Abstract
Dysfunction of mitochondrial respiration is an increasingly recognized cause of isolated hypertrophic cardiomyopathy. To gain insight into the genetic origin of this condition, we used next-generation exome sequencing to identify mutations in MTO1, which encodes mitochondrial translation optimization 1. Two affected siblings carried a maternal c.1858dup (p.Arg620Lysfs∗8) frameshift and a paternal c.1282G>A (p.Ala428Thr) missense mutation. A third unrelated individual was homozygous for the latter change. In both humans and yeast, MTO1 increases the accuracy and efficiency of mtDNA translation by catalyzing the 5-carboxymethylaminomethylation of the wobble uridine base in three mitochondrial tRNAs (mt-tRNAs). Accordingly, mutant muscle and fibroblasts showed variably combined reduction in mtDNA-dependent respiratory chain activities. Reduced respiration in mutant cells was corrected by expressing a wild-type MTO1 cDNA. Conversely, defective respiration of a yeast mto1Δ strain failed to be corrected by an Mto1Pro622∗ variant, equivalent to human MTO1Arg620Lysfs∗8, whereas incomplete correction was achieved by an Mto1Ala431Thr variant, corresponding to human MTO1Ala428Thr. The respiratory yeast phenotype was dramatically worsened in stress conditions and in the presence of a paromomycin-resistant (PR) mitochondrial rRNA mutation. Lastly, in vivo mtDNA translation was impaired in the mutant yeast strains.
Main Text
Infantile hypertrophic cardiomyopathy and lactic acidosis are key clinical features in an increasing number of mitochondrial disorders associated with severe dysfunction of oxidative phosphorylation (OXPHOS), the main energy-supply pathway of cardiomyocytes. The advent of exome analysis by next-generation sequencing (NGS) technology has begun to elucidate the genetic defects underpinning this condition. Recently, exome-NGS allowed us to identify mutations in ACAD9 (MIM 611103), which encodes mitochondrial flavin adenine dinucleotide (FAD)-dependent acyl-coenzyme-A dehydrogenase 9, in several children affected by early-onset, isolated hypertrophic cardiomyopathy (MIM 611126).1 The role of ACAD9 seems to be marginal for fatty-acid beta oxidation, but essential for the assembly of mitochondrial respiratory chain (MRC) complex I (CI).1–3 Another recent example is mutations in AGK (MIM 610345), which encodes acyl-glycerol kinase, a mitochondrial enzyme involved in the biosynthesis of cardiolipin; these mutations are responsible for hypertrophic cardiomyopathy and congenital cataracts (Sengers syndrome [MIM 212350]).4 Cardiolipin is an essential component of the lipid milieu of the inner mitochondrial membrane that participates in the integrity and optimization of the activity of both the MRC complexes and the skeletal-muscle- and heart-specific solute carrier family 25 (adenine nucleotide translocator 1), SLC25A4. Likewise, rare, recessive mutations in SLC25A4 (MIM 103220) also cause hypertrophic cardiomyopathy (MIM 192600), and yet another X-linked recessive condition, Barth syndrome, hallmarked by severe mitochondrial cardiomyopathy (MIM 302060), is caused by a mutation in TAZ (MIM 300394), which encodes Tafazzin, an acyl-transferase, specific to cardiolipin, that optimizes its fatty-acid composition to the structural and functional needs of the MRC. Other children with severe, isolated cardiomyopathy and lactic acidosis harbor recessive mutations in TMEM70 (MIM 612418), which encodes a bona fide assembly factor of MRC CV (ATP-synthase).5 Syndromic cardiomyopathy, in combination with encephalopathy, myopathy, or both, is also associated with a number of mutations of mtDNA or nuclear genes that affect MRC activities.6 Nevertheless, a substantial proportion of cases characterized by OXPHOS-related severe hypertrophic cardiomyopathy remains genetically undiagnosed.
Through exome-NGS analysis of a selected cohort of affected individuals, we identified pathogenic mutations in MTO1 (NC_000006.11), which encodes an enzyme involved in posttranscriptional modification of mitochondrial tRNAs (mt-tRNAs).
Affected person 1 (Pt1) was the first child of nonconsanguineous, healthy parents from northern Italy. He was born at 29 weeks of gestational age, by caesarean section because of oligohydramnios and reduced fetal growth. His birth weight was 790 g, his length was 34.5 cm, and his head circumference was 25.5 cm. Immediately after birth he had an episode of metabolic failure with severe hypoglycemia (25 mg%), metabolic acidosis (pH 7.17, base excess [BE] −11.7 mEq/l), and high blood lactate (13 mM, normal values [nv] < 2.0). In the subsequent days, blood glucose levels were corrected by the infusion of dextrose, whereas plasma lactate remained high (10–15 mM), with mild hyperammonemia (195 μg%, nv < 80). Electroencephalogram (EEG) and cerebral echography results were normal, as were those of a liver and spleen ultrasound examination. Interventricular septum hypertrophy was detected on the 15th day (6.4 mm, nv < 3). He died on the 19th day of sudden bradycardia unresponsive to resuscitation procedures. MRC activities in digitonin-permeabilized skin fibroblasts showed a reduction of CIII normalized to citrate synthase (CS) (CIII/CS = 60% of the controls' mean) and of CIV/CS (56%), whereas the other activities were within the controls' range (Table 1). Sequence analysis of muscle mtDNA revealed a normal H1t haplogroup common in Europeans.
Table 1.
