Background: Mitochondrial complex I deficiency is a common inherited metabolic disease.
Results: B2 transposable element insertion into Ndufs4 in mice causes loss of the “N assembly module” of complex I, alterations in cellular metabolites, and neurological symptoms.
Conclusion: NDUFS4 subunit is required for complex I stability.
Significance: Understanding the effects of oxidative phosphorylation defects is essential for the development of treatments.
Keywords: Mitochondrial Diseases, Mitochondrial Metabolism, Mouse, Mouse Genetics, Proteomics, B2 SINE, CI Deficiency, Leigh Syndrome, NDUFS4, Insertional Mutagenesis, Mouse Model
Abstract
Eukaryotic cells generate energy in the form of ATP, through a network of mitochondrial complexes and electron carriers known as the oxidative phosphorylation system. In mammals, mitochondrial complex I (CI) is the largest component of this system, comprising 45 different subunits encoded by mitochondrial and nuclear DNA. Humans diagnosed with mutations in the gene NDUFS4, encoding a nuclear DNA-encoded subunit of CI (NADH dehydrogenase ubiquinone Fe-S protein 4), typically suffer from Leigh syndrome, a neurodegenerative disease with onset in infancy or early childhood. Mitochondria from NDUFS4 patients usually lack detectable NDUFS4 protein and show a CI stability/assembly defect. Here, we describe a recessive mouse phenotype caused by the insertion of a transposable element into Ndufs4, identified by a novel combined linkage and expression analysis. Designated Ndufs4fky, the mutation leads to aberrant transcript splicing and absence of NDUFS4 protein in all tissues tested of homozygous mice. Physical and behavioral symptoms displayed by Ndufs4fky/fky mice include temporary fur loss, growth retardation, unsteady gait, and abnormal body posture when suspended by the tail. Analysis of CI in Ndufs4fky/fky mice using blue native PAGE revealed the presence of a faster migrating crippled complex. This crippled CI was shown to lack subunits of the “N assembly module”, which contains the NADH binding site, but contained two assembly factors not present in intact CI. Metabolomic analysis of the blood by tandem mass spectrometry showed increased hydroxyacylcarnitine species, implying that the CI defect leads to an imbalanced NADH/NAD+ ratio that inhibits mitochondrial fatty acid β-oxidation.
Introduction
The oxidative phosphorylation (OXPHOS)4 system in mitochondria consists of five protein complexes (CI–CV) and is responsible for the generation of the majority of the energy for the cell in the form of the energy-rich molecule ATP (1). Complex I (CI; NADH-ubiquinone oxidoreductase) is a major entry point of electrons into the OXPHOS system via the enzymatic conversion of NADH into NAD+, the transfer of the released electrons to ubiquinone, and the consequent enzymatic reduction of the latter. Simultaneous with these actions, CI pumps protons out of the mitochondrial matrix, thereby contributing to the generation of the mitochondrial membrane potential (ΔψM), which can be used by complex V for ATP synthesis or alternatively for the generation of heat. In mammals, CI requires the correct assembly of 45 subunits encoded by both the nuclear and mitochondrial DNA in order to function correctly, making it the largest of the OXPHOS complexes.
Defects in the OXPHOS system are the most common cause of inherited metabolic diseases affecting 1 in 5000 live births (2). CI deficiency is the most common of the OXPHOS disorders and has been shown to be caused by mutations in at least 20 subunit genes plus at least nine other genes encoding CI assembly factors (3), with many other affected individuals yet to have their genetic cause identified. As for any of the OXPHOS disorders, CI deficiency may affect any organ at any age, but most often organs with a high energy demand, such as the heart and brain, are affected. Accordingly, patients with isolated CI deficiency most often present with the early onset neurodegenerative disease Leigh syndrome (OMIM number 256000) (4, 5). There is currently no effective treatment available for patients with CI deficiency, as is generally the case for all patients with OXPHOS disorders. Up until now, patient management has largely been limited to the treatment of symptoms (6) and genetic counseling for those families where mutations have been identified with the possibility of prenatal testing.
One problem in developing new treatment approaches has been the lack of relevant animal models for OXPHOS disorders, in particular mouse models, to test promising drugs. Until recently, the only mouse models with CI deficiency were the spontaneous mutant “Harlequin” mouse with a proviral insertion in intron 1 of the Aif (apoptosis-inducing factor) gene, resulting in a reduced overall expression of AIF protein and the subsequently generated conditional Aif knockouts (7–10). These models have some limitations because the exact role of AIF in CI maintenance and activity is still unclear. This was emphasized by the recent discovery of patients with an isolated CIV deficiency caused by mutations in the orthologous AIF gene (11). Furthermore, the variability of the phenotype of Harlequin mice may complicate interpretation of the treatment outcomes.
The Ndufs4 gene encodes a matrix arm subunit of CI. Patients with mutations in the NDUFS4 gene develop Leigh syndrome and have a CI assembly/stability defect and a severe defect in CI activity. Homozygous expression of a truncated form of NDUFS4 results in embryonic lethality (12), whereas mice with a complete knock-out of Ndufs4 (Ndufs4−/− mice) develop severe neurological impairment, leading to death at ∼7 weeks (13). Ndufs4−/− mice develop severe neurological impairment, leading to death at ∼7 weeks. Additional studies showed that the majority of the symptoms of Ndufs4−/− mice were due to the CI defect in the brain (14). Our CI-deficient mouse model presented here arose due to a spontaneous B2 SINE retroviral insertion into the Ndufs4 gene (Ndufs4fky/fky mice). We used a novel strategy involving genetic linkage based on single nucleotide polymorphisms (SNPs) and microsatellites, together with expression exon arrays, which identified not only the affected gene but the affected exon. The phenotype of our Ndufs4fky/fky mice is similar to that described by Kruse et al. (13) for the conventional knockout. However, we show here that in Ndufs4fky/fky mice, certain metabolites in the blood are significantly different compared with unaffected controls, which may potentially be used as biomarkers for disease in the future. Furthermore, we demonstrate a clear CI assembly defect in Ndufs4fky/fky mice, which is similar to that found in patients with mutations in NDUFS4.
EXPERIMENTAL PROCEDURES
Animal Handling
The spontaneous Ndufs4fky mutation originated in a transgenic C57BL/6 (B6) strain. For genetic mapping, B6 Ndufs4fky/+ males were crossed with BALB/c (BC) females. Experiments were carried out on backcross (N2) and intercross (Fn) offspring, as well as progeny from the founding B6 colony. Ethics approval was granted by the Walter and Eliza Hall Institute Animal Ethics Committee (Project 2005.041) and an approved protocol by the Murdoch Childrens Research Institute Animal Ethics Committee (A526). Mice were housed on 12-h light/dark cycles and provided ad libitum access to food and water. Ndufs4fky/fky mice were euthanized around postnatal day 40, when they lost more than 10% of their body weight and/or showed hind limb clasping or forward curling when suspended by the tail according to our ethical guidelines.
Growth Curves and Organ Weights
Mice were weighed from the age of 5 to 45 days postnatal every afternoon. Differences between control and affected mice were assessed using a permutation test designed for growth curves (available on the Walter and Eliza Hall Institute of Medical Research Bioinformatics Web site) with 10,000 permutations. p values were computed as recommended by Phipson and Smyth (15).
Genetic Linkage
Genetic linkage to locate the fky locus was performed by outcrossing onto BALB/c, and there was complete penetrance in the resulting progeny (81 of 370 = 0.22, χ2 = 1.91, df = 1, p = 0.17) with no sex bias (χ2 = 0.62, df = 1, p = 0.43). Penetrance of the mutant phenotype in founding and outcross colonies was determined using Pearson's χ2 goodness of fit test. Sex bias was examined using Pearson's test of independence with Yates correction. A total of 99 offspring, comprising 20 backcross (N2) and 79 intercross (F2) animals, were analyzed at 309 SNP markers using the SEQUENOM® iPLEXTM assay and a custom panel of 15 microsatellites (document number 8876-006, R01, April 2005). Briefly, each SNP region was PCR-amplified, followed by a clean-up step to dephosphorylate unincorporated dNTPs. A second amplification was performed using primers that stop just before each SNP nucleotide, with the reaction terminating after a single base extension. Therefore, the final amplification products for each polymorphic marker would differ by only one nucleotide, which was the SNP itself. Amplicons were analyzed using mass spectrometry (SpectroCHIP® bioarrays) with a MassARRAY work station (version 3.3). Single point analysis was done on the SNP genotypes to check segregation distortion and determine a threshold for genome-wide significance. A custom binomial test implemented in R was then used to look for bias to B6 homozygosity.
Microsatellite genotyping was performed using markers from public databases (supplemental Table 1). Amplification was done under standard conditions using fluorescently labeled primers (Applied Biosystems), and PCR products were separated by capillary electrophoresis on the AB3730 DNA analyzer. Fluorescent signals were translated into fragment sizes using Genemapper (version 3.7). Microsatellite genotypes were examined by visual comparison of haplotypes to identify regions of B6 homozygosity.
Genotyping
The novel Ndufs4 mutation is genotyped using PCR under standard conditions with one forward primer in intron 3 (5′-TAGGAAGGGAGAGACGAGCA-3′) and two reverse primers located in the B2 SINE insert (5′-TTACCCACTGAGCCATCTCAC-3′) and exon 3 (5′-GATGCCCAACCCATCAAAG-3′), respectively.
