Background: Proteases activate ENaC by releasing inhibitory tracts.
Results: Inhibitory peptides cross-link to the finger and thumb domains of ENaC. Simply cross-linking these domains inhibits channel activity.
Conclusion: Inhibitory peptides bind at a finger-thumb interface, inhibiting the channel by maintaining the interface in a tight conformation.
Significance: These observations provide insights regarding the mechanisms of channel activation by proteases.
Keywords: Allosteric Regulation, Enac, Protease, Protein Cross-linking, Protein Structure
Abstract
Proteolysis plays an important role in the maturation and activation of epithelial Na+ channels (ENaCs). Non-cleaved channels are inactive at high extracellular Na+ concentrations and fully cleaved channels are constitutively active. Cleavage of the α and γ subunits at multiple sites activates the channel through the release of imbedded inhibitory tracts. Peptides derived from these released tracts are also inhibitory, likely through binding at the inhibitory tract sites. We recently reported a model of the α subunit. We have now cross-linked Cys derivatives of the inhibitory peptide to the channel, using our model to predict sites at a domain interface of the α subunit that is in proximity to the N terminus of the peptide. Furthermore, peptide inhibition was mimicked in the absence of peptide by cross-linking the channel across the domain interface. Our results suggest a dynamic domain interface that can be exploited by inhibitory peptides and provides a mechanism for peptide inhibition and proteolytic activation.
Introduction
Na+ is the predominant cation in the extracellular fluids. Na+ balance and extracellular fluid volume homeostasis are linked directly. The epithelial Na+ channel (ENaC)2 is crucial to extracellular fluid volume homeostasis through its role in transepithelial Na+ transport in the aldosterone-sensitive distal nephron (1, 2). ENaC also participates in the regulation of airway surface liquid volume by facilitating Na+ transport across airway epithelia. It is a member of the ENaC/degenerin family of ion channels, which are gated by ligands and/or mechanical stimuli, and share several structural and functional features.
Rare among ion channels, ENaC is activated by proteolysis (3). This processing event transitions channels to a high open probability (Po) state and tailors channel function to its biological role in bulk Na+ transport. We have suggested previously that proteolysis provided a mechanism that allowed ENaCs to evolve from other members of the ENaC/degenerin family that reside primarily in the closed state and transiently open in response to external factors (3). Cleavage likely occurs during channel maturation by proteases in the biosynthetic pathway, as well as at the cell surface (3). ENaC proteolysis has been suggested to play a role in disorders such as nephrotic syndrome and cystic fibrosis, where concentrations of specific protease are elevated in the extracellular milieu (4, 5). Activation requires cleavage at multiple sites within the α and/or γ subunits with the liberation of imbedded inhibitory tracts (3, 6, 7). Crucially, it is the release of inhibitory tracts and not cleavage itself that activates ENaC. Consequently, peptides that correspond to the released fragments are inhibitory (3, 6–9). We recently reported that introduction of Trp mutations at a large number of sites in the α subunit reduced the inhibition of channel activity by the α subunit-derived 8-mer peptide, Ac-LPHPLQRL-amide (10). A structural model of the α subunit extracellular region was constructed based on these data and on homology to ASIC1 (11), the only member of the ENaC/degenerin family whose structure has been resolved (12, 13). ASIC1 is homotrimeric, and the extracellular region of each subunit has three peripheral helical domains (termed the thumb, finger, and knuckle) surrounding two central β sheet domains (termed palm and β-ball) that form the core of the structure. Our model placed the bound peptide at the finger-thumb domain interface, suggesting that this domain interface changes conformation upon gating. Motions around this interface were correlated with twisting motions in the pore in normal mode analysis calculations performed on both ASIC1 (14) and our α ENaC model (11).
To test our model of inhibitory peptide binding, we attempted to cross-link Cys derivatives of the peptide to the channel using bifunctional methanethiosulfonate (MTS) compounds. We found that sites at the finger-thumb interface crosslink to the N terminus of the peptide but not the C terminus. We also found sites in the finger domain further from the finger-thumb interface that cross-link to the C terminus of the peptide but not the N terminus. In probing the mechanism of peptide inhibition, we found that cross-linking adjacent sites at the thumb-finger interface trapped the channel in a low activity state. Our data suggest a dynamic finger-thumb interface. Narrowing the interface transitions the channel to a low activity state, providing a mechanism for peptide inhibition and proteolytic ENaC activation.