Biochemical Analysis of OXPHOS Activities
| Muscle | CSa | CI | SDH | CII | CIII | CIV | CV |
|---|---|---|---|---|---|---|---|
| Ct values | 80–210 | 13–24 | 10.7–17,4 | 15–28 | 60–100 | 120–220 | 130–280 |
| Pt2 | 73 | 5 (27) | 16.8 (120) | 28.7 (133) | 95.3 (119) | 46.5 (27) | 351 (171) |
| Pt3 (3 mo) | 209 | 2.2 (12) | 13.1 (93) | nd b | nd b | 51 (30) | nd |
| Pt3 (17 yr) | 147 | 1.3 (7) | 13.6 (97) | 26.3 (122) | 75 (94) | 59 (35) | 144 (70) |
| Fibroblasts | CSa | CI | SDH | CII | CIII | CIV | CV |
| Ct values | 100–200 | 10.7–26 | 6.5–14,3 | 8.6–18,4 | 70–120 | 70–125 | 65–113 |
| Pt1 | 160 | 12.5 (68) | 14.3 (137) | 24.6 (182) | 57 (60) | 55 (56) | 93 (104) |
| Pt2 | 101 | 10.1 (55) | 11 (105) | 30.3 (224) | 92 (97) | 111 (114) | 126 (141) |
| Pt1 + riboflavin | 142 | 11.6 (63) | nd | nd | 53 (56) | 62 (64) | nd |
| Pt2 + riboflavin | 82 | 6.5 (35) | nd | nd | 71 (75) | 76 (78) | nd |
nd, not determined; + riboflavin, fibroblasts cultured for 1 week with Dulbecco's modified Eagle's medium (DMEM) supplemented with 5.3 μM riboflavin.
Parentheses indicate percentages relative to the mean control (Ct) value. Values below the control range are reported in bold. All enzymatic activities are normalized for CS activity. Two muscle biopsies were analyzed in Pt3; the first taken at 3 months of age (3 mo) and the second at 17 years of age (17 yr).
Citrate synthase (CS) activity, expressed as nmol min−1 mg−1.
In Pt3 muscle, the ratio CII+CIII (succinate cytochrome c reductase)/CS was 16.6 (105% of the Ct value; normal range 12.2–19.4).
Affected person 2 (Pt2), our index case, was the younger sister of Pt1. She also was born at 36 weeks of gestational age by caesarean section because of oligohydramnios and reduced fetal growth. Her birth weight was 1,380 g, her length was 42 cm, and her head circumference was 30 cm. At birth, she was mildly hypotonic and had severe metabolic acidosis (pH 7.21, BE −13 mEq/l), with high blood lactate (17.9 mM). She was immediately started on biotin (10 mg per day), Coenzyme Q10 (14 mg per day), thiamine (150 mg per day), and dichloroacetate (DCA; 30 mg per day). Plasma lactate stabilized to values between 6 and 10 mM. EEG and brain ultrasound results were normal. On the seventh day, she became tachycardic. Heart ultrasound findings were normal until the 38th day, when septum hypertrophy (7 mm, nv 3.5) and left-ventricular-wall hypertrophy (6 mm, nv 4) were found. She died on the 40th day of sudden bradycardia unresponsive to resuscitation procedures. An autopsy showed the presence of cardiomegaly, pleural effusion, and ascites. Biochemical assays, performed on the 800× g supernatant from the homogenate of a muscle biopsy, showed a reduction of the ratios of CI/CS and CIV/CS. MRC activities in digitonin-permeabilized fibroblasts showed only the reduction of CI/CS (Table 1). Sequence analysis of muscle mtDNA revealed a normal H1t haplogroup, and Sanger sequence analysis of ACAD9, TMEM70, NDUFS2 (MIM 602985), and NDUFV2 (MIM 600532) showed no mutation.
Affected person 3 (Pt3), a boy, was born at term to reportedly nonconsanguineous, healthy parents originating from a small village in the alpine region of northeastern Italy. His initial clinical history has been reported elsewhere.7 At the age of 1 month, he developed hyperpnea, difficulty feeding, weakness, and a lack of ocular fixation. His liver was 5 cm below the costal margin; he had severe metabolic acidosis, with high blood lactate (5.5 mM, nv < 2.0). An electrocardiogram (ECG) showed signs of ischemia, and a cardiac ultrasound examination revealed marked hypertrophic cardiomyopathy, particularly affecting the posterior wall of the left ventricle (8.5 mm, nv 4), reduced left-ventricular function, and mild pericardial effusion. Biochemical assays on the 800× g supernatant of the homogenate from a muscle biopsy taken at 3 months of age revealed severe reduction of CI/CS (12% of the controls' mean) and CIV/CS (30%) ratios, whereas succinate dehydrogenase (SDH)/CS and CII+III/CS ratios as well as CS (Table 1) and pyruvate dehydrogenase activities were normal. DCA treatment resulted in marked improvement of both metabolic acidosis and cardiomyopathy. After 9 months of DCA therapy, a cardiac ultrasound examination showed a normal-sized heart, with normal left-ventricular-wall thickness (5 mm) and function (ejection fraction 76%, systolic fraction 43%) and low blood lactate values, ranging from 1.6 to 3.1 mM. During his first years of life, Pt3 had no severe episode of metabolic acidosis, even if plasma lactate remained moderately high, ranging from 2.5 to 4 mM. His growth rate has been normal, with good neurological development and a normal brain anatomy, according to magnetic resonance imaging. He was put on a permanent treatment of DCA (200 mg per day), carnitine (1 g per day), and CoQ10 (100 mg per day). Because of the possible side effects of DCA, he was monitored regularly through the evaluation of visual and brainstem auditory evoked potentials as well as electromyography and nerve conduction velocities. At the age of 7 years, ultrasound examination showed a normal systolic ejection fraction in spite of a slight dilation of the left-ventricular chamber. At 12 years, DCA was stopped because of normalization of plasma lactate. A second muscle biopsy, taken at 17 years, again showed severe reduction of CI/CS (7% of the controls' mean) and CIV/CS (35%), whereas the other MRC activities were normal (Table 1). Sequence analysis of mtDNA showed a normal H2 haplogroup. He is now 19 years old with a normal scholastic performance. A recent ultrasound examination revealed the presence of hypertrophic cardiomyopathy with an ejection fraction of 60%. An ECG showed sinus bradycardia (45 beats per min) but a Holter ECG was otherwise normal. A neurological examination was normal, except for a reduction of skills in the execution of fine movements, more evident in the left hand. Ophthalmoscopic examination showed moderate bilateral optic atrophy. Visual-evoked potential showed an increased P100 latency, more marked in the left eye. Electromyography and nerve-conduction velocity were normal.
