Background: The low density lipoprotein receptor-related protein 1 (LRP1) is a transforming growth factor β (TGF-β) receptor in ovarian cells.
Results: GULP is an adapter to LRP1 and mediates TGF-β signaling in signaling-competent early endosomes.
Conclusion: GULP positively regulates TGF-β signaling in ovarian cells.
Significance: GULP is poorly expressed in ovarian cancer cells and is a target for TGF-β-mediated growth inhibition.
Keywords: Adapter Proteins, Cancer Biology, Cell Biology, Cell Growth, Cell Invasion, Cell Surface Receptor, Confocal Microscopy, Transforming Growth Factor β (TGFβ)
Abstract
Transforming growth factor β (TGF-β) is a key regulatory molecule with pleiotropic effects on cell growth, migration, and invasion. As a result, impairment of proper TGF-β signaling is central to tumorigenesis and metastasis. The TGF-β receptor V (TGFBRV or LRP1) has been shown to be responsible for TGF-β-mediated cell growth inhibition in Chinese hamster ovary (CHO) cells. The LRP1 adapter protein GULP mediates internalization of the various LRP1-specific ligands, and we hypothesize that GULP acts as a novel regulator of TGF-β signaling in ovarian cells. CHO cells that overexpress exogenous GULP (FL) demonstrate enhancement in growth inhibition, migration, and invasion from TGF-β treatment, whereas cells that lack GULP (AS) show impairment of growth inhibition and decreased migration and invasion. The enhanced TGF-β response in FL cells was confirmed by a prolonged TGF-β-induced SMAD3 phosphorylation, whereas a shortening of the phosphorylation event is observed in AS cells. Mechanistically, the presence of GULP retains the TGF-β in a signaling-competent early endosome for enhanced signaling. To address this mechanism in a physiological setting, TGF-β insensitive ovarian adenocarcinoma cells (HEY) have a very low GULP expression level, similar to the observation made in a wide selection of human ovarian adenocarcinomas. Transfection of GULP into the HEY cells restored the TGF-β responsiveness, as measured by SMAD3 phosphorylation and impairment of cell growth. Because GULP expression positively regulates TGF-β signaling leading to growth inhibition, this may represent an attractive target to achieve TGF-β responsiveness in ovarian cells.
Introduction
Transforming growth factor-β (TGF-β) is a family of structurally homologous dimeric polypeptides consisting of three mammalian isoforms: TGF-β1, TGF-β2, and TGF-β3 (1, 2). TGF-βs are involved in the regulation of multiple important biological processes, including cell proliferation, cell differentiation, and extracellular matrix modification, leading to epithelial-mesenchymal transition (EMT),3 migration, and invasion (3, 4). The regulatory roles of TGF-βs are critical to the progression of many vital human diseases, including fibrosis, atherosclerosis, and cancer (5–9). The role of TGF-βs in cancer is sophisticated. In normal cells, TGF-β acts as a tumor suppressor by promoting cell growth arrest and apoptosis (10) as well as by preventing cell immortalization (11). However, many cancer cells from various origins have gained resistance to the growth-inhibitory effect of TGF-β with mutations in the mediators of the TGF-β signaling pathway along with increased production of TGF-β. Moreover, TGF-β is capable of triggering EMT in various carcinomas making them more invasive by promoting extracellular matrix remodeling and eventually metastasis (3, 5). The TGF-β signaling pathway thus acts as a key regulator in tumorigenesis and, therefore, as a potential target for chemotherapy (7, 10).
The general mechanism of TGF-β signaling begins with the binding of ligand to the type III TGF-β receptor or type II receptor (TGF-β·RII). Upon binding of TGF-β, TGF-β·RII recruits, trans-phosphorylates, and activates the type I receptor (TGF-β·RI) (12–15). After activation, TGF-β·RI then transduces the signal through a class of signaling mediators known as SMADs (16). Receptor-regulated SMADs (R-SMADs), such as SMAD-2 and SMAD-3, are phosphorylated by TGF-β·RI and subsequently bind to SMAD-4 (co-SMAD) (16, 17). The heterodimerization of R-SMAD and co-SMAD forms a transcription activator complex, which is translocated to the nucleus and regulates the transcription of various target genes (18, 19). Also, SMAD-6 and -7 can inhibit the formation of a transcription activator complex by competing with co-SMAD for binding with R-SMADs (20).
The signal of TGF-β could also be down-regulated or terminated by numerous mechanisms other than the action of antagonistic SMAD-6 or -7. Phosphatases and ubiquitin ligases are capable of deactivating and down-regulating TGF-β·RI through dephosphorylation and ubiquitination (21, 22). Endocytic adapter proteins also regulate the signal by affecting the rate of internalization and/or expression of the receptors on the cell surface (23, 24). The discovery of the SMAD anchor for receptor activation protein (SARA) also provided another level of complexity to the mechanism of TGF-β signaling. SARA is a FYVE domain protein that localizes to the early endosome and recruits SMADs to the internalized receptor complex for phosphorylation and activation (25–27). Moreover, the SMAD complex is also susceptible to dephosphorylation and ubiquitin-mediated degradation, which ultimately terminates the signal (28–30). Other factors may also help transduce and regulate the signal of TGF-β (31, 32). The type V receptor (TGF-β-RV) was shown to be identical to the low density lipoprotein receptor-related protein-1 (LRP1) (33). LRP1 has been shown to affect the signaling of TGF-β in Chinese hamster ovary (CHO) cells, where LRP1-deficient CHO cells gained resistance toward TGF-β-induced cell growth arrest (34). Many other groups have also demonstrated the importance of LRP1 in TGF-β signaling events (35–39). These were important observations demonstrating that LRP1 facilitates TGF-β signaling because LRP1 was only originally classified as a scavenger receptor facilitating degradation through receptor-mediated ligand endocytosis (40).
We have previously described the effect of the phosphotyrosine binding domain (PTB)-containing engulfment adapter protein (GULP), an adapter protein of LRP1, on cellular lipid homeostasis by modulating the endocytosis of LRP1 and LRP ligands (41). In this study, we hypothesize that GULP is a key regulator of LRP1-mediated TGF-β signaling and thereby plays an important role in modulating TGF-β signals in ovarian cells. To support this, microarray studies have shown that GULP is significantly down-regulated in most ovarian adenocarcinomas (42, 43). Hereby, we present evidence that GULP is required in ovarian cells to maintain their sensitivity toward TGF-β-induced cell growth arrest, demonstrating the important role that GULP may perform in ovarian cancer progression.
MATERIALS AND METHODS
Cell Culture and Transfection
The CHO LR73 cells were a gift from Dr. Kodi Ravichandran (University of Virginia) and were described previously (41, 44). Multiple lines of stable transfection of GULP-overexpressing CHO cells (FL), cells overexpressing only the PTB of GULP, and GULP-knocked down CHO cells (AS) were generated and maintained as described previously (41). We deliberately chose moderate expression FL clones (between 1.7- and 3.8-fold endogenous protein expression), and the expression level of GULP in FL and AS cells was validated prior to each set of experiments by Western blot. CHO cells with a deficiency in LRP1 (13-5-1) were acquired from Dr. David Fitzgerald through Dr. Zemin Yao (45, 46). 13-5-1 cells were authenticated by Pseudomonas exotoxin treatment (46). Mouse embryonic fibroblasts (MEF-1), MEF cells genetically deficient in gulp (GULP KO), GULP KO MEF cells with GULP expression reconstituted, and LRP-deficient MEF (MEF-2) were obtained from Dr. Kodi Ravichandran (University of Virginia). HEY and SKOV3 ovarian adenocarcinomas were originally obtained from ATCC (Manassas, VA). Morphology, expression levels, and signaling mechanisms were routinely assessed to reconfirm the phenotype. 13-5-1, HEY, and SKOV3 cells were all cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% penicillin/streptomycin. For transient transfection in HEY cells, cells were grown to 60% confluence and transfected with expression vectors encoding yellow fluorescent protein (YFP)-conjugated GULP or the fluorescence protein only in antibiotic-free DMEM with 5% FBS using FuGENE HD (Roche Applied Science; according to the manufacturer's instructions). After 48 h of transfection, cells were used for MTT assays and Western blotting. For all quantitative assays, statistically significant differences were determined by using Student's t test.
Reverse Transcription-PCR
mRNA was isolated and reverse transcribed into cDNA using the RNeasy® minikit (Qiagen) according to the manufacturer's instructions. Transcribed cDNA were then amplified by PCR up to 35 cycles.
Western Blotting
Cells were cultured in various amounts of FBS and stimulated with or without 0.2 nm TGF-β for different periods of time as described in the figure legends. Cells were lysed in lysis buffer containing phosphatase inhibitors (Sigma, BioShop). 25 μg of protein from each cell lysate was separated using SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was then incubated with specific primary antibodies (phospho-SMAD3, SMAD3, GULP, GAPDH, LRP1, Dab2, fluorescent protein, p21, p15, TGF-β·RI, and anti-tubulin) and subsequently incubated with specific horseradish peroxidase-conjugated secondary antibodies. The immunoreactive proteins were detected using the ECL system (Thermo Scientific). Quantification was performed using the built-in software from the gel imager (Alpha Innotech). Scrambled RNA or siRNA (10 nmol) was transfected into cells with HiPerfect transfection reagent (Qiagen) for 16–24 h before treatment with TGF-β, Western blotting, and cell proliferation assays.
Cell Proliferation Assays
1 × 105 cells from each cell line were plated in 6-cm dishes with 5% FBS and stimulated with or without 0.2 nm TGF-β on the next day. After 48 h of stimulation, cells were resuspended with 0.25% trypsin (Wisent) and counted using 0.4% trypan blue (Invitrogen) under a light microscope (Leica) with a hemacytometer. The percentage of cells susceptible to TGF-β-induced growth inhibition was calculated by the formula, (1 − (TGF-β-treated/non-treated control)) × 100. Apoptosis of cells was measured by using the Annexin V-Cy3 Apoptosis kit from Sigma according to the manufacturer's instructions.
MTT Assays
CHO and 13-5-1 cells were resuspended and plated into 96-well plates at a concentration of 5,000 cells/100 μl in culture medium with 5% FBS. Cells were stimulated or not with 0.2 nm TGF-β for 3 days and then incubated with 0.5% MTT in PBS for 4 h. The dye was extracted with Sorenson's buffer and DMSO, and the intensity was measured using a Packard microplate reader at 590 nm (47). HEY cells were plated at a concentration of 3,000 cells/100 μl in culture medium with 5% FBS. After 48 h of transfection, HEY cells were stimulated or not with 0.2 nm TGF-β in serum-free DMEM for 3 days. The rest of the experiments were conducted in a similar manner as described above. The percentage of cells susceptible to TGF-β-induced growth inhibition was calculated by the formula, (1 − (TGF-β-treated/non-treated control)) × 100.
Cell Migration/Invasion Assays
Cells were resuspended in serum-free medium and plated into each of the upper wells of the 24-multiwell insert system (BD Biosciences) with 5 × 104 cells in 300 μl with or without 0.2 nm TGF-β. 500 μl of full growth medium was added to the lower well to allow migration. After 24 h of incubation, media were removed, and cotton-tipped swabs were used to remove the non-migratory cells on the insert. The cells were then fixed with 4% paraformaldehyde in PBS for 15 min and subsequently stained with 0.2% crystal violet in PBS for 10 min. After staining, the insert was washed three times with distilled water, and the crystal violet was extracted by incubating with 200 μl of 10% acetic acid. The amount of migration was quantified by measuring the optical density at 590 nm using a microplate reader (Packard). Invasion assays were conducted in a similar manner; however, Matrigel (BD Biosciences) was coated onto the insert 24 h prior to the experiment following the manufacturer's specifications.