EXPERIMENTAL PROCEDURES
ENaC Subunit Mutagenesis and Expression
cDNAs for wild type mouse ENaC α, β, and γ subunits in pBluescript SK− vector (Stratagene, La Jolla, CA) were used as templates for point mutations and to synthesize cRNAs. Site-directed mutagenesis was performed using the QuikChange II XL kit (Stratagene) according to the manufacturer's instructions. Targeted mutations were confirmed by direct sequencing. cRNAs were prepared from corresponding cDNAs using the mMESSAGE mMACHINE T3 kit (Ambion/Applied Biosystems, Austin, TX). cRNA concentrations were determined spectroscopically. cRNA size was determined by agarose gel electrophoresis. Oocytes were isolated from adult female Xenopus laevis (NASCO, Plant City, FL) using a protocol approved by the University of Pittsburgh Institutional Animal Care and Use Committee. ENaCs were expressed in stage V–VI X. laevis oocytes by injecting 1 ng of cRNA per subunit. Injected oocytes were maintained at 18 °C in modified Barth's saline (88 mm NaCl, 1 mm KCl, 2.4 mm NaHCO3, 15 mm HEPES, 0.3 mm Ca(NO3)2, 0.41 mm CaCl2, 0.82 mm MgSO4, 10 μg/ml sodium penicillin, 10 μg/ml streptomycin sulfate, 100 μg/ml gentamycin sulfate, pH 7.4). All experiments were performed at ambient temperatures (21–23 °C) 20–30 h following cRNA injection.
Peptides
All peptides were synthesized and HPLC-purified by GenScript Corp. (Piscataway, NJ), and were modified by N-terminal acetylation and C-terminal amidation.
ENaC Current Measurements by Two-electrode Voltage Clamp
Oocytes were mounted in a 20-μl recording chamber (AutoMate Scientific, Berkeley, CA) and perfused at a flow rate of 3–5 ml/min using a high Na+ bath solution (110 mm NaCl, 2 mm KCl, 2 mm CaCl2, 10 mm HEPES, pH 7.4). Electrophysiological measurements were performed using a GeneClamp 500B voltage clamp amplifier (Axon Instruments, Foster City, CA), Clampex software (Axon Instruments). Perfusion was controlled by an eight-channel pinch valve perfusion system (AutoMate Scientific). Stock solutions of peptide, MTS compounds, and amiloride were prepared for dilution into the high Na+ bath solution. Peptides were dissolved in water at 10 mm, whereas amiloride and uncharged MTS compounds were dissolved in dimethyl sulfoxide at 100 and 10 mm, respectively. Uncharged MTS stock solutions were diluted into the aqueous high Na+ bath solution immediately preceding each experiment and used within 5 min of mixing. Solutions of charged 2-(trimethylammonium)ethyl methanethiosulfonate (MTSET) and 2-sulfonatoethyl methanethiosulfonate (MTSES) at 1 mm in high Na+ bath solution were prepared from dry powder immediately before each experiment.
Statistical Analyses
p values were determined by a one-way ANOVA followed a Newman-Keuls post hoc test performed with Igor Pro (Wavemetrics, Lake Oswego, OR) or by a Student's t test performed with Excel (Microsoft Corp., Redmond, WA). Values of p < 0.01 were considered significant.
RESULTS
Cys Derivatives of LPHPLQRL Inhibit ENaC
To identify inhibitory peptides for use in cross-linking experiments, we assessed the ability of Cys derivatives of the inhibitory peptide Ac-LPHPLQRL-amide to inhibit ENaC. The parent peptide has an apparent affinity of 0.9 μm (8, 10). We tested several Cys derivatives: two 9-mers with a Cys added to either end of the parent sequence and five 8-mers with a Cys substitution at interior positions. We found that adding Cys to either the N terminus (N-Cys) or C terminus (C-Cys), or substituting position 6 with Cys had modest effects on peptide inhibition of ENaC currents (Table 1). We examined whether N-Cys or C-Cys could cross-link to ENaC with Cys substitutions at defined sites.
TABLE 1.
Peptide inhibition of wild type ENaC
Peptide inhibition of amiloride-sensitive currents was measured using two-electrode voltage clamp at −100 mV. Titrations were performed from 0.3 to 30 μm peptide, and all currents were normalized to current measured in the absence of peptide in the same experiment. Data were fitted to the Hill equation, with the value of the Hill coefficient fixed at 1, to determine IC50 values (mean ± S.D. of 4–6 experiments).
| Peptide | IC50 |
|---|---|
| μm | |
| LPHPLQRL | 0.9a |
| CLPHPLQRL (N-Cys) | 1.4 ± 0.2 |
| LCHPLQRL | >10 |
| LPCPLQRL | 10 ± 2 |
| LPHCLQRL | 7.5 ± 0.9 |
| LPHPCQRL | >10 |
| LPHPLCRL | 1.8 ± 0.3 |
| LPHPLQRLC (C-Cys) | 1.2 ± 0.2 |
Cross-linking N-Cys to ENaC
Based on our model for the ENaC α subunit (11), the bound inhibitory peptide assumes an extended conformation that places the N and C termini far apart (Fig. 1A). We hypothesized that selected sites near the N terminus of the peptide will cross-link to N-Cys but not to C-Cys. Our model places residues 170–175 and 281–285 of the finger and 473–477 of the thumb near the N terminus of the bound inhibitory peptide (Fig. 1A). We recently showed that N-Cys can be cross-linked to E174C but not E173C (11). We mutated each of the remaining sites to Cys and attempted to cross-link these mutants to both N-Cys and C-Cys using bifunctional MTS compounds with alkanediyl linkers of various lengths. We denote these compounds as MTS-n-MTS, where n indicates the number of carbon atoms between MTS groups.
