized by the SH3 domains of at least 2 proteins, IRSp53 and IRTKS (13, 14). Because activation of the native N-WASP/WIP complex by Cdc42 normally requires the SH3 domain-containing F-BAR protein TOCA1 (20), we explored whether TOCA1 and other closely-related factors play a role in EspFU-mediated actin pedestal assembly.
Through a combination of protein depletion and overexpression studies, we found that TOCA1 stimulates actin pedestal assembly, whereas surprisingly its homologs FBP17 and CIP4 inhibit this process. After 3-h infections, when pedestals have typically reached steady-state in quantity, length, and intensity, the frequency of pedestal formation decreased by approximately one-third when TOCA1 was depleted from cells, while the average intensity of F-actin staining within pedestals more than doubled when TOCA1 was overexpressed. In contrast, when either FBP17 or CIP4 were depleted, the efficiency of pedestal formation increased slightly, and the average intensity of F-actin staining more than doubled. Moreover, FBP17 overexpression caused a major decrease in the efficiency of actin pedestal formation. We do not yet understand the mechanistic basis for how FBP17 (and perhaps CIP4) normally inhibits pedestal assembly, but these results are consistent with previous observations suggesting that TOCA1, FBP17, and CIP4 have different abilities to promote plasma membrane remodeling (21, 24, 25). Given that TOCA1 was the only one of these factors to have a positive effect on pedestal assembly, we further explored its function during this process.
GFP-TOCA1 localized to actin pedestals generated by EspFU, and the SH3 domain of TOCA1 was required for this recruitment. TOCA1 also bound to EspFU in an SH3 domain-dependent manner in multiple protein-protein interaction assays. A W518K substitution that disrupts SH3 function abrogated both EspFU binding in vitro and the dominant negative activity of this isolated domain in cells. Collectively, these results suggest that EspFU is capable of directly engaging TOCA1 via its SH3 domain during actin pedestal assembly.
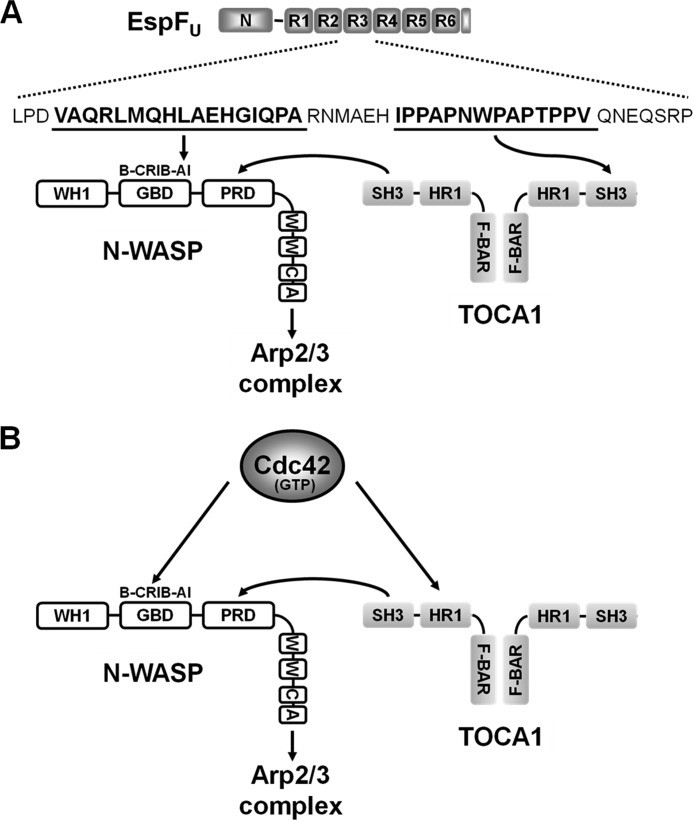
Importantly, although EspFU and Cdc42 each bind to both TOCA1 and N-WASP, they recognize different sequences in these proteins (Fig. 9, A and B). EspFU binds to the autoinhibitory portion of the N-WASP GBD, whereas Cdc42 binds to a nearby motif called the CRIB (32, 33). In addition, while EspFU interacts with the TOCA1 SH3 domain, Cdc42 recognizes its HR1 domain (20). Thus, EspFU has evolved strategies that are parallel to, but distinct from those used by Cdc42 to activate the N-WASP/WIP actin polymerization machinery.
FIGURE 9.