Gelatin Zymography
WT, FL, and AS cells were treated with 0.2 nm TGF-β or not for 24 h in AMEM with 2% FBS. The media were collected and concentrated down to 200 μl using an Amicon® Ultra-4 centrifugal unit (Millipore). Equal volumes of the concentrated media were loaded onto a 10% acrylamide gel co-polymerized with 1 mg/ml gelatin (Fisher) in reducing agent-free SDS-loading buffer. The gel was washed with buffer (2.5% Triton X-100, 50 mm Tris, 5 mm CaCl2, 1 μm ZnCl2, pH 7.4) for 1 h at room temperature and then incubated overnight at 37 °C with renaturing buffer (washing buffer without Triton X-100). The gel was stained with Coomassie Blue and destained. Matrix metallopeptidase-9 (MMP-9) activities were shown as opaque bands on a blue background. The gel was scanned using a gel imager from Alpha Innotech, and densitometry analyses were conducted using the built-in software.
Wound Healing Assays
Cells were grown in 6-well plates with 5% FBS until confluent. A scratch was created using a P-200 pipette tip, and then the cells were washed twice with serum-free medium and incubated with or without 0.2 nm TGF-β. After 24 h, three randomly selected fields were acquired using an inverted bright field microscope with a 4× objective. The scratch area was quantified by using the Image Pro-Plus software. The results were presented as percentage wound healing with the equation, % wound healing = (1 − (wound area at t24 h/wound area at t0 h)) × 100 (48).
Luciferase Assays
Cells were transiently cotransfected with 0.5 μg of 3TP-Luc and β-galactosidase reporter for 24 h and then serum-starved for 24 h. Cells were then stimulated with 0.2 nm TGF-β or not until the next morning. The luciferase activity was measured using a luminometer (EG&G Berthold) and normalized to the β-galactosidase activity by measuring the optical density at 420 nm with a plate reader (BioTek).
Pseudomonas Exotoxin A Cytotoxicity Assay
1 × 106 cells from each cell line were plated in 6-cm dishes with full growth medium and stimulated with or without 100 ng/ml Pseudomonas exotoxin A (PEA) (Sigma) on the next day. After 24 h of stimulation, cells were resuspended with 0.25% trypsin and counted using 0.4% trypan blue under a light microscope.
GST-RAP Expression, Purification, and Cy5 Labeling
The expression vector of GST-RAP fusion protein was expressed and purified from Escherichia coli (DH5α) as described by Herz et al. (49). The purified GST-RAP proteins were labeled with Cy5 (GE Healthcare) according to the manufacturer's protocol.
Fluorescent Labeling of Lipoproteins
βVLDL and LDL were prepared by ultracentrifugation. Both lipoproteins were diluted to 3 mg/ml with PBS containing 10% lipoprotein-deficient serum and 1% penicillin/streptomycin. Prepared lipoprotein samples were labeled with fluorescent lipophilic tracer 1,1′-dioctadecyl-3,3,3′,3′,-tetramethyindodicarbocyanine perchlorate (DiD; Invitrogen) by microinjection using a insulin syringe.
Ligand Binding and Ligand Degradation Assays
LRP ligands (α2M (Sigma) and TGF-β (Leinco)) were labeled with Na125I according to Kiss et al. (41). Cell surface ligand binding experiments and ligand degradation assays were performed as described previously (41).
PIP Strips and Efflux Assays
Commercial lipid blots on nitrocellulose are available (PIP Strips, Invitrogen) and were used similarly to a Western blot (blocking, incubation with GULP protein, washing, detection of bound GULP by primary/secondary antibodies, and ECL detection). Efflux assays were performed as described previously (41).
Fluorescence Microscopy
AS and FL cells were seeded on 35-mm glass bottom MatTek dishes overnight. The cells were transfected with YFP-Rab5 or YFP-Rab7 with FuGENE HD for 24 h. After a 15-min incubation with fluorescently labeled LRP1 ligands (Cy5-RAP, Cy5-TGF-β, DiD-βVLDL, and DiD-LDL), the samples were washed three times with PBS and replaced with HEPES-buffered medium, pH 7.4. Images were collected on a Zeiss LSM-510 Meta laser-scanning microscope with either a 63× or a 40× oil immersion lens. Cy5 and DiD-labeled ligands were excited at 633 nm, and YFP was excited at 514 nm.
RESULTS
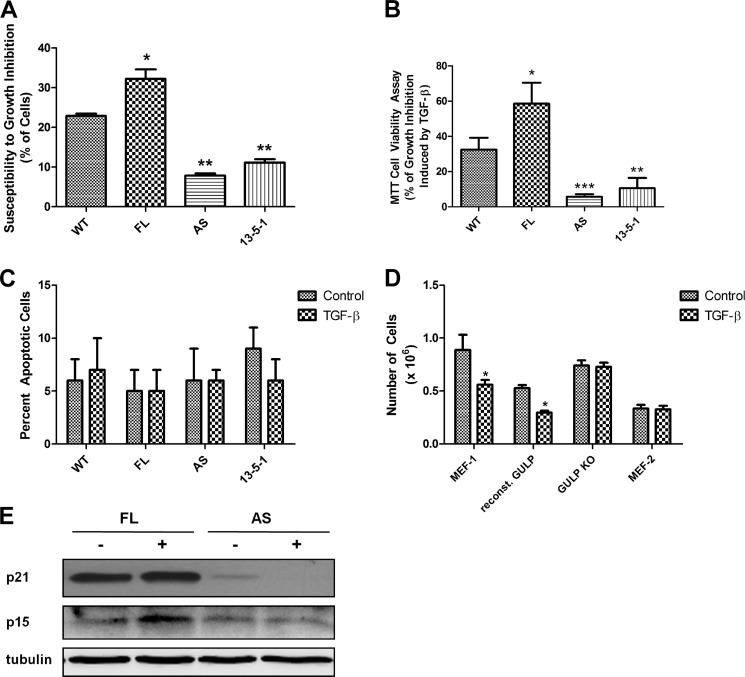
One of the most important downstream effects of TGF-β signaling is the promotion of cell growth arrest by means of inhibition of cell proliferation and the induction of apoptosis. The ability of TGF-β to promote growth arrest has drawn vast attention from researchers because it implies a potential target for cancer treatment. Moreover, many carcinomas have been shown to gain resistance to the TGF-β effect, making it an important pathway to study for carcinogenesis. To assess the effect of differential GULP expression level on TGF-β-induced cell growth arrest, we utilized four different cell lines: wild-type CHO cells (WT), CHO with overexpressed full-length GULP (FL), CHO with knocked down GULP (AS), and LRP1 null CHO cells (13-5-1). Of all the cells treated with TGF-β, FL cells had the strongest inhibitory effect (>30%), and WT cells had a moderate inhibition (>20%) (Fig. 1A). On the other hand, 13-5-1 cells and AS cells both had a weak growth inhibition effect (∼10% each). To further assess the effect of GULP on TGF-β-induced cell growth arrest, we performed an MTT cell viability assay. The chemical MTT is reduced to a purple color in living cells, proportional to the number of cells, and this can be quantified by a colorimetric assay. FL cells were growth-inhibited by ∼59%, compared with WT cells at ∼33% (Fig. 1B). 13-5-1 (10%) and AS (7%) cells showed little growth inhibition in comparison. The results show that GULP-overexpressing cells were more sensitive to TGF-β-induced cell growth arrest, whereas the GULP-knocked down cells were more resistant. To determine if the reduced cell numbers were due to increased apoptosis, we quantified the number of apoptotic cells. All cell types were seeded at the same density and allowed to grow in the absence or presence of TGF-β. Then cells were stained by a vital fluorescent dye (which stains live cells) and Annexin V (which binds apoptotic cells). There was no significant increase in apoptotic cells in any cell type (Fig. 1C), suggesting that TGF-β-induced apoptosis is not a major contributor to reduced cell number. Thus, our data clearly indicate that GULP acts as a positive regulator of TGF-β-mediated cell growth arrest in CHO cells. To confirm that this effect also occurs in other cell types, we performed the growth assay in MEFs. In parallel with the CHO experiment, the experiment was performed with parental MEFs (MEF-1), MEF cells with GULP knocked out (GULP KO), GULP KO MEFs with reconstituted GULP, and LRP1-deficient MEFs (MEF2). The results show that decreased GULP prevents a TGF-β response, whereas GULP expression promotes a TGF-β response (Fig. 1D), corroborating the CHO experiments and demonstrating that GULP is a requirement for TGF-β-mediated growth inhibition. To dissect the mechanism of the GULP effect on cell growth inhibition, we performed Western blots of p21Cip1 and p15ink4b. These cell cycle regulators have previously been shown to play a critical role in relaying TGF-β family member cell proliferation inhibition in human keratinocytes, astrocytes, and hepatocatcinomas (50–54). Western blot analysis revealed in FL cells that TGF-β induced p21 and p15 expression, whereas in AS cells, p15 and p21 expression levels were very low and not induced by TGF-β (Fig. 1E), indicating that TGF-β does induce p15 and p21 expression and cell growth inhibition in a GULP-dependent manner in our model system.
FIGURE 1.
The growth-inhibitory effect of TGF-β in CHO cells correlates to the expression level of GULP. A, control CHO cells (WT), CHO cells overexpressing GULP (FL), CHO cells with reduced GULP expression (AS), and LRP1-deficient CHO cells (13-5-1) were treated with TGF-β or not for 48 h. Cells were resuspended and counted using a hemacytometer. Values are expressed as the percentage decrease of the number of cells between the conditions with and without TGF-β. Results presented here are the mean ± S.D. (error bars) of three separate experiments. *, p < 0.003; **, p < 0.0001 in comparison with WT. B, WT, FL, AS, and 13-5-1 cells were stimulated without or with TGF-β for 72 h before cell growth was measured by a cell viability assay (MTT). Values were computed and presented as percentage of growth inhibition induced by TGF-β as described under “Materials and Methods.” Results shown here are the mean ± S.D. of two separate experiments, each performed in quadruplicate. *, p < 0.03; **, p < 0.01; ***, p < 0.002 in comparison with WT. C, WT, FL, AS, and 13-5-1 cells were stimulated without or with TGF-β for 24 h, and then an apoptosis kit assay (Sigma) was performed according to manufacturer's instructions. Results are presented as the mean percentage of total cells ± S.D. that stain positive for Annexin V of five separate plates of at least 200 cells/plate for each condition. D, control MEF cells (MEF-1), MEF cells with a genetic deficiency of GULP (GULP KO), GULP KO cells with reconstituted GULP expression (reconst. GULP), and LRP1-deficient MEF cells (MEF-2) were treated without or with TGF-β for 48 h, and then cells were counted as in A. *, p < 0.01 comparing TGF-β-treated and -untreated conditions. E, FL or AS cells were treated without or with TGF-β for 16 h; cell lysates were harvested; and Western blots of p21Cip1, p15ink4b, and tubulin were performed.