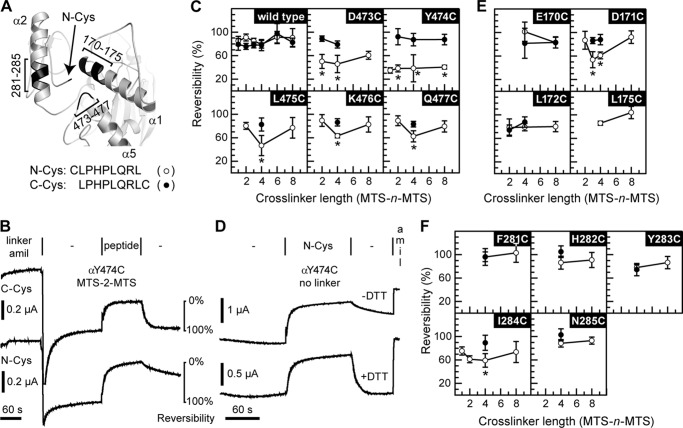
FIGURE 1.
Selected sites in the finger and thumb domains cross-link to N-Cys. A, model of ENaC α subunit finger thumb-domain interface. Position of Cys on the N terminus of the bound inhibitory peptide is indicated by an arrow. Areas predicted to be close to the N-terminal Cys are highlighted (black) and labeled. B, representative example of experiments performed in c, e, and f. Oocytes were injected with 1 ng of mutant or wild type α subunit, and 1 ng each of wild type β and γ subunits. Currents were measured by two-electrode voltage clamp at −60 mV. Channels were labeled by perfusing oocytes with 10 μm MTS-n-MTS (linker) in the presence of 20 μm amiloride (amil) for 2 min, followed by a 3-min wash (−). Channels were then inhibited by either N-Cys or C-Cys (peptide) for 2 min, followed by a 2-min wash to assess reversibility of peptide inhibition. Species tested with either N-Cys (○) or C-Cys (●) include wild type (C), mutants of the loop connecting helices α4 and α5 in the thumb (C), and mutants of helices α1 (E) and α2 (F) in the finger. Values are mean ± S.D. (n = 5–10). An asterisk indicates p < 0.01 versus the same mutant with the alternate peptide and wild type with either peptide at a given cross-linker length using ANOVA and Newman-Keuls post hoc test. D, αY474C was tested for the reversibility of N-Cys inhibition in the absence of MTS compounds. N-Cys was added to oocytes expressing the αY474C mutant for 2 min after a 2-min period establishing the base-line current. The reversibility of N-Cys inhibition after washout ± DTT was then assessed, followed by amiloride addition to determine amiloride-sensitive currents.
We co-expressed wild type and mutant ENaC α subunits with wild type β and γ subunits and measured their currents in a 110 mm Na+ bath using the two-electrode voltage clamp technique. We found that MTS-n-MTS compounds irreversibly inhibited ENaC currents in the absence of inhibitory peptide and that the ENaC pore blocker amiloride attenuated this effect. We therefore labeled channels with 10 μm MTS-n-MTS in the presence of 20 μm amiloride (Fig. 1B). After a wash to establish base-line current, we added either 10 μm N-Cys or 10 μm C-Cys and allowed sufficient time (2 min) for peptide inhibition and reaction with MTS labeled channels. Peptides were then removed from the bath, and the reversibility of peptide inhibition was determined. Wild type channels exhibited an 80–90% reversibility of inhibition with either peptide after labeling with each of the MTS reagents tested (Fig. 1C). Mutant channels showed similar near-complete reversibility for inhibition with C-Cys after treatment with MTS compounds (Fig. 1, C, E, and F). In contrast, N-Cys inhibition was less reversible for several mutants. This effect was dependent on the ENaC Cys mutant and the length of the cross-linker. We interpreted a reduction of inhibition reversibility with a given MTS-n-MTS compound as a specific N-Cys/ENaC mutant crosslink if reversibility was lower than that for C-Cys with the same mutant and for either peptide with wild type ENaC. Of the sites tested in the thumb α4–α5 loop, Cys substitutions at 473–477 each cross-linked to N-Cys (Fig. 1C). Except for Y474C, successful cross-linking depended on the length of the alkanediyl linker of the MTS compound. Of the sites 170–175 at the N terminus of helix α1, we previously examined MTS-4-MTS cross-linking of N-Cys and C-Cys to Cys introduced at Glu-173 and Glu-174 (11). These data provided evidence for cross-linking between E174C and N-Cys but not cross-linking involving E173C. Here, we also found that D171C cross-linked to N-Cys in a linker length-dependent manner (Fig. 1E). This is consistent with residues at positions 171 and 174 facing the same side on a helix near the N terminus of the bound peptide. We also tested Cys substitutions at sites 281–285 (Fig. 1F). Of these, only I284C cross-linked N-Cys.