Model for EspFU-mediated recruitment of TOCA1 and enhancement of N-WASP activation. A, EspFU contains an N-terminal sequence (N) important for entry into mammalian cells and multiple 47-residue peptide repeats (R1-R6) that include an N-WASP-binding amphipathic helix and a proline-rich SH3-binding sequence. Interactions between an EspFU repeat and the N-WASP autoinhibitory (AI) motif within the GBD exposes the WCA region to promote Arp2/3 complex activation. The EspFU repeats also bind to the SH3 domain of TOCA1. In a simple model, one subunit of a TOCA1 homodimer may interact with EspFU, while the SH3 domain of the other subunit may enhance N-WASP activation by binding to the N-WASP proline-rich domain (PRD). Other N-WASP sequences include a WASP-homology-1 (WH1) domain, a basic (B) peptide, and a C-terminal WCA region. B, in contrast to EspFU, Cdc42 binds to the N-WASP Cdc42/Rac-interactive-binding (CRIB) motif within the GBD and the TOCA1 HR1 domain to promote actin assembly. The TOCA1 SH3 domain can facilitate N-WASP activation by binding to the PRD.
Interestingly, pulldown assays, Far-Western blots, and peptide-binding ELISAs all indicated that multiple 47-residue repeats are necessary for the physical association of EspFU with TOCA1, implying that repeat cooperativity is required for recruitment of TOCA1 during pedestal biogenesis. Cooperativity among the EspFU repeats was previously shown to result in simultaneous engagement of multiple N-WASP molecules and a dramatic enhancement of subsequent Arp2/3 binding and activation (9, 34). Taken together, these findings suggest that, in addition to their synergistic effects on N-WASP activation, the tandem EspFU repeats may provide a scaffold for recruiting additional host proteins like TOCA1 that coalesce into a multisubunit complex capable of robust actin polymerization. Notably, the number of EspFU repeat units varies among pathogenic E. coli isolates, but all known EspFU proteins contain at least two repeats (45), implying that multivalent interactions, likely with multiple binding partners, are important for actin pedestal assembly in vivo.
Consistent with previous observations indicating that N-WASP has higher levels of activity when it is engaged by multiple signaling molecules (19, 20, 37, 44), pyrene-actin assembly assays demonstrated that EspFU and TOCA1 activate the N-WASP/WIP complex in an additive manner when present at low concentrations. However, EspFU and TOCA1 are each independently capable of potently stimulating assembly when used in higher quantities. Interestingly, our observation that TOCA1 alone activates the N-WASP/WIP complex in an SH3-dependent manner indicates that it does not always require coordination with other endogenous N-WASP activators such as Cdc42 to promote actin polymerization.
The findings that the SH3 domain of TOCA1 can bind EspFU as well as activate N-WASP might be interpreted to mean that simultaneous interactions of TOCA1 with EspFU (to promote recruitment) and N-WASP (to promote activation) does not occur. However, F-BAR domains like the one present in TOCA1 dimerize, so native TOCA1 might harbor two (or more) SH3 domains that could promote coincident binding to EspFU and N-WASP (Fig. 9A). It is also possible that different cellular conditions dictate whether TOCA1 is bound to EspFU, N-WASP, both proteins, or neither protein within the pedestal. Finally, it is important to note that the proline-rich sequences within EspFU are additionally recognized by the SH3 domains of IRSp53 and IRTKS, I-BAR proteins that mediate the recruitment of EspFU to the EHEC transmembrane receptor, Tir (13, 14). EspFU has also been reported to bind the SH3 domain of cortactin (46), another actin-associated factor. Thus, with numerous potential host cell targets, the mechanisms by which the EspFU repeat region organizes a complex signaling platform during actin pedestal formation are only beginning to be elucidated.
Our preliminary investigations into how the SH3 domains from TOCA1 and FBP17, as well as IRSp53 and IRTKS, bind to EspFU and activate the N-WASP/WIP complex have begun to reveal some of the similarities and differences between the F-BAR and I-BAR subclasses of the BAR superfamily of membrane-deforming proteins. Interestingly, a monomeric form of the proline-rich EspFU peptide binds to the recombinant SH3 domains of IRTKS and (to an even greater extent) IRSp53, but not to TOCA1 or FBP17. Thus, there appears to be a fundamental difference in the way in which the SH3 domains from IRSp53/IRTKS and TOCA1 are recognized by EspFU. The SH3 domains from the I-BAR proteins also are slightly better than the SH3 domains from the F-BAR proteins at activating the N-WASP/WIP complex in vitro. However, it is not yet clear if such subtle differences are manifested in any significant effects on actin assembly in cells.
