Abstract
In HIV type 1 (HIV-1), the dimerization initiation site (DIS) is the sequence primarily responsible for initiating the noncovalent linkage of two homologous strands of genomic RNA during viral assembly. The DIS loop contains an autocomplementary hexanucleotide sequence and forms a symmetric homodimer through a loop–loop kissing interaction. In a structural rearrangement catalyzed by the HIV-1 nucleocapsid protein (NCp7) and suggested to be associated with maturation of the budded viral particle, the DIS converts from a metastable kissing dimer to an extended duplex. Here, we demonstrate that the DIS kissing dimer displays localized conformational dynamics that result from the specific protonation of the N1 base nitrogen of the DIS loop residue A272 at near-physiological pH. The rate of NCp7-catalyzed maturation of the DIS kissing dimer is also shown to directly correlate with the observed proton-coupled conformational dynamics, where NCp7 is found to convert the dynamic A272 protonated state with a faster rate. Taken together, these results reveal a previously undescribed role for base protonation in modulating local RNA structure and demonstrate a mechanism for promoting the chaperone-mediated structural rearrangement of a kinetically trapped RNA conformational state.
Keywords: NCp7, RNA, NMR, fluorescence
Two copies of genomic RNA, linked noncovalently through a 5′ leader sequence termed the dimer linkage structure (DLS) (1, 2), are packaged into budding retroviral particles. In HIV type-1 (HIV-1), dimerization is initiated by a highly conserved 35-nt stem–loop within the DLS, called the dimerization initiation site (DIS) (3). The DIS loop contains a hexanucleotide palindromic sequence that is most often either GUGCAC (subtype A, Mal isolate) or GCGCGC (subtype B, Lai isolate), together with highly conserved 5′ and 3′ flanking purine nucleotides (Fig. 1A). Deletion or mutation of the DIS sequence dramatically decreases viral replication rates and reduces infectivity (3–6), with specific defects shown to affect genomic RNA dimerization and encapsidation, as well as proviral DNA synthesis (4–9). In vitro experiments have shown that RNA sequences derived from the DLS, including isolated DIS stem–loops, can spontaneously form homodimers through loop–loop kissing interactions (3, 10–16). These studies suggest that the initial association of HIV-1 genomic RNA in vivo occurs through an RNA kissing interaction.
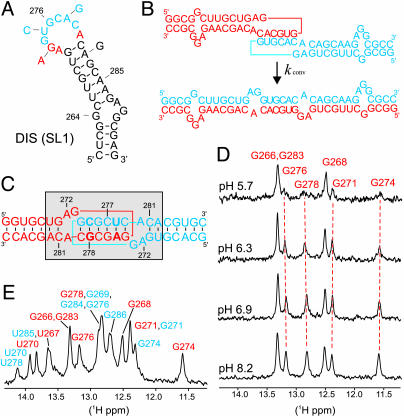
Fig. 1.
Secondary structure and pH-dependent DIS dynamics. (A) Stem–loop sequence and secondary structure for DIS (SL1) from HIV-1 subtype A. The autocomplementary hexanucleotide loop sequence is shown in blue, and conserved purines are bolded in red. Arrows indicate differences in sequence from the HIV-1 subtype B strain. (B) Representation of the DIS homodimer conversion from kissing dimer to extended duplex, with DIS stem–loops shown in blue and red; purine junction bases are boxed. (C) Sequence and secondary structure of the DIS23(GA)·DIS23(HxUC) heterodimeric kissing complex formed by DIS23(GA) in red and DIS23(HxUC) designed with stem sequences noncomplementary to the stems of DIS23(GA) in blue. Point mutation palindromic hexanucleotide DIS loop sequences used to generate heterodimeric complexes are bolded. The conformationally dynamic region is shown by a shaded box. (D)1D 15N-filtered 1H-NMR spectra of the imino proton region of a 13C,15N-A,G-labeled DIS23(GA)·DIS23(HxUC) kissing complex in NMR buffer and 300 mM NaCl as a function of pH. Imino assignments are indicated and the subset of pH-dependent resonances followed by dashed red lines. (E) Assigned imino proton resonances [DIS23(GA) in red and DIS23(HxUC) in blue] of the DIS kissing dimer at pH 8.2.
In the postbudded viral particle, retroviral genomic RNA undergoes further conformational changes that result in the formation of a more compact thermodynamically stable genomic RNA dimer (17, 18). This maturation process requires active viral protease and correlates with cleavage of the nucleocapsid protein (NCp7) from gag in the budded particle. In viral maturation, NCp7 has been proposed to activate RNA refolding events, acting as a nucleic acid chaperone (19). In vitro studies have demonstrated that NCp7 can activate conversion of metastable homodimers formed by RNA transcripts derived from DLS sequences to more thermodynamically stable dimers (20, 21). The isolated DIS stem–loop kissing dimers have also been shown to be converted by NCp7 to the mature dimer form (11, 20, 22–24) and maturation demonstrated to occur through a mechanism in which the loop–loop helix is maintained, while strands of the stem helices are melted and exchanged (Fig. 1B) (25). Here, we demonstrate that the DIS kissing dimer displays localized conformational dynamics associated with specific protonation of the N1 nitrogen of loop residue A272 at nearphysiological pH. The rate of NCp7-catalyzed maturation of the DIS kissing dimer is also shown to directly correlate with the protonation state of A272.