Microarray Analysis
Total RNA was isolated from whole brain of two unaffected and two affected P37 mice using the Qiagen RNeasy® Lipid Tissue Midi Kit according to the manufacturer's instructions, including DNase treatment. RNA was processed and hybridized onto an Affymetrix GeneChip® Mouse Exon 1.0ST Array according to the manufacturer's instructions (GeneChip® Whole Transcript Sense Target Labeling Assay Manual 701880 Rev.2). Fluorescent signals were detected using a GeneChip® Scanner 3000.
Intensity values were background corrected, log2-transformed, and quantile-normalized using the robust multiarray average algorithm (16). Expression values were summarized and analyzed at both the gene and the exon levels. In each case, differential expression was assessed using empirical Bayes moderated t tests (17). Benjamini and Hochberg adjustment was applied to control the false discovery rate for the 30 genes in the linkage region. A custom RefSeq-based CDF file was used for the gene level analysis (18). Pathway analysis was run on the genome-wide differential expression results using ROAST (19), Camera,5 and gene sets from the Broad Institute's Molecular Signatures Database (MSigDB) (20) mapped to mouse orthologs (see the Walter and Eliza Hall Institute of Medical Research Bioinformatics Web site).
Cloning and Sequencing
Genomic Ndufs4 primers for each exon were designed to include 200–300 bp of adjacent splice boundaries, whereas primers for cDNA amplification were designed to the middle of each exon (primer sequence available on request). Cloning of cDNA amplicons was carried out using the Promega pGEM®-T Vector System with JM109 competent cells according to the manufacturer's instructions. Sequencing reactions were prepared using the Applied Biosystems Prism® BigDyeTM Terminator (version 3.1) as recommended by the Australian Genome Research Facility. Sequencing of clones was done using M13 primers 5′-GGAAACAGCTATGACCATG-3′ and 5′-GTAAAACGACGGCCAGT-3′. Fluorescently labeled amplicons were analyzed using capillary separation, and sequence reads were examined using SeqMan software (DNASTAR® Lasergene). Ndufs4 reference sequences were downloaded from the University of California, Santa Cruz, Genome Browser Mouse Feb 2006 assembly.
SDS-PAGE, BN-PAGE, and Immunoblotting
For SDS-PAGE, fresh frozen tissues were homogenized in lysis buffer (100 mm Tris/HCl pH 8, 100 mm NaCl, 0.1% Triton X-100, 2 mm EDTA, and protease inhibitor mixture), briefly sonicated, and centrifuged at 4 °C. The resulting supernatant was collected, and 20 μg of protein extract was resolved using SDS-PAGE on a 15% acrylamide gel. NDUFS4 was detected using a monoclonal antibody (MitoSciences, MS104).
For BN-PAGE, mitochondria were isolated from mouse heart as described previously for cultured cells (21) and solubilized in either 1% n-dodecyl-β-d-maltoside or 1% Triton X-100 before separation of complexes by BN-PAGE (22). Proteins were either stained with 0.5% Coomassie Blue R (Sigma) in 40% ethanol, 7% acetic acid or transferred to PVDF membrane (Immobilon, Millipore) for immunodecoration (22). Membranes were probed with antibodies against the 70-kDa subunit of CII (Invitrogen) or against the CI subunit NDUFA9 (23).
For mass spectrometric analysis of holo-CI and “crippled” CI, mitochondria were solubilized in 1% Triton X-100 followed by separation on a 4–10% BN-PAGE gel. Complex I was further purified by another round of BN-PAGE. To this end, strips of the first dimension BN-polyacrylamide gel were excised and placed and run on a second BN-polyacrylamide gel.
The resolved gel was fixed in 50% methanol, 2% phosphoric acid; washed in distilled H2O; incubated in 34% methanol, 2% phosphoric acid, 17% ammonium sulfate; and stained with Brilliant Blue G colloidal Coomassie. Complexes were then excised from the gel for mass spectrometric analysis.
Nano-LC-MS/MS Analysis
Excised gel bands were washed, reduced, carbamidomethylated, and subjected to tryptic in-gel digestion as described previously (24). Generated peptides were extracted with 15 μl of 0.1% trifluoroacetic acid (TFA). Nano-LC-MS/MS analyses were conducted using an LTQ-Orbitrap XL (Thermo Scientific, Bremen, Germany) mass spectrometer, online-coupled to an inert U3000 nano-HPLC system (Dionex, Amsterdam, The Netherlands).
Briefly, peptides were preconcentrated in 0.1% TFA on a 100-μm inner diameter reverse phase trapping column (Synergi HydroRP, 4-μm particle size, 80-Å pore size, 2-cm length, Phenomenex) and afterward separated on a 75-μm inner diameter reverse phase column (Synergi HydroRP, 2-μm particle size, 80-Å pore size, 30-cm length, Phenomenex) main column by applying a 40-min binary gradient (solvent A, 0.1% formic acid; solvent B, 0.1% formic acid, 84% acetontrile) ranging from 5 to 50% solvent B in 40 min and from 50 to 95% B in 1 min, at a flow rate of 270 nl/min and 60 °C.
MS survey scans from 350 to 2000 m/z were acquired in the Orbitrap with a resolution of 60,000 using the polysiloxane at m/z 445.120030 as lock mass. The five most intense signals were subjected to collision-induced dissociation-based MS/MS in the LTQ using a dynamic exclusion. AGC target values were set to 106 and 104 for the Orbitrap and the LTQ, respectively.
Raw data were transformed into mgf (Mascot generic file) format, and generated peak lists were searched against the IPI mouse database version 3.52 (55,303 sequences) using Mascot 2.2.2 with the following settings: trypsin as enzyme with a maximum of one missed cleavage, carbamidomethylation of Cys (+57.0214 Da) as fixed and oxidation of Met (+15.9949 Da) as well as phosphorylation of Ser, Thr, and Tyr (+79.9663 Da) as variable modifications, a peptide mass tolerance of 10 ppm, and an MS/MS tolerance of ±0.5 Da. Only proteins with at least two different, manually checked peptide MS/MS spectra above a Mascot score of 35 (p < 0.05 at 28) were taken into account.
Mitochondrial CI Activity
OXPHOS enzyme activities were determined spectrophotometrically as described previously (5), except that assays were performed at 25 °C. Activities of respiratory chain complexes of tissues from Ndufs4fky/fky and wild type mice were compared by a t test. Differences were considered significant at p < 0.05.
Mitochondrial ATP Synthesis Assay
Mitochondria were immediately isolated from tissues of mice euthanized by cervical dislocation according to Pallotti and Lenaz (25) for heart and according to Sims (26) for brain. ATP synthesis capacity of isolated mitochondria (6.25 μg/ml end concentration) was measured as described previously with digitonin omitted (27). CI-dependent ATP synthesis was achieved with the substrates malate (10 mm) and glutamate (10 mm), whereas CII-dependent ATP synthesis was achieved with succinate (10 mm) in the presence of rotenone (2.5 μm). ATP synthesis capacity was expressed as the ratio between CI-dependent ATP synthesis normalized against CII-dependent ATP synthesis. Data obtained with mitochondria from Ndufs4fky/fky and control mice were compared by t test. Differences were considered significant at p < 0.05.
Histology
Skeletal muscle (quadriceps) was cryosectioned to 8 μm and stained with oil red O. Oil red O stock solution was prepared by solubilizing 300 mg of dye in 100 ml of isopropyl alcohol. Cryosections were fixed in paraformaldehyde for 10 min and incubated in 60% oil red O stock solution for 10 min. Slides were washed three times with distilled water, mounted, and examined. For the analysis of the optic nerve the mice were perfused with 1% paraformaldehyde and 2% glutaraldehyde in 0.1 m cacodylate buffer (pH 7.3) for 5 min. Optic nerves were accurately dissected and postfixed by immersion in the fixative solution for 2 h at 4 °C, washed in 0.1 m cacodylate buffer, and osmicated for 4 h in 1% OsO4 (Fluka). Nerves were rinsed in water, dehydrated, and embedded in Epon 812 substitute (Fluka). One-micrometer semithin sections were stained with 1% toluidine blue and examined by light microscopy.
Mouse brains were cryosectioned in a 1:4 series at 20 μm. Spinal cords were sectioned in a 1:24 series at 20 μm. All cut sections were mounted onto gelatin-coated slides. One series from each cut tissue was stained with cresyl violet. Images (×10) of sections were obtained using an Olympus Bx51 microscope and then analyzed using Neurolucida software. ImagePro Plus software was used to measure the motor cortex, sensory cortex, corpus collosum, ventricle, striatum, hippocampus (dentate gyrus and CA1), substantia nigra, pontine nucleus, ventral posterior thalamic nucleus (VP), cerebellar layers, and the areas of entire hemispheres.
Metabolic Analysis of Blood from Dried Bloodspot Cards
Blood drawn from the heart was placed on Whatman 903 paper and analyzed by electrospray tandem mass spectrometry as described previously with minor modifications (28). Multiple reaction monitoring was used to target a selected panel of amino acids and acylcarnitines. Quantitation was performed using added stable isotope internal standards. Significant changes between Ndufs4fky/fky mice and control were determined by t test. Differences were considered significant at p < 0.05.
