Abstract
Duplication (dup7q11.23) and deletion (Williams syndrome) of chromosomal region 7q11.23 cause neurodevelopmental disorders with contrasting anxiety phenotypes. We found that 30% of 4- to 12-year-olds with dup7q11.23 but fewer than 5% of children with WS or in the general population met diagnostic criteria for a separation-anxiety disorder. To address the role of one commonly duplicated or deleted gene in separation anxiety, we compared mice that had varying numbers of Gtf2i copies. Relative to mouse pups with one or two Gtf2i copies, pups with additional Gtf2i copies showed significantly increased maternal separation-induced anxiety as measured by ultrasonic vocalizations. This study links the copy number of a single gene from 7q11.23 to separation anxiety in both mice and humans, highlighting the utility of mouse models in dissecting specific gene functions for genomic disorders that span many genes. This study also offers insight into molecular separation-anxiety pathways that might enable the development of targeted therapeutics.
Main Text
Anxiety disorders are the most common form of psychopathology in childhood, and separation-anxiety disorder (SAD) is among the most frequent anxiety diagnoses.1 Based on diagnostic interviews conducted with parents, the prevalence of SAD in children has been estimated as ∼2.5%.1,2 Anxiety disorders show strong heritability, but despite researchers' considerable efforts to identify genetic factors, no robust molecular genetic linkages have yet been demonstrated in humans.3
Williams syndrome4 (WS [MIM 194050]) and duplication of chromosomal region 7q11.235 (dup7q11.23 [MIM 609757]) are genomic disorders caused by the deletion and duplication, respectively, of a common 1,500,000 bp segment spanning 26 genes on human chromosome 7. These syndromes are both associated with neurocognitive and behavioral features. Specifically, intellectual disability, relative strength in language and considerable weakness in visuospatial construction, social disinhibition, and nonsocial anxiety are associated with WS.6–8 In contrast, dup7q11.23 is associated with speech disorder, language delay, and both social and nonsocial anxiety.8,9 The genomic overlap and penetrant but contrasting social-anxiety phenotypes associated with these rare neurodevelopmental disorders might help establish direct links with genes and pathways that play a role in particular anxiety disorders.
Anecdotal reports of separation difficulties in young children with dup7q11.23 (C.B.M., unpublished data) led us to focus our investigation on separation anxiety. We measured separation anxiety in children with dup7q11.23 or WS by using the Anxiety Disorders Interview Schedule: Parent Version10 (ADIS-P) for the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) and the Child Behavior Checklist for Ages 1.5–5 (CBCL 1.5–5).11 All procedures were approved by the institutional review board at the University of Louisville, and written informed consent was obtained from the parents of all participants. 7q11.23 deletion or duplication size was determined by fluorescence in situ hybridization with labeled probes (purified from bacterial artificial chromosomes [BACs], plasmid artificial chromosomes [PACs], and cosmids) that bound to metaphase or interphase chromosomes prepared from established lymphoblastoid cell lines, as previously described.12 Only children with classic deletions or duplications were included. Demographic characteristics of the child participants are presented in Table 1.
Table 1.
Participant Characteristics for ADIS-P and CBCL 1.5–5 Samples
| Sample Characteristics |
Group |
|
|---|---|---|
| Dup7q11.23 | Williams Syndrome | |
| ADIS-P | ||
| Sample size | 27 (12 girls, 15 boys) | 214 (107 girls, 107 boys) |
| Chronological age | mean: 7.78 years (SD = 2.78, range: 4.03–12.93) | mean: 8.19 years (SD = 2.74, range: 4.07–12.96) |
| CBCL 1.5–5 | ||
| Sample size | 18 (11 girls, 7 boys) | 181 (92 girls, 89 boys) |
| Chronological age | mean: 3.87 years (SD = 1.19, range: 2.03–5.81) | mean: 4.31 years (SD = 1.11, range: 2.08–5.81) |
The ADIS-P,10 a semistructured interview designed to assess anxiety and related disorders in children aged 4–16 years, has been used in several studies of children with WS6,13–15. As part of a larger study of the development of children with 7q11.23 deletion or duplication, parents completed the ADIS-P,10 which was used for determining diagnoses of SAD. The ADIS-P has excellent reliability for SAD as well as excellent test-retest reliability for the interview.16 Interviewers were licensed clinical psychologists or advanced clinical-psychology doctoral students who had completed a rigorous training procedure (see Woodruff-Borden et al.15). All interview protocols were reviewed with the supervising clinical psychologist, who concurred on all diagnoses of SAD. A child was diagnosed with SAD if he or she met the DSM-IV diagnostic criteria, including significant distress or impairment in functioning.