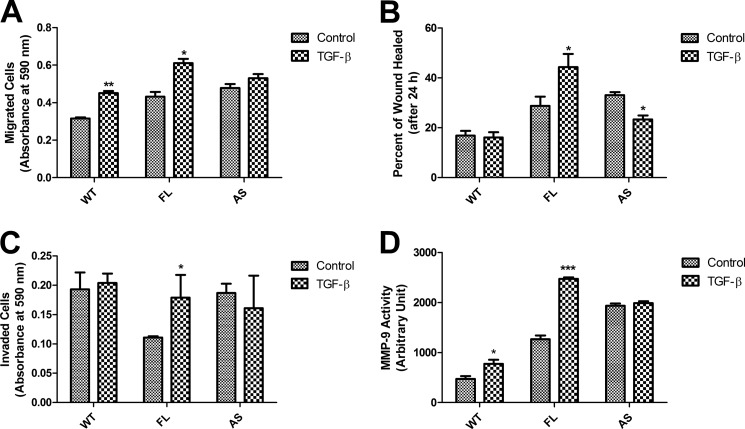
TGF-β also regulates the motility of cells. It has been shown that CHO cells adopted a more metastatic phenotype upon the presence of excessive TGF-β (55). We examined the migration ability of the different cell lines in multiwell inserts in response to TGF-β. FL cells showed the most migration in response to TGF-β, with AS cells showing the least TGF-β-induced cell migration, in proportion to the level of expressed GULP (Fig. 2A). We also assessed the GULP effect on TGF-β-mediated cell migration using the wound healing assay (48). The changes in the area of the wound were measured, computed, and represented as percentage wound healing described under “Materials and Methods.” Results were plotted showing the TGF-β-treated and untreated control cells. FL cells showed the most wound healing in response to TGF-β treatment, whereas WT cells showed a somewhat neutral response toward TGF-β treatment (Fig. 2B, supplemental Fig. 1). However, AS cells showed a strong decrease in percentage of wound healing in response to TGF-β compared with WT cells. Further, we examined the invasion ability of the different cell lines in multiwell inserts that had been filled with Matrigel in response to TGF-β. Cells must secrete enzymes to digest the Matrigel in order to penetrate it and access the well. FL cells showed the most invasion in response to TGF-β, with AS cells showing the least invasion, in proportion to the level of expressed GULP (Fig. 2C). As a secondary measure, we performed a gelatin zymography assay. The medium from cells grown in the absence or presence of TGF-β was collected and then run on a native polyacrylamide gel with gelatin. After a period of incubation, the cell-secreted MMP-9 digests the gelatin and produces a cloudy white area at 50 kDa (the molecular mass of MMP-9) on the polyacrylamide gel. For the purposes of quantification, we scanned the gels, inverted the color, and determined the intensity and area of digestion by densitometry. FL cells showed the most induction of MMP-9 activity with TGF-β treatment, whereas AS cells showed no difference, as compared with WT cells (Fig. 2D).
FIGURE 2.
Overexpression of GULP in CHO cells leads to enhanced cell motility and invasion in response to TGF-β treatment. A, transwell migration assays were conducted using WT, FL, and AS cells. Results show migration (as measured by crystal violet color intensity) for TGF-β-untreated (Control) and -treated cells (TGF-β). Data presented here are the mean ± S.D. (error bars) of three experiments, each done in triplicate. *, p < 0.003; **, p < 0.0003 in comparison with the untreated condition. B, wound healing assays were conducted using WT, FL, and AS cells. Cells were grown to confluence and serum-starved for 24 h, and then a scratch of ∼1 mm was introduced through the middle of the plate. Cells were then treated without or with TGF-β for 24 h, and the percentage of area that TGF-β-induced cells grow into the scratch (wound healing) was measured and computed as described under “Materials and Methods.” Results presented here are the mean ± S.D. of three experiments, each done in triplicate. *, p ≤ 0.001 in comparison with the untreated condition. C, Transwell invasion assays were conducted using WT, FL, and AS cells without and with TGF-β treatment for 24 h. Cell invasion (the number of cells that crossed the Matrigel barrier) was measured by the intensity of crystal violet staining in the well. Data presented here were the mean ± S.D. of two experiments, each done in triplicate. *, p ≤ 0.004 in comparison with the untreated condition. D, gelatin zymography experiments were performed on media from WT, FL, and AS cells treated without or with TGF-β for 24 h. To quantify the extent of active MMP-9 secreted, densitometry was performed on the MMP-9 digestion band in the absence and presence of TGF-β and presented as the mean ± S.D. of three separate gelatin zymography experiments. *, p < 0.03; ***, p < 0.0003.
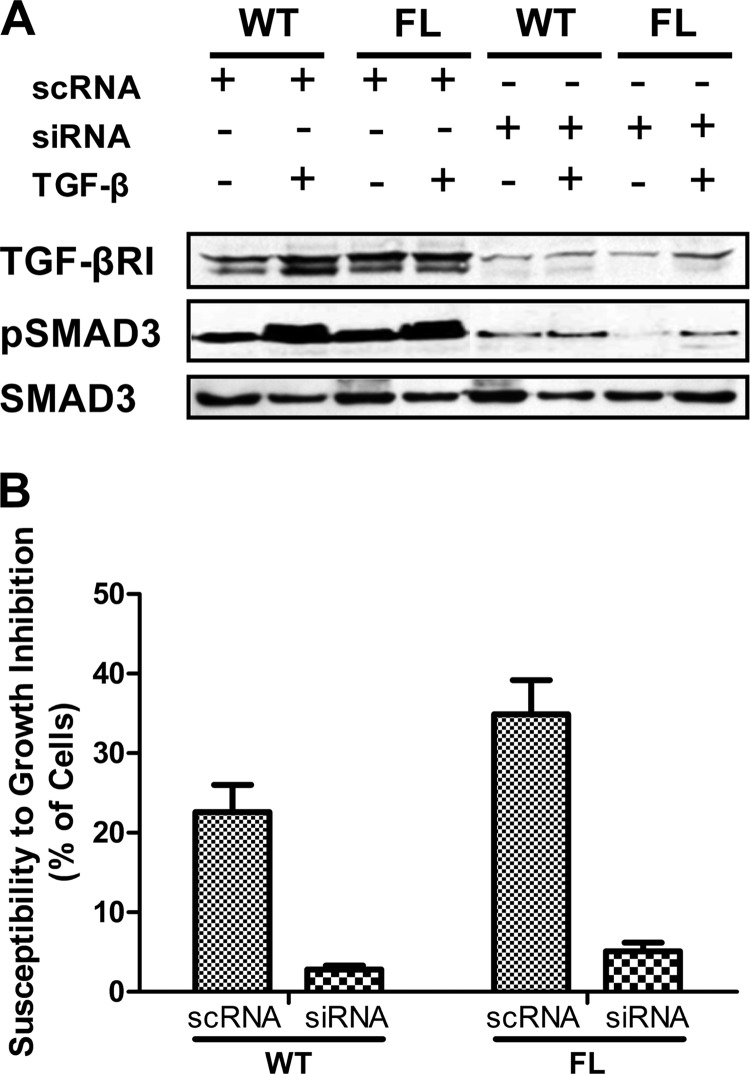
TGF-β signaling is transduced through the phosphorylation of R-SMADs by the TGF-β·RI. To determine the involvement of TGF-β·RI, we used siRNA to silence TGF-β·RI and then detect if TGF-β and GULP can still signal. The results show that knockdown of TGF-β·RI (Fig. 3A) prevents GULP from mediating TGF-β-induced phospho-SMAD3 formation in WT and FL cells (Fig. 3A). Furthermore, silencing of TGF-β·RI prevents TGF-β-dependent growth inhibition in both WT and FL cells (Fig. 3B). These results demonstrate that LRP1 is probably acting as a co-receptor to the TGF-β·RI.
FIGURE 3.
GULP signaling involves TGF-β·RI. A, WT and FL cells were transfected with scrambled RNA or siRNA specific for TGF-β·RI for 16 h and then were treated without or with TGF-β for 60 min. Total cell lysates were collected and analyzed by Western blot using anti- TGF-β·RI, anti-pSMAD3, and anti-SMAD antibodies. TGF-β·RI expression is efficiently prevented by siRNA, which also prevents pSMAD3 formation. B, a growth inhibition experiment (similar to Fig. 1) was performed. WT and FL cells were transfected with scrambled RNA or siRNA specific for TGF-β·RI for 16 h and then treated without or with TGF-β for 24 h. Cells were resuspended and counted using a hemacytometer. Values are expressed as the percentage decrease of the number of cells between the conditions with and without TGF-β. Results presented here are the mean ± S.D. (error bars) of three separate experiments.
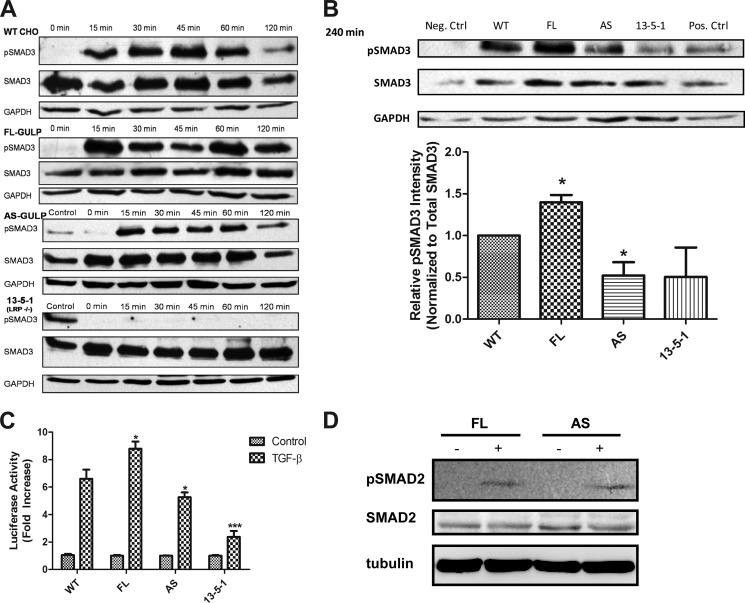
The amount of phosphorylation on SMAD3 can be used as a measure of the intensity of the TGF-β response. We quantified the amount of phosphorylation of SMAD3 in cells with differential GULP expression by Western blot. In WT cells, we see a typical TGF-β response, where the phosphorylation of SMAD3 peaks at 45 min and is back to background by 120 min (Fig. 4A). In FL cells, phosphorylation of SMAD3 remained at a high level even at 120 min. AS cells showed a time course of phosphorylation similar to WT, but SMAD3 phosphorylation was attenuated more than both FL and WT cells. In 13-5-1 cells, the phosphorylation of SMAD3 remained low. To validate this result, we ran the different samples of the same time points on a single blot and normalized to total SMAD protein expression. At the 240 min time point, AS had clearly a strongly diminished phospho-SMAD3 in comparison with WT (Fig. 4B). FL cells, on the other hand, had the strongest expression at the 240 min time point. Our observations are that overexpression of GULP sustained the phosphorylation of SMAD3, and this effect is lost when GULP was absent.
FIGURE 4.
Enhanced GULP expression results in prolonged SMAD3 phosphorylation and increased TGF-β activity. A, WT, FL, AS, and 13-5-1 cells were treated with TGF-β for the time periods indicated. Cell lysates were analyzed by Western blot using anti-pSMAD3 antibody (Cell Signaling), SMAD3 antibody (Cell Signaling), and GAPDH antibody (Santa Cruz Biotechnology, Inc.). Positive controls were included for AS and 13-5-1 cells to prove that the Western blots were detecting properly. B, WT, FL, AS, and 13-5-1 cells were stimulated with TGF-β for 240 min, and the cell lysates were analyzed by Western blot using anti-phospho-SMAD3 antibody, SMAD3 antibody, and GAPDH antibody. A representative blot is shown at the top, including a positive control. Densitometric scaling was performed to normalize the intensity of pSMAD3 to SMAD3 expression. The results were plotted as relative intensity of pSMAD3, with the intensity of WT being normalized to 1, and three independent Western blot analyses generated the mean ± S.D. (error bars). *, p ≤ 0.05. C, WT, FL, AS, and 13-5-1 cells were co-transfected with a 3TP-Lux reporter construct and a β-galactosidase expression plasmid. Cells were treated with TGF-β or not for 16 h, and luciferase assays were performed. Data were presented as -fold increase in activity of TGF-β-treated samples compared with the non-treated WT control. The experiments were repeated three times, each done in triplicate, and results presented here are the mean ± S.D. from three experiments. In comparison with the -fold increase in TGF-β activity in WT, significance is shown. *, p < 0.05; ***, p < 0.001. D, FL and AS cells were stimulated without or with TGF-β for 60 min, and the cell lysates were analyzed by Western blot using anti-pSMAD2 antibody, SMAD2 antibody, and tubulin antibody. The experiment was repeated three times, and a representative blot is shown.