Acylcarnitine Analysis of Tissues
Approximately 10 mg of heart or liver tissue was analyzed according to van Vlies et al. (29) with minor modifications. The amounts of acylcarnitine species were semiquantified using the following internal standards: [2H9]carnitine for C0–C2 carnitines, [2H3]isovalerylcarnitine for C3–C6 carnitines, [2H3]octanoylcarnitine for C8–C10 carnitines, and [2H3]tetradecanoylcarnitine for C12–C20 carnitines. Butylated acylcarnitine species were measured in duplicate by electrospray tandem mass spectrometry using a Waters Quattro LC instrument.
RESULTS
Spontaneous B2 SINE Insertion (fky) into Ndufs4 Results in Progressive Neurological Disorder
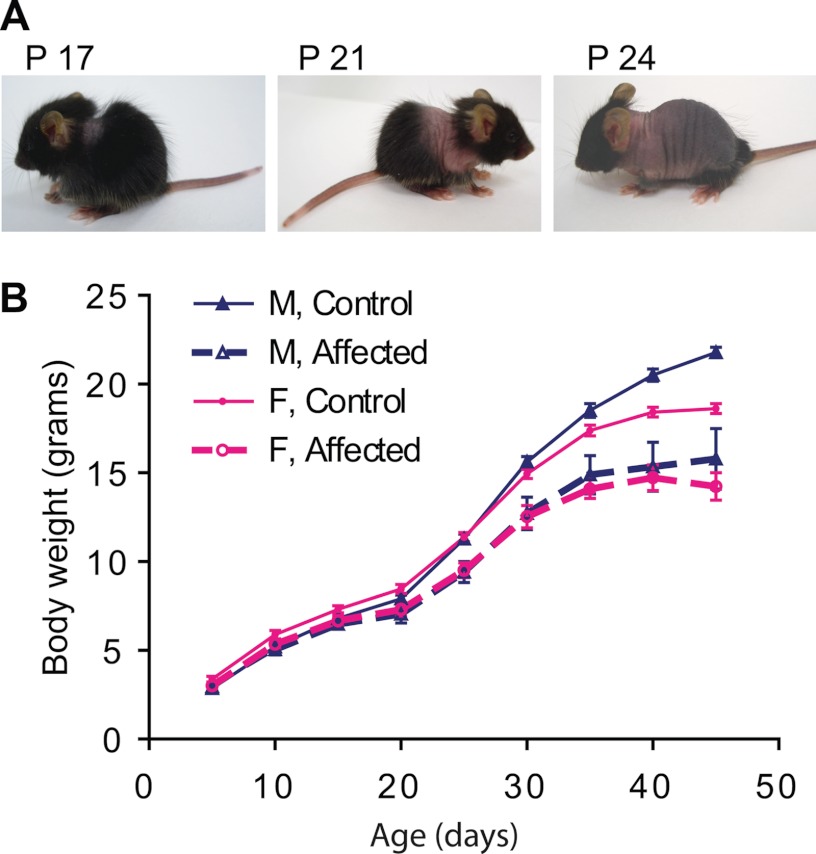
During a breeding strategy with heterozygous male Dnmt3L mice (30) and female RIP-mOVA transgenic C57BL/6 mice (31) a spontaneous “funky” mutation (fky) occurred. This mutation segregates independently of the parental transgenes, and inheritance is consistent with an autosomal recessive locus. The fky/fky phenotype presents from around postnatal day 16 (P16) with fur loss in the neck area, which subsequently spreads caudally along the trunk toward the tail (Fig. 1A). This fur loss is transient and regrows after a week in a similar pattern from the neck down the trunk. Homozygous mice show a failure to thrive from postnatal week 2 and by P40 are, on average, 26% smaller than their normal littermates (p = 10−5 for males; p = 10−5 for females; Fig. 1B). From week 5 onward, the affected mice develop neurological symptoms, including tilting of the head (16 of 75 = 0.21), walking in circles (17 of 75 = 0.23), and forward curling in combination with twisting of the body when suspended by the tail (51 of 75 = 0.68). From P40 onward, the symptoms progress rapidly, resulting in euthanasia according to ethical guidelines. Despite the severity of the mutation in the homozygous state, there was no evidence of prenatal lethality, as determined for a cohort of 465 progeny, of which 107 displayed the recessive phenotype (107 of 465 = 0.23, χ2 = 0.98, df = 1, p = 0.32) with no sex bias (51 males and 56 females, df = 1, χ2 = 0.48, p = 0.49). We performed a novel combination of both linkage and expression analysis to rapidly identify the gene responsible for the fky/fky phenotype. Genetic linkage to locate the fky locus was initiated by outcrossing the C57BL/6 fky with BALB/c wild-type mice and performing a genome-wide interrogation of 309 SNPs in 20 backcross and 79 intercross offspring. Analysis of these SNPs led to a subregion of chromosome 13, where a custom panel of 15 microsatellites further fine-mapped the recessive locus to a 6-megabase interval on mouse chromosome 13 between D13Mit196 and rs3708633 (supplemental Fig. 1A and Table 2), which contained 30 annotated RefSeq genes (Ensembl Genome Browser; mouse build 35). Expression analysis of these 30 genes using whole brain RNA on the Affymetrix GeneChip® Mouse Exon 1.0ST Array found Ndufs4 to be down-regulated in homozygous mice (p = 4 × 10−24; supplemental Table 2). Moreover, the expression of Ndufs4 showed relatively uniform down-regulation of all five exons with the exception of exon 3, which was at least 2.5-fold more down-regulated than the other exons (supplemental Fig. 1C and Table 3).
FIGURE 1.
Mice with a homozygous “funky” mutation have transient hair loss (A) and a failure to thrive (B). A, funky mice present with baldness behind the neck around P17, which progresses down the trunk (P21) toward the tail (P24) after which the hair grows back in the same order. B, growth curves comparing homozygous funky males and females to normal littermates. Homozygous mice show a reduction in growth rate from the age of 15 days. n = 6 for affected males (M, Affected); n = 33 for control males (M, Control); n = 10 for affected females (F, Affected); n = 20 for control females (F, Control); p = 10−5 for males; p = 10−5 for females. Error bars, S.E.
Sequencing of all five Ndufs4 exons and neighboring splice boundaries revealed a 258-nucleotide (nt) insertion within exon 3 (Fig. 2A). The insert sequence was queried against Repbase using RepeatMasker (A. F. A. Smit, R. Hubley, and P. Green, RepeatMasker Open-3.0, 1996–2004; available on the World Wide Web), and a 97% match (171 of 176 nt excluding the 69-nt poly(A) tail) to a known B2 short interspersed nuclear element (SINE) was found (Fig. 2, B and C). Genetic screening of our mouse colony found that this mutant Ndufs4 variant, Ndufs4fky, segregates with the recessive phenotype.
FIGURE 2.
The “funky” mutation is a B2 SINE insertion into exon 3 of Ndufs4. A, sequencing chromatograph comparing Ndufs4 exon 3 in wild type and homozygous mice; the inserted sequence begins 52 nt after the 5′ start of the exon. B, RepeatMasker output showing alignment of insert sequence with B2_Mm1a; 171 of 176 nt = 97% match. Transition and transversion substitutions are indicated by i and v, respectively. C, Ndufs4fky locus showing the B2 insert and target site duplication (TSD) sequences.
B2 SINE Insertion Results in Depletion of NDUFS4 Protein in Ndufs4fky/fky Mice
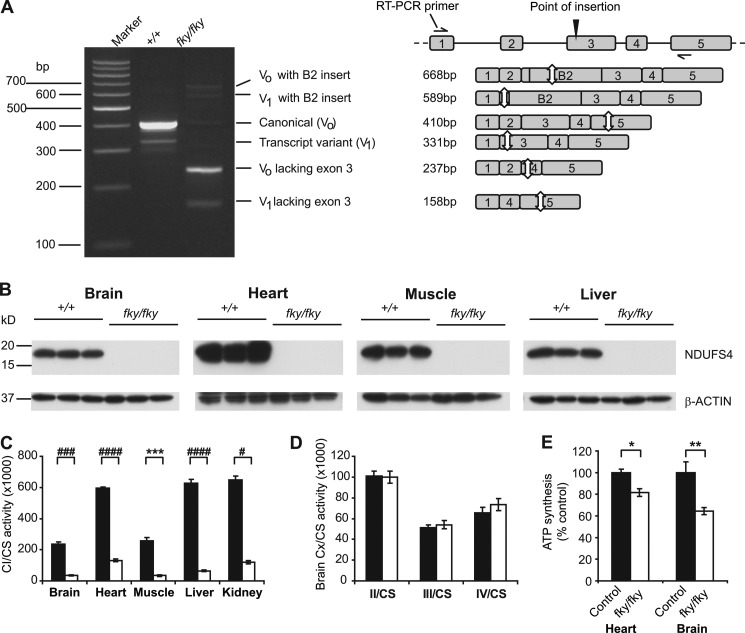
RT-PCR analysis of whole brain RNA showed a truncated canonical Ndufs4 transcript in Ndufs4fky/fky mice, and sequencing revealed that exon 3 was absent (Fig. 3A), which is concordant with the exon level microarray analysis. The absence of exon 3 from the canonical Ndufs4 transcript is predicted to cause a frameshift resulting in the formation of premature termination codons within exons 4 and 5. Canonical transcripts containing both exon 3 and the B2 insert were present in lower levels, but positioning of the B2 element in exon 3 also results in premature termination codon formation within the exon. Western blot analysis of brain, heart, liver, and skeletal muscle failed to detect NDUFS4 protein in Ndufs4fky/fky mouse tissues compared with wild type and heterozygous littermates (n = 3) (Fig. 3B), consistent with post-transcriptional degradation of the Ndufs4 product due to premature termination codon-induced nonsense-mediated decay (32).
