Abstract
The heat shock cognate protein 70 (Hsc70) is a member of a 70-kDa heat shock protein (HSP70) family that functions as molecular chaperones. In this study, a novel Hsc70 gene from Chinese soft-shelled turtle (Pelodiscus sinensis) (tHsc70) was identified. The tHsc70 full-length complementary DNA (cDNA) is 2 272 bp long with a 1 941-bp open reading frame (ORF) encoding 646 amino acids. Three characteristic signature regions of the HSP70 family, two major domains of an adenosine triphosphate (ATP)/guanosine triphosphate (GTP) binding domain (ABD), and a substrate-binding domain (SBD) were present in the predicted tHsc70 amino acid sequence. The tHsc70 gene was expressed in Escherichia coli BL21 and the expression product reacted with the anti-Hsc70 mouse monoclonal antibody by Western blotting. Homology analysis revealed that tHsc70 shared identity from 53.9% to 87.7% at the nucleotide level, and 49.1% to 99.5% at the amino acid level with the known Hsc70s. Phylogenetic analysis showed that tHsc70 was clustered together with the Hsc70 gene of another reptile species (Alligator mississippiensis). The tHsc70 was expressed in the liver, lung, heart, and skeletal muscle. The expression patterns of tHsc70 messenger RNA (mRNA) differed among different tissues under different durations of heat stress at 40 °C. Adaptation at 25 °C for 1 h after heat stress was also different among tissues and length of heat stress. Irrespective of different profiles of expression under heat stress, tHsc70 may play roles in protecting turtles from thermal stress.
Keywords: Pelodiscus sinensis, Heat shock cognate protein 70 (Hsc70), Phylogenetic analysis, Heat stress
1. Introduction
Heat shock proteins (HSPs) serve as molecular chaperone proteins and play critical roles in cell protection and protein folding in almost all organisms, from prokaryotes to eukaryotes (Lindquist and Craig, 1988). In addition, HSPs have attracted considerable interest among immunologists in recent years, because they have been implicated as chaperokines in the pathogenesis of a number of autoimmune diseases and in antigen presentation, cross-presentation, and tumor immunity (Pockley et al., 2008; Tsan and Gao, 2009). However, there are controversies over their roles in maintaining the balance between pro- and anti-inflammatory responses in mammalian cellular models (de Maio, 2011).
HSPs are well-conserved proteins that are named by their approximate molecular weights, Hsp60, Hsp70, and Hsp90, referring to heat shock proteins of sizes 60, 70, and 90 kDa, respectively (Lindquist and Craig, 1988; Kampinga et al., 2009). The Hsp70 family, one of the most abundant HSP families, has been extensively studied and consists of cytosolic Hsp70, including stress-inducible Hsp70 and constitutive heat shock cognate protein 70 (Hsc70), glucose-regulated protein 78 (Grp78), and mitochondrion Hsp70 (Wang et al., 2008; Luan et al., 2010). Hsp70, known to be activated by heat shock, is also induced by a variety of stressors, such as microbial infection, hypoxic condition, osmotic stress, ultraviolet (UV) irradiation, and heavy metals. It functions to protect organisms from damages (Pockley, 2003). Hsc70 is expressed under normal growth conditions such as development and cellular differentiation (Karouna-Renier et al., 2003; Yamashita et al., 2004; Uenishi et al., 2006; Chuang et al., 2007). It is also involved in regulating innate and adaptive immune responses and modulating hepatitis C virus infectivity (Parent et al., 2009; Su et al., 2010). Recent evidence indicates that Hsc70 gene expression pattern varies in response to different stresses and development (Shim et al., 2006). A number of Hsc70s have been identified from such species as human, mouse, two-spotted spider mite, zebrafish, and tiger shrimp (Santacruz et al., 1997; Hunt et al., 1999; Lo et al., 2004; Shim et al., 2006; Brocchieri et al., 2008) that have helped us to better understand their phylogenetic relationship and explore their biological functions. However, there are still many Hsc70 molecules that remain unidentified so far in less popular animal species, such as turtles and snakes.
Chinese soft-shell turtle Pelodiscus sinensis is an ectothermic amniotes reptile with a prominent evolutionary link between ectothermic anamniotic fishes and amphibians and endothermic amniotic birds and mammals (Zimmerman et al., 2010). Turtles are popular as a health-promoting food in Asian countries, including China, Japan, and Korea. The turtle industry has been growing steadily by large-scale artificial breeding in southeastern Chinese provinces Zhejiang, Jiangsu, and Fujian (Li et al., 2008). However, growth performance of the turtles can be affected by the aquatic environments where temperature, salinity, pollutant content, and oxygen can be greatly affected by season, weather, or even human activity. Turtles have to regulate expression of many of their genes to cope with varying environments and even microbial infection as part of their survival strategies (Yamashita et al., 2004; Luan et al., 2010). However, the HSPs in turtles have not been characterized. It remains unknown how the Hsp70s are expressed in turtles in response to stressors such as elevated temperatures. The main objectives of this study were to characterize the genetic structure of the tHsc70 gene from Chinese soft-shelled turtles and to analyze its expression at messenger RNA (mRNA) and protein levels in different tissues under heat shock.