Our model places Tyr-474 adjacent to the N terminus of the bound peptide. As Y474C/N-Cys cross-linking was not linker length-dependent, we hypothesized that Y474C and N-Cys may be able to form a disulfide bond. We added N-Cys to oocytes expressing Y474C in the absence of pretreatment with MTS compounds (Fig. 1D). Upon removal of peptide from the bath, reversal of inhibition was incomplete (29 ± 8%, n = 7). We repeated the experiment but supplemented the post-peptide wash with the reducing agent DTT. This treatment resulted in complete reversal of N-Cys inhibition of Y474C (99 ± 19%, n = 6, p < 10−5 versus absence of DTT by unpaired Student's t test). In the absence of peptide inhibition, treatment with DTT alone modestly stimulated ENaC currents (19 ± 11%, n = 4). These results suggest that Y474C and N-Cys spontaneously form a disulfide bond.
Cross-linking C-Cys to ENaC
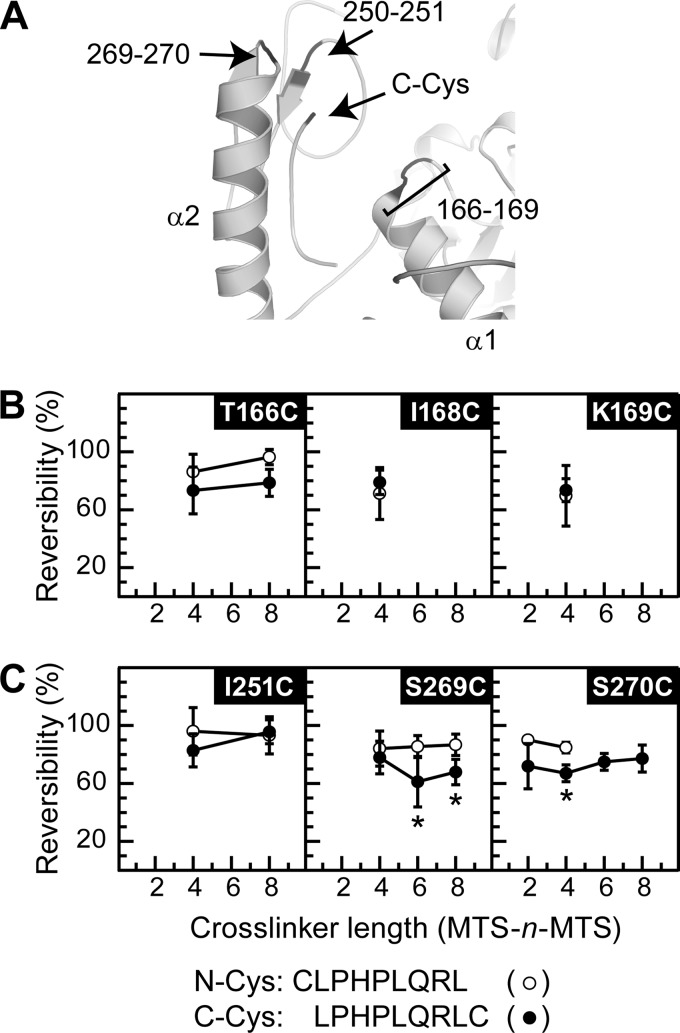
Using the same logic as above, we hypothesized that sites near the C terminus of the peptide may cross-link to C-Cys but not to N-Cys. Our model positions residues 166–169, 250–251, and 269–270 of the finger near the C terminus of the bound inhibitory peptide (Fig. 2A). We mutated each site to Cys and assessed the ability of MTS-n-MTS compounds to cross-link C-Cys and N-Cys to the channel using the protocol described above (Fig. 1B). We interpreted our data as evidence of a specific C-Cys/ENaC mutant cross-link in a manner analogous to that for Fig. 1. We found linker length-dependent C-Cys cross-linking to residues S269C and S270C preceding helix α2 (Fig. 2C). We did not report results for E167C and K250C due to poor functional expression in oocytes. These data are consistent with the bound peptide depicted in our α ENaC model (11). Namely, (i) that the peptide assumes an extended conformation in the bound state, (ii) that the N terminus of the bound peptide lies at the interface of the finger and thumb domains comprised of the 171/174 face of helix α1, the α4–α5 loop of the thumb, and the C-terminal half of helix α2 (Fig. 1), and (iii) that the C terminus of the bound peptide lies near the N terminus of helix α2 (Fig. 2A).
FIGURE 2.
Selected sites in the finger domain cross-link to C-Cys. A, model of ENaC α subunit finger domain focused on the C terminus of the bound inhibitory peptide. Position of Cys on the C terminus of the bound inhibitory peptide is indicated by an arrow. Areas predicted to be close to the C-terminal Cys are highlighted (black) and labeled. Experiments were performed as described in Fig. 1. Mutants tested with either N-Cys (○) or C-Cys (●) include residues preceding helices α1 (B) and α2 (C). Values are mean ± S.D. (n = 5–7). Neither αE167C nor αK250C could be tested due to poor functional expression. *, p < 0.01 versus the same mutant with the alternate peptide and wild type with either peptide (Fig. 1C) at a given cross-linker length using ANOVA and a Newman-Keuls post hoc test.