FIGURE 3.
B2 SINE insertion into Ndufs4 results in isolated CI deficiency in all tissues tested. A, RT-PCR showing truncated Ndufs4 transcripts in whole brain RNA of Ndufs4fky/fky mice; wild type transcript variant (V1) was identified through the sequencing of clones; the schematic on the right describes the different amplicons visible on the gel; white arrows indicate the locations of stop codons. B, NDUFS4 protein was not detected in whole brain, heart, liver, and skeletal muscle of Ndufs4fky/fky mice via Western blot. C, CI activity, normalized against citrate synthase (CS), was reduced in all tissues tested of Ndufs4fky/fky mice (n = 3–5). D, mitochondrial CII, CIII, and CIV activities were normal in brain tissues of Ndufs4fky/fky mice (n = 4). E, ATP synthesis capacity of heart and brain mitochondria was determined as the CI-dependent ATP production (with substrate combination malate and glutamate) normalized against CII-dependent ATP production (with substrate succinate in the presence of CI inhibitor rotenone) and is shown as a percentage of control mitochondria (age range P30–P41, heart n = 13, brain n = 5). C and D, black bars, wild type mice; white bars, Ndufs4fky/fky mice. E, control, Ndufs4+/f+ and Ndufs4+/fky; fky/fky, Ndufs4fky/fky. *, p < 0.05; **, p < 0.01; ***, p < 0.005; #, p < 0.001; ##, p < 0.0005; ###, p < 0.0001; ####, p < 0.00005. Error bars, S.E.
Ndufs4 encodes the 18-kDa structural subunit of mitochondrial CI. In humans mutations in NDUFS4 cause mitochondrial CI deficiency (OMIM number 252010) (33–35), whereas a conventional knockout of the Ndufs4 gene in mouse was recently also shown to cause CI deficiency (13). Measurement of CI activity in brain, heart, muscle, liver, and kidney tissues from Ndufs4fky/fky mice, normalized against citrate synthase activity, showed decreased CI activity levels compared with wild type (Fig. 3C). The highest residual activity was detected in heart tissue (22.1 ± 1.3% of wild type level), whereas liver had the lowest residual activity compared with wild type (10.4% ± 1.1%). Additionally, no difference in CI activity was found between wild type and heterozygous mice (data not shown). This corresponds with the phenotype of heterozygous animals, which we found indistinguishable from wild type animals.
The activity of the other OXPHOS complexes (CII, CIII, and CIV) was not affected by the depletion of NDUFS4, as shown for brain, indicating an isolated effect on CI in Ndufs4fky/fky mice (n = 4, p ≫ 0.05, unpaired t test; Fig. 3D).
In order to determine the effect of the NDUFS4 depletion on the functionality of the OXPHOS system, we studied the capacity of isolated mitochondria to produce ATP when respiring on the substrates malate and glutamate, which feed electrons into the system at the level of CI. As a control, we measured ATP synthesis with succinate, which feeds electrons in via CII, plus rotenone to inhibit CI activity. Because no difference in CI activity and phenotype was found between the wild type and heterozygous genotypes, they were combined and referred to as the “control” group. As a result of the defect in CI activity, mitochondria isolated from heart and brain tissue of Ndufs4fky/fky show a decreased capacity to generate ATP compared with control mitochondria (ratio of ATP generated by respiring on malate and glutamate versus succinate and rotenone; Fig. 3E). Despite the large reduction measured in the CI activity assays for both the heart and brain, there was only a small reduction of 18% in the capacity to generate ATP in the heart, with a somewhat greater reduction of 36% in the brain (Fig. 3E).
Depletion of NDUFS4 Protein Leads to Loss of N Module of CI
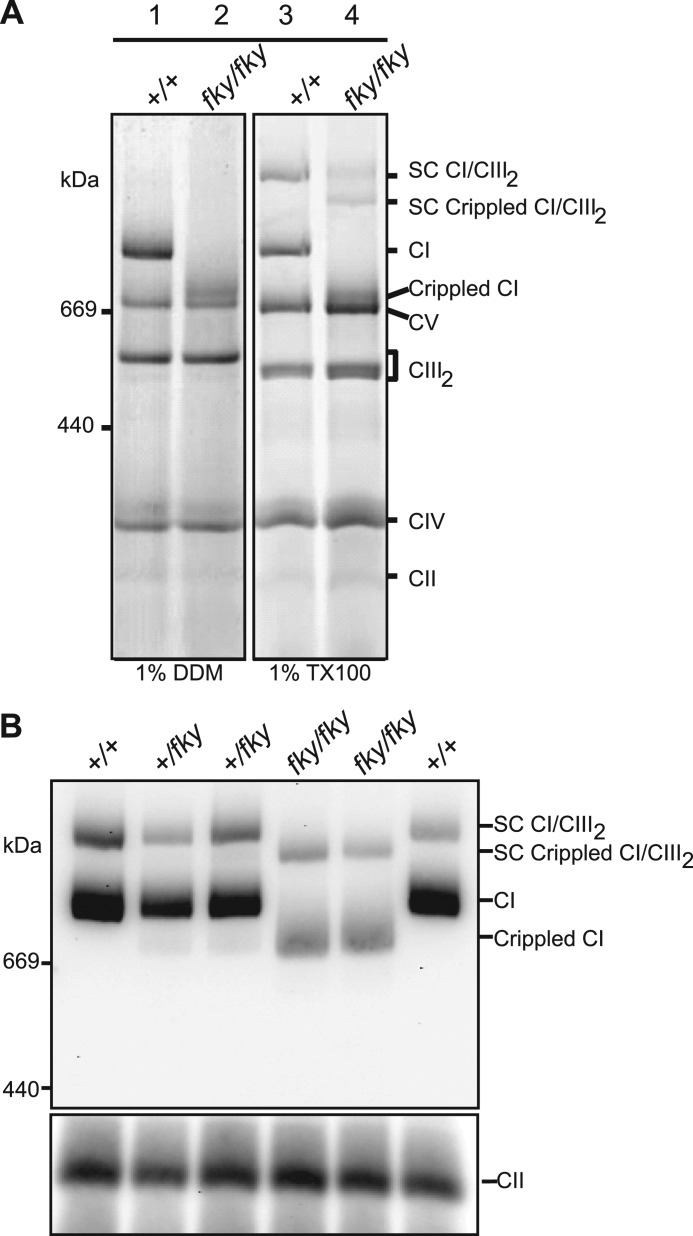
In humans, nonsense mutations, deletions, and duplications within NDUFS4 have been demonstrated to disrupt NDUFS4 protein synthesis, leading to abnormal mitochondrial CI formation and causing reduced activity (33–37). We examined mitochondrial CI assembly state in Ndufs4fky/fky mice using BN-PAGE separation followed by Coomassie staining. Ndufs4fky/fky heart mitochondria solubilized in 1% n-dodecyl-β-d-maltoside for BN-PAGE have a clear defect in CI assembly/stability, as shown by disappearance of the band corresponding to the correct molecular weight for CI as visible in the control lane (Fig. 4A, lanes 1 and 2). Instead, there is a band with a lower molecular weight corresponding to a protein complex of ∼800 kDa appearing in Ndufs4fky/fky heart mitochondria (lane 2), indicating a crippled CI. We next used milder solubilization conditions to allow for detection of the supercomplex of CI and CIII (CI-CIII2) on BN-PAGE (Fig. 4A, lanes 3 and 4). The CI-CIII2 supercomplexes present as a lower molecular weight species, indicating a supercomplex of crippled CI with CIII in the Ndufs4fky/fky mitochondria. Immunoblotting with antibody against the CI subunit NDUFA9 subsequent to BN-PAGE of 1% Triton X-100-solubilized mitochondria confirmed the crippled state of CI and the CI-CIII2 supercomplex from Ndufs4fky/fky mitochondria (Fig. 4B). The amount of the crippled CI and CI-CIII2 supercomplex was also decreased compared with wild type mice. Furthermore, the assembly state and amount of CI from heterozygous Ndufs4+/fky mitochondria did not differ from wild type mitochondria, consistent with the results obtained for the CI activity measurements (data not shown).
FIGURE 4.
Depletion of NDUFS4 changes the assembly status of CI. A, representative BN-polyacrylamide gel showing crippled CI monomer when Ndufs4fky/fky heart mitochondria are solubilized in 1% n-dodecyl β-maltoside (DDM; lane 2). When Ndufs4fky/fky heart mitochondria are solubilized in 1% Triton X-100 (TX100), the CI-CIII2 supercomplex (SC) can be observed (lanes 3 and 4). A large proportion of this CI-CIII2 SC is detected in the crippled state in Ndufs4fky/fky mitochondria (lane 4). B, BN-PAGE followed by Western blot with antibody against CI subunit NDUFA9 shows weaker staining and crippled status of CI (CI) monomer and CI-CIII2 (CI/CIII2) supercomplex in 1% Triton X-100-solubilized mitochondria from heart tissues of Ndufs4fky/fky mice.