Informed consent for participation in this study was obtained from the parents of all individuals involved, in agreement with the Declaration of Helsinki and approved by the ethics committees of the Fondazione IRCCS (Istituto di Ricovero e Cura a Carattere Scientifico) Istituto Neurologico (Milan, Italy).
We first carried out exome-NGS in the index case. The DNA extracted from fibroblasts was processed with the SureSelect Human All Exon 50 Mb kit (Agilent) and subsequently sequenced as 76 base-pair (bp) paired-end runs to an average coverage of 120×, corresponding to 9–12 Gb of sequence data. Read alignment was performed with the use of the Burrows-Wheeler Aligner (version 0.5.8) applied to the human genome assembly hg19. Single-nucleotide variants and small insertions and deletions were detected with SAMtools (version 0.1.7). Given that mitochondrial disorders are rare conditions, we excluded variants with a frequency > 0.2% in “in house” control exomes and public databases. Assuming an autosomal-recessive mode of inheritance, we searched for homozygous or compound-heterozygous variants, which were filtered against (1) variants known to be associated with MRC defects and (2) novel homozygous or compound-heterozygous variants affecting genes that encode mitochondrial proteins listed in MitoP2 (score > 0.5). This filtering procedure (Table S1 available online) revealed that our index case was compoundheterozygous for mutations in MTO1: a maternal c.1858dup (p.Arg620Lysfs∗8) and a paternal c.1282G>A (p.Ala428Thr) (Figure 1A). The same mutations were also found in her affected brother.
Figure 1.
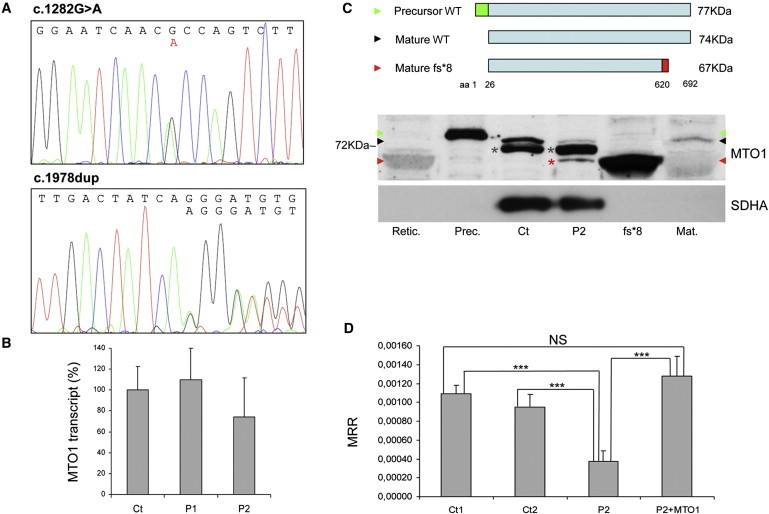
MTO1 Mutations
(A) Electropherograms of MTO1 of Pt2 showing the c.1282G>A (left) and c.1858dup (right), both in heterozygosis.
(B) Real-time PCR on retrotranscribed cDNA from fibroblasts of individuals 1 (P1) and 2 (P2). The amount of MTO1 transcript (normalized to GAPDH levels) is comparable in mutant versus WT control samples (Ct), indicating no mRNA decay. Data are represented as mean ± SD.
(C) Western-blot analysis of MTO1. Top: schematic representation of the precursor WT MTO1 (isoform a), its mature species after cleavage of a predicted 25 aa mitochondrial targeting sequence in the N terminus, and the mature p.Arg620Lysfs∗8 mutant species. Bottom: Western-blot analysis on isolated mitochondria. Retic.: reticulocyte lysate used for in-vitro protein synthesis; Prec.: in-vitro-translated 77 kDa MTO1 precursor protein (green arrowhead); Ct: isolated mitochondria from control fibroblasts; P2: isolated mitochondria from individual 2 fibroblasts; fs∗8: in-vitro-translated 67 kDa mature protein carrying the truncating p.Arg620Lysfs∗8 variant (red arrowhead); Mat.: in-vitro-translated 74 kDa WT mature MTO1 (black arrowhead). A faint crossreacting band is visualized in mt P2 sample (red asterisk), corresponding to the mature p.Arg620Lysfs∗8 truncated protein. An unspecific signal is present in mt samples (gray asterisks). The position of the 72 kDa molecular weight marker protein is also indicated. SDHA was used as loading control.
(D) MRR measured in immortalized fibroblasts from Pt 2, in naive condition (P2) or overexpressing MTO1 (P2+MTO1), and in control subjects (Ct1, Ct2). MRR values are expressed as pMolesO2/min/cells. Data are represented as mean ± SD. Two-tail, unpaired Student's t test was applied for statistical significance. ∗∗∗: p < 0.001.