Materials and Methods
Sample Preparation. RNA samples were prepared by in vitro T7 polymerase transcription by using synthetic DNA templates (26). Uniformly 15N-labeled NTPs were harvested from Escherichia coli grown on M9 minimal media by using 15NH4Cl as the sole nitrogen source, prepared by using standard protocols (27), and used to synthesize uniformly 15N-labeled DIS23(GA) and DIS23(HxUC). Similarly, commercial 13C15N-rATP and 13C15N-rGTP nucleotides (Spectra Stable Isotopes, Columbia, MD) were used to synthesize a 13C15N-A,G-labeled DIS23(GA) stem–loop. The 2-aminopurine (2-AP)-containing stem–loops (25), DIS24(GA)-4ap, and DIS24(GA-A272C)-4ap were chemically synthesized by using standard protocols. Complementary DIS24(UC) and DIS24(UC-A272C) stem–loops were prepared by in vitro T7 polymerase synthesis. All RNAs were purified by using denaturing polyacrylamide gel electrophoresis, recovered by electrophoretic elution, and exchanged into NMR buffer (1 mM cacodylate/25 mM NaCl, pH 6.5). NCp7 protein was expressed and purified as described (28).
NMR Spectroscopy. Several different isotopically labeled DIS kissing complexes (Fig. 1C) were formed for NMR studies: DIS23(GA)·DIS23(HxUC), 15N-DIS23(GA)·DIS23(HxUC), DIS23(GA)·15N-DIS23(HxUC), and 13C15N-A,G-labeled DIS23(GA)·DIS23(HxUC). For all dimer complexes, corresponding DIS23(GA) and DIS23(HxUC) stem–loops were mixed in a 1:1 ratio followed by snap-cooling. The DIS kissing complexes were stabilized by using either 300 mM NaCl or 3.3 mM MgCl2, which was added from concentrated stock solutions. NMR samples contained RNA at concentrations from 0.6 to 0.8 mM and were brought to a final volume of ≈280 μl in Shigemi susceptibility matched tubes (Shigemi, Allison Park, PA). The pH of the samples was adjusted by using concentrated stocks of NaOH and HCl. NMR spectra were recorded on either Bruker (Billerica, MA) AVANCE 500 MHz or 600 MHz spectrometers equipped with 1H,13C,15N triple-axis-gradient probes and processed by using nmrpipe (29). NMR experiments were collected at 25°C unless otherwise indicated. 1D proton spectra were collected by using either a jump-and-return pulse sequence or a 15N-selected jump-and-return pulse sequence with a sweep width of 12,500 Hz, 8 K complex points, and 128 scans and processed by using a 5-Hz line-broadening in xwinnmr. Assignment of the imino proton resonances was made by using 2D 15N-1H heteronuclear single quantum correlation spectroscopy (HSQC) spectra, as well as 2D 15N/14N edited/filtered NOESY spectra. The assignments of the DIS23(GA) purine base C2, H2, C8, and H8 resonances were made by using a 3D 13C-edited NOESY experiment collected at 35°C. For the pKa determinations, 2D 13C-1H HSQC spectra were recorded by using pH values ranging from 5.1 to 8.2. The 13C-1H HSQC spectra were collected with sweep widths of 6,000 Hz in the t2 and 5,000 Hz in the t1 dimensions, 2 K by 240 complex data points in t2 and t1, respectively, and 120 scans per increment.
Rates of NCp7 Chaperone Activity. 2-AP was inserted as a single bulged nucleotide to form construct DIS24(GA)-4ap. This construct allowed conversion from kissing to duplex dimer to be monitored directly by fluorescence quenching by using the complementary DIS24(UC) stem–loop (25). Mutant constructs DIS24(GA-A272C)-4ap and DIS24(UC-A272C), together with their complements, were used to determine the extent to which the DIS maturation rate is influenced by the specific nucleotide at position 272.
NCp7-catalyzed structural isomerization of the DIS was measured at 25°C by using a Spex Fluoromax-3 (Spex Industries, Metuchen, NJ), by following the decrease in fluorescence emission over time as the DIS24(GA)-4ap·DIS24(UC) kissing dimer is converted to the extended duplex. The DIS24(GA)-4ap·DIS24(UC) kissing dimer was first formed in NMR buffer solution (at pH 6.0 and 7.2, respectively) in the presence of 5 mM MgCl2, then mixed manually with NCp7 and rapidly inserted into the spectrofluorometer for fluorescence measurements. A similar procedure was used to determine the conversion rates of the kissing complexes formed with mutated junctions. In all experiments, the concentration of the kissing complex was 100 nM, and that of the NCp7 protein was 225 nM. The time course of the conversion of the DIS kissing dimer was fit by using the equation, Ft = F1 exp(–kconv t) + F2 exp(–karr t) + C, where kconv is the observed isomerization rate for conversion of the DIS kissing to extended mature duplex dimer, and karr is attributed to the rate of rearrangement of the 2-AP probe subsequent to its stacking in the duplex conformation. In all experiments, both rates showed similar pH-dependent behavior.