Cross-linking Finger and Thumb Domains Inhibits ENaC
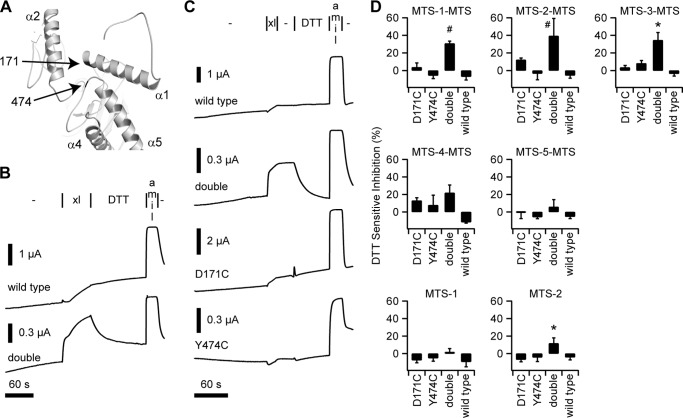
We previously proposed that the 8-mer peptide inhibits ENaC current by damping motions near the finger-thumb interface that are associated with potential gating motions in the pore (11). This hypothesis of peptide inhibition builds upon the proposal by Jasti and colleagues (12), which speculated that relative motions between the finger and thumb domains induced by ligand binding transduce conformational changes through the thumb domain to the pore. Residues D171C of the finger and Y474C of the thumb clearly form cross-links to N-Cys (Fig. 1, C and E), suggesting that these are neighboring residues (Fig. 3A). We hypothesized that we could recapitulate the inhibitory effect of peptide binding, in the absence of peptide, by cross-linking D171C and Y474C.
FIGURE 3.
Conformational trapping using sites in the finger and thumb domains. A, model of the α subunit finger domain highlighting interfacing residues 171 and 474 in the finger and thumb, respectively. B, preliminary experiments with MTS-2-MTS (xl) show the effect of a 1-min addition of the reagent, followed by a 2-min wash with DTT to wild type and αD171C,Y474C (double) channels. C, representative recordings of the effect of adding MTS-2-MTS (xl) for 15 s, followed by a 30-s wash (−), and then a 1-min wash with DTT. Amiloride (amil) was added at the end of each experiment to measure the background current in each experiment. D, DTT-sensitive component of inhibition by various MTS compounds of wild type ENaC, each of the single mutants (D171C and Y474C) and the double mutant. DTT-sensitive inhibition was calculated by dividing the change in current upon DTT addition by the total amiloride-sensitive current. Values are mean ± S.D. (n = 5–10). *, p < 0.01 versus wild type and single mutants using the same MTS reagent, as determined by ANOVA and a Newman-Keuls post hoc test. #, p < 0.01 versus wild type and single mutants using the same MTS reagent and all channels using the analogous monofunctional MTS reagent, as determined by ANOVA and a Newman-Keuls post hoc test.
Preliminary experiments examining the effect of MTS-2-MTS on currents from wild type channels showed a slow time-dependent inhibition of ENaC current that was DTT-insensitive (Fig. 3B). When we examined the effect of MTS-2-MTS on currents from the D171C,Y474C double mutant, we observed a biphasic (rapid followed by a slow phase) inhibition of ENaC current that was partially DTT-sensitive. The slow phase of inhibition for the double mutant was similar to the inhibition time course for wild type. Furthermore, the rapid phase of double mutant inhibition was paralleled by a lag phase for wild type.
To kinetically separate these effects, we performed experiments in which we added MTS-2-MTS for 15 s, corresponding to the fast inhibitory phase, followed by a 30-s wash and subsequent DTT addition (Fig. 3C). Under these conditions, MTS-2-MTS induced a DTT-sensitive inhibition of ENaC currents for the double mutant that was greater than that for wild type or for either the D171C or Y474C single mutants (Fig. 3, C and D). This effect required a bifunctional compound, as DTT-sensitive inhibition was less when using the comparable monofunctional MTS-2 (Fig. 3D). We also observed a length requirement, as we observed a significant effect with the methanediyl, ethanediyl, and propanediyl bis-MTS compounds but not with the bifunctional butanediyl and pentanediyl compounds. These data suggest that alkanediyl linkers with three or fewer carbons sufficiently constrain the distance between D171C and Y474C at the finger-thumb interface to induce channel closure, whereas longer linkers do not. However, we cannot discriminate between the possibilities that the longer linkers insufficiently constrain this interface to influence channel Po or that the longer linkers do no react with both introduced Cys residues. The longer linkers were active in our assays as DTT-insensitive inhibition generally increased with the length of the linker (see supplemental Table 1).