Along with activating N-WASP/WIP, TOCA1 can also bind and deform membranes via its F-BAR domain, which recognizes charged phospholipids (21–23). In fact, membrane binding by TOCA1 may contribute to its ability to activate N-WASP (30). Interestingly, the concave-shaped F-BAR dimer induces membrane invagination, in contrast to the convex membrane binding I-BAR domains of IRTKS and IRSp53 that induce protrusive structures (47). How these opposing activities might influence the morphology of the actin pedestal also remains to be determined.
Bacterial comandeering of multiple host factors to promote actin-based motility has also been demonstrated for Shigella, which recruits both N-WASP and TOCA1 during actin tail assembly (31). In that experimental system, TOCA1 depletion reduces the average length of the actin tail, suggesting that TOCA1 facilitates tail elongation (31). However, the Shigella N-WASP activator, IcsA, does not bind TOCA1. Instead, TOCA1 recruitment requires a second unidentified Shigella effector protein that functions by an unknown mechanism. Thus, in contrast to a multifaceted strategy for N-WASP activation by Shigella, our results indicate that the mechanism of actin pedestal assembly driven by N-WASP and TOCA1 relies solely upon the versatile EHEC effector protein EspFU. Further characterizations of EspFU and its host protein targets will undoubtedly continue to shed light on how cells and pathogens control the actin assembly machinery.
Acknowledgments
We thank Henry Ho and Marc Kirschner for TOCA1 plasmids and Pietro DeCamilli for the GFP-FBP17 plasmid.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grants R01-AI46454 (to J. M. L.) and R01-GM59609 (to M. D. W.).
- F-actin
- filamentous-actin
- EHEC
- enterohemorrhagic Escherichia coli
- I-BAR
- inverse-BAR
- GBD
- GTPase binding domain
- WCA
- WH2-connector-acidic
- TOCA
- transducer of Cdc42-dependent actin assembly
- F-BAR
- FCH-Bin-Amphiphysin-Rvs.
REFERENCES
- 1. Stevens J. M., Galyov E. E., Stevens M. P. (2006) Actin-dependent movement of bacterial pathogens. Nat. Rev. Microbiol. 4, 91–101 [DOI] [PubMed] [Google Scholar]
- 2. Haglund C. M., Welch M. D. (2011) Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J. Cell Biol. 195, 7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hayward R. D., Leong J. M., Koronakis V., Campellone K. G. (2006) Exploiting pathogenic Escherichia coli to model transmembrane receptor signaling. Nat. Rev. Microbiol. 4, 358–370 [DOI] [PubMed] [Google Scholar]
- 4. Kaper J. B., Nataro J. P., Mobley H. L. (2004) Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140 [DOI] [PubMed] [Google Scholar]
- 5. Spears K. J., Roe A. J., Gally D. L. (2006) A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol. Lett. 255, 187–202 [DOI] [PubMed] [Google Scholar]
- 6. Dean P., Maresca M., Kenny B. (2005) EPEC's weapons of mass subversion. Curr. Opin. Microbiol. 8, 28–34 [DOI] [PubMed] [Google Scholar]
- 7. Garmendia J., Frankel G., Crepin V. F. (2005) Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73, 2573–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Campellone K. G. (2010) Cytoskeleton-modulating effectors of enteropathogenic and enterohaemorrhagic Escherichia coli: Tir, EspFU, and actin pedestal assembly. FEBS J. 277, 2390–2402 [DOI] [PubMed] [Google Scholar]
- 9. Campellone K. G., Cheng H. C., Robbins D., Siripala A. D., McGhie E. J., Hayward R. D., Welch M. D., Rosen M. K., Koronakis V., Leong J. M. (2008) Repetitive N-WASP-binding elements of the enterohemorrhagic Escherichia coli effector EspF(U) synergistically activate actin assembly. PLoS Pathog. 4, e1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allen-Vercoe E., Waddell B., Toh M. C., DeVinney R. (2006) Amino acid residues within enterohemorrhagic Escherichia coli O157:H7 Tir involved in phosphorylation, α-actinin recruitment, and Nck-independent pedestal formation. Infect Immun. 74, 6196–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brady M. J., Campellone K. G., Ghildiyal M., Leong J. M. (2007) Enterohaemorrhagic and enteropathogenic Escherichia coli Tir proteins trigger a common Nck-independent actin assembly pathway. Cell Microbiol. 9, 2242–2253 [DOI] [PubMed] [Google Scholar]