The frameshift mutation is predicted to introduce a stop codon after an aberrant sequence of seven amino acids (aa) (Figure S1), causing the loss of the C-terminal 73 aa residues (≈10% of the protein size); the missense mutation affects an amino-acid residue, Ala428, which is invariant in all available animal, plant, and yeast species, including Saccharomyces cerevisiae (Figures S2A and S2B). In addition, the Ala428Thr change scored very highly for likelihood to be deleterious according to ad-hoc softwares for pathogenicity prediction (deleterious for Polyphen2: p = 0.999; Panther: 0.88041; MutPred: 0.903). The exome variant server (EVS) of the NHLBI GO Exome Sequencing Project reports a frequency of 0.028% (2/7,020) in the American population of European origin for the c.1282G>A change, whereas the c.1858dup change was not present in the database. We also excluded the missense mutation from our exome database, which contains 973 genomes from Europeans (=1,946 alleles), and from a database of 300 alleles from consecutive control DNA samples from subjects originating from northern Italy.
To evaluate a possible influence of the two mutations on the stability of the transcript, we extracted mRNA from mutant fibroblasts of affected individuals 1 and 2 and retrotranscribed it into cDNA. Quantitative real-time PCR showed that the content of mutant MTO1 transcripts was similar to that of wild-type (WT) control samples (Figure 1B), and sequence analysis revealed the presence of comparable amounts of either mutant transcript (data not shown), indicating no RNA decay.
Next, we analyzed MTO1 on isolated mitochondria and on total cell lysates obtained from both mutant and control immortalized fibroblasts, using a polyclonal MTO1 antibody (Proteintech). MTO1 is predicted by bioinformatic tools (MitoProt, TargetP) to be imported inside mitochondria after the cleavage of a 25-aa-long mitochondrial targeting sequence. We synthesized the polypeptides corresponding to the precursor, the mature WT, and the mature p.Arg620Lysfs∗8 truncated MTO1 species (TNT Transcription-Translation System kit, Promega). After performing SDS-polyacrylamide electrophoresis and electroblotting, we immunovisualized a band reacting with an MTO1 antibody in mitochondria (Figure 1C) and in fibroblast lysates (Figure S3). This band, which comigrates with a band corresponding to the 74 kDa full-length, mature in-vitro-synthesized MTO1 polypeptide (NM_012123, NP_036255), was markedly increased in MTO1-overexpressing fibroblasts (Figure S3). In mutant mitochondria, the amount of the 74 kDa protein was clearly reduced, whereas an additional band was detected, with the same electrophoretic mobility of the in-vitro-synthesized mature p.Arg620Lysfs∗8 MTO1 species, predicted to have a molecular weight of ≈67 kDa. This result suggests that the p.Arg620Lysfs∗8 truncated MTO1 is relatively stable (Figure 1C).
In order to prove the causative role of the MTO1 variants found in the affected siblings of family 1, we first tested whether the expression of WT MTO1 cDNA could rescue the biochemical phenotype of mutant cells. Given that the MRC defects in Pt1 and Pt2 fibroblasts were relatively mild and variable, we immortalized the Pt2 fibroblasts using pBABE-puro SV40 and evaluated the oxygen consumption through microscale oxygraphy (Seahorse Bioscience XF-96). This assay, which depends upon and reflects the cumulative proficiency of the whole set of MRC complexes, is more sensitive than individual assays of each complex.8 We demonstrated a clear reduction of the maximal respiration rate (MRR) in immortalized Pt2 cells compared to immortalized control fibroblasts, which returned to normal after transduction with a MTO1WT-expressing lentivirus (pLenti6 Gateway Vector kit, Invitrogen) (Figures 1E and S3). This result indicates a pathogenic role of the MTO1 variants found in Pt2 mutant cells.
Second, we sequenced the exons and exon-intron boundaries of MTO1 in DNA samples from 17 individuals with early-onset hypertrophic cardiomyopathy, lactic acidosis, and defective MRC activities. We found a single individual, Pt3, homozygous for the c.1282G>A (p.Ala428Thr) mutation, identical to that found in the paternal allele of family 1.
In both yeast and humans, MTO1 encodes the enzyme that catalyzes the 5-carboxymethylaminomethylation (mnm5s2U34) of the wobble uridine base in mt-tRNAGln, mt-tRNAGlu, and mt-tRNALys. 9 This modification is usually coupled to the 2-thiolation of the same uridine moiety, a reaction catalyzed by 2-thiouridylase, encoded by MTO2 (TRMU in humans [MIM 610230]); both these posttranscriptional modifications increase accuracy and efficiency of mtDNA translation.10 In order to further test the pathogenic role of the MTO1 mutations, we used the yeast Saccharomyces cerevisiae. We first showed that the absence of MTO1 (mto1Δ) was associated with decreased respiration rate in yeast incubated at 28°C (Figure 2A). Second, we demonstrated that this phenotype failed to be corrected by expression of a recombinant yeast MTO1 cDNA encoding protein variant Pro622∗, corresponding to the human Arg620Lysfs∗8 change (Figure S2, Table S2). This result suggests that, although present in human mitochondria, the Arg620Lysfs∗8 MTO1 variant is functionally inactive. The respiratory phenotype of the mto1Δ strain was only partially corrected by expression of a yeast recombinant MTO1 encoding protein variant Ala431Thr, corresponding to the human Ala428Thr change (Figure S2, Table S2), whereas the expression of yeast MTO1WT led to full recovery (Figure 2A). These results were qualitatively unchanged, but dramatically amplified, in experiments carried out under temperature-induced stress conditions; i.e., at 37°C (Figure 2B). However, neither the growth of mto1Δ nor that of mto1Pro622∗ or mto1Ala431Thr strains on oxidative carbon sources was significantly impaired. The OXPHOS-negative growth phenotype of mto1 mutants is in fact contingent on the presence of a C>G transversion at nucleotide 1477 of the 15S rRNA in mtDNA.11 The mutation disrupts the C1477-G1583 base pairing in a functionally relevant hairpin structure, which is part of the decoding site (site A) of the ribosome, where the codon-anticodon recognition occurs.12 This mutation confers resistance to the antibiotic paromomycin (Figure 3) by destabilizing the hairpin and results in a synthetic phenotype with MTO1 disruption,11 most likely due to impaired interaction between unprocessed MTO1-dependent mt-tRNAs with ribosomal site A.13
Figure 2.