Nature of Inhibition via Finger-thumb Domain Interface
Based on our finding that constraining the finger and thumb domains to a tight conformation resulted in channel inhibition, we predicted that pushing them apart would increase currents. We tested our hypothesis using electrostatics. Helix α1 in our model spans residues 170–187 and hosts seven residues bearing carboxylate groups and one Arg. Using the Y474C mutant, we tested whether adding a positively or negatively charged group to the α4–α5 loop in the thumb domain would change channel current. Upon addition of positively charged MTSET, currents decreased rapidly (Fig. 4A). The loss of current was sustained after MTSET was removed, suggesting a covalent modification. Finally, DTT reversed the effect of MTSET, consistent with a covalent modification at the introduced Cys. These data reinforce the notion that a tight finger-thumb domain interface is synonymous with channel inhibition. Upon addition of negatively charged MTSES, currents also decreased. Current reduction was sustained following MTSES removal, suggesting covalent modification. But in contrast to MTSET, DTT did not reverse the effect of MTSES. As reported by others, we found that neither MTSET nor MTSES affected wild type ENaC currents.
FIGURE 4.
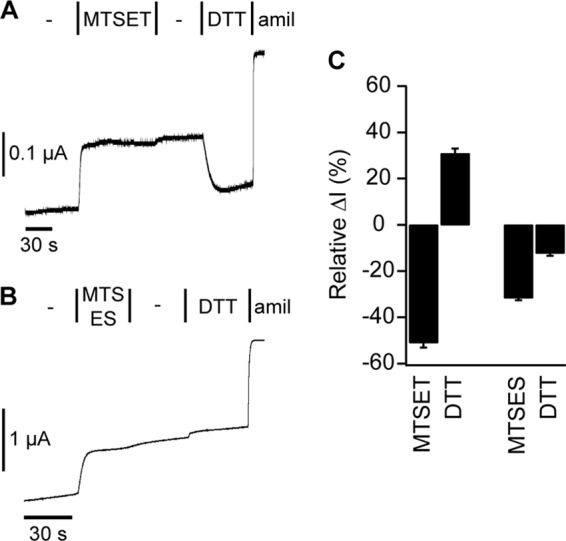
Chemical modification adding a charged group to αY474C is inhibitory. 1 mm solutions of positively charged MTSET (A) or negatively charged MTSES (B) in high Na+ buffer were added to Y474C channels for a sufficient amount of time to achieve a saturated effect. After a wash to remove MTS compounds, DTT was added. Amiloride (amil) was added at the end of each experiment to determine the ENaC-specific current. C, the change in current upon MTS compound or DTT addition was measured relative to the total amiloride-sensitive current immediately prior to MTS or DTT addition. Values are mean ± S.D. (n = 7).
We were able to inhibit channel currents using MTS reagents to cross-link the finger and thumb domains. We also were able to reproduce this effect using electrostatics. In each case, we found that currents were reduced by 50% or less. It is possible that either a fraction of the channels were cross-linked resulting in channels unable to open or that nearly all of the channels were cross-linked, resulting in channels with a reduced Po. To distinguish between these two possibilities, we added MTS-2-MTS in three 15-s pulses separated by 30-s intervals to D171C,Y474C mutant channels. In the former case, we predict that additional pulses of MTS-2-MTS would inhibit channels further. In the latter case, we predict that additional MTS-2-MTS pulses would have little effect. We found that the first pulse reduced ENaC currents by 35%, and that the second and third pulses reduced ENaC currents by a further 4 to 6% each (Fig. 5A). Similar experiments performed with wild type ENaC resulted in a 6–7% change in current with each MTS-2-MTS pulse. These data are consistent with the notion that a tight conformation at finger-thumb interface is associated with a lower Po but not exclusively associated with the closed state of the channel.
FIGURE 5.
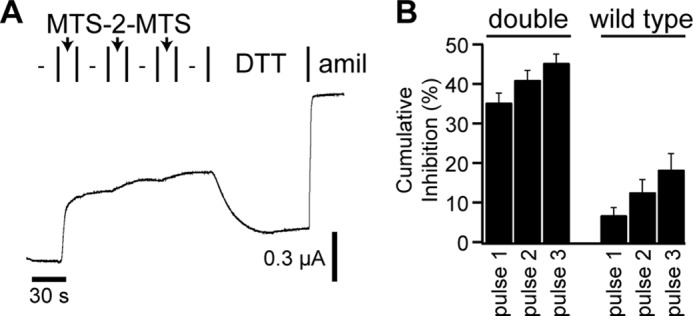
Saturated cross-linking across the α subunit finger-thumb interface does not fully inhibit ENaC. MTS-2-MTS was added in three 15-s pulses (arrows) to wild type or αD171C,Y474C double mutants, with 30-s washes between pulses. DTT was then added followed by amiloride (amil). A, representative recording. B, the cumulative inhibition following each MTS-2-MTS pulse, normalized to the total amiloride-sensitive current immediately prior to the first MTS-2-MTS pulse. Values are mean ± S.D. (n = 5 (wild type) or n = 8 (double)).