Having shown that SMAD3 phosphorylation was potentiated by GULP, we next investigated whether GULP could enhance SMAD3 activity. For this, we transfected a synthetic TGF-β-responsive gene promoter construct fused to the luciferase gene (3TP-Lux) with a β-galactosidase-expressing vector (12) in the different CHO cell lines described above. The β-gal reporter was co-transfected to normalize for the difference in transfection efficiency across the four cell lines (Fig. 4C). After normalizing to the base line of each cell line, WT cells have a 6.6 ± 0.66-fold induction in reporter activity, whereas FL and AS cells have an 8.8 ± 0.53- and 5.3 ± 0.35-fold induction, respectively (Fig. 4C). 13-5-1 cells have only 2.4 ± 0.43-fold induction, which confirms previously published data showing how LRP1 is required for TGF-β induced cell growth arrest in CHO cells. After normalization to WT, FL cells showed about 133% increase in luciferase activity, whereas AS cells showed a moderate but significant (p < 0.009) decrease in luciferase activity. Collectively, our results indicate that GULP acts as positive regulator of SMAD3 phosphorylation and TGF-β transcriptional control and that blocking GULP expression significantly impairs these effects. To identify a possible role of SMAD2, we performed a similar experiment to Fig. 4A and detected phospho-SMAD2 in response to TGF-β. Both FL and AS cells showed a low but similar amount of phospho-SMAD2 (Fig. 4D), suggesting that SMAD2 does not play a major role in GULP-mediated TGF-β signaling in these cells.
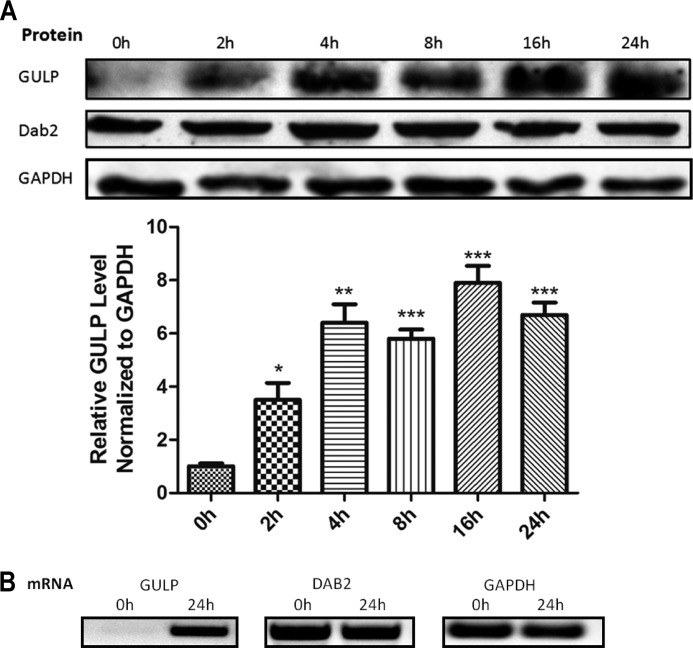
As one mechanism of how GULP may respond to TGF-β treatment, we quantified GULP protein and mRNA in response to TGF-β. WT cells were serum-starved for 24 h and then treated with TGF-β for the time period as indicated and harvested for Western blot or RT-PCR. After normalizing for GAPDH levels, the protein expression level of GULP increased with time, up to 24 h with TGF-β treatment (Fig. 5). However, the expression level of another PTB-containing adapter protein Dab2 was not stimulated by TGF-β despite its pivotal role in TGF-β signaling (56–58). Moreover, the RT-PCR also showed an induction of GULP mRNA level at the 24 h time point (Fig. 5). As a control experiment, we show that TGF-β does not significantly increase GULP expression levels in AS cells (supplemental Fig. 2A) because this might have confounded experimental observations with AS cells. Thus, our data highlight GULP as a novel TGF-β downstream target gene that acts in a positive feedback loop to promote and potentiate TGF-β signaling, leading to cell growth inhibition, cell migration, and cell invasion.
FIGURE 5.
GULP expression is inducible by TGF-β. A (top), WT cells were stimulated with TGF-β for a time course as indicated, and total cell lysates were analyzed by Western blot using anti-GULP, anti-Dab2, and anti-GAPDH antibodies. This experiment was performed three times, and a representative Western blot is shown here. A (bottom), to quantify the bands using densitometry, we measured the relative intensity of the GULP band after normalization to the intensity of GAPDH and then calculated the average ± S.D. (error bars). *, p < 0.05; **, p < 0.005; ***, p < 0.001. B, WT cells were stimulated with TGF-β for 24 h, and then mRNAs were extracted, and reverse transcription PCR was performed. Resulting cDNAs were then amplified using the primers specific for GULP, Dab2, and GAPDH for 25 cycles.
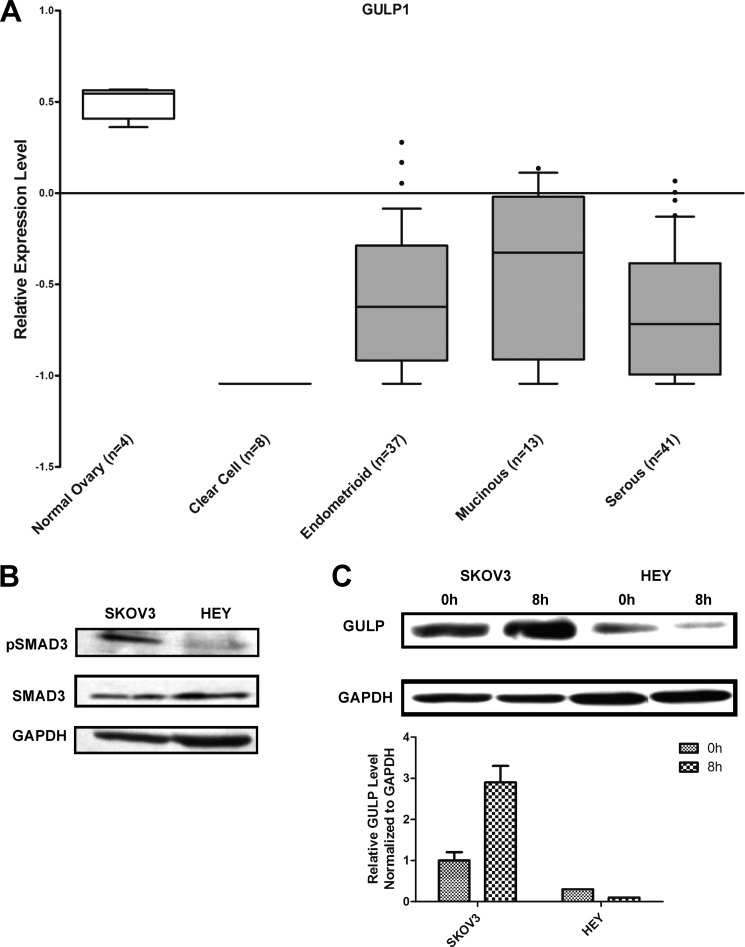
Based on our observations on cell growth inhibition, phospho-SMAD3 Western blots, and the reporter assay, we have demonstrated that GULP plays a role in the regulation of SMAD-dependent TGF-β signaling. As a result, we hypothesized that a decrease in GULP expression results in impaired TGF-β signaling, including within human cancerous tumors. To demonstrate this directly, we analyzed microarray data from a previously published data set (43) from human ovarian adenocarcinomas: clear cell, endometrioid, mucinous, and serous types. All adenocarcinoma types showed a significant decrease in GULP expression level of ∼2-fold when compared with normal tissue (Fig. 6A). We then analyzed five other data sets of ovarian adenocarcinoma and found that GULP mRNA expression was significantly reduced in all but one of them (supplemental Fig. 3). These results suggest that decreased GULP expression may be a direct or indirect cause of tumorigenesis.
FIGURE 6.
Ovarian adenocarcinoma cells have a low expression level of GULP. A, microarray data on the relative expression level of GULP from normal ovary and several different ovarian adenocarcinomas was analyzed and plotted into box plots (number of samples is shown below the bar; original data are from Schwartz et al. (43), with permission from authors). The values are displayed at a base 2 logarithmic transformation. B, SKOV3 and HEY cells were characterized in terms of TGF-β sensitivity. Total cell lysates of SKOV3 and HEY were collected after 45 min of TGF-β treatment and were analyzed by Western blot using anti-pSMAD3, SMAD3, and anti-GAPDH antibodies. C, SKOV3 and HEY cells were stimulated with TGF-β for 0 and 8 h. Total cell lysates were analyzed by Western blot using anti-GULP and anti-GAPDH antibodies. Sample blots are displayed here. The experiment was replicated three times, and densitometry was used to measure the band intensities (normalized to GAPDH), to generate the average intensity ± S.D. (error bars).
To confirm these results in human ovarian cells, ovarian adenocarcinomas (SKOV3 and HEY) were utilized. We first characterized the TGF-β responsiveness of SKOV3 and HEY cells by performing a phospho-SMAD3 Western blot after 45 min of TGF-β treatment. Phosphorylation of SMAD3 in HEY cells is clearly lacking in comparison with SKOV3 cells (Fig. 6B). Therefore, SKOV3 is a TGF-β-sensitive cell line, and HEY is a TGF-β-insensitive cell line. As a result, SKOV3 and HEY cells were utilized to examine the relationship between GULP protein expression and TGF-β signaling. Further, the GULP protein expression levels in SKOV3 and HEY cells were quantified by Western blot after treatment with TGF-β for 8 h. In SKOV3 cells, GULP protein was expressed and was induced upon TGF-β treatment (Fig. 6C), similar to that observed in CHO cells (Fig. 5). On the other hand, our result shows that HEY cells had a much lower GULP expression level to begin with and failed to elevate the GULP level in response to TGF-β treatment. This defect in induction of GULP expression may be one of the causes that contribute to the TGF-β-insensitive phenotype in HEY cells. On the other hand, LRP1 expression levels were similar between SKOV3 and HEY cells and were not induced by TGF-β treatment (supplemental Fig. 2B), suggesting that the effect is primarily governed by GULP expression levels.