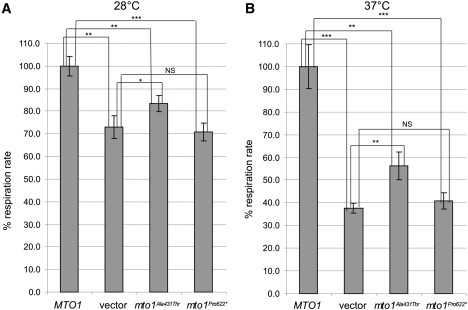
Respiratory Phenotypes of Yeast Mutant Strains
(A and B) Respiratory activity of yeast mto1Δ strains transformed with MTO1WT recombinant vector, (empty) vector, and mto1Ala431Thrand mto1Pro622∗ recombinant vectors at 28°C (A) and 37°C (B). Respiratory rates were normalized to the WT strain, for which the respiratory rate was 38.2 nmol min−1 mg−1 at 28°C and 28.9 nmol min−1 mg−1 at 37°C. Values are the mean of at least three independent experiments. Two-tail, unpaired t test was applied for statistical significance. ∗: p < 0.05; ∗∗p < 0.01; ∗∗∗: p < 0.001. Data are represented as mean ± SD.
Figure 3.
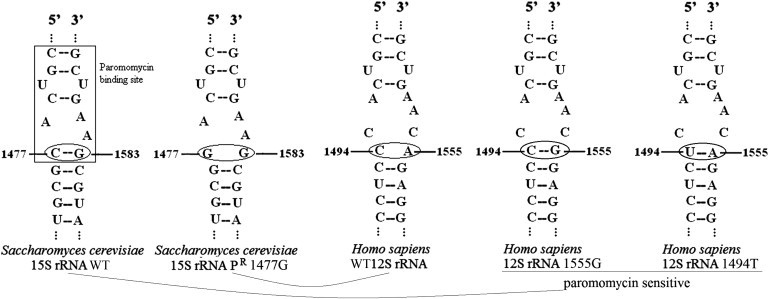
A-Site Structures of Yeast 15S and Human 12S rRNAs
Secondary structure of site A of the WT yeast strain 15S rRNA, of the PR yeast strain 15S rRNA, of the WT human 12S rRNA, and of the mutant human 12S rRNA—the last two carrying either the 1555G or the 1494T mtDNA mutation, both associated with PS. The pairing of 1477–1583 nucleotides in WT yeast rRNA corresponds to that of 1494-1555 nucleotides in mutant human rRNA and confers aminoglycoside susceptibility.
The normal human mitochondrial 12S rRNA contains a hairpin structure that corresponds to the paromomycin-resistant (PR) variant in yeast, because C1494 and A1555, which are equivalent to yeast C1477 and G1583, cannot form a pair. Incidentally, the well-known pathogenic m.1555A>G mutation of human mtDNA as well as the m.1494C>T can establish a C1494-G1555 pairing or the equivalent U1494-A1555 pairing, respectively, both of which increase the length of the hairpin structure, thus letting paromomycin (and other aminoglycosides) bind to site A (Figure 3).14 As a consequence, the m.1555A>G and m.1494C>T both confer aminoglycoside susceptibility to human mtDNA, being associated with a specific phenotype, aminoglycoside-induced nonsyndromic deafness (MIM 580000).15,16
Given that human WT 12S RNA site A is structurally similar to the yeast PR variant of the 15S RNA site A (Figure 3), we extended our complementation analysis to a m.1477C>G mutant PR yeast strain.
We showed that, in contrast to the mto1Δ paromomycin-sensitive (PS) strain, the mto1Δ PR strain was unable to grow on oxidative carbon sources such as glycerol (Figure 4A). The oxidative growth remained abolished with the expression of a cDNA encoding Mto1Pro622∗, clearly reduced with the expression of a cDNA encoding Mto1Ala431Thr, and fully restored with the expression of a cDNA encoding MTO1WT(Figure 4A).
Figure 4.