Results and Discussion
pH-Dependent Conformational Dynamics in the DIS Kissing Dimer. The DISMal kissing dimers formed by RNA hairpins with noncomplementary stems and point mutations in the loop palindromic sequence [so-called DIS23(GA) and DIS23(HxUC)] have been used in this study (Fig. 1C). These altered sequences exhibit wild-type kissing dimer properties (3, 16, 24, 25) while exclusively forming kissing heterocomplexes at NMR concentrations and maintaining pseudosymmetry in the complex. Formation of the DIS kissing heterodimer was monitored by using changes in 1D imino proton NMR spectra that are observed on mixture of equimolar amounts of DIS23(GA) with DIS23(HxUC) and addition of either NaCl or MgCl2 to the NMR buffer at pH 6.5. Additional imino proton resonances associated with new Watson–Crick base pairs in the loop–loop helix, as well as shifts of the resonances associated with some of the Watson–Crick base pairs in the stems, are expected on DIS kissing dimer formation. Surprisingly, initial imino spectra collected on an unlabeled DIS kissing heterodimer formed under standard NMR conditions at pH 6.5 showed not only the anticipated spectral changes but also a selective broadening of a subset of resonances, which was independent of the buffer, salt, or temperature used in the experiments (data not shown). To investigate a possible pH dependence of the observed broadening, the imino spectra of different selectively isotope-labeled DIS kissing heterodimers, designed to facilitate NMR resonance assignment, were acquired over a varied pH range. As an example, Fig. 1D shows the G imino proton resonances from DIS23(GA), which are selectively observed by using a 15N-filtered NMR experiment applied to a complex formed by 13C15N-A,G-labeled DIS23(GA) and DIS23(HxUC). In these experiments, the line widths of the imino proton resonances originally observed to be broadened at pH 6.5 showed pronounced pH dependencies over a near-physiological range, with complete exchange broadening observed at low pH and relatively sharp lines observed at high pH.
From the complete assignment of the imino proton spectra obtained at high pH (Fig. 1E), it becomes evident that the imino resonances belonging to residues that display pH-dependent line widths, like G276, G278, G271, and G274 seen in Fig. 1D, are associated with base pairs in the loop–loop and stem helices that flank the purine junctions (A272, G273, and A280) of the DIS kissing dimer (these base pairs and DIS purine junctions are highlighted in Fig. 1C by a shaded box). In addition, the pH effect is observed for imino proton resonances from both stem–loops in the kissing dimer to approximately the same degree, consistent with the pseudosymmetry of the kissing heterodimer complex. In contrast, those imino proton resonances associated with base pairs in the stems, like G268, that are further from the junctions do not display pH dependence. The selective exchange broadening of imino protons involved in Watson–Crick base pairs in the loop–loop helix and in the apical Watson–Crick base pairs of the stem helices indicates that this region of the DIS dimer structure becomes dynamic on protonation at low pH. In addition, the observation of exchange broadening of the NMR resonances at low pH defines a relatively slow, micro- to millisecond, time scale for these associated dynamic motions in the DIS kissing dimer. It should be noted that imino protons are in exchange with the bulk water, and the line widths of these resonances contain contributions from this exchange rate. However, the observed trend for the DIS kissing dimer in the pH range used here is opposite what would be expected if the increase of the line widths was a result of an increase in the rate of exchange with bulk H2O. In addition, no evidence for dissociation of the kissing dimer is found in the pH range investigated, ruling out the possibility that the broadening of the line widths is due to exchange between individual hairpins and kissing dimer.
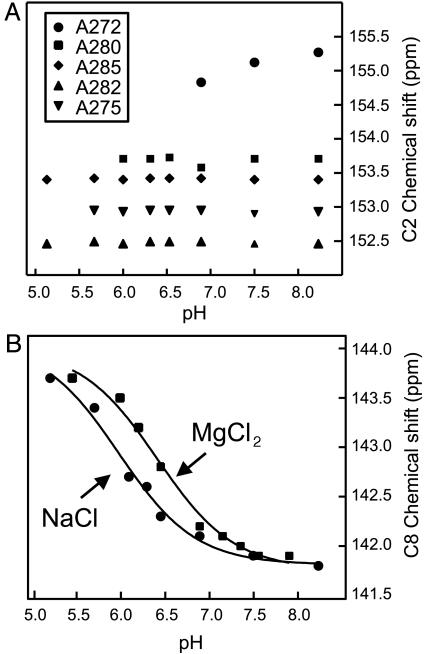
To directly probe whether the pH dependence of the DIS imino proton spectra is coupled to an RNA protonation event(s), a series of 1H,13C HSQC spectra were collected over a pH range by using the complex formed by 13C15N-A,G-labeled DIS23(GA) and DIS23(HxUC), which allowed purine base resonances from DIS23(GA) to be selectively observed. Base carbon chemical shifts, in particular adenosine C2 chemical shifts, are very sensitive to the protonation state of a base ring and can be used to determine in situ the pKa of these bases in nucleic acid structures (30, 31). In addition, pH-dependent dynamics may also be manifested in these experiments in the form of cross-peak broadening. In the 1H,13C HSQC spectra, the C2 chemical shift of A272 is observed to shift upfield as the pH is decreased from 8.2, as would be expected if N1 of this adenosine base becomes protonated over the titrated pH range, whereas the chemical