2. Materials and methods
2.1. Treatments and tissue sample collection of turtles
Chinese soft-shelled turtles P. sinensis (CSST, two years old with weight at (550±20) g) were obtained from Hangzhou Zhongde Aquatic Culture Co., Ltd., China. The turtles were raised individually in 30 cm×23 cm×18 cm tanks placed in a room at (25±1) °C (as normal temperature), and acclimatized for one week before experiments. The animal experiments were approved by the Zhejiang University Committee for Experimental Animal Management.
For cloning of the tHsc70 gene, the turtle was subjected to a 40 °C environment for 4 h and a liver sample was taken for RNA extraction.
To evaluate heat shock stress and tHsc70 expression profiling, 21 turtles were divided into seven groups, with three in each group. Turtles in Group I were raised at 25 °C as a control. Groups II, III, and IV were exposed to 40 °C for 1, 2, and 4 h, respectively (Wang et al., 2008). To evaluate heat shock recovery, Groups V, VI, and VII were first exposed to 40 °C for 1, 2, and 4 h, respectively, consistent with Groups II–IV, and then moved to tanks with water temperature at 25 °C for 1 h for adaptation. After the treatments, the liver, skeletal muscle, lung, and heart were quickly dissected and immediately placed in liquid nitrogen, and stored at −80 °C until analysis.
2.2. Total RNA extraction
Total RNA was extracted from the liver, skeletal muscle, lung, and heart of CSST and purified as described by Li et al. (2011). Purity and quantity of RNA were measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). Integrity of RNA was evaluated by agarose gel electrophoresis. Only intact RNA samples with 260/280 nm absorbance between 1.8 and 2.0 were used for complementary DNA (cDNA) synthesis.
2.3. Amplification and cloning of tHsc70 cDNA
One microgram of total RNA containing the tHsc70 gene was used for reverse transcription using a SuperScript™ III First-Strand Synthesis SuperMix kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. cDNA synthesized was denatured and diluted to 100 μl. The polymerase chain reaction (PCR) fragment of tHsc70 was amplified using the primer pair Hsc70-dF and Hsc70-dR (Table 1) on a thermal cycler S1000™ (Bio-Rad, Hercules, CA, USA). PCR was carried out using 2 μl of cDNA as a template in a 30-μl reaction mixture with a denaturation step of 95 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 50 °C for 50 s and 72 °C for 30 s, and completed with a 10-min extension at 72 °C.
Table 1.
Primers for PCR and real-time PCR
| Primer name* | Sequence (5′–3′) |
| Hsc70-dF | CARCAYGGCAARGTGGAGATCAT |
| Hsc70-dR | TCAATACTGGCYTGDGTGCT |
| Q-RT | CCAGTGAGCAGAGTGACGAGGACTCGAGCTCAAGC(T)17 |
| Q-O | CCAGTGAGCAGAGTGACG |
| Q-I | GAGGACTCGAGCTCAAGC |
| Hsc70-3O | AGAAGGTTGGAGCGGAAAGGAATGT |
| Hsc70-3I | TCCCACCAAGCAGACTCAGACATTCA |
| Hsc70-5RT | TGCCACGGAACAGATCAGCATTTAA |
| Hsc70-5O | CCATACGGTTATCAAAGTCCTCCCCA |
| Hsc70-5I | ACATTCCTTTCCGCTCCAACCTT |
| Universal primer mix (UPM) | Long: CTAATACGACTCACTATAGCAAGCAGTGGTATCAACGCAGAGT |
| Short: CTAATACGACTCACTATAGC | |
| Nested universal primer (NUP) | AAGCAGTGGTATCAACGCAGAGT |
| Hsc70-ORF-F# | CCCAAGCTTATGTCTAAGGGACCAGCA |
| Hsc70-ORF-R§ | CCGCTCGAGATCTACTTCCTCAATGGTT |
| β-actin-qF | CTACAGCTTCACCACCACAG |
| β-actin-qR | GACCATCAGGGAGTTCGTA |
| Hsc70-qF | ATGAAGCTGTGGCTTACGGTGCA |
| Hsc70-qR | TGTTCCGCTTGATCAGGACTGTCA |
Last letters in the primer names: F, forward; R, reverse; RT, reverse transcription; O, outer; I, inner; qF, forward primers for quantitative PCR; qR, reverse primers for quantitative PCR
Underline means HindIII restriction endonuclease site
Underline means XhoI restriction endonuclease site
For 3′-end rapid amplification of cDNA ends (RACE), Q-RT, Q-O and Q-I primers (Table 1) were used for nested PCR according to Scotto-Lavino et al. (2006). The first-strand cDNA was synthesized with Q-RT primer. The 1st round PCR was performed using the synthesized cDNA and the primer pair Q-O/Hsc70-3O (Table 1). The 2nd round PCR products were further amplified using primer pair Q-I/Hsc70-3I (Table 1) as described by Li et al. (2011).