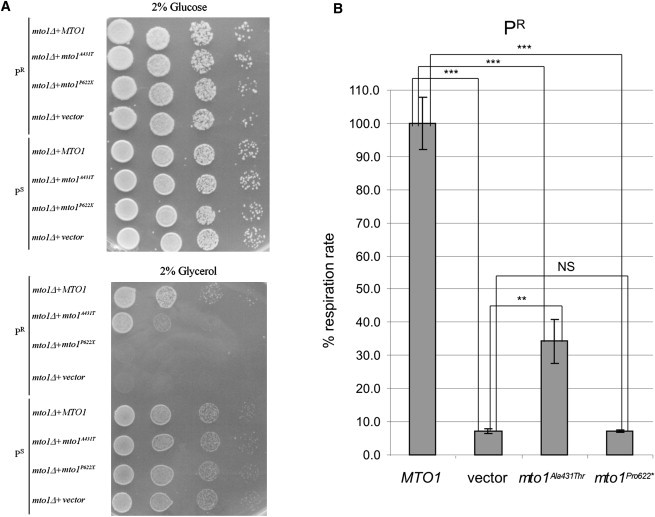
PR and PS Yeast Phenotypes
(A) Spot assay of mto1Δ PR and PS strains, transformed with MTO1WT recombinant vector, (empty) vector, and mto1Ala431Thrand mto1Pro622∗ recombinant vectors. The assay was performed by spotting decreasing concentrations of yeast cells (105, 104, 103, and 102) on a medium supplemented with either 2% glucose (upper panel) or 2% glycerol (lower panel). See text for details.
(B) Respiratory activity of yeast MTO1WT and mutant PR strains at 28°C. Respiratory rates were normalized to the WT strain, for which the respiratory rate was 26.6 nmol min−1 mg−1. Values are the mean of at least four independent experiments. Two-tail, unpaired t test was applied for statistical significance. ∗: p < 0.05; ∗∗p < 0.01; ∗∗∗: p < 0.001.
Data are represented as mean ± SD.
Likewise, the respiration rate, which was nearly abolished in the mto1Δ PR strain, was not corrected by transformation with the Mto1Pro622∗-encoding cDNA and only partially corrected by the Mto1Ala431Thr-encoding cDNA, in contrast to the full recovery obtained by expressing a MTO1WT cDNA (Figure 4B). We observed no difference between MTO1WT and mto1 mutant strains in the frequency of “petite” colonies; i.e., respiration-defective clones caused by large deletions or loss of mtDNA (data not shown), which indicates that mutations in MTO1 did not affect mtDNA stability.
Analysis in yeast clearly shows that while both mutations are detrimental for respiratory activity, the deleterious effects of the Pro622∗ protein variant is more severe than the Ala431Thr replacement. These results are concordant with the clinical phenotype associated with the equivalent mutations in humans: the third affected individual, homozygous for the MTO1 mutation encoding the Ala428Thr protein variant, equivalent to the yeast Ala431Thr, is now 19 years old, and in relatively well-compensated condition, whereas individuals 1 and 2, who were compound heterozygous for the MTO1 mutations encoding the Ala428Thr and Arg620Lysfs∗8 mutations, the latter being equivalent to the yeast Pro622∗, died a few days after birth of intractable congestive heart failure and severe lactic acidosis.
MTO1 encodes a FAD-containing enzyme involved in posttranscriptional modification of specific mt-tRNAs, thus contributing to the optimization of mtDNA-dependent protein synthesis. However, similar to a recent report on the effects on in-vivo mtDNA translation of TRMU (MTO2) mutations,17 we failed to show consistent alterations of mtDNA-dependent protein synthesis in Pt2 mutant fibroblasts assayed in standard conditions18 (Figure S4). Hence, we evaluated the effect on mtDNA translation of the expression of cDNAs encoding the Mto1Pro622∗ and Mto1Ala431Thr variants, versus MTO1WT, in the highly sensitive mto1Δ PR yeast model.19 Interestingly, the mtDNA protein synthesis in the mto1Δ strain expressing a cDNA encoding Mto1Pro622∗ was markedly reduced, particularly for cytochrome c oxidase (Cox) subunits 1 and 2 and for cytochrome b, similar to, albeit lesser than, the null mto1Δ strain. The mtDNA translation pattern in the mto1Δ strain expressing a cDNA encoding Mto1Ala431Thr was similar to that of the MTO1WT strain, although some bands, such as those corresponding to the Var1 and adenosine triphosphatase (ATPase) 6 polypeptides, were slightly reduced in the mutant versus WT strains (Figure 5). As expected, these results confirm that the effects of the Mto1Pro622∗ truncating mutation are more deleterious on mtDNA translation than those of the Mto1Ala431Thr missense mutation, in agreement with the results of the respiratory and oxidative-growth phenotypes in yeast and of the greater severity of the clinical phenotype in individuals 1 and 2 versus that of individual 3.
Figure 5.
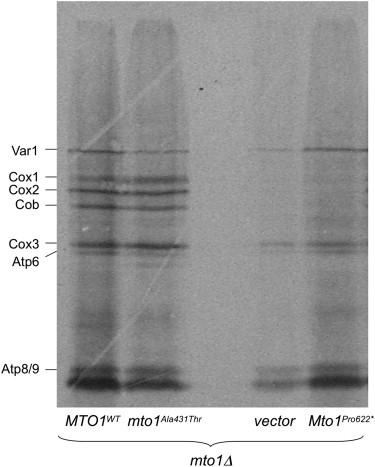
In-Vivo Mitochondrial Translation of PRmto1Δ Strain Transformed with WT or Mutant MTO1 Alleles
Mitochondrial gene products were labeled with [35S] methionine in whole cells at 30°C in the presence of cycloheximide for 7 min.19 Equivalent amounts of total cellular proteins were separated by SDS-PAGE on a 17.5% polyacrylamide gel, transferred to a nitrocellulose membrane, and exposed to X-ray film. The mto1Δ strain transformed with the empty vector (vector) contained 40% of petite cells, thus reducing the overall signal. Cox: cytochrome c oxidase; Cob: cytochrome b; Atp: ATP synthase; Var1: small mitochondrial ribosome subunit. In the mto1Pro622∗ strain, several bands, particularly Cox1, Cox2, and Cob, are virtually missing, and the overall pattern is similar to that of the (empty) vector. In the mto1Ala431Thr, the intensity of some bands, including Var1 and Atp6, is slightly reduced compared to the MTO1WT strain, but the two patterns are similar, suggesting mild impairment.