shifts of the C2 resonances belonging to the other DIS23(GA) adenosine bases are unchanged over the pH range investigated, indicating that the pKas of these bases are below pH 4.5 (Fig. 2A). Determination of the pKa of protonation for A272 by using changes in the C2 chemical shift, however, was not possible, because the C2-H2 correlation of A272 was also observed to be significantly exchange broadened as a function of pH and is not detectable below pH 6.9. A pKa for A272 in the kissing dimer complex was therefore determined by monitoring changes in the C8 chemical shift because the H8–C8 correlation of A272 could be detected over the entire pH range. The magnitude and direction of the change in the A272 C8 chemical shift is consistent with what would be expected to result from protonation of the N1 position of this base and could be fit to a Henderson–Hasselbach equation, yielding an apparent pKa of 6.0 ± 0.1 in NMR buffer with 300 mM NaCl and 6.4 ± 0.1 in NMR buffer with 3.3 mM MgCl2 (Fig. 2B). Taken together, these results indicate that the pH-dependent dynamics observed for the DIS kissing dimer is directly coupled to the protonation state of A272, which titrates over a near-physiological pH range, with the slight difference in the A272 pKa for complexes formed in NaCl vs. MgCl2, suggestive of subtle cation-dependent difference in the structure of the DIS kissing dimer junctions. Moreover, although the pKa in the kissing dimer of residue A272 from DIS23(HxUC) was not determined explicitly, the generally observed pH dependence of resonances from this base and surrounding residues, together with the pseudosymmetry of the complex, strongly indicates that this symmetrically related residue has a similarly shifted pKa.
Fig. 2.
In situ determination of a pKa for A272. (A) Plot of the C2 carbon chemical shifts vs. pH of the five adenosines in the DIS23(GA) stem–loop in the 13C,15N-A,G-labeled DIS23(GA)·DIS23(HxUC) kissing complex formed in the presence of 300 mM NaCl. Note that resonances from residue A272, A280, and A275 become exchange broadened beyond detection as the pH is lowered, and, therefore, the pH-dependent correlation of the C2 chemical shifts for these bases is incomplete. (B) Plot of the C8 carbon chemical shift of A272 from DIS23(GA) vs. pH in the 13C,15N-A,G-labeled DIS23(GA)·DIS23(HxUC) kissing complex formed in the presence of either 300 mM NaCl or 3.3 mM MgCl2. Solid lines are best fits using a Henderson–Hasselbach equation.
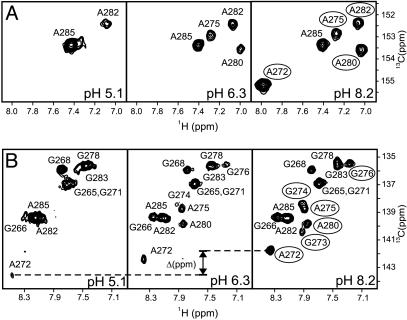
In addition to pH-dependent exchange broadening of the A272 resonances, nonexchangeable resonances assigned to A280 and G273, as well as those assigned to bases in the stems and loop–loop helix flanking A272 in the DIS kissing dimer, are observed in the 1H,13C HSQC spectra to display pH-dependent broadening (Fig. 3). At low pH, most of these resonances are exchange broadened beyond detection. Thus, as indicated by the pH dependency of the imino proton resonances, the pH-dependent exchange broadening of the nonexchangeable resonances provides further evidence that the A272 protonated state of DIS kissing dimer is dynamic. For residue G273, the C8–H8 correlation is relatively broad even at high pH, indicating that this base is somewhat dynamic even when A272 is deprotonated. The imino proton of G273 is also not observed at any pH investigated, which may be the result of rapid exchange with water and/or dynamic motions of the DIS junctions. Note that the observed exchange broadening in the DIS kissing dimer cannot be attributed solely to an intermediate rate of exchange between protonated/deprotonated forms of A272, because such kinetics would predict a sharpening of resonances at low pH (30, 31), which is not observed. In addition, a quantitative description of the two exchange processes is not possible because resonances needed for this analysis are broadened beyond detection at low pH. Interestingly, although the constellation and relative positions of the purine junction bases are identical in the DIS kissing and duplex dimers (Fig. 1B), pH-dependent conformation dynamics associated with protonation of A272 are not observed in the mature duplex dimer or the individual stem–loops (data not shown), indicating that these purines adopt a unique conformation in the kissing dimer that results in the shifted pKa.
Fig. 3.
Selected regions of a 600-MHz 2D 1H-13C HSQC spectrum of the 13C,15N-A,G-labeled DIS23(GA)·DIS23(HxUC) kissing complex formed in NMR buffer supplemented with 300 mM NaCl showing purine C2-H2 (A) and C8-H8 (B) correlations at pH 5.1, 6.3, and 8.2. Note that, although the C2 chemical shift is expected to shift upfield by as much as 8 ppm on protonation of an adenosine base, no C2–H2 crosspeaks are observed outside the plotted region over the entire pH range. The change in C8 carbon chemical shift for A272 is indicated; other resolved crosspeaks observed to broadened at low pH are circled in the pH 8.2 spectrum.