Parents also completed the CBCL 1.5–5,11 a standardized questionnaire composed of 99 items describing behavioral, emotional, and social problems. For this questionnaire, parents rate each item on a 3 point scale—0 (not true), 1 (somewhat or sometimes true), or 2 (very true or often true)—on the basis of their child's behavior during the preceding 2 months. Parental response to item 37 (“Gets too upset when separated from parents”) was used as a measure of children's separation difficulties.
Statistical analyses were performed with SPSS version 20. Confidence intervals (CIs) for proportions were calculated with the adjusted Wald method on a CI calculator.
On the basis of the ADIS-P interview, 8 of 27 children with dup7q11.23 and 9 of 214 children with WS met DSM-IV criteria for SAD, including the presence of interference or distress. Binomial tests comparing these proportions to the proportion of children in the general population (0.0232) indicated that the proportion of dup7q11.23-affected children who had SAD (0.296) was significantly higher than the general-population proportion (p < .0001, dup7q11.23 CI.95 = [0.157, 0.487]). The proportion of WS-affected children who had SAD (0.042) did not differ significantly from the general-population proportion (p = 0.102, WS CI.95 = [0.021, 0.079]). A comparison of the proportions of children with SAD in the dup7q11.23 group and the WS group—with the use of the WS group's proportion as an estimate of the WS population value—indicated that SAD was significantly more common among children with dup7q11.23 than among children with WS (p < 0.0001).
To provide a more conservative estimate of possible differences in prevalence of SAD between children with dup7q11.23 or WS and children in the general population, we compared the prevalence rates for the participants in our study to the prevalence rate reported by Leyfer and her colleagues6 for 84 typically developing siblings of children with WS (0.036). SAD was determined on the basis of parental interviews with the use of the ADIS-P, the same diagnostic interview that was used for assessing SAD for the children with dup7q11.23 or WS in the present study. A Fisher exact test indicated that the prevalence of SAD was significantly higher for children with dup7q11.23 than for Leyfer et al.'s typically developing sample (p = 0.005). However, there was no difference in prevalence of SAD between children with WS and the children in Leyfer et al.'s typically developing sample (p = 1.00).
Parental responses to item 37 (addressing separation difficulty) on the CBCL 1.5–511 are shown in Table 2. A binomial test indicated that when difficulty with separation was defined as a score of 2 on item 37, the proportion of dup7q11.23-affected children who had difficulty separating from their parents (0.333) was significantly higher than the proportion of children with WS (0.011) (p < .0001, dup7q11.23 CI.95 = [0.161, 0.564]). Similarly, when difficulty with separation was defined as a score of 1 or 2 on item 37, the dup7q11.23 proportion (0.611) was significantly higher than the WS proportion (0.182) (p < .0001, dup7q11.23 CI.95 = [0.384, 0.798]). In both these analyses, the WS group's proportion was used as an estimate of the WS population value.
Table 2.
Proportion of Parental Responses to Item 37a as a Function of CBCL 1.5–5 Response Category
| CBCL 1.5–5 Response Category |
Group |
|
|---|---|---|
| Dup7q11.23 (n = 18) | Williams Syndrome (n = 181) | |
| 0 (not true) | 0.389 | 0.818 |
| 1 (somewhat or sometimes true) | 0.278 | 0.171 |
| 2 (very true or often true) | 0.333 | 0.011 |
“Gets too upset when separated from parents.”