DISCUSSION
Our α ENaC model places the N terminus of the inhibitory 8-mer peptide at the finger-thumb interface, and its C terminus near the beginning of helix α2, far from the finger-thumb interface (11). Cross-linking data are clearly consistent with this model of inhibitory peptide binding. We hypothesized that the peptide draws the finger and thumb together in the peptide bound state, which is inhibitory. Peptide release frees the finger and thumb to move, allowing the channel to more readily assume an open conformation. This idea is supported by our demonstration that cross-linking D171C and Y474C across the finger-thumb interface reduces channel activity. These sites were selected because they each cross-linked with the N-Cys peptide and were in close proximity in our α ENaC model. The inhibitory effect of cross-linking D171C and Y474C demonstrated a length requirement, suggesting that linkers with three or fewer carbons sufficiently constrained the finger-thumb interface to favor the closed channel conformation. Longer linkers either did not cross-link or allowed sufficient mobility at the interface so as to have little effect. We also reproduced the cross-linking effect by placing opposing charges at this interface. Taken together, these data are the first experimental evidence of movement between the finger and thumb domains of ENaC and show that such rearrangements affect channel gating. The stabilization of a specific conformation at a dynamic domain interface provides a mechanism for the allosteric inhibition of ENaC by peptides. We propose that proteolytic ENaC activation functions in an analogous manner by releasing a constraint at the finger-thumb interface.
In all of our experiments designed to constrain movements between the α4–α5 loop of the thumb domain and helix α1 of the finger domain, we never observed >50% inhibition despite saturating the effect of the MTS reagent. We draw two important conclusions from this observation. First, the gate does not change its conformation in a lockstep mechanism with the finger-thumb interface. Cross-linked channels still transition between closed and open states but likely exhibit a lower Po than their non-cross-linked counterparts. Second, the 26-mer and 8-mer peptides derived from the 206–231 inhibitory tract also inhibit ENaC through additional mechanisms. We conclude this based on the fact that each peptide can achieve 80–90% inhibition at saturation. Our model postulates extensive interactions between the peptide and other parts of the finger domain, which suggests additional structures have a role in peptide inhibition.
Our data link dynamic changes at the finger-thumb interface to functional changes at the channel gate. Although we were able to inhibit the channel by exploiting this interface, we were not able to stimulate the channel. We attempted to stimulate ENaC currents by introducing like charges at the interface. Modification of Y474C channels with MTSES led to reduced channel activity, which was an unexpected result. Furthermore, DTT did not reverse the effect of MTSES, in contrast to its effect on MTSET-modified Y474C channels. It is possible that MTSES modification led to a change in the conformation of the α4–α5 loop that prevented DTT access to the modified Cys at position 474. Alternatively, MTSES modification led to misfolding of the α4–α5 loop that could not be reversed even after MTSES modification was reversed by DTT. We speculate that rather than pushing helix α1 and the α4–α5 loop apart, MTSES modification of Y474C led to a non-reversible thumb loop conformational change that reduced channel currents.
The thumb domains of the ENaC/degenerin family of ion channels are poorly conserved yet share two important features: two predicted helices and 10 absolutely conserved Cys that form a five-disulfide bond ladder (12, 15). These hallmark features of ENaC/degenerin channels suggest that the antiparallel arrangement of the two thumb helices in ASIC1 is widely conserved. In the ASIC1 structure, the thumb connects to the palm near the opening of the pore and extends up from the membrane toward the finger. The thumb also interacts with the β-ball and finger of the same subunit and the palm of a neighboring subunit. Our data suggests that the 8-mer inhibitory peptide exploits the finger-thumb interface to stabilize the channel in the closed conformation. Recent data suggest that Cl−, which inhibits ENaC, binds at the thumb-palm intersubunit interface (16, 17). It remains to be determined whether the relative finger-thumb movements also involve the thumb-palm intersubunit interface. Furthermore, it is unclear whether inhibitory peptides, Cl−, and other extracellular allosteric inhibitors (e.g. Na+) recruit the same allosteric machinery but use different sites to trigger conformational changes. The rigidity implied by the presence of five disulfide bonds in the thumb suggests that movement at any thumb interdomain interface will induce movement at other thumb interdomain interfaces. We reported recently that mutations in the thumb near the outer mouth of the pore modulated the efficacy of multiple extracellular stimuli, suggesting mechanistic convergence (18). The remarkably similar kinetics of peptide, Cl−, and Na+ inhibition hint at a common rate-limiting step (10, 16, 19, 20). As ENaC/degenerin channels have evolved to respond to numerous extracellular stimuli, the basic architecture of the thumb domain seems to have been preserved. In this work, we found evidence for movement at a finger-thumb interface where an inhibitory peptide binds. Influencing interdomain interfaces that involve the thumb may be a general mechanism for extracellular ligand gating among ENaC/degenerin channels.
Acknowledgment
We thank Michael Grabe for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK078734 (to O. B. K.), DK065161 (to T. R. K.), DK084060 (to M. D. C.), and DK079307 (the Pittsburgh Center for Kidney Research).

This article contains supplemental Table 1.