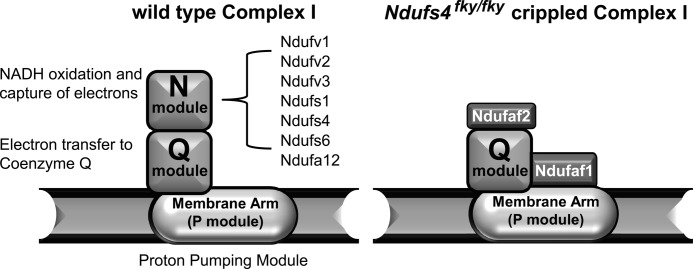
The composition of the crippled CI found in the BN-PAGE experiments with Ndufs4fky/fky mitochondria was subsequently determined using nano-liquid chromatography tandem mass spectrometry (nano-LC-MS/MS) with CI from wild type and Ndufs4fky/fky mitochondria. This resulted in the detection of 38 of the 45 subunits in wild type CI, whereas in the crippled complex, 30 subunits were detected (Table 1 and supplemental Table 4). Five subunits remained undetected in both wild type and crippled CI, indicating that these subunits may be difficult to detect. Strikingly, in addition to NDUFS4, the subunits NDUFA12, NDUFS1, NDUFS6, NDUFV1, and NDUFV2 were not detected in the crippled CI. These subunits, including NDUFS4, make up the N module responsible for NADH oxidation and transfer of electrons to iron sulfur clusters (Fig. 5). These data confirm that NDUFS4 is essential for the assembly and/or stability of the N module component of the CI matrix arm.
TABLE 1.
Representative MALDI-TOF results of isolated wild type and “crippled” Ndufs4fky/fky complex I
| Wild type holocomplex | Ndufs4fky/fky crippled complex | |
|---|---|---|
| Detected subunits | 38 | 30 |
| Not detected subunits | 7a | 15a |
| N module subunits detected | 6 | 0b |
| Detected assembly factors | 0 | 2 |
a Includes 5 subunit that were not detected in both.
b None of the N module subunits were detected.
FIGURE 5.
Schematic representation of the results from nano-LC-MS/MS analysis of the crippled CI in mitochondria from Ndufs4fky/fky mice. Left, the different modules present in CI in wild type mice. Right, crippled complex I from Ndufs4fky/fky mice misses all subunits that make up the N module, whereas the assembly factors Ndufaf1 and Ndufaf2 were found associated with the crippled complex I. Representation of the outcome of nano-LC-MS/MS analysis was based on the assembly model described by McKenzie and Ryan (64).
Not only is the crippled CI missing the N module, there were additional proteins detected that specifically associated with the crippled complex, namely NDUFAF1 (CIA30) and NDUFAF2 (B17.2L) (supplmental Table 4 and Fig. 5). These proteins are known to be CI assembly factors and have previously been found to associate with assembly intermediates in multiple CI-deficient patients (38–40). Moreover, NDUFAF1 was found associated with the crippled CI in patients with pathogenic NDUFS4 mutations (39, 41).
Metabolomic Analysis of Ndufs4fky/fky Mice Shows Increased Hydroxyacylcarnitines
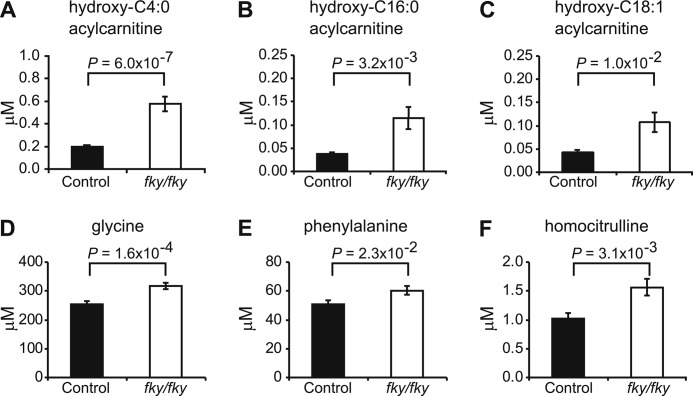
Having characterized the effect of NDUFS4 loss on CI assembly, we next sought to assess its impact on broader aspects of cellular metabolism. Tandem mass spectrometric targeted analysis of acylcarnitines and amino acids in the blood of Ndufs4fky/fky mice (mean age 40.1 ± 4.4 days for Ndufs4fky/fky mice and 40.1 ± 4.4 days for control mice, mean ± S.D.) showed only a few specific differences in metabolite levels when compared with control animals (Fig. 6 and supplemental Table 5). Interestingly, hydroxyacylcarnitines with a carbon chain of C4:0, C16:0, and C18:1 were all increased compared with control animals (Fig. 6, A–C). Although the absolute levels of these metabolites in the blood are low and probably will not directly influence disease outcome, the increased hydroxyacylcarnitines may indicate a block in the third NAD+-dependent step of the mitochondrial fatty acid β-oxidation pathway caused by the CI defect. However, additional acylcarnitine analysis of liver and heart of Ndufs4fky/fky mice revealed no large differences in carnitine species compared with control mice (supplemental Tables 6 and 7). Furthermore, oil red O staining of skeletal muscle failed to detect lipid accumulation in Ndufs4fky/fky mice (supplemental Fig. 2), which is occasionally found in patients with CI deficiency (5). These results suggest that the defect may result in altered ratios of fatty acyl species but does not cause a gross defect in fatty acid oxidation.
FIGURE 6.
NDUFS4 depletion results in significant changes in blood metabolite levels as measured by tandem MS. A–C, tandem MS analysis of blood from dried bloodspot cards showed significant increases in hydroxyacylcarnitine species with a mass corresponding to hydroxy-C4:0 acylcarnitine, hydroxy-C16:0 acylcarnitine, and hydroxy-C18:1 acylcarnitine in Ndufs4fky/fky mice. D–F, the only amino acid levels that were significantly changed were those of glycine, phenylalanine, and homocitrulline (all increased). n = 27 for controls; n = 26 for Ndufs4fky/fky. Error bars, S.E.
Tandem MS analysis of the blood amino acid levels showed only significant changes in the levels of glycine, phenylalanine, and homocitrulline, which were all elevated in Ndufs4fky/fky blood (Fig. 6, D–F).
DISCUSSION
Linkage in combination with expression profiling has been successful in identifying autosomal recessive disease genes in both mice and humans (42, 43). Here we performed an analysis of expression data focused solely on the critical interval identified by the customized linkage analysis and fine mapping of the susceptibility locus. This reduces the multiple testing penalty for the differential expression analysis. Additionally, as far as we are aware, this is the only example of using exon arrays, where in hindsight, due to the nature of the mutation (see below), the expression analyses identified not only the gene but the exon of the gene mutated in the fky/fky phenotype. SINEs belong to a family of endogenous retrovirus-like sequences known as transposable elements, which are known to cause disease through insertional mutagenesis (44–46). The spontaneous occurrence of the fky mutation not only highlights the detrimental effects of insertional mutagenesis but also prompts us to question if either of the parental transgenes Dnmt3L (DNA methyltransferase 3-like gene) or RIP-mOVA (membrane-bound ovalbumin under the control of the rat insulin promoter) had played a part in promoting mutagenic activity. Although RIP-mOVA is an unlikely candidate (31), DNMT3L has been found to be required for DNA methylation and genetic repression of transposable elements in spermatogonia (30). Dnmt3L−/− male mice are sterile due to spermatogenic arrest, whereas Dnmt3L+/− males are phenotypically normal and fertile. Considering that the fky/fky phenotype arose from breeding Dnmt3L+/− males, this suggests that the loss of one functional Dnmt3L allele may be sufficient to increase the frequency of transposable element-related mutagenesis in the male germ line.
The Ndufs4fky locus has a 13-nt target site duplication 5′-AAAACCAGAAAGG-3′ and a putative target sequence 5′-TCAAAA-3′, which are characteristic of documented insertions (47, 48) mediated by long interspersed nuclear elements (LINEs) (49, 50) (Fig. 2C). The B2 SINE insertion introduced a premature termination codon in Ndufs4, which most likely triggered nonsense-mediated decay of mutant mRNA, resulting in a complete depletion of NDUFS4 protein as shown in Fig. 3, A and B (32).
Ndufs4 encodes an 18-kDa subunit of mitochondrial CI and is highly expressed in energy-demanding tissues, such as the heart, skeletal muscle, and various components of the central nervous system, such as the amygdala, cerebellum, and dorsal root ganglion (51). Patients diagnosed with homozygous NDUFS4 mutations suffer from mitochondrial CI deficiency (OMIM number 252010). Symptomatically, these patients suffer from Leigh syndrome (OMIM number 256000), an early onset progressive neurodegenerative disorder often seen in congenital mitochondrial disorders, and exhibit symptoms such as failure to thrive, lethargy, hypotonia, and neurodegenerative disease (4, 52). This spectrum of symptoms was recapitulated in a conventional knock-out mouse of Ndufs4 in which exon 2 was excised using the Cre recombinase-loxP (Cre-loxP) system (13). The Ndufs4fky/fky mice described here show essentially the same phenotype as the conventional knockout, including the transient hair loss, failure to thrive, and development of neurological defects from P35, which usually resulted in euthanasia of affected mice by P40 according to our ethical guidelines.