To test the hypothesis that SAD and separation difficulties in dup7q11.23 might be linked with dosage of a single gene, we generated mice with decreased or increased genomic copy number of general transcription factor 2I (Gtf2i [MIM 601679]), a gene from the commonly deleted or duplicated region. Gtf2i is highly expressed in the prenatal and postnatal developing brain17,18 and acts as both a transcriptional activator19 and a regulator of intracellular calcium levels.20 In mouse pups, separation anxiety can be assessed through ultrasonic vocalizations (USVs).21 Mouse pups emit 50–120 kHz USVs in response to maternal separation, as well as to other stressful stimuli.21 These pup USVs are believed to increase mother-infant social contact after separation and prompt retrieval by the mother. Maternal separation-induced USVs are strongly associated with anxiety and are an ethologically validated measure for preclinical characterization of anxiolytic drugs.22
Mice with a genomic duplication of Gtf2i were generated with Cre-loxP technology in combination with an in vivo breeding strategy (Figure 1A). Independent mouse lines with loxP sites at the desired endpoints were generated: Gtf2ird1Gt(XS0608)Wtsi was created as described previously,23 and Gtf2i3′UTRloxP was created by gap-repair targeting with clones from a 5′/3′ phage library.24 The targeting vector included a 9 kb Gtf2i fragment spanning from exon 25 to the 3′UTR, resulting in the duplication of these exons downstream of Gtf2i and the insertion of a loxP site after recombination between the vector and the endogenous locus. Aggregation of correctly targeted embryonic stem cells with morula-stage embryos generated chimeras, which were bred to CD1 females for the production of Gtf2i+/3′UTRloxP mice.
Figure 1.
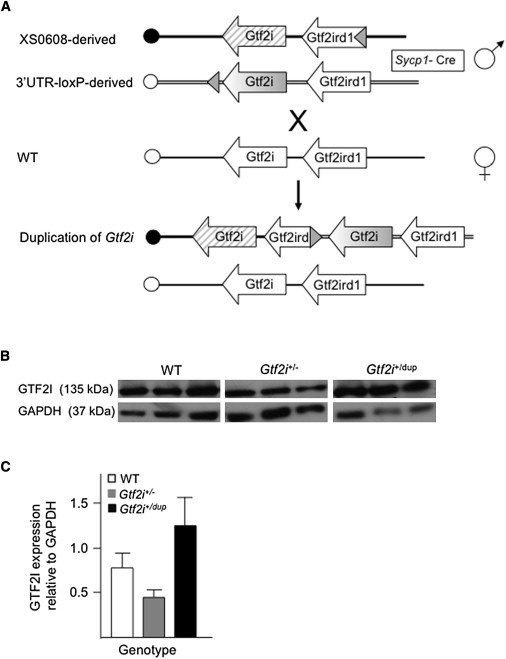
Generation of Mice with Decreased or Increased Gtf2i Dosage
(A) Schematic representation of the generation of mice with duplication of Gtf2i. XS0608 contains a loxP site inserted into intron 4 of Gtf2ird1, whereas the 3′UTR-loxP line contains a loxP site downstream of the last coding exon of Gtf2i. Recombination between the loxP sites in vivo resulted in duplication of the entire Gtf2i gene in the male gamete of transloxer male mice carrying both loxP sites in trans, as well as a Cre transgene under the control of the Sycp1 promoter. Duplicated chromosomes were transmitted to offspring upon crossing with wild-type (WT) females. The centromere of mouse chromosome 5 is represented by the circle at the left end of each diagram. LoxP sites are represented by a shaded triangle.
(B) Immunoblot analysis of GTF2I from whole brain of P8 mice with the use of a polyclonal antibody (610943, BD Biosciences, Franklin Lakes, NJ). GAPDH (ab36840, Abcam, Cambridge, MA) was used as a control for protein loading (50 μg per lane). Representative blots from three independent pups are shown.
(C) A graph (created with Image J [National Institutes of Health]) comparing the densitometric measurement of GAPDH with that of GTF2I in whole brain of P8 mice. Data are represented as mean ± SEM (n = 3 for each group).
To generate the Gtf2i duplication, Gtf2ird1Gt(XS0608)Wtsi mice were crossed with Gtf2i3′UTRloxP mice that also carried a Cre transgene under the control of the Sycp1 promoter (Sycp1-Cre).25 “Transloxer” males carrying both the Gtf2ird1Gt(XS0608)Wts and Gtf2i3′UTRloxP alleles as well as the Sycp1-Cre transgene were crossed with CD1 females (Figure 1A), and the offspring were screened by PCR with primer set GtDup, which generated a 250 bp amplicon specific for the duplication. We used quantitative PCR (qPCR) to characterize mice carrying the duplication (Gtf2i+/Dup) in order to identify changes in copy number of Gtf2i exons 5 and 30. Primer sequences are shown in Table S1, available online. We generated Gtf2i+/Dup mice, Gtf2iDup/Dup mice, and wild-type (WT) littermates through the intercrossing of Gtf2i+/Dup mice to minimize variation of the maternal environment and provide within-litter controls.