The near-physiological pKa observed for A272 in the kissing dimer represents a shift of ≈2.5 pH units relative to what is observed for 5′-AMP (31). Adenosines with similarly shifted pKas have been found in other RNA structures; however, in these cases, protonation of adenosine (A+) is associated with stabilization of structure, either because A+ becomes the donor in a hydrogen bond or because the stacking interactions are enhanced when one member of the stacking pair becomes protonated (refs. 31–34 and references therein). In this respect, the DIS kissing dimer is unique in that formation of A272+ state of DIS results in a conformationally dynamic state and an apparent destabilization of local RNA structure.
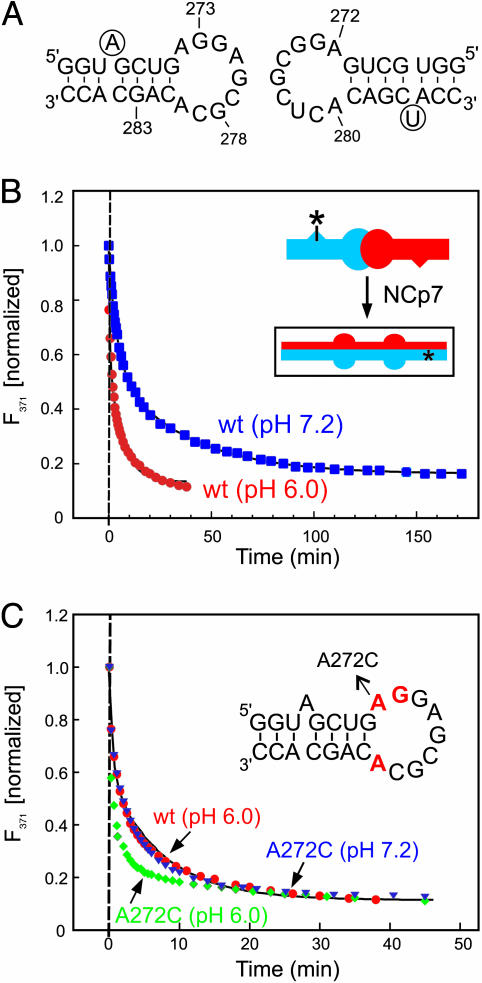
pH Dependence of the NCp7-Catalyzed DIS Structural Rearrangement. Rates of conversion from DIS kissing to extended duplex conformation were measured as a function of pH to determine whether the observed proton-coupled dynamics of the DIS kissing dimer effected NCp7-catalyzed DIS maturation. The conversion rates for the DIS isomerization from the kissing to duplex conformation at pH 6.0 and 7.2 were determined by monitoring the change in the steady-state fluorescence after addition of NCp7 to a DIS kissing complex formed between stem–loops DIS24(GA)-4ap and DIS24(UC), which contain complementary stems and can adopt the mature dimer form (Fig. 4A). In these experiments (25), the fluorescence of the bulged 2-AP reporter is observed to quench on forming a 2-AP-U base pair in the mature DIS dimer. Structural maturation rates determined with these experiments (Fig. 4B) are found to be approximately five times faster at pH 6.0, where 72% of A272 is protonated, than at pH 7.2, where only 20% of A272 is protonated.
Fig. 4.
pH dependence of NCp7-catalyzed DIS maturation. (A) Sequence of the DIS24(GA)-4ap and DIS24(UC) stem–loops used in the fluorescence experiments. 2-AP label at position 4 of DIS24(GA)-4ap and the added U-bulge in DIS24(UC) are circled. (B) Plot of the normalized fluorescence decay as a function of time after NCp7 protein was added to a DIS24(GA)-4ap·DIS24(UC) kissing complex preformed in NMR buffer with 5 mM MgCl2 at pH 6.0 (red circles) and 7.2 (blue squares). Structural isomerization rates (pH 6.0: kconv = 1.11 ± 0.12 min–1, karr = 0.11 ± 0.01 min–1; pH 7.2: kconv = 0.23 ± 0.01 min–1, karr = 0.026 ± 0.001 min–1) were fit by using a double exponential rate equation. (Inset) A schematic of the NCp7-catalyzed structural isomerization of the kissing dimer. Asterisks indicate the position of 2-AP in the DIS24(GA)-4ap stem-loop. (C) Plot of the normalized fluorescence decay as a function of time after NCp7 protein was added to kissing complexes formed with A272C mutant DIS sequences DIS24(GA,A272C)-4ap·DIS24(UC,A272C) by using conditions as in A, at pH 6.0 (green diamonds) and 7.2 (blue triangles). Structural isomerization rates (pH 6.0: kconv = 4.92 ± 0.59 min–1, karr = 0.28 ± 0.03 min–1; pH 7.2: kconv = 3.94 ± 0.49 min–1, karr = 0.22 ± 0.01 min–1) were fit by using a double exponential rate equation. The DIS wild-type sequence data (red circles) is shown for comparison. (Inset) A schematic of the DIS24(GA)-4ap stem–loop with junction bases shown in red and the A272C mutation indicated.