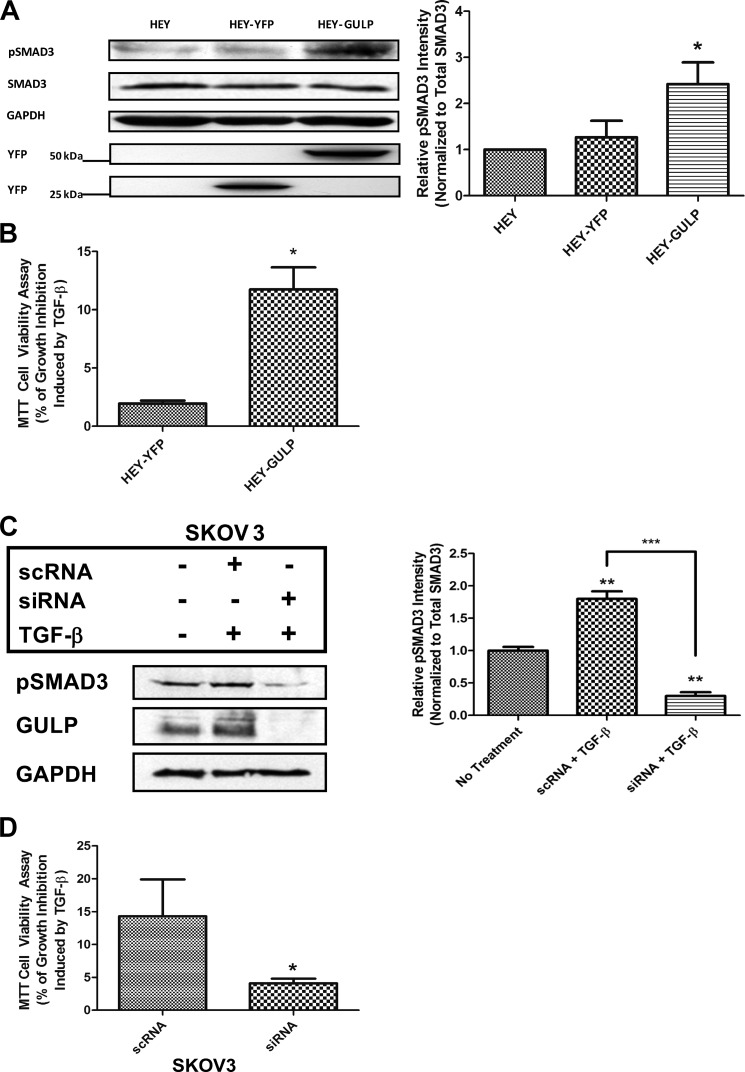
To verify our observations, we hypothesized that reintroduction of GULP into HEY cells may restore the TGF-β response. HEY cells were transiently transfected with either YFP alone (HEY-YFP) or YFP-GULP (HEY-GULP), as confirmed by Western blotting for YFP (Fig. 7A), showing comparable expression levels. Phospho-SMAD3 Western blots were performed after 120 min of TGF-β treatment, and the phosphorylation of SMAD3 in HEY-GULP cells was significantly stronger than in both the non-transfected and YFP transfected controls (Fig. 7A). These results demonstrate that the GULP expression level is low in TGF-β-unresponsive cells, and TGF-β responsiveness can be recovered upon GULP expression. Furthermore, these results confirm the observations in CHO cells, where higher GULP expression level led to a prolonged SMAD3 phosphorylation period and thus enhanced the TGF-β signaling in cells.
FIGURE 7.
Enhanced GULP expression in TGF-β-unresponsive HEY cells results in increased TGF-β activity. A, HEY cells, HEY cells transiently expressing YFP (HEY-YFP), and HEY cells transiently expressing YFP-GULP (HEY-GULP) were stimulated with TGF-β for 120 min. Total cell lysates were collected and analyzed by Western blot using anti-pSMAD3, anti-SMAD3, anti-GAPDH, and anti-YFP antibodies. Densitometric scaling was performed to normalize the intensity of pSMAD3 to total SMAD3 levels. The results are plotted below as relative intensity of pSMAD3 normalized to untransfected HEY cells. Data presented here are the mean ± S.D. (error bars) of three independent analyses, with a representative blot shown at the top. *, p < 0.03. B, HEY cells were transfected with a YFP expression plasmid or transfected with a plasmid expressing YFP-GULP. Transfected cells were treated without or with TGF-β for 48 h, and cell growth was measured by the cell viability MTT assay. Values were computed and presented as percentage of growth inhibition induced by TGF-β as described under “Materials and Methods.” Results shown here are the mean ± S.D. of two independent experiments, each performed with six replicates. *, p < 0.0001. C, SKOV3 cells were transfected with scrambled RNA (scRNA) or siRNA to GULP for 24 h, and then they were treated with TGF-β for 60 min. Cell lysates were prepared, and Western blots of pSMAD3, SMAD3, and GULP were performed. Relative intensities of pSMAD3 as measured by densitometry (normalized to total SMAD3 levels) were measured, and the average ± S.D. is presented here. **, p < 0.01 in comparison with the no treatment condition; ***, p < 0.002 between the scrambled RNA and siRNA conditions. D, SKOV3 cells were transfected with GULP siRNA or a scrambled control (scRNA). Transfected cells were treated without or with TGF-β for 48 h, and cell growth was measured by the cell viability MTT assay. Values were computed and presented as percentage of growth inhibition induced by TGF-β as described under “Materials and Methods.” Results shown here are the mean ± S.D. of two independent experiments, each performed with six replicates. *, p < 0.05.
Although we showed biochemically that the TGF-β-insensitive phenotype can be partially corrected by restoring the GULP expression level, it is important to see whether this effect has physiological importance. We thus assessed whether we could restore TGF-β growth-inhibitory responses in HEY cells by overexpressing GULP. Control HEY cells transfected with YFP alone or with a YFP-tagged GULP were stimulated or not with TGF-β for 72 h, and cell growth was measured using the cell viability MTT assay. Interestingly, overexpression of GULP significantly enhanced the TGF-β growth-inhibitory response in these cells from 2.0 ± 0.3 to 12.0 ± 1.9% (Fig. 7B).
To confirm this effect, we took the opposite approach by knocking down GULP expression in SKOV3 cells. Transfection of siRNA was effective in knocking down GULP (Fig. 7C; scrambled RNA (scRNA) was used as a transfection control), and as a result, phospho-SMAD3 was significantly decreased. Functionally, a decrease in GULP expression resulted in significantly less cell growth inhibition (Fig. 7D). Together, these results highlight GULP as a central signaling molecule, required for TGF-β/SMAD growth-inhibitory responses.
So far, we have demonstrated the impact of GULP expression level on TGF-β responsiveness in ovarian cells and ovarian carcinoma. Moreover, GULP expression level mediates its effect on TGF-β sensitivity through the modulation of SMAD3 phosphorylation. The exact mechanism of how GULP prolongs the phosphorylation state of SMAD3 is unknown. Previous evidence has suggested that GULP functions by stabilizing an early endosome complex containing the LRP ligand, preventing its maturation and conversion to a late endosome for ligand degradation. Although it is still controversial whether TGF-β can signal from the cell surface or if it needs to be internalized to signal (25, 26, 59, 60), we hypothesize that a GULP-mediated stabilization of a “signaling-competent” TGF-β-containing early endosome may promote TGF-β signaling.
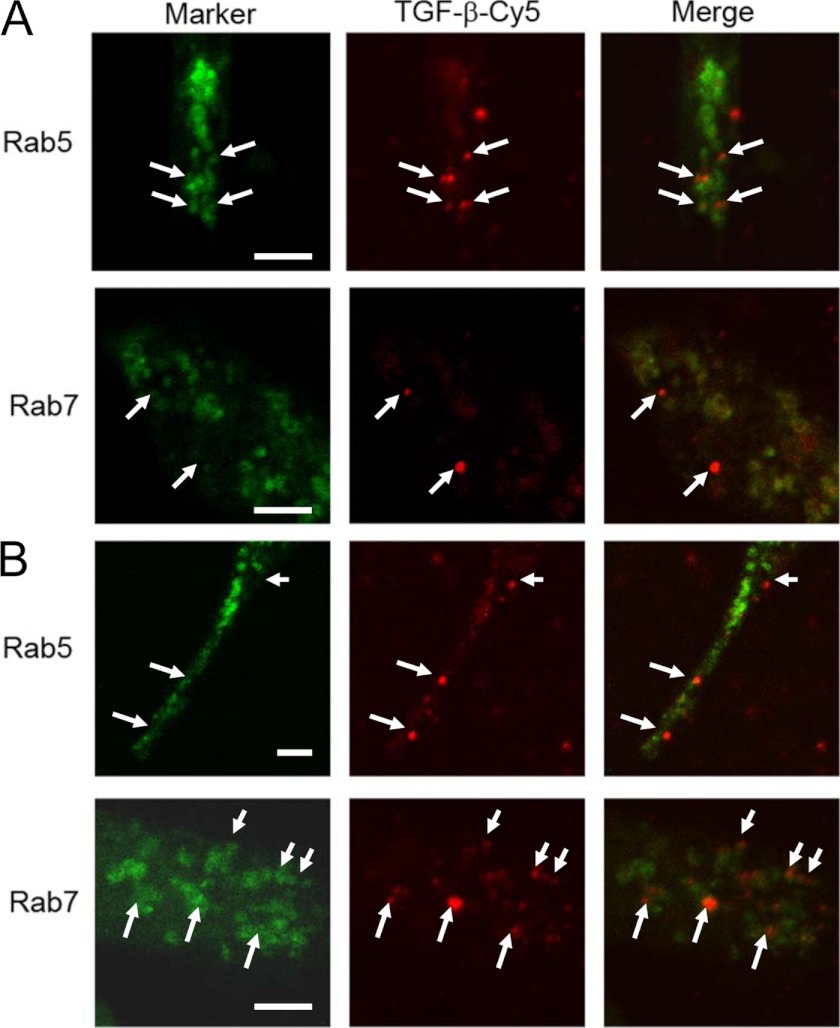
To address a possible mechanism, we performed a number of experiments. GULP is primarily cytosolic and not localized to the plasma membrane but to an early endosome compartment (41). We provide evidence that GULP binds PI 3-phosphate (and other monophosphate PI species) and PI 3,5-diphosphate but not PI-4,5-diphosphate (supplemental Fig. 4A), suggesting that GULP is not targeted to the plasma membrane, like other adapter proteins. Furthermore, disruption of the polybasic binding site of the PI species at the N terminus of GULP results in impairment of GULP function (both receptor activity and effect on cholesterol efflux; supplemental Fig. 4, B and C). Interestingly, knockdown of GULP does not prevent uptake of LRP ligands (41) (supplemental Fig. 5). We previously showed that GULP partially localizes to an early endosome compartment, and expression of GULP traps LRP ligands in an early endosome, preventing trafficking to a late endosome (41). Therefore, we propose that GULP maintains a “signaling-competent” early endosome compartment that contributes to enhanced TGF-β signaling. To address this, we performed fluorescence microscopy of Cy5-labeled TGF-β in FL and AS cells, colocalized with an early endosome marker (Rab5) and a late endosome marker (Rab7). In FL cells, TGF-β colocalized with Rab5 after a 1-h incubation (Fig. 8A), whereas in AS cells, TGF-β colocalized with Rab7 (Fig. 8B). Therefore, we hypothesized that GULP overexpression led to the stabilization of the early endosome complex, preventing the early to late endosome transition and therefore prolonging the event of TGF-β signaling and phosphorylation of SMAD3 (Fig. 3) at the early endosomes.
FIGURE 8.
TGF-β is trapped in the early endosome in FL cells. FL cells (A) or AS cells (B) were transfected with Rab5 (early endosome marker) or Rab7 (late endosome marker) to label endosome compartments. These cells were incubated with Cy5-labeled TGF-β (red fluorescence) for 1 h and then analyzed by fluorescence microscopy. Overlap of TGF-β in an endosome compartment can be determined by examining the white arrow in the separate frames (as an example). FL cells show considerable overlap of Rab5 endosomes and TGF-β but no overlap of Rab7 endosomes and TGF-β. AS cells show little overlap of Rab5 endosomes and TGF-β but significant overlap of Rab7 endosomes and TGF-β. The white arrows indicate endosomal TGF-β. White bar, 10 μm.