Leigh syndrome is characterized by lesions in various parts of the central nervous system, such as the brainstem, basal ganglia, cerebellum, and spinal cord (4). Preliminary neuropathology revealed no gross morphological abnormalities in the brain of 41- and 48-day-old Ndufs4fky/fky mice (supplemental Table 8). In agreement with this observation, genome-wide microarray analysis of the total RNA between whole brain of unaffected and affected P37 mice showed only two differentially expressed genes, both down-regulated, Ndufs4 and Ppdpf. Intensive and sophisticated bioinformatic attempts failed to find even subtly altered relevant biological pathways both with (e.g. keywords brain, neuro, oxidative, ampk, and fatty) and without (MSigDB) prior knowledge.
However, in agreement with the visual loss observed in Ndufs4−/− mice (13), axonal damage was present in optic nerve of Ndufs4fky/fky mice by P58 (supplemental Fig. 3). Initial light microscopic studies in brain regions of Ndufs4−/− mice also did not show differences between wild type and knock-out mice (13). However, a more detailed study published subsequently by the same group revealed neuronal loss in particular brain regions of mice with a late stage disease phenotype, which was suggested to result from progressive glial activation (14). Due to our ethical guidelines, our mice were typically euthanized before the onset of the late stage of disease as defined by Quintana et al. (14). This probably explains why we did not detect significant neuropathological changes in the brain.
Measurement of CI activity, normalized against citrate synthase, detected a 78–90% reduction in brain, heart, liver, and muscle tissues from Ndufs4fky/fky mice (Fig. 3C), which is similar to the levels found in muscle tissue of patients and initially reported in tissues of the conventional Ndufs4−/− mice (13, 52). A subsequent study found somewhat higher activity in the heart, where CI activity was only 56% reduced (53), which is probably due to different sample preparation and assay methods. Similar assays of the other major mitochondrial complexes (CII, CIII, and CIV) showed no differences in activity in Ndufs4fky/fky mice (Fig. 3, C and D). These results provide convincing evidence that the homozygous Ndufs4 mutation has an isolated effect on CI. Reduced CIII activity, which has been found in some NDUFS4 patients (33), was not detected in Ndufs4fky/fky mice. Ndufs4fky/fky mice survive to adulthood at 6 weeks of age, whereas most patients with homozygous Ndufs4 mutations die in infancy (33). This could suggest some functional redundancy of NDUFS4 in mice. However, a more likely explanation is that because mice mature to adulthood in just 6 weeks, their longer survival in developmental terms and milder neuropathology are due to the time required for pathology to result from accumulated tissue damage.
At first glance, the predominantly neurological phenotype of Ndufs4fky/fky mice may seem surprising, considering that the Ndufs4 subunit is expressed ubiquitously and that CI activity was decreased substantially in all tissues studied. However, the selective vulnerability of the nervous system to mitochondrial dysfunction has been noted previously (54). This is partly explained by its high energy requirement, with the brain accounting for ∼20% of oxygen consumption and ∼25% of glucose consumption despite representing only ∼2% of body weight (55). In Ndufs4fky/fky mice, a contributing factor may lie in the observation that CI-linked ATP synthesis capacity was also decreased more markedly in the brain than in the heart (64 and 82% of control values, respectively). Our studies of brain homogenate also probably underestimate the functional consequences of decreased CI activity in some neural cells. Previous studies found that ATP synthesis in rat synaptic mitochondria began to fall when CI activity was decreased by 25%, whereas in non-synaptic mitochondria, there was no effect until CI activity was decreased by 72% (56). Hence, the neurological susceptibility of Ndufs4fky/fky mice probably relates to heterogeneity at the level of CI activity within and between tissues, differing functional thresholds for CI activity loss between and within tissues, and variation in downstream pathways related to cellular function and cell death (57).
Although the phenotypes of Ndufs4−/− (13) and Ndufs4fky/fky mice bear many similarities to the clinical features seen in patients with CI deficiency, there are also differences. Distinctly, the transient fur loss seen in both models does not have an equivalent human symptom, although hair and skin abnormalities have been observed in patients with mitochondrial disease (58–61). Progressive fur loss has been reported in a different mouse model of mitochondrial disease, the Polg “mutator” mouse, which accumulate mitochondrial DNA mutations with increasing age (62, 63). Ndufs4 does not appear to be highly expressed in the skin, but age-specific onset of fur loss suggests temporal regulation of Ndufs4. Future expression analysis of Ndufs4 in skin samples before, during, and after the onset of fur loss may provide further insight.
Analysis of the assembly status of Ndufs4fky/fky CI using BN-PAGE revealed the presence of an ∼830-kDa crippled CI species that could associate into supercomplexes (Fig. 4, A and B). Earlier studies using fibroblasts of patients with Ndufs4 mutations showed the presence of a crippled species that is believed to be an accumulated assembly intermediate of CI (36, 37, 41). As with the low complex I activity found in all tissues, the small amounts of assembled complex I and CI-CIII2 contrast with the relatively high level of ATP synthesis. The latter is measured in fibroblasts whose plasma membrane has been permeabilized with a low concentration of digitonin and which retain an intact mitochondrial inner membrane. CI activity is measured in sonicated tissue homogenates, and BN-PAGE studies use relatively mild detergent conditions but nonetheless solubilize the OXPHOS complexes from the mitochondrial inner membrane. Given that NDUFS4 is involved in CI assembly and stability, these observations together suggest that the amount of fully assembled CI in the intact inner membrane may be higher than that predicted by enzyme assay and BN-PAGE analyses.
Nano-LC-MS/MS analysis was performed to identify the subunits that make up the crippled CI and showed the complete lack of subunits belonging to the NADH oxidation (N) module. This module is believed to be attached as a whole to an intermediate containing all other subunits at the final stage of the assembly into mature CI (Fig. 5) (for a review, see Ref. 64). A CI species lacking the N module has been identified previously in in vitro subunit import studies using control and patient mitochondria (lacking NDUFS4 or NDUFS6) and is considered to be a genuine CI assembly intermediate (41, 64). Supporting this, our study here showed the association of CI assembly factors NDUFAF1 and NDUFAF2 with the crippled complex species, suggesting that the complex does not dissociate under the conditions of BN-PAGE. Although NDUFAF2 has been found associated with the 830-kDa assembly intermediate present in patients with mutations in NDUFS4, this has not been shown previously for NDUFAF1 (39, 40). However, NDUFAF1 has been detected in the 830-kDa assembly intermediate of control mitochondria (38). We note that crippled CI was not detected in the initial report on Ndufs4−/− mice (13) but was described recently in a study that also identified a small complex of ∼200 kDa retaining NADH oxidation activity (53). Those observations are consistent with our results of mass spectrometric analysis of crippled CI.
Measurement of acylcarnitine species is a well characterized method of detecting defects in fatty acid oxidation in humans and mice (65). Our metabolic screen of the blood of Ndufs4fky/fky mice showed an increase in multiple hydroxyacylcarnitine species (Fig. 6, A–C). The exact structure of these hydroxyacylcarnitine species cannot be determined via the tandem mass spectrometry method used in this study. However, 3-hydroxyacylcoenzyme A species are formed during mitochondrial β-oxidation and converted into β-ketoacylcoenzyme A in the NAD+-dependent step of the β-oxidation spiral (for a review, see Ref. 66). Hence, the increase of hydroxyacylcarnitines suggests a block in β-oxidation at the conversion of 3-hydroxyacylcoenzyme A into the corresponding acylcarnitine species. In Ndufs4fky/fky mice, CI appears to lack the module responsible for the conversion of NADH into NAD+. This may cause an imbalance in the NAD+/NADH ratio inside mitochondria and be the reason for the block in fatty acid β-oxidation. Studies in patients suffering from OXPHOS disorders have shown the presence of increased serum alanine levels and/or decreases in citrulline levels (67–70). This was not found in the blood of Ndufs4fky/fky mice, which only had levels of glycine, phenylalanine, and homocitrulline significantly elevated (Fig. 6, D–F). However, these levels were not dramatically changed, and they probably do not impact greatly on the phenotype.
In summary, we have characterized an animal model of CI deficiency that spontaneously arose due to the insertion of a B2 SINE transposable element, using a novel combination of linkage and exon arrays. Biochemically, the Ndufs4fky/fky mice have a CI defect similar to that observed in fibroblasts of patients with mutations in NDUFS4. CI is unstable, resulting in the detachment of the NADH oxidation module with the remaining crippled CI associated with the assembly factors NDUFAF1 and NDUFAF2.
Acknowledgments
Efficient animal management and data collection were facilitated by the Walter and Eliza Hall Institute of Medical Research (WEHI) and Murdoch Childrens Research Institute animal technicians. Processing of histological samples was aided by WEHI Histology and the Integrative Neuroscience Facility. Affymetrix exon arrays and linkage genotyping were performed by the Australian Genome Research Facility, which was established through the Commonwealth-funded Major National Research Facilities Program.
This work was supported by an Australian Postgraduate Award (to S. A. K.), National Health and Medical Research Council (NHMRC) fellowships (to H. H. S, D. R. T., G. K. S., and A. K. V.), NHMRC career development awards (to M. M. and M. B.), NHMRC Program Grant 257501, and NHMRC Project Grants 607403 and 602530, the Australian Research Council (to M. T. R.), the Victorian Government's Operational Infrastructure Support Program, and the Nossal Leadership Award from the Walter and Eliza Hall Institute of Medical Research (to H. S. S.).