Gtf2i+/− mice contain a genetrap insertion within intron 3 of Gtf2i, and this insertion creates a null allele (Gtf2iGt(YTA365)Byg/+).26 Gtf2i−/− mice died during embryogenesis. Mice were generated by injection of Gt(YTA365)Byg embryonic stem cells into C57BL/6 blastocysts, were maintained on a CD1 background, and had reached the third generation of backcrossing at the time of these experiments. We generated Gtf2i+/− mice and WT littermates through the intercrossing of Gtf2i+/− mice to minimize variation of the maternal environment and provide within-litter controls. All procedures were approved by the University of Toronto Animal Care Committee and were carried out in compliance with the Canadian Council on Animal Care guidelines.
An approximate 50% reduction in Gtf2i mRNA expression and protein levels was seen in whole adult brain tissue from Gtf2i+/− mice, and an approximate 50% increase in Gtf2i mRNA expression and protein levels was seen in brain tissue from Gtf2i+/Dup mice (Figures 1B and 1C). Expression of flanking genes Gtf2ird1 (MIM 604318) and Clip2 (MIM 603432) was assessed by real-time PCR as described previously27 and was found to be unchanged (data not shown).
We recorded ultrasonic vocalization in mouse pups with one (Gtf2i+/−, n = 11), two (WT, n = 49), three (Gtf2i+/Dup, n = 30), or four (Gtf2iDup/Dup, n = 18) Gtf2i copies during a 4 min separation from their mother. Mice were housed with access to food and water ad libitum and were on a 12 hr light/dark cycle throughout the experiments. Pups came from litters of between 5 and 13 animals and lived with both parents in their home cage. Date of birth was considered postnatal day (P) 0, and pups were tattooed for identification on the hind paw at P6 by the subcutaneous insertion of dye near the end of the toe with the use of capillary action from a 20G needle. Data were pooled across all male and female pups within genotype groups and were collapsed across litters as in previously reported studies.28
Maternal separation-induced USVs were recorded on P8 between 10 am and 6 pm. The family cage containing the litter of pups and their parents was transported to the procedure room, illuminated by two 40 W red bulbs. Each pup was separated from the litter one at a time in random order and placed in a shallow plastic beaker (height = 6 cm, diameter = 4 cm) in a sound-attenuating chamber (40 × 25 × 30 cm). The ultrasound detector D1000X (Pettersson Elektronik AB, Uppsala, Sweden) was suspended 12 cm above the floor of the beaker. After a 5 s habituation period, USV emissions for the next 4 min at a sampling frequency of 250 kHz were recorded. The primary dependent variable was the total number of USVs. Immediately after USV recording, the pups were weighed and returned to the home cage. Mean weight was 5.45 g (standard deviation [SD] = 0.69) for the Gtf2i+/− group, 5.29 g (SD = 0.87) for the Gtf2i+/+ group, 5.67 g (SD = 0.99) for the Gtf2i+/Dup group, and 5.56 g (SD = 0.98) for the Gtf2iDup/Dup group, indicating a healthy nutritional status for each group. The results of a Kruskal-Wallis test indicated that body weight did not differ significantly as a function of genotype (p = 0.36). Accordingly, body weight was not included as a factor in subsequent analyses.
Approximately half of the pups were tested in beakers with clean materials (cotton pads), and half were tested with soiled cotton nest material in counterbalanced orders. A preliminary Mann-Whitney U-test indicated that the distributions of the median number of USVs produced by mouse pups during the 4 min separation period did not differ significantly as a function of bedding type (nest or clean) (p = 0.82). Accordingly, bedding type was not included as a factor in subsequent analyses. Spectrographs (20–125 kHz) were generated by discrete Fourier transformation (256 bins) and analyzed with Avisoft SASLab Pro Software v.4.39 (Avisoft Bioacoustics, Berlin, Germany). Sonograms were analyzed by a trained coder who was blind to genotype. A second trained coder, also blind to genotype, independently analyzed the sonograms of two litters (two Gtf2i+/−, six Gtf2i+/+, three Gtf2i+/Dup, and four Gtf2iDup/Dup). The means and SDs for the total number of USVs counted by the two coders for the 4 min separation period were very similar (coder 1: mean = 305.87, SD = 197.03; coder 2: mean = 307.60, SD = 195.98), and reliability was excellent (r = 0.999, p < 0.0001).