- ENaC
- epithelial Na+ channel
- Po
- open probability
- MTS
- methanethiosulfonate
- MTSET
- 2-(trimethylammonium)ethyl methanethiosulfonate
- MTSES
- 2-sulfonatoethyl methanethiosulfonate
- ANOVA
- analysis of variance
- N-Cys
- CLPHPLQRL
- C-Cys
- LPHPLQRLC.
REFERENCES
- 1. Kashlan O. B., Kleyman T. R. (2011) ENaC structure and function in the wake of a resolved structure of a family member. Am. J. Physiol. Renal Physiol. 301, F684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhalla V., Hallows K. R. (2008) Mechanisms of ENaC regulation and clinical implications. J. Am. Soc. Nephrol. 19, 1845–1854 [DOI] [PubMed] [Google Scholar]
- 3. Kleyman T. R., Carattino M. D., Hughey R. P. (2009) ENaC at the cutting edge: regulation of epithelial sodium channels by proteases. J. Biol. Chem. 284, 20447–20451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Passero C. J., Mueller G. M., Rondon-Berrios H., Tofovic S. P., Hughey R. P., Kleyman T. R. (2008) Plasmin activates epithelial Na+ channels by cleaving the γ subunit. J. Biol. Chem. 283, 36586–36591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caldwell R. A., Boucher R. C., Stutts M. J. (2005) Neutrophil elastase activates near-silent epithelial Na+-channels and increases airway epithelial Na+-transport. Am. J. Physiol. Lung Cell Mol. Physiol. 288, L813–819 [DOI] [PubMed] [Google Scholar]
- 6. Carattino M. D., Sheng S., Bruns J. B., Pilewski J. M., Hughey R. P., Kleyman T. R. (2006) The epithelial Na+ channel is inhibited by a peptide derived from proteolytic processing of its a subunit. J. Biol. Chem. 281, 18901–18907 [DOI] [PubMed] [Google Scholar]
- 7. Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., Kleyman T. R. (2007) Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the γ subunit. J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 8. Carattino M. D., Passero C. J., Steren C. A., Maarouf A. B., Pilewski J. M., Myerburg M. M., Hughey R. P., Kleyman T. R. (2008) Defining an inhibitory domain in the α subunit of the epithelial sodium channel. Am. J. Physiol. Renal Physiol. 294, F47–52 [DOI] [PubMed] [Google Scholar]
- 9. Passero C. J., Carattino M. D., Kashlan O. B., Myerburg M. M., Hughey R. P., Kleyman T. R. (2010) Defining an inhibitory domain in the γ subunit of the epithelial sodium channel. Am. J. Physiol. Renal Physiol. 299, F854-F861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kashlan O. B., Boyd C. R., Argyropoulos C., Okumura S., Hughey R. P., Grabe M., Kleyman T. R. (2010) Allosteric Inhibition of the Epithelial Na+ Channel (ENaC) through Peptide Binding at Peripheral Finger and Thumb Domains. J. Biol. Chem. 285, 35216–35223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kashlan O. B., Adelman J. L., Okumura S., Blobner B. M., Zuzek Z., Hughey R. P., Kleyman T. R., Grabe M. (2011) Constraint-based, homology model of the extracellular domain of the epithelial Na+ channel α subunit reveals a mechanism of channel activation by proteases. J. Biol. Chem. 286, 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 13. Gonzales E. B., Kawate T., Gouaux E. (2009) Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 460, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang H., Yu Y., Li W. G., Yu F., Cao H., Xu T. L., Jiang H. (2009) Inherent dynamics of the acid-sensing ion channel 1 correlates with the gating mechanism. PLoS Biol. 7, e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheng S., Maarouf A. B., Bruns J. B., Hughey R. P., Kleyman T. R. (2007) Functional role of extracellular loop cysteine residues of the epithelial Na+ channel in Na+ self-inhibition. J. Biol. Chem. 282, 20180–20190 [DOI] [PubMed] [Google Scholar]
- 16. Collier D. M., Snyder P. M. (2009) Extracellular chloride regulates the epithelial sodium channel. J. Biol. Chem. 284, 29320–29325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Collier D. M., Snyder P. M. (2011) Identification of epithelial Na+ channel (ENaC) intersubunit Cl− inhibitory residues suggests a trimeric αγβ channel architecture. J. Biol. Chem. 286, 6027–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shi S., Ghosh D. D., Okumura S., Carattino M. D., Kashlan O. B., Sheng S., Kleyman T. R. (2011) Base of the thumb domain modulates epithelial sodium channel gating. J. Biol. Chem. 286, 14753–14761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sheng S., Bruns J. B., Kleyman T. R. (2004) Extracellular histidine residues crucial for Na+ self-inhibition of epithelial Na+ channels. J. Biol. Chem. 279, 9743–9749 [DOI] [PubMed] [Google Scholar]
- 20. Chraïbi A., Horisberger J. D. (2002) Na self inhibition of human epithelial Na channel: Temperature dependence and effect of extracellular proteases. J. Gen. Physiol. 120, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]