This article contains supplemental Tables 1–8 and Figs. 1–3.
D. Wu and G. K. Smyth, manuscript in preparation.
- OXPHOS
- oxidative phosphorylation
- CI–CV
- complex I–V, respectively
- OMIM
- Online Mendelian Inheritance in Man®
- df
- degree of freedom
- SNP
- single nucleotide polymorphism
- BN
- blue native
- Pn
- postnatal day n
- nt
- nucleotide(s)
- SINE
- short interspersed nuclear element
- LINE
- long interspersed nuclear element.
REFERENCES
- 1. Smeitink J. A., Zeviani M., Turnbull D. M., Jacobs H. T. (2006) Mitochondrial medicine. A metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 3, 9–13 [DOI] [PubMed] [Google Scholar]
- 2. Skladal D., Halliday J., Thorburn D. R. (2003) Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain 126, 1905–1912 [DOI] [PubMed] [Google Scholar]
- 3. Tucker E. J., Compton A. G., Calvo S. E., Thorburn D. R. (2011) The molecular basis of human complex I deficiency. IUBMB Life 63, 669–777 [DOI] [PubMed] [Google Scholar]
- 4. Rahman S., Blok R. B., Dahl H. H., Danks D. M., Kirby D. M., Chow C. W., Christodoulou J., Thorburn D. R. (1996) Leigh syndrome. Clinical features and biochemical and DNA abnormalities. Ann. Neurol. 39, 343–351 [DOI] [PubMed] [Google Scholar]
- 5. Kirby D. M., Crawford M., Cleary M. A., Dahl H. H., Dennett X., Thorburn D. R. (1999) Respiratory chain complex I deficiency. An underdiagnosed energy generation disorder. Neurology 52, 1255–1264 [DOI] [PubMed] [Google Scholar]
- 6. Chinnery P., Majamaa K., Turnbull D., Thorburn D. (2006) Treatment for mitochondrial disorders. Cochrane Database Syst. Rev. CD004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klein J. A., Longo-Guess C. M., Rossmann M. P., Seburn K. L., Hurd R. E., Frankel W. N., Bronson R. T., Ackerman S. L. (2002) The harlequin mouse mutation down-regulates apoptosis-inducing factor. Nature 419, 367–374 [DOI] [PubMed] [Google Scholar]
- 8. Joza N., Oudit G. Y., Brown D., Bénit P., Kassiri Z., Vahsen N., Benoit L., Patel M. M., Nowikovsky K., Vassault A., Backx P. H., Wada T., Kroemer G., Rustin P., Penninger J. M. (2005) Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol. Cell Biol. 25, 10261–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pospisilik J. A., Knauf C., Joza N., Benit P., Orthofer M., Cani P. D., Ebersberger I., Nakashima T., Sarao R., Neely G., Esterbauer H., Kozlov A., Kahn C. R., Kroemer G., Rustin P., Burcelin R., Penninger J. M. (2007) Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell 131, 476–491 [DOI] [PubMed] [Google Scholar]
- 10. Ishimura R., Martin G. R., Ackerman S. L. (2008) Loss of apoptosis-inducing factor results in cell type-specific neurogenesis defects. J. Neurosci. 28, 4938–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghezzi D., Sevrioukova I., Invernizzi F., Lamperti C., Mora M., D'Adamo P., Novara F., Zuffardi O., Uziel G., Zeviani M. (2010) Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am. J. Hum. Genet. 86, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ingraham C. A., Burwell L. S., Skalska J., Brookes P. S., Howell R. L., Sheu S. S., Pinkert C. A. (2009) NDUFS4. Creation of a mouse model mimicking a complex I disorder. Mitochondrion 9, 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kruse S. E., Watt W. C., Marcinek D. J., Kapur R. P., Schenkman K. A., Palmiter R. D. (2008) Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab. 7, 312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quintana A., Kruse S. E., Kapur R. P., Sanz E., Palmiter R. D. (2010) Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc. Natl. Acad. Sci. U.S.A. 107, 10996–11001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Phipson B., Smyth G. K. (2010) Permutation P-values should never be zero. Calculating exact P-values when permutations are randomly drawn. Stat. Appl. Genet. Mol. Biol. 9, Article 39 [DOI] [PubMed] [Google Scholar]
- 16. Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., Speed T. P. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 [DOI] [PubMed] [Google Scholar]
- 17. Smyth G. K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article 3 [DOI] [PubMed] [Google Scholar]
- 18. Dai M., Wang P., Boyd A. D., Kostov G., Athey B., Jones E. G., Bunney W. E., Myers R. M., Speed T. P., Akil H., Watson S. J., Meng F. (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33, e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu D., Lim E., Vaillant F., Asselin-Labat M. L., Visvader J. E., Smyth G. K. (2010) ROAST. Rotation gene set tests for complex microarray experiments. Bioinformatics 26, 2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis. A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McKenzie M., Lazarou M., Thorburn D. R., Ryan M. T. (2006) Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients. J. Mol. Biol. 361, 462–469 [DOI] [PubMed] [Google Scholar]
- 22. McKenzie M., Lazarou M., Thorburn D. R., Ryan M. T. (2007) Analysis of mitochondrial subunit assembly into respiratory chain complexes using blue native polyacrylamide gel electrophoresis. Anal. Biochem. 364, 128–137 [DOI] [PubMed] [Google Scholar]
- 23. Johnston A. J., Hoogenraad J., Dougan D. A., Truscott K. N., Yano M., Mori M., Hoogenraad N. J., Ryan M. T. (2002) Insertion and assembly of human tom7 into the preprotein translocase complex of the outer mitochondrial membrane. J. Biol. Chem. 277, 42197–42204 [DOI] [PubMed] [Google Scholar]
- 24. Winkler C., Denker K., Wortelkamp S., Sickmann A. (2007) Silver- and Coomassie-staining protocols. Detection limits and compatibility with ESI MS. Electrophoresis 28, 2095–2099 [DOI] [PubMed] [Google Scholar]
- 25. Pallotti F., Lenaz G. (2001) Isolation and subfractionation of mitochondria from animal cells and tissue culture lines. Methods Cell Biol. 65, 1–35 [DOI] [PubMed] [Google Scholar]
- 26. Sims N. R., Anderson M. F. (2008) Isolation of mitochondria from rat brain using Percoll density gradient centrifugation. Nat. Protoc. 3, 1228–1239 [DOI] [PubMed] [Google Scholar]
- 27. Wanders R. J., Ruiter J. P., Wijburg F. A. (1993) Studies on mitochondrial oxidative phosphorylation in permeabilized human skin fibroblasts. Application to mitochondrial encephalomyopathies. Biochim. Biophys. Acta 1181, 219–222 [DOI] [PubMed] [Google Scholar]
- 28. Rashed M. S., Bucknall M. P., Little D., Awad A., Jacob M., Alamoudi M., Alwattar M., Ozand P. T. (1997) Screening blood spots for inborn errors of metabolism by electrospray tandem mass spectrometry with a microplate batch process and a computer algorithm for automated flagging of abnormal profiles. Clin. Chem. 43, 1129–1141 [PubMed] [Google Scholar]
- 29. van Vlies N., Tian L., Overmars H., Bootsma A. H., Kulik W., Wanders R. J., Wood P. A., Vaz F. M. (2005) Characterization of carnitine and fatty acid metabolism in the long-chain acyl-CoA dehydrogenase-deficient mouse. Biochem. J. 387, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Webster K. E., O'Bryan M. K., Fletcher S., Crewther P. E., Aapola U., Craig J., Harrison D. K., Aung H., Phutikanit N., Lyle R., Meachem S. J., Antonarakis S. E., de Kretser D. M., Hedger M. P., Peterson P., Carroll B. J., Scott H. S. (2005) Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kurts C., Heath W. R., Carbone F. R., Allison J., Miller J. F., Kosaka H. (1996) Constitutive class I-restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 184, 923–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang Y. F., Imam J. S., Wilkinson M. F. (2007) The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76, 51–74 [DOI] [PubMed] [Google Scholar]
- 33. Budde S. M., van den Heuvel L. P., Janssen A. J., Smeets R. J., Buskens C. A., DeMeirleir L., Van Coster R., Baethmann M., Voit T., Trijbels J. M., Smeitink J. A. (2000) Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem. Biophys. Res. Commun. 275, 63–68 [DOI] [PubMed] [Google Scholar]
- 34. Petruzzella V., Vergari R., Puzziferri I., Boffoli D., Lamantea E., Zeviani M., Papa S. (2001) A nonsense mutation in the NDUFS4 gene encoding the 18 kDa (AQDQ) subunit of complex I abolishes assembly and activity of the complex in a patient with Leigh-like syndrome. Hum. Mol. Genet. 10, 529–535 [DOI] [PubMed] [Google Scholar]