Because of violations of the assumptions of normality and homogeneity of variance and because of unequal numbers of mouse pups across genotypes, nonparametric statistics were used for the analysis of the mouse USV data. The distributions of the numbers of USVs produced during the 4 min separation period are indicated separately for each genotype in Figure 2A. The median number of USVs produced in each minute of the separation period as a function of genotype is shown in Figure 2B. A Kruskal-Wallis test indicated a significant difference among genotypes in the median number of USVs produced over the 4 min period (standardized test statistic = 15.95, p < 0.001). Follow-up stepwise-stepdown tests identified two homogeneous subsets. Subset 2 (Gtf2i+/Dup and Gtf2iDup/Dup genotypes) produced significantly more USVs than did subset 1 (Gtf2i+/− and Gtf2+/+ genotypes) (p < 0.05).
Figure 2.
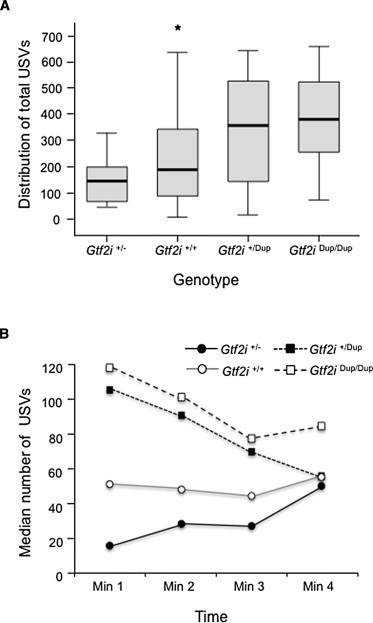
Number of USVs Produced by Mouse Pups during Maternal-Separation Period
(A) Box plots show the distribution of USVs produced over the 4 min testing period as a function of genotype. Data are represented as the median plus the upper and lower quartiles. The lowest and highest observations are indicated by whiskers. The outlier is indicated by an asterisk.
(B) Median number of USVs produced by P8 mouse pups during each minute of the 4 min maternal-separation period as a function of genotype.
The production of USVs in mouse pups can be influenced by both genetic and environmental factors, including cold temperatures, odors, handling, and maternal-pup interactions.21 The results from our study argue strongly in favor of a genetic cause of separation-induced USVs in our mice given that maternal genotype, handling, and odors were controlled across genotypes. As shown in Figure 2B, USVs in the Gtf2i+/Dup and Gtf2iDup/Dup mice decline over the time of separation (excluding temperature as the basis for genotype differences in the number of USVs produced), demonstrating that the initial separation is key to the increased production of USVs.
USVs can be altered in mice through the modulation of several different systems, including the serotonergic, cholinergic, GABAergic, and opiate systems. In particular, selective serotonin inhibitors (SSRIs) have been shown to attenuate production of USVs in mouse pups.28 Interestingly, SSRIs are the first-line medication for SAD, and indeed, for most anxiety disorders in children.1,29 Although SSRIs are generally well tolerated, the response rate in children with SAD has not been well studied, and the available data from randomized placebo-controlled trials suggest that it is quite variable.29 Comorbid diagnoses are also very common in children with anxiety disorders, and it is not clear which groups benefit most from specific medications.29 SSRI treatment in children with dup7q11.23 has not been formally tested, but identification of GTF2I downstream molecular pathways that mediate separation anxiety will allow the development and use of alternative, pathway-directed medications.