To confirm that the pH dependence of the NCp7-catalyzed DIS isomerization is directly correlated to the protonation state of A272, maturation rates for DIS kissing complexes formed between DIS24(GA)-4ap and DIS24(UC) stem–loops in which A272 was mutated to C were determined (Fig. 4C). The A272C mutation is a nonpurine substitution that does not introduce a possible new Watson–Crick base pair in the loop. For the A272C mutant kissing complex, the pH dependence of the NCp7-catalyzed isomerization rate is almost completely lost (the difference between pHs is ≈25%), indicating that the protonation state of A272 is primarily responsible for the pH effect. In addition, the pH dependence of conversion rates measured for the DIS kissing complexes, in which only one hairpin was mutated, were found to differ by only about half that observed for kissing complexes with the mutation in both junctions. As suggested by the NMR data, this result indicates that residue A272 in both junctions of the kissing complex modulates the local RNA structure to a similar extent through a proton-coupled effect. It also reveals that the proton-coupled conformational change experienced by the individual junctions is noncooperative with respect to NCp7-catalyzed DIS maturation. It should also be noted that for the kissing complexes in which A272 was mutated, measured DIS conversion rates at comparable pH are accelerated relative to the wild-type sequences (Fig. 4C). This is likely due to a more significant destabilization of the DIS junctions by the mutations than by A272 protonation.
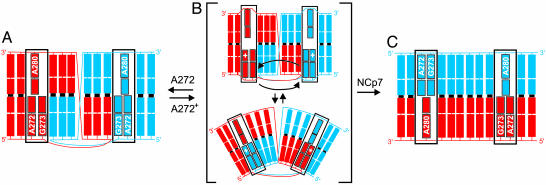
A Conformationally Dynamic Switch in the DIS Kissing Dimer. Proton-coupled switches between RNA conformational states have previously been observed in E. coli 5S RNA (35) and a pseudoknot structure surrounding the first ribosome initiation site in E. coli α mRNA (36). Here, the observed exchange broadening of the protonated state of the DIS kissing dimer suggests a dynamic model for the DIS kissing dimer that likely involves motions of the 5′ purine junction bases A272 and G273 (Fig. 5). In this model, the unprotonated A272 state of the DIS kissing dimer, which adopts a defined conformation based on the line widths and nuclear Overhauser effects (NOEs) observed for the heterodimeric DIS kissing complex at pH 8.2 (data not shown) becomes destabilized on A272 protonation. As shown schematically in Fig. 5A, NOEs observed in 3D 13C-edited-NOESY experiments applied to selectively 15N,13C-labeled heterodimeric DIS kissing complexes at pH 8.2 (data not shown) are consistent with a single conformation of the kissing dimer junctions that involves 5′ stacking of A272 and G273 and 3′ stacking of A280 between the stem helix of the same stem–loop and the kissing loop–loop helix. NMR data collected on the DIS kissing complex at pH 8.2, however, did not provide evidence for formation of any stable base pairing between the junction bases in the kissing complex. These bases therefore appear to adopt a base pair independent but unique conformation, resulting in a chemical environment around A272 that shifts its pKa.
Fig. 5.
Model for the pH-dependent structural interconversion of the DIS dimer. DIS stem–loops are shown in blue and red to the second base in the stem. Junction bases are labeled and boxed. (A) The A272 nonprotonated state of the DIS kissing dimer observed at high pH. (B) A two-state representation of the dynamic ensemble of states that results on protonation of A272. Black arrows indicate possible 5′ base motions of A272 and G273 that could result in the observed exchange broadening of the NMR resonances belonging to these and flanking residues. Note that the ensemble may also be comprised of states in which only one of the 5′ bases flips between stacked positions or states in which the purine base motions are more localized. (C) The mature duplex dimer state, showing a model of the NMR-determined structure (39, 40) with all junction bases stacked in toward the helix.
Protonation of residue A272 in the DIS kissing dimer introduces a positive charge and changes the hydrogen-bonding potential of this base. These changes could serve to disrupt the junction structure of the nonprotonated state of the DIS kissing dimer and allow bases G273, A272+, and A280 to adopt and exchange between two or more alternative conformational states (Fig. 5B). An intermediate exchange, on the NMR time scale, between these states not only would result in the observed broadening of resonances associated with the junction purines but also could cause broadening of resonances belonging to flanking residues in the loop–loop helix and the apical base pairs in the stem helices, as observed experimentally. Note that exchange between even slightly different conformations of the 5′ purine bases could result in significant broadening if resonances experience different ring current effects in these conformations due to changes in stacking geometry. Among the conformations accessible in this ensemble may be a state(s) more favorably predisposed structurally for NCp7-activated conversion (Fig. 5B). In this model, the pH dependence of the NCp7-catalyzed rate of DIS maturation is explained by a proton-coupled shift in the conformational equilibrium between DIS kissing dimer states that results in an increase in the population of such a favorable state(s).