The function of MTO1 could explain the variability of the biochemical defects, ranging from isolated CI deficiency, as in Pt2 fibroblasts, to combined CI-CIV deficiency in Pt2 and Pt3 skeletal muscle, or to combined CIII-CIV deficiency in Pt1 fibroblasts. Among the 13 mtDNA-encoded proteins, seven are subunits of CI, three of CIV, two of CV, and one of CIII. This gene distribution can explain why mtDNA translation defects such as those associated with mutations of MTO1 can predominantly impair the activity of CI, but also that of CIII and CIV. The two mtDNA-encoded subunits of CV are part of the F0 component of this complex, the function of which is not directly measured by the standard CV assay that is based on ATP-hydrolysis, a function carried out by the F1 component of CV. This could explain, at least in part, why CV activity was essentially normal in MTO1 mutant samples.
The presence of a FAD moiety in MTO1 opens the possibility that, as observed for other mitochondrial flavo-enzymes,1,3,20 riboflavin supplementation may be beneficial for correction, at least in part, of the biochemical defect and improvement of the clinical course. However, we observed neither correction of MRC biochemical activities (Table 1) nor improvement of oxygen consumption (data not shown) by growing Pt1 and Pt2 mutant fibroblasts in 5.3 μM riboflavin for 1 week. Likewise, addition of different amounts of riboflavin (0.53, 2.6, 5.3, 13.3, and 26.6 μM) to a glycerol medium had no effect on either growth or respiration of mto1 mutant yeast strains (data not shown). These results indicate that riboflavin supplementation is ineffective, possibly because the truncating mutation is too drastic and the missense mutation does not affect the N-terminal, FAD-binding domain of the protein.21
An additional source of complexity stems from the existence of transcript variants encoding at least three different MTO1 isoforms. Although isoform a is prevalent, being in fact the only one that we could detect in fibroblasts, the presence of isoforms b (NM_133645, NP_598400) and c (NM_001123226, NP_001116698) is also predicted, each retaining a different extra-exon resulting in protein sequences longer than those of isoform a.22 The existence and functional significance of these longer variants are presently unknown. Notably, both mutations found in this study affect the protein sequence common to all three isoforms, predicting overall impairment of the MTO1 function.
MTO1 and MTO2 (TRMU) take part in the same pathway involved in posttranscriptional modification of specific mt-tRNAs.10 Interestingly, specific TRMU mutations cause severe and sometimes fatal liver failure (MIM 613070),23 and other mutations in the same gene have been associated with reversible mitochondrial myopathy.24 Here we report mutations of MTO1 associated with impairment of yet another target organ, the heart, the severity of which seems to depend on the potential deleteriousness of the mutations. The mechanisms underlying such diverse tissue and organ specificity in the same enzyme or in the same enzymatic pathway is a challenge for future work in this rapidly expanding field of mitochondrial medicine.
Acknowledgments
We thank Alexander Tzagoloff for the generous gift of the PR yeast strain. We are grateful to Erika Fernandez-Vizarra for help with the in vivo translation assay, to Alessia Nasca for the screening of the Italian DNA control samples, and to Ilaria D'Amato for technical support with microscale oxygraphy. This work was supported by the Pierfranco and Luisa Mariani Foundation of Italy; Fondazione Telethon grants GGP11011 and GPP10005; CARIPLO grant 2011/0526; the Italian Association of Mitochondrial Disease Patients and Families (Mitocon); the Helmholtz Alliance for Mental Health in an Ageing Society (HA-215) and the German Federal Ministry of Education and Research (BMBF)-funded Systems Biology of Metabotypes grant (SysMBo 0315494A); the German Network for Mitochondrial Disorders (mitoNET 01GM0867 and 01GM0862); and E-Rare grant GenoMit JTC2011.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Exome Variant Server (EVS), http://evs.gs.washington.edu/EVS
MitoP2, http://www.mitop.de
MitoProt, http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html
MutPred, http://mutpred.mutdb.org
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Panther, http://www.pantherdb.org
Polyphen2, http://genetics.bwh.harvard.edu/pph2
References
- 1.Haack T.B., Danhauser K., Haberberger B., Hoser J., Strecker V., Boehm D., Uziel G., Lamantea E., Invernizzi F., Poulton J. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat. Genet. 2010;42:1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- 2.Nouws J., Nijtmans L., Houten S.M., van den Brand M., Huynen M., Venselaar H., Hoefs S., Gloerich J., Kronick J., Hutchin T. Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell Metab. 2010;12:283–294. doi: 10.1016/j.cmet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Gerards M., van den Bosch B.J., Danhauser K., Serre V., van Weeghel M., Wanders R.J., Nicolaes G.A., Sluiter W., Schoonderwoerd K., Scholte H.R. Riboflavin-responsive oxidative phosphorylation complex I deficiency caused by defective ACAD9: new function for an old gene. Brain. 2011;134:210–219. doi: 10.1093/brain/awq273. [DOI] [PubMed] [Google Scholar]