- 35. van den Heuvel L., Ruitenbeek W., Smeets R., Gelman-Kohan Z., Elpeleg O., Loeffen J., Trijbels F., Mariman E., de Bruijn D., Smeitink J. (1998) Demonstration of a new pathogenic mutation in human complex I deficiency. A 5-bp duplication in the nuclear gene encoding the 18-kDa (AQDQ) subunit. Am. J. Hum. Genet. 62, 262–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scacco S., Petruzzella V., Budde S., Vergari R., Tamborra R., Panelli D., van den Heuvel L. P., Smeitink J. A., Papa S. (2003) Pathological mutations of the human NDUFS4 gene of the 18-kDa (AQDQ) subunit of complex I affect the expression of the protein and the assembly and function of the complex. J. Biol. Chem. 278, 44161–44167 [DOI] [PubMed] [Google Scholar]
- 37. Iuso A., Scacco S., Piccoli C., Bellomo F., Petruzzella V., Trentadue R., Minuto M., Ripoli M., Capitanio N., Zeviani M., Papa S. (2006) Dysfunctions of cellular oxidative metabolism in patients with mutations in the NDUFS1 and NDUFS4 genes of complex I. J. Biol. Chem. 281, 10374–10380 [DOI] [PubMed] [Google Scholar]
- 38. Dunning C. J., McKenzie M., Sugiana C., Lazarou M., Silke J., Connelly A., Fletcher J. M., Kirby D. M., Thorburn D. R., Ryan M. T. (2007) Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 26, 3227–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ogilvie I., Kennaway N. G., Shoubridge E. A. (2005) A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Invest. 115, 2784–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vogel R. O., van den Brand M. A., Rodenburg R. J., van den Heuvel L. P., Tsuneoka M., Smeitink J. A., Nijtmans L. G. (2007) Investigation of the complex I assembly chaperones B17.2L and NDUFAF1 in a cohort of CI-deficient patients. Mol. Genet. Metab. 91, 176–182 [DOI] [PubMed] [Google Scholar]
- 41. Lazarou M., McKenzie M., Ohtake A., Thorburn D. R., Ryan M. T. (2007) Analysis of the assembly profiles for mitochondrial and nuclear DNA-encoded subunits into complex I. Mol. Cell Biol. 27, 4228–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lawn R. M., Wade D. P., Garvin M. R., Wang X., Schwartz K., Porter J. G., Seilhamer J. J., Vaughan A. M., Oram J. F. (1999) The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J. Clin. Invest. 104, R25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berge K. E., Tian H., Graf G. A., Yu L., Grishin N. V., Schultz J., Kwiterovich P., Shan B., Barnes R., Hobbs H. H. (2000) Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 290, 1771–1775 [DOI] [PubMed] [Google Scholar]
- 44. Gilbert N., Bomar J. M., Burmeister M., Moran J. V. (2004) Characterization of a mutagenic B1 retrotransposon insertion in the jittery mouse. Hum. Mutat. 24, 9–13 [DOI] [PubMed] [Google Scholar]
- 45. Druker R., Whitelaw E. (2004) Retrotransposon-derived elements in the mammalian genome. A potential source of disease. J. Inherit. Metab. Dis. 27, 319–330 [DOI] [PubMed] [Google Scholar]
- 46. Gu Y., Kodama H., Watanabe S., Kikuchi N., Ishitsuka I., Ozawa H., Fujisawa C., Shiga K. (2007) The first reported case of Menkes disease caused by an Alu insertion mutation. Brain Dev. 29, 105–108 [DOI] [PubMed] [Google Scholar]
- 47. Jurka J. (1997) Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. U.S.A. 94, 1872–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cost G. J., Boeke J. D. (1998) Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37, 18081–18093 [DOI] [PubMed] [Google Scholar]
- 49. Dewannieux M., Esnault C., Heidmann T. (2003) LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35, 41–48 [DOI] [PubMed] [Google Scholar]
- 50. Dewannieux M., Heidmann T. (2005) L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J. Mol. Biol. 349, 241–247 [DOI] [PubMed] [Google Scholar]
- 51. Su A. I., Cooke M. P., Ching K. A., Hakak Y., Walker J. R., Wiltshire T., Orth A. P., Vega R. G., Sapinoso L. M., Moqrich A., Patapoutian A., Hampton G. M., Schultz P. G., Hogenesch J. B. (2002) Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 99, 4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Budde S. M., van den Heuvel L. P., Smeets R. J., Skladal D., Mayr J. A., Boelen C., Petruzzella V., Papa S., Smeitink J. A. (2003) Clinical heterogeneity in patients with mutations in the NDUFS4 gene of mitochondrial complex I. J. Inherit. Metab. Dis. 26, 813–815 [DOI] [PubMed] [Google Scholar]
- 53. Calvaruso M. A., Willems P., van den Brand M., Valsecchi F., Kruse S., Palmiter R., Smeitink J., Nijtmans L. (2012) Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Genet. 21, 115–120 [DOI] [PubMed] [Google Scholar]
- 54. DiMauro S., Schon E. A. (2008) Mitochondrial disorders in the nervous system. Annu. Rev. Neurosci. 31, 91–123 [DOI] [PubMed] [Google Scholar]
- 55. Bélanger M., Allaman I., Magistretti P. J. (2011) Brain energy metabolism. Focus on astrocyte-neuron metabolic cooperation. Cell Metab. 14, 724–738 [DOI] [PubMed] [Google Scholar]
- 56. Davey G. P., Peuchen S., Clark J. B. (1998) Energy thresholds in brain mitochondria. Potential involvement in neurodegeneration. J. Biol. Chem. 273, 12753–12757 [DOI] [PubMed] [Google Scholar]
- 57. Dubinsky J. M. (2009) Heterogeneity of nervous system mitochondria. Location, location, location! Exp. Neurol. 218, 293–307 [DOI] [PubMed] [Google Scholar]
- 58. Ostergaard E., Bradinova I., Ravn S. H., Hansen F. J., Simeonov E., Christensen E., Wibrand F., Schwartz M. (2005) Hypertrichosis in patients with SURF1 mutations. Am. J. Med. Genet. A 138, 384–388 [DOI] [PubMed] [Google Scholar]
- 59. Kubota Y., Ishii T., Sugihara H., Goto Y., Mizoguchi M. (1999) Skin manifestations of a patient with mitochondrial encephalomyopathy with lactic acidosis and strokelike episodes (MELAS syndrome). J. Am. Acad. Dermatol. 41, 469–473 [DOI] [PubMed] [Google Scholar]
- 60. Bodemer C., Rötig A., Rustin P., Cormier V., Niaudet P., Saudubray J. M., Rabier D., Munnich A., de Prost Y. (1999) Hair and skin disorders as signs of mitochondrial disease. Pediatrics 103, 428–433 [DOI] [PubMed] [Google Scholar]
- 61. Silengo M., Valenzise M., Spada M., Ferrero G. B., Ferraris S., Dassi P., Jarre L. (2003) Hair anomalies as a sign of mitochondrial disease. Eur. J. Pediatr. 162, 459–461 [DOI] [PubMed] [Google Scholar]
- 62. Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J. N., Rovio A. T., Bruder C. E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R., Törnell J., Jacobs H. T., Larsson N. G. (2004) Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423 [DOI] [PubMed] [Google Scholar]
- 63. Kujoth G. C., Hiona A., Pugh T. D., Someya S., Panzer K., Wohlgemuth S. E., Hofer T., Seo A. Y., Sullivan R., Jobling W. A., Morrow J. D., Van Remmen H., Sedivy J. M., Yamasoba T., Tanokura M., Weindruch R., Leeuwenburgh C., Prolla T. A. (2005) Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309, 481–484 [DOI] [PubMed] [Google Scholar]
- 64. Mckenzie M., Ryan M. T. (2010) Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life 62, 497–502 [DOI] [PubMed] [Google Scholar]
- 65. Matern D. (2008) 3.2 Acylcarnitines, Including in Vitro Loading Tests. in Laboratory Guide to the Methods in Biochemical Genetics, 26th Ed (Blau N., Duran M., Gibson K. M. eds.) pp. 171–206, Springer Verlag, Berlin [Google Scholar]
- 66. Houten S. M., Wanders R. J. (2010) A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis. 33, 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rubio-Gozalbo M. E., Sengers R. C., Trijbels J. M., Doesburg W. H., Janssen A. J., Verbeek A. L., Smeitink J. A. (2000) A prognostic index as diagnostic strategy in children suspected of mitochondriocytopathy. Neuropediatrics 31, 114–121 [DOI] [PubMed] [Google Scholar]
- 68. Naini A., Kaufmann P., Shanske S., Engelstad K., De Vivo D. C., Schon E. A. (2005) Hypocitrullinemia in patients with MELAS. An insight into the “MELAS paradox.” J. Neurol. Sci. 229, 187–193 [DOI] [PubMed] [Google Scholar]
- 69. Parfait B., de Lonlay P., von Kleist-Retzow J. C., Cormier-Daire V., Chrétien D., Rötig A., Rabier D., Saudubray J. M., Rustin P., Munnich A. (1999) The neurogenic weakness, ataxia and retinitis pigmentosa (NARP) syndrome mtDNA mutation (T8993G) triggers muscle ATPase deficiency and hypocitrullinemia. Eur. J. Pediatr. 158, 55–58 [DOI] [PubMed] [Google Scholar]
- 70. Morava E., Hogeveen M., De Vries M., Ruitenbeek W., de Boode W. P., Smeitink J. (2006) Normal serum alanine concentration differentiates transient neonatal lactic acidemia from an inborn error of energy metabolism. Biol. Neonate 90, 207–209 [DOI] [PubMed] [Google Scholar]