Analysis of a pH-Dependent Dynamic RNA Structure. Our results indicate that the DIS kissing dimer does not adopt a single conformational state at near-physiological pH but rather appears to be dynamic in its functional form. The pH-dependent conformational dynamics identified for the DIS kissing dimer provide a possible physical explanation for previously reported differences in interpretation of chemical probing data (13, 14), as well as differences observed among our structural data, the NMR structure of the Lai isolate (37), and the crystal structures of the Mal and Lai isolates (38) of the DIS kissing dimer. In particular, accessibility of the N1 position of A272 to chemical probes appears to vary with changes in the protonated state of A272 and subsequent changes in its conformation under different pH conditions (13, 14). In addition, different stacked conformations observed for 5′ A272 and R273 may represent different conformations of the DIS kissing dimer observed under different experimental conditions with varied pHs close to the pKa of A272. It should be noted, however, that the stacked-out conformation of the 5′ purine bases observed in the crystal structure is stabilized by the interdimer stacking of these bases among symmetrically related kissing complexes (38). The relevance of this structure relative to the solution observations, which indicate that these bases are stacked in and adopt a conformation that results in a shifted pKa for A272, is not clear. Differences have also been observed in the NMR- (39, 40) and x-ray- (41) determined conformations of the purine junctions in the mature DIS dimer, indicating that even in this dimeric form, the junctions remain quite plastic.
A Possible Regulatory Mechanism for HIV-1 Genomic RNA Dimer Maturation. Previous studies have shown that the purine junction bases contribute to the stability and association kinetics of the DIS kissing dimer (8, 12, 13). In vivo, these bases may therefore act first in the role of promoting and kinetically trapping formation of genomic RNA homodimers in infected eukaryotic cells that are then packaged into new retroviral particles. In the postbudded particle, protonation of A272 could then act, as it is observed to function in vitro, to accelerate NCp7-catalyzed DIS maturation. In vivo, the pKa of A272 could also be further shifted on interaction of the DIS kissing dimer with other retroviral or cellular factors, like NCp7, thus making protonation the dominant state at physiological pH. Accordingly, proton-coupled modulation of the DIS purine junctions could allow these highly conserved nucleotides to serve dual functional roles in the HIV-1 replication life cycle, switching from enhancers of DIS kissing dimer formation to promoters of NCp7-catalyzed HIV-1 viral RNA maturation.
Conclusion
A proton-coupled dynamic conformational switch has been identified in the DIS kissing complex at near-physiological pH, which directly modulates local RNA structure and the rate of NCp7-catalyzed DIS dimer structural maturation. This proton-coupled switch demonstrates a specific mechanism by which modulation of local RNA structure could promote the chaperone-mediated structural rearrangement of a kinetically trapped RNA conformation. Although these observations are highly suggestive of a biological role for proton-coupled modulation of the HIV-1 DIS kissing dimer conformation and dynamics, further in vivo analysis will be required to determine its physiological significance in HIV-1 genomic RNA dimerization and viral particle maturation.
Acknowledgments
This work was supported by National Institutes of Health Grant GM59107 (to J.P.M.). NMR instrumentation is supported by the National Institutes of Health, the National Institute of Standards and Technology, and the W. M. Keck Foundation. We thank F. Song (Center for Advanced Research in Biotechnology) for oligonucleotide synthesis and M. Rist (Technische Universität München, Munich) for preparation of mutant DIS samples.
Abbreviations: HIV-1, HIV type 1; DIS, dimerization initiation site; DLS, dimer linkage structure; 2-AP, 2-aminopurine; HSQC, heteronuclear single quantum correlation spectroscopy.
References
- 1.Skripkin, E., Paillart, J. C., Marquet, R., Ehresmann, B. & Ehresmann, C. (1994) Proc. Natl. Acad. Sci. USA 91, 4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laughrea, M. & Jette, L. (1994) Biochemistry 33, 13464–13474. [DOI] [PubMed] [Google Scholar]
- 3.Paillart, J. C., Marquet, R., Skripkin, E., Ehresmann, C. & Ehresmann, B. (1996) Biochimie 78, 639–653. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout, B. & vanWamel, J. L. B. (1996) J. Virol. 70, 6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clever, J. L. & Parslow, T. G. (1997) J. Virol. 71, 3407–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laughrea, M., Jette, L., Mak, J., Kleiman, L., Liang, C. & Wainberg, M. A. (1997) J. Virol. 71, 3397–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haddrick, M., Lear, A. L., Cann, A. J. & Heaphy, S. (1996) J. Mol. Biol. 259, 58–68. [DOI] [PubMed] [Google Scholar]