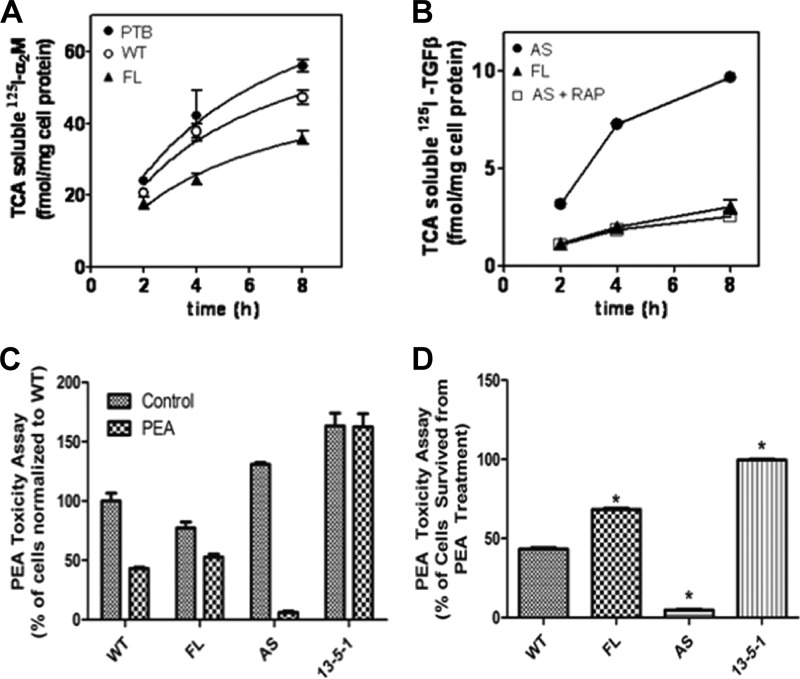
The ultimate fate of ligand endocytosis is the degradation of ligands in the lysosome. Therefore, we labeled the LRP1 ligands α2M and TGF-β with 125I and monitored their degradation in WT, FL, and AS cells by measuring TCA-soluble 125I radioactivity. Over the time course, the radioactivity was strongest in AS cells and weakest in FL cells, suggesting a more efficient degradation of α2M (Fig. 9A) and TGF-β (Fig. 9B) in AS cells. However, the degradation of transferrin is not affected by GULP, suggesting that GULP has specificity to LRP1 ligands (data not shown). PEA is a toxin and a ligand for LRP1-mediated endocytosis, utilizing a lysosome escape mechanism to poison mammalian hosts. In this case, it is important to point out that the PEA must get to the lysosome in order to escape and become toxic. A PEA cytotoxicity assay was performed on WT, FL, AS, and 13-5-1 cells to validate GULP function. The LRP1 null 13-5-1 cells showed complete resistance toward PEA toxicity as described previously (Fig. 9C) (61). The toxicity effect is strongest in AS cells, where only 4.69 ± 0.9% of cells survived from PEA treatment. In FL cells, 69.31 ± 1.23% of the cells survived in comparison with 43.21 ± 1.77% in WT cells, showing a significantly enhanced resistance toward PEA with higher GULP expression level (Fig. 9D). Overall, these results provided solid evidence that GULP acts as a stabilizer of the early endosome, preventing the early endosome-late endosome transition/maturation, whereas the transition toward the late endosome was enhanced when GULP was absent. It is this mechanism that explains the effect of GULP on TGF-β signaling.
FIGURE 9.
GULP affects trafficking and degradation of LRP ligands. A, WT-, FL-, or PTB (the dominant negative mutant that behaves similarly to AS cells)-expressing cells were incubated with 125I-labeled methylamine-activated α2M at 37 °C for various times. At each time point, cell medium was collected, and TCA-soluble 125I was detected (corresponding to degraded ligand). The most radioactivity was observed in PTB-expressing cells, demonstrating that LRP ligands are internalized and preferentially delivered to the lysosome in PTB-expressing cells. B, FL- and AS-expressing cells were incubated with 125I-labeled TGF-β at 37 °C for various times in the absence or presence of competing amounts of RAP. At each time point, cell medium was collected, and TCA-soluble 125I was detected. AS cells had the most radioactivity, indicating the most degradation of ligand, which could be effectively competed by the addition of RAP. C and D, PEA is an LRP ligand that, after internalization and trafficking to the late endosome, escapes the late endosome and becomes cytotoxic. WT, FL, AS, and 13-5-1 cells were incubated without (PEA Null) or with PEA (PEA), for 24 h and then cells were counted. C, actual cell counts normalized to WT at 100 cells. D, ratio of PEA/PEA null for each cell type. FL cells are only partially affected by PEA, whereas AS cells are extremely sensitive to PEA. *, p < 0.01. Error bars, S.D.
DISCUSSION
Cancer is an insidious disease which researchers have yet to master. It is difficult to obtain mechanistic molecular evidence on overlapping, interacting, redundant signaling pathways, and the role of TGF-β in normal cell biology and cancer is one such example. Under normal conditions, TGF-β can mediate a growth-inhibitory signal or apoptosis, which prevents cancer growth, but in some instances of cancer, TGF-β promotes EMT and angiogenesis and suppresses an immune response, which promotes cancer growth (7, 10, 62–65). This is the main reason for choosing CHO cells as an ovarian model. In this case, LRP1 (TGFBRV) has been documented as the primary TGF-β receptor in CHO cells that mediates the growth-inhibitory effect of TGF-β (33, 34). It is in this context that we can define the potential role of GULP (an LRP1 adapter protein) in TGF-β signaling. Our data, in addition to the microarray data showing that human ovarian adenocarcinoma cells have significantly less expressed GULP mRNA than normal ovary tissue, demonstrate a role of GULP in TGF-β signaling and a potential role of GULP in TGF-β signaling in a cancer setting.
In this study, we have presented data demonstrating the correlation between GULP expression level and TGF-β responsiveness using the model cell lines CHO cells, MEF cells, and two ovarian carcinomas: the TGF-β-sensitive SKOV3 cells and the TGF-β-insensitive HEY cells. At the level of physiological importance, the GULP expression level had a significant impact on TGF-β-induced cell growth arrest. Cells overexpressing GULP become more sensitive to TGF-β, leading to an increase in cell growth inhibition compared with cells with a normal level of GULP. On the other hand, cells with low GULP expression showed resistance toward TGF-β treatment. This effect was illustrated by comparing the endogenous GULP level of SKOV3 and HEY cells, where HEY cells had a negligible level of GULP compared with the TGF-β-sensitive SKOV3 cells.
We were also interested in whether GULP plays a role in TGF-β-induced metastasis. We did see a significant effect of GULP on cell motility and invasion in our CHO cells. First of all, CHO cells and ovarian cells in general are known to be less migratory and invasive (66); thus, TGF-β may be promoting its effect more on the cell division cycle instead of cell motility and invasion. However, that was not evident from our studies where GULP manipulation was effective at modulating TGF-β-induced migration and invasion. Some studies had shown that LRP1 plays an important role in cell migration/invasion (67). In order to promote invasion, cells secrete metalloproteinases (such as MMP-2 and MMP-9) to digest the extracellular matrix. We performed gelatin zymography to measure the activity of MMP-9 secreted by the WT, FL, and AS cells in response to TGF-β signaling. FL cells had the highest TGF-β-induced increase in gelatin digestion, and AS cells had the lowest. These results complement the data for invasion, where FL cells demonstrated the highest TGF-β-induced invasion.
In addition, to examine the physiological importance of GULP on tumor cells, we also elucidated the effect of GULP on TGF-β signaling at the biochemical level. Several key observations were made. First of all, an increase in GULP expression level led to a prolonged SMAD3 phosphorylation, where a decrease in GULP expression level had the opposite effect. This observation was further confirmed using the 3TP-Lux reporter, where GULP expression level had a positive correlation with the reporter activity. Interestingly, we also observed that GULP expression was induced by TGF-β treatment (up to 24 h), further suggesting the role of GULP in modulating the TGF-β signal. The induction of GULP expression by TGF-β treatment was also observed in other cancer cell lines, namely MCF-7, NMuMG, HUH7, and HepG2 cells,4 which suggested that the response of GULP expression is present in a wide selection of tissues. More importantly, this mechanism is absent in the TGF-β-insensitive HEY cells, which further strengthens the importance of GULP on TGF-β signaling. We show that HEY cells lack SMAD3 phosphorylation, but the TGF-β response can be rescued by introducing GULP through transfection. Conversely, SKOV3 cells have normal SMAD3 phosphorylation, which can be abrogated by silencing GULP. These observations suggest that GULP indeed plays a regulatory role in the TGF-β signaling pathway. It is also possible that the enhanced expression level of GULP induced by TGF-β is required to sustain the TGF-β signaling by maintaining the phosphorylation state of SMAD3.
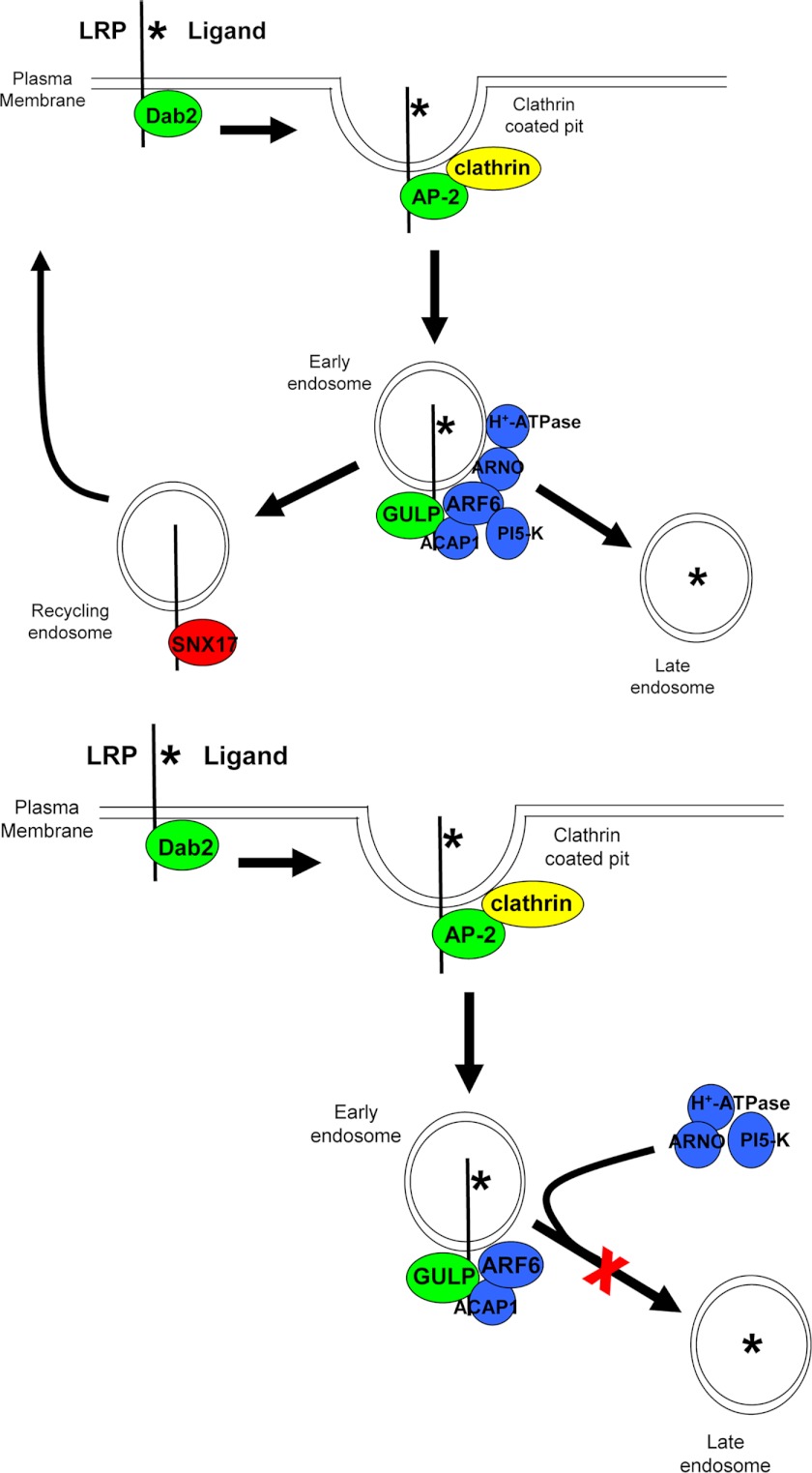
Moreover, we evaluated the mechanism of how GULP affects the phosphorylation of SMAD3 and TGF-β signaling in general. Together with our previous observations, we had hypothesized that GULP may enhance TGF-β signaling by stabilizing the early endosome (41). To provide evidence toward our hypothesis, fluorescence microscopy data showed trapping of TGF-β in the early endosome complex in FL cells with elevated GULP level. To further elucidate GULP mechanism, 125I-labeled TGF-β was utilized to monitor TGF-β trafficking and degradation. Similar to another LRP ligand, α2M, the degradation of TGF-β was increased in cells expressing a low level of GULP and decreased in cells expressing higher levels of GULP. Experiments using the cytotoxin PEA independently confirmed that low levels of GULP promote LRP ligand trafficking to the late endosome/lysosome, and increased levels of GULP restrict LRP ligands to the early endosome. Confocal imaging of LRP1-specific and nonspecific ligands also confirmed these observations. As a result, we propose the following mechanism of GULP function in TGF-β signaling. Upon the internalization of LRP1, GULP binds to LRP1 (Fig. 10). It has been shown previously that GULP binds ARF6 (preferentially in its GDP-bound inactive form) and ACAP1 (the GTPase-activating protein specific for ARF6) (68). Although the activity of ARF6 under conditions of high and low GULP expression has not been fully elucidated, it is plausible that direct interaction of GULP with ARF6(GDP) and ACAP1 may prevent ARF6 from interacting with its guanine nucleotide exchange factor ARF nucleotide binding site opener (ARNO), Grp1 or EFA6 (69–71). ARNO has been shown to interact with and recruit vacuolar-type H+-ATPase (V-ATPase) (71–73), and disruption of ARNO binding or recruitment of V-ATPase results in an impairment of maturation of early to late endosomes (71). This resembles closely the phenotype that we observe upon GULP expression. Therefore, we postulate that GULP, by virtue of its binding to ARF6(GDP) and ACAP1, prevents ARNO interacting with ARF6 and therefore recruitment of the V-ATPase. The net result is the prevention of maturation of early to late endosomes. The effect on TGF-β signaling is enhanced because the signaling complex (presumably at the early endosome) remains active and is not degraded. This correlates with enhanced phosphorylation of SMAD3 by sustaining the complex in the early endosome stage. When GULP finally falls off from the complex, receptor-ligand disassociation would occur, allowing TGF-β to enter the lysosome for degradation (Fig. 10). One other possibility that we have not addressed is raised in the report by Barrios-Rodiles et al. (74), in which, using the Lumier technique, GULP was found to interact with TGFBR-I, TGFBR-II, SMURF-1, and SMURF-2. As a result, GULP may very well enhance TGF-β signaling through possible receptor interaction or by preventing the ubiquitination of phospho-SMAD3 by binding the SMURF ubiquitin ligases, a finding that we will address in future experiments.