GTF2I has been shown to act as both a basal transcriptional factor and a transcriptional coactivator through binding to elements within the promoter of a number of genes.19 GTF2I is able to promote transcription upon phosphorylation by Src and subsequent translocation to the nucleus30 and might provide a link between signal transduction and gene expression. Of genes shown to be regulated by GTF2I, only c-FOS (MIM 164810) is known to function in the brain, where it combines with c-JUN (MIM 165160) to form the activity-dependent transcription factor activator protein 1.31
GTF2I is also able to negatively regulate agonist-induced calcium entry into cells through interfering with the transport of transient receptor potential channel 3 (TRPC3 [MIM 602345]) to the plasma membrane by phospholipase C-γ20. Although no direct link between calcium and anxiety has been demonstrated, aberrant regulation of intracellular calcium levels might affect downstream signaling pathways such as those that depend on Ca2+/calmodulin-dependent protein kinases (CaMKs). In mice, increased levels of CaMK2A (MIM 114078), which is activated in response to an increase in the intracellular concentration of Ca2+, lead to increased anxiety.32 Deletion of CaMK4 (MIM 114080), which phosphorylates cyclic-AMP response element-binding protein (CREB [MIM 123810]) and CRE modulator (CREM [MIM 123812]), causes reduced anxiety in mice.33 CREB and CREM are both activity-dependent regulators of the transcription of multiple genes, including those involved in emotional behavior.34,35 Interestingly, SSRIs increased expression of CREB protein in rats, but other medications such as antipsychotics and opioids did not.36
Anxiety disorders are common, disabling, and show comorbidity with depression, substance-abuse disorders, and other serious psychiatric disorders.29 Despite evidence of a strong genetic component underlying the etiology of many anxiety disorders, targeted and genome-wide association studies have identified only a few linked chromosomal regions, most of which have not been easily replicated. Rare disorders, such as dup7q11.23, provide more easily identifiable genetic causes that can be used as a starting point for identifying the neuromolecular underpinnings of anxiety disorders. Even if rare and common forms of anxiety are caused by different genetic alterations, these alterations might produce downstream effects in specific biological pathways or neural circuits that are shared. Thus, identifying the genes strongly associated with specific anxiety phenotypes in rare disorders should lead to a better understanding of pathophysiology and provide a starting point for the identification of genes and pathways that cause these common disorders in the general population.
In summary, the association between a major anxiety disorder and change in copy number of a single gene provides a starting point for understanding the molecular basis of anxiety. In doing so, the research reported in this paper demonstrates the promise of the dissection of rare genomic disorders for unraveling the genetic complexity of common disease.
Acknowledgments
We thank the children and their parents for their enthusiastic participation in our research, Elaine Tam for her assistance in maintaining the mouse colony, Lap-Chee Tsui for supporting the generation of the Gtf2i+/3′UTRloxP mice, and Doris J. Kistler for statistical consultation. This work was funded by a grant from the Canadian Institutes of Health Research (MOP77720) to L.R.O. and by grants from the National Institute of Neurological Disorders and Stroke (R01 NS35102) and the National Institute of Child Health and Development (R37 HD29957) to C.B.M.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Confidence-Interval Calculator (CIC), http://www.measuringusability.com/wald.htm
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
References
- 1.Beesdo K., Knappe S., Pine D.S. Anxiety and anxiety disorders in children and adolescents: Developmental issues and implications for DSM-V. Psychiatr. Clin. North Am. 2009;32:483–524. doi: 10.1016/j.psc.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaffer D., Fisher P., Dulcan M.K., Davies M., Piacentini J., Schwab-Stone M.E., Lahey B.B., Bourdon K., Jensen P.S., Bird H.R. The NIMH Diagnostic Interview Schedule for Children Version 2.3 (DISC-2.3): Description, acceptability, prevalence rates, and performance in the MECA Study. Methods for the Epidemiology of Child and Adolescent Mental Disorders Study. J. Am. Acad. Child Adolesc. Psychiatry. 1996;35:865–877. doi: 10.1097/00004583-199607000-00012. [DOI] [PubMed] [Google Scholar]