- 12. Campellone K. G., Brady M. J., Alamares J. G., Rowe D. C., Skehan B. M., Tipper D. J., Leong J. M. (2006) Enterohaemorrhagic Escherichia coli Tir requires a C-terminal 12-residue peptide to initiate EspF-mediated actin assembly and harbours N-terminal sequences that influence pedestal length. Cell Microbiol. 8, 1488–1503 [DOI] [PubMed] [Google Scholar]
- 13. Vingadassalom D., Kazlauskas A., Skehan B., Cheng H. C., Magoun L., Robbins D., Rosen M. K., Saksela K., Leong J. M. (2009) Insulin receptor tyrosine kinase substrate links the E. coli O157:H7 actin assembly effectors Tir and EspF(U) during pedestal formation. Proc. Natl. Acad. Sci. U.S.A. 106, 6754–6759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weiss S. M., Ladwein M., Schmidt D., Ehinger J., Lommel S., Städing K., Beutling U., Disanza A., Frank R., Jänsch L., Scita G., Gunzer F., Rottner K., Stradal T. E. (2009) IRSp53 links the enterohemorrhagic E. coli effectors Tir and EspFU for actin pedestal formation. Cell Host Microbe. 5, 244–258 [DOI] [PubMed] [Google Scholar]
- 15. Campellone K. G., Robbins D., Leong J. M. (2004) EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7, 217–228 [DOI] [PubMed] [Google Scholar]
- 16. Garmendia J., Phillips A. D., Carlier M. F., Chong Y., Schüller S., Marches O., Dahan S., Oswald E., Shaw R. K., Knutton S., Frankel G. (2004) TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell Microbiol. 6, 1167–1183 [DOI] [PubMed] [Google Scholar]
- 17. Campellone K. G., Welch M. D. (2010) A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rottner K., Hänisch J., Campellone K. G. (2010) WASH, WHAMM, and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 20, 650–661 [DOI] [PubMed] [Google Scholar]
- 19. Padrick S. B., Rosen M. K. (2010) Physical mechanisms of signal integration by WASP family proteins. Annu. Rev. Biochem. 79, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ho H. Y., Rohatgi R., Lebensohn A. M., Le M., Li J., Gygi S. P., Kirschner M. W. (2004) Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell 118, 203–216 [DOI] [PubMed] [Google Scholar]
- 21. Itoh T., Erdmann K. S., Roux A., Habermann B., Werner H., De Camilli P. (2005) Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell 9, 791–804 [DOI] [PubMed] [Google Scholar]
- 22. Kamioka Y., Fukuhara S., Sawa H., Nagashima K., Masuda M., Matsuda M., Mochizuki N. (2004) A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J. Biol. Chem. 279, 40091–40099 [DOI] [PubMed] [Google Scholar]
- 23. Tsujita K., Suetsugu S., Sasaki N., Furutani M., Oikawa T., Takenawa T. (2006) Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 172, 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Toguchi M., Richnau N., Ruusala A., Aspenström P. (2010) Members of the CIP4 family of proteins participate in the regulation of platelet-derived growth factor receptor-beta-dependent actin reorganization and migration. Biol. Cell 102, 215–230 [DOI] [PubMed] [Google Scholar]
- 25. Bu W., Lim K. B., Yu Y. H., Chou A. M., Sudhaharan T., Ahmed S. (2010) Cdc42 interaction with N-WASP and Toca-1 regulates membrane tubulation, vesicle formation, and vesicle motility: implications for endocytosis. PLoS One 5, e12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kakimoto T., Katoh H., Negishi M. (2006) Regulation of neuronal morphology by Toca-1, an F-BAR/EFC protein that induces plasma membrane invagination. J. Biol. Chem. 281, 29042–29053 [DOI] [PubMed] [Google Scholar]
- 27. Bu W., Chou A. M., Lim K. B., Sudhaharan T., Ahmed S. (2009) The Toca-1-N-WASP complex links filopodial formation to endocytosis. J. Biol. Chem. 284, 11622–11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu J., Mukhopadhyay A., Craig A. W. (2011) Transducer of Cdc42-dependent actin assembly promotes epidermal growth factor-induced cell motility and invasiveness. J. Biol. Chem. 286, 2261–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee K., Gallop J. L., Rambani K., Kirschner M. W. (2010) Self-assembly of filopodia-like structures on supported lipid bilayers. Science 329, 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takano K., Toyooka K., Suetsugu S. (2008) EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 27, 2817–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leung Y., Ally S., Goldberg M. B. (2008) Bacterial