- 4.Mayr J.A., Haack T.B., Graf E., Zimmermann F.A., Wieland T., Haberberger B., Superti-Furga A., Kirschner J., Steinmann B., Baumgartner M.R. Lack of the mitochondrial protein acylglycerol kinase causes Sengers syndrome. Am. J. Hum. Genet. 2012;90:314–320. doi: 10.1016/j.ajhg.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cízková A., Stránecký V., Mayr J.A., Tesarová M., Havlícková V., Paul J., Ivánek R., Kuss A.W., Hansíková H., Kaplanová V. TMEM70 mutations cause isolated ATP synthase deficiency and neonatal mitochondrial encephalocardiomyopathy. Nat. Genet. 2008;40:1288–1290. doi: 10.1038/ng.246. [DOI] [PubMed] [Google Scholar]
- 6.Uziel G., Ghezzi D., Zeviani M. Infantile mitochondrial encephalopathy. Semin. Fetal Neonatal Med. 2011;16:205–215. doi: 10.1016/j.siny.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Burlina A.B., Milanesi O., Biban P., Bordugo A., Garavaglia B., Zacchello F., DiMauro S. Beneficial effect of sodium dichloroacetate in muscle cytochrome C oxidase deficiency. Eur. J. Pediatr. 1993;152:537. doi: 10.1007/BF01955069. [DOI] [PubMed] [Google Scholar]
- 8.Invernizzi F., D'Amato I., Jensen P.B., Ravaglia S., Zeviani M., Tiranti V. Microscale oxygraphy reveals OXPHOS impairment in MRC mutant cells. Mitochondrion. 2012;12:328–335. doi: 10.1016/j.mito.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Yan Q., Guan M.X. Combination of the loss of cmnm5U34 with the lack of s2U34 modifications of tRNALys, tRNAGlu, and tRNAGln altered mitochondrial biogenesis and respiration. J. Mol. Biol. 2010;395:1038–1048. doi: 10.1016/j.jmb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umeda N., Suzuki T., Yukawa M., Ohya Y., Shindo H., Watanabe K., Suzuki T. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005;280:1613–1624. doi: 10.1074/jbc.M409306200. [DOI] [PubMed] [Google Scholar]
- 11.Colby G., Wu M., Tzagoloff A. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:27945–27952. doi: 10.1074/jbc.273.43.27945. [DOI] [PubMed] [Google Scholar]
- 12.Yan Q., Li X., Faye G., Guan M.X. Mutations in MTO2 related to tRNA modification impair mitochondrial gene expression and protein synthesis in the presence of a paromomycin resistance mutation in mitochondrial 15 S rRNA. J. Biol. Chem. 2005;280:29151–29157. doi: 10.1074/jbc.M504247200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Yan Q., Guan M.X. Mutation in MTO1 involved in tRNA modification impairs mitochondrial RNA metabolism in the yeast Saccharomyces cerevisiae. Mitochondrion. 2009;9:180–185. doi: 10.1016/j.mito.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian Y., Guan M.X. Interaction of aminoglycosides with human mitochondrial 12S rRNA carrying the deafness-associated mutation. Antimicrob. Agents Chemother. 2009;53:4612–4618. doi: 10.1128/AAC.00965-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prezant T.R., Agapian J.V., Bohlman M.C., Bu X., Oztas S., Qiu W.Q., Arnos K.S., Cortopassi G.A., Jaber L., Rotter J.I. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat. Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- 16.Zhao H., Li R., Wang Q., Yan Q., Deng J.H., Han D., Bai Y., Young W.Y., Guan M.X. Maternally inherited aminoglycoside-induced and nonsyndromic deafness is associated with the novel C1494T mutation in the mitochondrial 12S rRNA gene in a large Chinese family. Am. J. Hum. Genet. 2004;74:139–152. doi: 10.1086/381133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasarman F., Antonicka H., Horvath R., Shoubridge E.A. The 2-thiouridylase function of the human MTU1 (TRMU) enzyme is dispensable for mitochondrial translation. Hum. Mol. Genet. 2011;20:4634–4643. doi: 10.1093/hmg/ddr397. [DOI] [PubMed] [Google Scholar]
- 18.Fernández-Silva P., Acín-Pérez R., Fernández-Vizarra E., Pérez-Martos A., Enriquez J.A. In vivo and in organello analyses of mitochondrial translation. Methods Cell Biol. 2007;80:571–588. doi: 10.1016/S0091-679X(06)80028-2. [DOI] [PubMed] [Google Scholar]
- 19.Barrientos A., Korr D., Tzagoloff A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J. 2002;21:43–52. doi: 10.1093/emboj/21.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghezzi D., Sevrioukova I., Invernizzi F., Lamperti C., Mora M., D'Adamo P., Novara F., Zuffardi O., Uziel G., Zeviani M. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am. J. Hum. Genet. 2010;86:639–649. doi: 10.1016/j.ajhg.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi R., Villarroya M., Ruiz-Partida R., Li Y., Proteau A., Prado S., Moukadiri I., Benítez-Páez A., Lomas R., Wagner J. Structure-function analysis of Escherichia coli MnmG (GidA), a highly conserved tRNA-modifying enzyme. J. Bacteriol. 2009;191:7614–7619. doi: 10.1128/JB.00650-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., Li R., Lin X., Guan M.X. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12 S rRNA A1555G mutation. J. Biol. Chem. 2002;277:27256–27264. doi: 10.1074/jbc.M203267200. [DOI] [PubMed] [Google Scholar]
- 23.Zeharia A., Shaag A., Pappo O., Mager-Heckel A.M., Saada A., Beinat M., Karicheva O., Mandel H., Ofek N., Segel R. Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 2009;85:401–407. doi: 10.1016/j.ajhg.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uusimaa J., Jungbluth H., Fratter C., Crisponi G., Feng L., Zeviani M., Hughes I., Treacy E.P., Birks J., Brown G.K. Reversible infantile respiratory chain deficiency is a unique, genetically heterogenous mitochondrial disease. J. Med. Genet. 2011;48:660–668. doi: 10.1136/jmg.2011.089995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