- 3.Smoller J.W., Gardner-Schuster E., Covino J. The genetic basis of panic and phobic anxiety disorders. Am. J. Med. Genet. C. Semin. Med. Genet. 2008;148C:118–126. doi: 10.1002/ajmg.c.30174. [DOI] [PubMed] [Google Scholar]
- 4.Bayés M., Magano L.F., Rivera N., Flores R., Pérez Jurado L.A. Mutational mechanisms of Williams-Beuren syndrome deletions. Am. J. Hum. Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somerville M.J., Mervis C.B., Young E.J., Seo E.J., del Campo M., Bamforth S., Peregrine E., Loo W., Lilley M., Pérez-Jurado L.A. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N. Engl. J. Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leyfer O., Woodruff-Borden J., Mervis C.B. Anxiety disorders in children with williams syndrome, their mothers, and their siblings: Implications for the etiology of anxiety disorders. J. Neurodev. Disord. 2009;1:4–14. doi: 10.1007/s11689-009-9003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mervis C.B., John A.E. Cognitive and behavioral characteristics of children with Williams syndrome: Implications for intervention approaches. Am. J. Med. Genet. C. Semin. Med. Genet. 2010;154C:229–248. doi: 10.1002/ajmg.c.30263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osborne L.R., Mervis C.B. Rearrangements of the Williams-Beuren syndrome locus: Molecular basis and implications for speech and language development. Expert Rev. Mol. Med. 2007;9:1–16. doi: 10.1017/S146239940700035X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velleman S.L., Mervis C.B. Children with 7q11.23 duplication syndrome: Speech, language, cognitive, and behavioral characteristics and implications for intervention. Perspect. Lang. Learn. Educ. 2011;18:108–116. doi: 10.1044/lle18.3.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silverman W., Albano A. Graywind Publications; San Antonio, TX: 1996. The Anxiety Disorders Interview Schedule for DSM-IV: Parent interview schedule. [Google Scholar]
- 11.Achenbach T., Rescorla L. University of Vermont Research Center for Children, Youth & Families; Burlington, VT: 2000. Manual for the Child Behavior Checklist for ages 1.5–5. [Google Scholar]
- 12.Hobart H.H., Morris C.A., Mervis C.B., Pani A.M., Kistler D.J., Rios C.M., Kimberley K.W., Gregg R.G., Bray-Ward P. Inversion of the Williams syndrome region is a common polymorphism found more frequently in parents of children with Williams syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 2010;154C:220–228. doi: 10.1002/ajmg.c.30258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy J.C., Kaye D.L., Sadler L.S. Psychiatric diagnoses in patients with Williams syndrome and their families. Jeff. J. Psychiatry. 2006;20:22–31. [Google Scholar]
- 14.Leyfer O.T., Woodruff-Borden J., Klein-Tasman B.P., Fricke J.S., Mervis C.B. Prevalence of psychiatric disorders in 4 to 16-year-olds with Williams syndrome. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2006;141B:615–622. doi: 10.1002/ajmg.b.30344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodruff-Borden J., Kistler D.J., Henderson D.R., Crawford N.A., Mervis C.B. Longitudinal course of anxiety in children and adolescents with Williams syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 2010;154C:277–290. doi: 10.1002/ajmg.c.30259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman W.K., Saavedra L.M., Pina A.A. Test-retest reliability of anxiety symptoms and diagnoses with the Anxiety Disorders Interview Schedule for DSM-IV: Child and parent versions. J. Am. Acad. Child Adolesc. Psychiatry. 2001;40:937–944. doi: 10.1097/00004583-200108000-00016. [DOI] [PubMed] [Google Scholar]
- 17.Enkhmandakh B., Bitchevaia N., Ruddle F., Bayarsaihan D. The early embryonic expression of TFII-I during mouse preimplantation development. Gene Expr. Patterns. 2004;4:25–28. doi: 10.1016/s1567-133x(03)00155-8. [DOI] [PubMed] [Google Scholar]
- 18.Fijalkowska I., Sharma D., Bult C.J., Danoff S.K. Expression of the transcription factor, TFII-I, during post-implantation mouse embryonic development. BMC Res. Notes. 2010;3:203. doi: 10.1186/1756-0500-3-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy A.L. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I. Gene. 2001;274:1–13. doi: 10.1016/s0378-1119(01)00625-4. [DOI] [PubMed] [Google Scholar]