FIGURE 10.
Schematic model of the role of GULP in the trafficking of LRP ligands. Degradation of LRP ligands requires the timely and coordinated recruitment of adapter and effector proteins to mediate internalization and trafficking of the LRP ligands to the lysosome (top). Dab2 is positioned at the plasma membrane to recruit LRP to clathrin-coated pits. AP-2 binds clathrin and mediates inward curvature to generate an endosome. At the early endosome, GULP binds LRP and recruits ARF6 (GDP-bound) and ACAP1 (GAP specific to ARF6), thereby excluding ARNO (guanine nucleotide exchange factor specific to ARF6), vacuolar H+-ATPase, and the PI 5-kinase (PI-5-K). Exclusion of ARNO and PI 5-kinase prevents maturation of the early endosome to a late endosome (bottom) and prevents efficient sorting of the receptor (to be recycled) and ligand (to be degraded). Thus, in the presence of GULP, TGF-β is trapped in the early endosome and consequently promotes enhanced TGF-β signaling.
Further study is needed to elucidate the exact mechanism. All in all, our results suggest that the GULP expression level is critical to TGF-β sensitivity and that its importance in TGF-β sensitivity may have an effect on tumorigenesis of ovarian carcinomas.
Acknowledgment
We thank Dr. Kathleen Cho for generously providing microarray data.

This article contains supplemental Figs. 1–5.
C.-I. Ma, C. Martin, Z. Ma, A. Hafiane, M. Dai, J.-J. Lebrun, and R. S. Kiss, unpublished data.
- EMT
- epithelial-mesenchymal transition
- R-SMAD
- receptor-regulated SMAD
- PTB
- phosphotyrosine binding domain
- FL
- full-length
- MEF
- mouse embryo fibroblast
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- MMP
- matrix metallopeptidase
- PI
- phosphatidylinositol
- PEA
- Pseudomonas exotoxin A
- ARF
- ADP-ribosylation factor
- ARNO
- ARF nucleotide binding site opener
- V-ATPase
- vacuolar-type H+-ATPase
- pSMAD3
- phospho-SMAD3
- α2M
- α2-macroglobulin.
REFERENCES
- 1. Roberts A. B., Sporn M. B. (1985) Transforming growth factors. Cancer Surv. 4, 683–705 [PubMed] [Google Scholar]
- 2. Sporn M. B., Roberts A. B., Wakefield L. M., Assoian R. K. (1986) Transforming growth factor-β. Biological function and chemical structure. Science 233, 532–534 [DOI] [PubMed] [Google Scholar]
- 3. Zhou L., Leung B. S. (1992) Growth regulation of ovarian cancer cells by epidermal growth factor and transforming growth factors α and β1. Biochim. Biophys. Acta 1180, 130–136 [DOI] [PubMed] [Google Scholar]
- 4. Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem. 67, 753–791 [DOI] [PubMed] [Google Scholar]
- 5. Blobe G. C., Schiemann W. P., Lodish H. F. (2000) Role of transforming growth factor β in human disease. N. Engl. J. Med. 342, 1350–1358 [DOI] [PubMed] [Google Scholar]
- 6. Chatterjee S., Van Marck E. (2005) A word on the possible role of the circulating transforming growth factor β-1 in hypertension, diabetes, obesity, smoking, and human disease involving fibrosis. Med. Sci. Monit. 11, LE10–LE11 [PubMed] [Google Scholar]
- 7. Massagué J. (2008) TGFβ in cancer. Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gordon K. J., Blobe G. C. (2008) Role of transforming growth factor-β superfamily signaling pathways in human disease. Biochim. Biophys. Acta 1782, 197–228 [DOI] [PubMed] [Google Scholar]
- 9. Goumans M. J., Liu Z., ten Dijke P. (2009) TGF-β signaling in vascular biology and dysfunction. Cell Res. 19, 116–127 [DOI] [PubMed] [Google Scholar]
- 10. Derynck R., Akhurst R. J., Balmain A. (2001) TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29, 117–129 [DOI] [PubMed] [Google Scholar]
- 11. Lacerte A., Korah J., Roy M., Yang X. J., Lemay S., Lebrun J. J. (2008) Transforming growth factor-β inhibits telomerase through SMAD3 and E2F transcription factors. Cell. Signal. 20, 50–59 [DOI] [PubMed] [Google Scholar]
- 12. Wrana J. L., Attisano L., Cárcamo J., Zentella A., Doody J., Laiho M., Wang X. F., Massagué J. (1992) TGF β signals through a heteromeric protein kinase receptor complex. Cell 71, 1003–1014 [DOI] [PubMed] [Google Scholar]
- 13. Massagué J. (1992) Receptors for the TGF-β family. Cell 69, 1067–1070 [DOI] [PubMed] [Google Scholar]
- 14. Lewis K. A., Gray P. C., Blount A. L., MacConell L. A., Wiater E., Bilezikjian L. M., Vale W. (2000) Betaglycan binds inhibin and can mediate functional antagonism of activin signaling. Nature 404, 411–414 [DOI] [PubMed] [Google Scholar]
- 15. McLean S., Di Guglielmo G. M. (2010) TGF β (transforming growth factor β) receptor type III directs clathrin-mediated endocytosis of TGF β receptor types I and II. Biochem. J. 429, 137–145 [DOI] [PubMed] [Google Scholar]
- 16. Nakao A., Imamura T., Souchelnytskyi S., Kawabata M., Ishisaki A., Oeda E., Tamaki K., Hanai J., Heldin C. H., Miyazono K., ten Dijke P. (1997) TGF-β receptor-mediated signaling through Smad2, Smad3, and Smad4. EMBO J. 16, 5353–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lagna G., Hata A., Hemmati-Brivanlou A., Massagué J. (1996) Partnership between DPC4 and SMAD proteins in TGF-β signaling pathways. Nature 383, 832–836 [DOI] [PubMed] [Google Scholar]
- 18. Wotton D., Lo R. S., Lee S., Massagué J. (1999) A Smad transcriptional corepressor. Cell 97, 29–39 [DOI] [PubMed] [Google Scholar]
- 19. Xu L., Kang Y., Cöl S., Massagué J. (2002) Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell 10, 271–282 [DOI] [PubMed] [Google Scholar]
- 20. Hayashi H., Abdollah S., Qiu Y., Cai J., Xu Y. Y., Grinnell B. W., Richardson M. A., Topper J. N., Gimbrone M. A., Jr., Wrana J. L., Falb D. (1997) The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell 89, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 21. Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., Miyazono K. (2001) Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276, 12477–12480 [DOI] [PubMed] [Google Scholar]
- 22. Shi W., Sun C., He B., Xiong W., Shi X., Yao D., Cao X. (2004) GADD34-PP1c recruited by Smad7 dephosphorylates TGFβ type I receptor. J. Cell Biol. 164, 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finger E. C., Lee N. Y., You H. J., Blobe G. C. (2008) Endocytosis of the type III transforming growth factor-β (TGF-β) receptor through the clathrin-independent/lipid raft pathway regulates TGF-β signaling and receptor down-regulation. J. Biol. Chem. 283, 34808–34818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yao D., Ehrlich M., Henis Y. I., Leof E. B. (2002) Transforming growth factor-beta receptors interact with AP2 by direct binding to β2 subunit. Mol. Biol. Cell 13, 4001–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Panopoulou E., Gillooly D. J., Wrana J. L., Zerial M., Stenmark H., Murphy C., Fotsis T. (2002) Early endosomal regulation of Smad-dependent signaling in endothelial cells. J. Biol. Chem. 277, 18046–18052 [DOI] [PubMed] [Google Scholar]
- 26. Tsukazaki T., Chiang T. A., Davison A. F., Attisano L., Wrana J. L. (1998) SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell 95, 779–791 [DOI] [PubMed] [Google Scholar]
- 27. Runyan C. E., Schnaper H. W., Poncelet A. C. (2005) The role of internalization in transforming growth factor β1-induced Smad2 association with Smad anchor for receptor activation (SARA) and Smad2-dependent signaling in human mesangial cells. J. Biol. Chem. 280, 8300–8308 [DOI] [PubMed] [Google Scholar]
- 28. Lo R. S., Massagué J. (1999) Ubiquitin-dependent degradation of TGF-β-activated smad2. Nat. Cell Biol. 1, 472–478 [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y., Chang C., Gehling D. J., Hemmati-Brivanlou A., Derynck R. (2001) Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. U.S.A. 98, 974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seo S. R., Lallemand F., Ferrand N., Pessah M., L'Hoste S., Camonis J., Atfi A. (2004) The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J. 23, 3780–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Komuro A., Imamura T., Saitoh M., Yoshida Y., Yamori T., Miyazono K., Miyazawa K. (2004) Negative regulation of transforming growth factor-beta (TGF-β) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23, 6914–6923 [DOI] [PubMed] [Google Scholar]
- 32. Matsuura I., Denissova N. G., Wang G., He D., Long J., Liu F. (2004) Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature 430, 226–231 [DOI] [PubMed] [Google Scholar]
- 33. Huang S. S., Ling T. Y., Tseng W. F., Huang Y. H., Tang F. M., Leal S. M., Huang J. S. (2003) Cellular growth inhibition by IGFBP-3 and TGF-β1 requires LRP-1. FASEB J. 17, 2068–2081 [DOI] [PubMed] [Google Scholar]
- 34. Tseng W. F., Huang S. S., Huang J. S. (2004) LRP-1/TβR-V mediates TGF-β1-induced growth inhibition in CHO cells. FEBS Lett. 562, 71–78 [DOI] [PubMed] [Google Scholar]
- 35. Cabello-Verrugio C., Brandan E. (2007) A novel modulatory mechanism of transforming growth factor-β signaling through decorin and LRP-1. J. Biol. Chem. 282, 18842–18850 [DOI] [PubMed] [Google Scholar]