actin assembly requires toca-1 to relieve N-wasp autoinhibition. Cell Host Microbe 3, 39–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng H. C., Skehan B. M., Campellone K. G., Leong J. M., Rosen M. K. (2008) Structural mechanism of WASP activation by the enterohaemorrhagic E. coli effector EspF(U). Nature 454, 1009–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sallee N. A., Rivera G. M., Dueber J. E., Vasilescu D., Mullins R. D., Mayer B. J., Lim W. A. (2008) The pathogen protein EspF(U) hijacks actin polymerization using mimicry and multivalency. Nature 454, 1005–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Padrick S. B., Cheng H. C., Ismail A. M., Panchal S. C., Doolittle L. K., Kim S., Skehan B. M., Umetani J., Brautigam C. A., Leong J. M., Rosen M. K. (2008) Hierarchical regulation of WASP/WAVE proteins. Mol. Cell 32, 426–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Campellone K. G., Leong J. M. (2005) Nck-independent actin assembly is mediated by two phosphorylated tyrosines within enteropathogenic Escherichia coli Tir. Mol. Microbiol. 56, 416–432 [DOI] [PubMed] [Google Scholar]
- 36. Campellone K. G., Webb N. J., Znameroski E. A., Welch M. D. (2008) WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell 134, 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rohatgi R., Nollau P., Ho H. Y., Kirschner M. W., Mayer B. J. (2001) Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 276, 26448–26452 [DOI] [PubMed] [Google Scholar]
- 38. Vingadassalom D., Campellone K. G., Brady M. J., Skehan B., Battle S. E., Robbins D., Kapoor A., Hecht G., Snapper S. B., Leong J. M. (2010) PLoS Pathog 6, e1001056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cameron L. A., Footer M. J., van Oudenaarden A., Theriot J. A. (1999) Motility of ActA protein-coated microspheres driven by actin polymerization. Proc. Natl. Acad. Sci. U.S.A. 96, 4908–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goldberg M. B., Theriot J. A. (1995) Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc. Natl. Acad. Sci. U.S.A. 92, 6572–6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loisel T. P., Boujemaa R., Pantaloni D., Carlier M. F. (1999) Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401, 613–616 [DOI] [PubMed] [Google Scholar]
- 42. Alto N. M., Weflen A. W., Rardin M. J., Yarar D., Lazar C. S., Tonikian R., Koller A., Taylor S. S., Boone C., Sidhu S. S., Schmid S. L., Hecht G. A., Dixon J. E. (2007) The type III effector EspF coordinates membrane trafficking by the spatiotemporal activation of two eukaryotic signaling pathways. J. Cell Biol. 178, 1265–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aitio O., Hellman M., Kazlauskas A., Vingadassalom D. F., Leong J. M., Saksela K., Permi P. (2010) Recognition of tandem PXXP motifs as a unique Src homology 3-binding mode triggers pathogen-driven actin assembly. Proc. Natl. Acad. Sci. U.S.A. 107, 21743–21748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carlier M. F., Nioche P., Broutin-L'Hermite I., Boujemaa R., Le Clainche C., Egile C., Garbay C., Ducruix A., Sansonetti P., Pantaloni D. (2000) GRB2 links signaling to actin assembly by enhancing interaction of neural Wiskott-Aldrich syndrome protein (N-WASp) with actin-related protein (ARP2/3) complex. J. Biol. Chem. 275, 21946–21952 [DOI] [PubMed] [Google Scholar]
- 45. Garmendia J., Ren Z., Tennant S., Midolli Viera M. A., Chong Y., Whale A., Azzopardi K., Dahan S., Sircili M. P., Franzolin M. R., Trabulsi L. R., Phillips A., Gomes T. A., Xu J., Robins-Browne R., Frankel G. (2005) Distribution of tccP in clinical enterohemorrhagic and enteropathogenic Escherichia coli isolates. J. Clin. Microbiol. 43, 5715–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cantarelli V. V., Kodama T., Nijstad N., Abolghait S. K., Nada S., Okada M., Iida T., Honda T. (2007) Tyrosine phosphorylation controls cortactin binding to two enterohaemorrhagic Escherichia coli effectors: Tir and EspFu/TccP. Cell Microbiol. 9, 1782–1795 [DOI] [PubMed] [Google Scholar]
- 47. Saarikangas J., Zhao H., Pykäläinen A., Laurinmäki P., Mattila P. K., Kinnunen P. K., Butcher S. J., Lappalainen P. (2009) Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 19, 95–107 [DOI] [PubMed] [Google Scholar]