- 20.Caraveo G., van Rossum D.B., Patterson R.L., Snyder S.H., Desiderio S. Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science. 2006;314:122–125. doi: 10.1126/science.1127815. [DOI] [PubMed] [Google Scholar]
- 21.Scattoni M.L., Crawley J., Ricceri L. Ultrasonic vocalizations: A tool for behavioural phenotyping of mouse models of neurodevelopmental disorders. Neurosci. Biobehav. Rev. 2009;33:508–515. doi: 10.1016/j.neubiorev.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodgson R.A., Guthrie D.H., Varty G.B. Duration of ultrasonic vocalizations in the isolated rat pup as a behavioral measure: Sensitivity to anxiolytic and antidepressant drugs. Pharmacol. Biochem. Behav. 2008;88:341–348. doi: 10.1016/j.pbb.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Proulx E., Young E.J., Osborne L.R., Lambe E.K. Enhanced prefrontal serotonin 5-HT(1A) currents in a mouse model of Williams-Beuren syndrome with low innate anxiety. J. Neurodev. Disord. 2010;2:99–108. doi: 10.1007/s11689-010-9044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng B., Mills A.A., Bradley A. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 1999;27:2354–2360. doi: 10.1093/nar/27.11.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vidal F., Sage J., Cuzin F., Rassoulzadegan M. Cre expression in primary spermatocytes: A tool for genetic engineering of the germ line. Mol. Reprod. Dev. 1998;51:274–280. doi: 10.1002/(SICI)1098-2795(199811)51:3<274::AID-MRD6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 26.Stryke D., Kawamoto M., Huang C.C., Johns S.J., King L.A., Harper C.A., Meng E.C., Lee R.E., Yee A., L'Italien L. BayGenomics: A resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31:278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young E.J., Lipina T., Tam E., Mandel A., Clapcote S.J., Bechard A.R., Chambers J., Mount H.T., Fletcher P.J., Roder J.C., Osborne L.R. Reduced fear and aggression and altered serotonin metabolism in Gtf2ird1-targeted mice. Genes Brain Behav. 2008;7:224–234. doi: 10.1111/j.1601-183X.2007.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fish E.W., Faccidomo S., Gupta S., Miczek K.A. Anxiolytic-like effects of escitalopram, citalopram, and R-citalopram in maternally separated mouse pups. J. Pharmacol. Exp. Ther. 2004;308:474–480. doi: 10.1124/jpet.103.058206. [DOI] [PubMed] [Google Scholar]
- 29.Connolly S.D., Suarez L., Sylvester C. Assessment and treatment of anxiety disorders in children and adolescents. Curr. Psychiatry Rep. 2011;13:99–110. doi: 10.1007/s11920-010-0173-z. [DOI] [PubMed] [Google Scholar]
- 30.Cheriyath V., Desgranges Z.P., Roy A.L. c-Src-dependent transcriptional activation of TFII-I. J. Biol. Chem. 2002;277:22798–22805. doi: 10.1074/jbc.M202956200. [DOI] [PubMed] [Google Scholar]
- 31.Pérez-Cadahía B., Drobic B., Davie J.R. Activation and function of immediate-early genes in the nervous system. Biochem. Cell Biol. 2011;89:61–73. doi: 10.1139/O10-138. [DOI] [PubMed] [Google Scholar]
- 32.Hasegawa S., Furuichi T., Yoshida T., Endoh K., Kato K., Sado M., Maeda R., Kitamoto A., Miyao T., Suzuki R. Transgenic up-regulation of alpha-CaMKII in forebrain leads to increased anxiety-like behaviors and aggression. Mol. Brain. 2009;2:6–16. doi: 10.1186/1756-6606-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shum F.W., Ko S.W., Lee Y.S., Kaang B.K., Zhuo M. Genetic alteration of anxiety and stress-like behavior in mice lacking CaMKIV. Mol. Pain. 2005;1:22–30. doi: 10.1186/1744-8069-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maldonado R., Smadja C., Mazzucchelli C., Sassone-Corsi P. Altered emotional and locomotor responses in mice deficient in the transcription factor CREM. Proc. Natl. Acad. Sci. USA. 1999;96:14094–14099. doi: 10.1073/pnas.96.24.14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valverde O., Mantamadiotis T., Torrecilla M., Ugedo L., Pineda J., Bleckmann S., Gass P., Kretz O., Mitchell J.M., Schütz G. Modulation of anxiety-like behavior and morphine dependence in CREB-deficient mice. Neuropsychopharmacology. 2004;29:1122–1133. doi: 10.1038/sj.npp.1300416. [DOI] [PubMed] [Google Scholar]
- 36.Nibuya M., Nestler E.J., Duman R.S. Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. J. Neurosci. 1996;16:2365–2372. doi: 10.1523/JNEUROSCI.16-07-02365.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