- 36. Boucher P., Li W. P., Matz R. L., Takayama Y., Auwerx J., Anderson R. G., Herz J. (2007) LRP1 functions as an atheroprotective integrator of TGFβ and PDFG signals in the vascular wall. Implications for Marfan syndrome. PLoS One 2, e448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eslami P., Johnson M. F., Terzakaryan E., Chew C., Harris-White M. E. (2008) TGF β2-induced changes in LRP-1/T β R-V and the impact on lysosomal A β uptake and neurotoxicity. Brain Res. 1241, 176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou L., Takayama Y., Boucher P., Tallquist M. D., Herz J. (2009) LRP1 regulates architecture of the vascular wall by controlling PDGFRβ-dependent phosphatidylinositol 3-kinase activation. PLoS One 4, e6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng C. F., Fan J., Fedesco M., Guan S., Li Y., Bandyopadhyay B., Bright A. M., Yerushalmi D., Liang M., Chen M., Han Y. P., Woodley D. T., Li W. (2008) Transforming growth factor α (TGFα)-stimulated secretion of HSP90α. Using the receptor LRP-1/CD91 to promote human skin cell migration against a TGFβ-rich environment during wound healing. Mol. Cell Biol. 28, 3344–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herz J., Strickland D. K. (2001) LRP. A multifunctional scavenger and signaling receptor. J. Clin. Invest. 108, 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kiss R. S., Ma Z., Nakada-Tsukui K., Brugnera E., Vassiliou G., McBride H. M., Ravichandran K. S., Marcel Y. L. (2006) The lipoprotein receptor-related protein-1 (LRP) adapter protein GULP mediates trafficking of the LRP ligand prosaposin, leading to sphingolipid and free cholesterol accumulation in late endosomes and impaired efflux. J. Biol. Chem. 281, 12081–12092 [DOI] [PubMed] [Google Scholar]
- 42. Hendrix N. D., Wu R., Kuick R., Schwartz D. R., Fearon E. R., Cho K. R. (2006) Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 66, 1354–1362 [DOI] [PubMed] [Google Scholar]
- 43. Schwartz D. R., Wu R., Kardia S. L., Levin A. M., Huang C. C., Shedden K. A., Kuick R., Misek D. E., Hanash S. M., Taylor J. M., Reed H., Hendrix N., Zhai Y., Fearon E. R., Cho K. R. (2003) Novel candidate targets of β-catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 63, 2913–2922 [PubMed] [Google Scholar]
- 44. Su H. P., Nakada-Tsukui K., Tosello-Trampont A. C., Li Y., Bu G., Henson P. M., Ravichandran K. S. (2002) Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP). J. Biol. Chem. 277, 11772–11779 [DOI] [PubMed] [Google Scholar]
- 45. Zhang H., Lee J. M., Wang Y., Dong L., Ko K. W., Pelletier L., Yao Z. (2008) Mutational analysis of the FXNPXY motif within LDL receptor-related protein 1 (LRP1) reveals the functional importance of the tyrosine residues in cell growth regulation and signal transduction. Biochem. J. 409, 53–64 [DOI] [PubMed] [Google Scholar]
- 46. FitzGerald D. J., Fryling C. M., Zdanovsky A., Saelinger C. B., Kounnas M., Winkles J. A., Strickland D., Leppla S. (1995) Pseudomonas exotoxin-mediated selection yields cells with altered expression of low density lipoprotein receptor-related protein. J. Cell Biol. 129, 1533–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dawson C. W., Young L. S. (2001) In vitro assays to study epithelial cell growth. Methods Mol. Biol. 174, 165–172 [DOI] [PubMed] [Google Scholar]
- 48. Harfouche R., Hussain S. N. (2006) Signaling and regulation of endothelial cell survival by angiopoietin-2. Am. J. Physiol. Heart Circ. Physiol 291, H1635–H1645 [DOI] [PubMed] [Google Scholar]
- 49. Herz J., Goldstein J. L., Strickland D. K., Ho Y. K., Brown M. S. (1991) 39-kDa protein modulates binding of ligands to low density lipoprotein receptor-related protein/α2-macroglobulin receptor. J. Biol. Chem. 266, 21232–21238 [PubMed] [Google Scholar]
- 50. Datto M. B., Li Y., Panus J. F., Howe D. J., Xiong Y., Wang X. F. (1995) Transforming growth factor β induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc. Natl. Acad. Sci. U.S.A. 92, 5545–5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rich J. N., Zhang M., Datto M. B., Bigner D. D., Wang X. F. (1999) Transforming growth factor-β-mediated p15INK4B induction and growth inhibition in astrocytes is SMAD3-dependent and a pathway prominently altered in human glioma cell lines. J. Biol. Chem. 274, 35053–35058 [DOI] [PubMed] [Google Scholar]
- 52. Li R., Waga S., Hannon G. J., Beach D., Stillman B. (1994) Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature 371, 534–537 [DOI] [PubMed] [Google Scholar]
- 53. Ho J., Cocolakis E., Dumas V. M., Posner B. I., Laporte S. A., Lebrun J. J. (2005) The G protein-coupled receptor kinase-2 is a TGFβ-inducible antagonist of TGFβ signal transduction. EMBO J. 24, 3247–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ho J., de Guise C., Kim C., Lemay S., Wang X. F., Lebrun J. J. (2004) Activin induces hepatocyte cell growth arrest through induction of the cyclin-dependent kinase inhibitor p15INK4B and Sp1. Cell. Signal. 16, 693–701 [DOI] [PubMed] [Google Scholar]
- 55. Ueki N., Ohkawa T., Yokoyama Y., Maeda J., Kawai Y., Ikeda T., Amuro Y., Hada T., Higashino K. (1993) Potentiation of metastatic capacity by transforming growth factor-β1 gene transfection. Jpn. J. Cancer Res. 84, 589–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hocevar B. A., Prunier C., Howe P. H. (2005) Disabled-2 (Dab2) mediates transforming growth factor β (TGFβ)-stimulated fibronectin synthesis through TGFβ-activated kinase 1 and activation of the JNK pathway. J. Biol. Chem. 280, 25920–25927 [DOI] [PubMed] [Google Scholar]
- 57. Prunier C., Howe P. H. (2005) Disabled-2 (Dab2) is required for transforming growth factor β-induced epithelial to mesenchymal transition (EMT). J. Biol. Chem. 280, 17540–17548 [DOI] [PubMed] [Google Scholar]
- 58. Penheiter S. G., Singh R. D., Deep S. R., Repellin C. E., Wilkes M. C., Edens M., Howe P. H., Pagano R. E., Leof E. B. (2010) Type II transforming growth factor-β receptor recycling is dependent upon the clathrin adaptor protein Dab2. Mol. Biol. Cell 21, 4009–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Di Guglielmo G. M., Le Roy C., Goodfellow A. F., Wrana J. L. (2003) Distinct endocytic pathways regulate TGF-beta receptor signaling and turnover. Nat. Cell Biol. 5, 410–421 [DOI] [PubMed] [Google Scholar]
- 60. Chen C. L., Hou W. H., Liu I. H., Hsiao G., Huang S. S., Huang J. S. (2009) Inhibitors of clathrin-dependent endocytosis enhance TGFβ signaling and responses. J. Cell Sci. 122, 1863–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Avramoglu R. K., Nimpf J., McLeod R. S., Ko K. W., Wang Y., FitzGerald D., Yao Z. (1998) Functional expression of the chicken low density lipoprotein receptor-related protein in a mutant chinese hamster ovary cell line restores toxicity of Pseudomonas exotoxin A and degradation of α2-macroglobulin. J. Biol. Chem. 273, 6057–6065 [DOI] [PubMed] [Google Scholar]
- 62. Kuilman T., Michaloglou C., Mooi W. J., Peeper D. S. (2010) The essence of senescence. Genes Dev. 24, 2463–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oft M., Heider K. H., Beug H. (1998) TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Curr. Biol. 8, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 64. Wakefield L. M., Roberts A. B. (2002) TGF-β signaling. Positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 12, 22–29 [DOI] [PubMed] [Google Scholar]
- 65. Zhu Y., Nilsson M., Sundfeldt K. (2010) Phenotypic plasticity of the ovarian surface epithelium. TGF-β1 induction of epithelial to mesenchymal transition (EMT) in vitro. Endocrinology 151, 5497–5505 [DOI] [PubMed] [Google Scholar]
- 66. Wang H., Quah S. Y., Dong J. M., Manser E., Tang J. P., Zeng Q. (2007) PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res. 67, 2922–2926 [DOI] [PubMed] [Google Scholar]
- 67. Song H., Li Y., Lee J., Schwartz A. L., Bu G. (2009) Low density lipoprotein receptor-related protein 1 promotes cancer cell migration and invasion by inducing the expression of matrix metalloproteinases 2 and 9. Cancer Res. 69, 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ma Z., Nie Z., Luo R., Casanova J. E., Ravichandran K. S. (2007) Regulation of Arf6 and ACAP1 signaling by the PTB domain-containing adaptor protein GULP. Curr. Biol. 17, 722–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cohen L. A., Honda A., Varnai P., Brown F. D., Balla T., Donaldson J. G. (2007) Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol. Biol. Cell 18, 2244–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shultz T., Nash-Livni N., Shmuel M., Altschuler Y. (2006) EFA6 regulates endosomal trafficking and affects early endosomes in polarized MDCK cells. Biochem. Biophys. Res. Commun. 351, 106–112 [DOI] [PubMed] [Google Scholar]
- 71. Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G. H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D. A., Brown D., Marshansky V. (2006) V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat. Cell Biol. 8, 124–136 [DOI] [PubMed] [Google Scholar]
- 72. Várnai P., Bondeva T., Tamás P., Tóth B., Buday L., Hunyady L., Balla T. (2005) Selective cellular effects of overexpressed pleckstrin homology domains that recognize PtdIns(3,4,5)P3 suggest their interaction with protein binding partners. J. Cell Sci. 118, 4879–4888 [DOI] [PubMed] [Google Scholar]
- 73. Hernández-Deviez D., Mackay-Sim A., Wilson J. M. (2007) A role for ARF6 and ARNO in the regulation of endosomal dynamics in neurons. Traffic 8, 1750–1764 [DOI] [PubMed] [Google Scholar]
- 74. Barrios-Rodiles M., Brown K. R., Ozdamar B., Bose R., Liu Z., Donovan R. S., Shinjo F., Liu Y., Dembowy J., Taylor I. W., Luga V., Przulj N., Robinson M., Suzuki H., Hayashizaki Y., Jurisica I., Wrana J. L. (2005) High throughput mapping of a dynamic signaling network in mammalian cells. Science 307, 1621–1625 [DOI] [PubMed] [Google Scholar]