- 8.Paillart, J. C., Berthoux, L., Ottmann, M., Darlix, J. L., Marquet, R., Ehresmann, B. & Ehresmann, C. (1996) J. Virol. 70, 8348–8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen, N., Jette, L., Liang, C., Wainberg, M. A. & Laughrea, M. (2000) J. Virol. 74, 5729–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paillart, J. C., Skripkin, E., Ehresmann, B., Ehresmann, C. & Marquet, R. (1996) Proc. Natl. Acad. Sci. USA 93, 5572–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laughrea, M. & Jette, L. (1996) Biochemistry 35, 1589–1598. [DOI] [PubMed] [Google Scholar]
- 12.Skripkin, E., Paillart, J. C., Marquet, R., Blumenfeld, M., Ehresmann, B. & Ehresmann, C. (1996) J. Biol. Chem. 271, 28812–28817. [DOI] [PubMed] [Google Scholar]
- 13.Paillart, J. C., Westhof, E., Ehresmann, C., Ehresmann, B. & Marquet, R. (1997) J. Mol. Biol. 270, 36–49. [DOI] [PubMed] [Google Scholar]
- 14.Jossinet, F., Paillart, J. C., Westhof, E., Hermann, T., Skripkin, E., Lodmell, J. S., Ehresmann, C., Ehresmann, B. & Marquet, R. (1999) RNA 5, 1222–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodmell, J. S., Ehresmann, C., Ehresmann, B. & Marquet, R. (2000) RNA 6, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lodmell, J. S., Ehresmann, C., Ehresmann, B. & Marquet, R. (2001) J. Mol. Biol. 311, 475–490. [DOI] [PubMed] [Google Scholar]
- 17.Fu, W. & Rein, A. (1993) J. Virol. 67, 5443–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu, W., Gorelick, R. J. & Rein, A. (1994) J. Virol. 68, 5013–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rein, A., Henderson, L. E. & Levin, J. G. (1998) Trends Biochem. Sci. 23, 297–301. [DOI] [PubMed] [Google Scholar]
- 20.Muriaux, D., DeRocquigny, H., Roques, B. P. & Paoletti, J. (1996) J. Biol. Chem. 271, 33686–33692. [DOI] [PubMed] [Google Scholar]
- 21.Feng, Y. X., Copeland, T. D., Henderson, L. E., Gorelick, R. J., Bosche, W. J., Levin, J. G. & Rein, A. (1996) Proc. Natl. Acad. Sci. USA 93, 7577–7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muriaux, D., Fosse, P. & Paoletti, J. (1996) Biochemistry 35, 5075–5082. [DOI] [PubMed] [Google Scholar]
- 23.Theilleux-Delalande, V., Girard, F., Huynh-Dinh, T., Lancelot, G. & Paoletti, J. (2000) Eur. J. Biochem. 267, 2711–2719. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi, K., Baba, S., Koyanagi, Y., Yamamoto, N., Takaku, H. & Kawai, G. (2001) J. Biol. Chem. 276, 31274–31278. [DOI] [PubMed] [Google Scholar]
- 25.Rist, M. J. & Marino, J. P. (2002) Biochemistry 41, 14762–14770. [DOI] [PubMed] [Google Scholar]
- 26.Milligan, J. F. & Uhlenbeck, O. C. (1989) Methods Enzymol. 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 27.Batey, R. T., Inada, M., Kujawinski, E., Puglisi, J. D. & Williamson, J. R. (1992) Nucleic Acids Res. 20, 4515–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amarasinghe, G. K., De Guzman, R. N., Turner, R. B., Chancellor, K. J., Wu, Z. R. & Summers, M. F. (2000) J. Mol. Biol. 301, 491–511. [DOI] [PubMed] [Google Scholar]
- 29.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 30.Legault, P. & Pardi, A. (1994) J. Am. Chem. Soc. 116, 8390–8391. [Google Scholar]
- 31.Legault, P. & Pardi, A. (1997) J. Am. Chem. Soc. 119, 6621–6628. [Google Scholar]
- 32.Cai, Z. P. & Tinoco, I. (1996) Biochemistry 35, 6026–6036. [DOI] [PubMed] [Google Scholar]
- 33.Ravindranathan, S., Butcher, S. E. & Feigon, J. (2000) Biochemistry 39, 16026–16032. [DOI] [PubMed] [Google Scholar]
- 34.Huppler, A., Nikstad, L. J., Allmann, A. M., Brow, D. A. & Butcher, S. E. (2002) Nat. Struct. Biol. 9, 431–435. [DOI] [PubMed] [Google Scholar]
- 35.Kao, T. & Crothers, D. M. (1980) Proc. Natl. Acad. Sci. USA 77, 3360–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gluick, T. C., Gerstner, R. B. & Draper, D. E. (1997) J. Mol. Biol. 270, 451–463. [DOI] [PubMed] [Google Scholar]
- 37.Mujeeb, A., Clever, J. L., Billeci, T. M., James, T. L. & Parslow, T. G. (1998) Nat. Struct. Biol. 5, 432–436. [DOI] [PubMed] [Google Scholar]
- 38.Ennifar, E., Walter, P., Ehresmann, B., Ehresmann, C. & Dumas, P. (2001) Nat. Struct. Biol. 8, 1064–1068. [DOI] [PubMed] [Google Scholar]
- 39.Girard, F., Barbault, F., Gouyette, C., Huynh-Dinh, T., Paoletti, J. & Lancelot, G. (1999) J. Biomol. Struct. Dyn. 16, 1145–1157. [DOI] [PubMed] [Google Scholar]
- 40.Mujeeb, A., Parslow, T. G., Zarrinpar, A., Das, C. & James, T. L. (1999) FEBS Lett. 458, 387–392. [DOI] [PubMed] [Google Scholar]
- 41.Ennifar, E., Yusupov, M., Walter, P., Marquet, R., Ehresmann, B., Ehresmann, C. & Dumas, P. (1999) Struct. Fold. Des. 7, 1439–1449. [DOI] [PubMed] [Google Scholar]