For 5′-end RACE amplification, cDNA was synthesized using the primer Hsc70-5RT (Table 1) to obtain the first-strand cDNA as described by Li et al. (2011). The cDNA containing CSST tHsc70 was used for PCR amplification with gene specific primers and adapter primers contained in the 5′-end RACE kit (D315, TaKaRa Biotechnology Co., Ltd., Dalian, China) (Table 1). The primer pair, Hsc70-5O and universal primer mix (UPM), was used in the 1st round PCR. The programs were as follows: 5 cycles at 94 °C for 30 s and 72 °C for 3 min, 5 cycles at 94 °C for 30 s, 70 °C for 30 s and 72 °C for 3 min, 5 cycles at 94 °C for 30 s, 68 °C for 30 s and 72 °C for 3 min, 20 cycles at 94 °C for 30 s, 66 °C for 30 s and 72 °C for 3 min. The 1st round PCR products were further amplified using the primer pair Hsc70-5I and nested universal primer (NUP) with cycling conditions as follows: 95 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 66 °C for 40 s and 72 °C for 3 min, and completed with a 12-min extension at 72 °C. The CSST tHsc70 open reading frame (ORF) was amplified using primer pair Hsc70-ORF-F/Hsc70-ORF-R (Table 1), gel-purified, and cloned into pMD18-T vector. The insertion was confirmed by sequencing.
2.4. Sequence analysis of turtle tHsc70
The corresponding sequences of CSST tHsc70 gene and encoding protein were blasted using ClustalW program and the phylogenetic tree was generated accordingly by MEGA 4.0 software (http://www.megasoftware.net).
2.5. Prokaryotic expression and purification of recombinant protein turtle Hsc70
The CSST tHsc70 ORF was amplified by the primer pair Hsc70-ORF-F and Hsc70-ORF-R (Table 1) containing HindIII and XhoI restriction sites, respectively, and then inserted into pET30a(+) vector. The recombinant pET30-tHsc70 was used to transform Escherichia coli BL21 cells. The transformation mixture was plated on Luria-Bertani (LB) agar containing kanamycin at 50 μg/ml. Insert orientation was further confirmed by restriction digestion and sequencing. One positive clone was cultured in LB medium containing kanamycin 50 μg/ml until the culture reached an optical density at 600 nm (OD600) at about 0.5, and the expression of the recombinant protein was induced with isopropy-β-D-thiogalactoside (IPTG) at 1 mmol/L for 4 h. The recombinant protein CSST Hsc70 (tHsc70) was then purified using Ni-NTA protein purification column (QIAGEN®, Hilden, Germany).
2.6. Tissue preparation and protein assay
Protein was extracted from CSST liver using cell lysis buffer for Western and IP kit (P0013, Beyotime® Biotech Institute, Jiangsu, China) according to the manufacturer’s instructions. Protein concentration was quantified using BCA protein assay kit (P0010, Beyotime® Biotech Institute).
2.7. Western blotting analysis
The protein samples were mixed with 4×protein loading buffer (containing 10% dodecyl sulfate, sodium salt (SDS) and 10% β-mercaptoethanol) and treated on a boiling water bath for 5 min before loading into gel. The proteins were separated on 12% SDS-polyacrylamide gel electrophoresis (PAGE), and transferred onto polyvinylidene fluoride (PVDF) membrane. The membrane was blocked with 5% non-fat milk, incubated with mouse anti-Hsc70 monoclonal antibody (sc-7298, Santa Cruz Biotechnol. Inc., CA, USA) and peroxidase conjugated goat anti-mouse IgG (H+L) (Multisciences, Shanghai, China), respectively. Blot was revealed on a ChampChemi™ professional machine (SageCreation Science Co., Ltd., Beijing, China) in the presence of Supersignal® west pico chemiluminescent substrate system (Thermo, Boston, USA).
2.8. Turtle tHsc70 mRNA expression in the liver, lung, muscle, and heart under normal temperature
First-strand cDNA was synthesized from total RNAs of the liver, lung, muscle, and heart as described above with a SuperScript™ III First-Strand Synthesis SuperMix kit. Semi-quantitative PCR was used to detect tHsc70 mRNA expression levels in four different tissues relative to untreated control turtles on a thermal cycler S1000™ (Bio-Rad). The cDNA fragments were amplified using primer pair Hsc70-qF/Hsc70-qR, and β-actin gene amplified as an internal control using primer pair β-actin-qF/β-actin-qR (Table 1). The cycle numbers were pre-examined to determine the linear reaction range for both tHsc70 and β-actin. The final cycling program used was: 94 °C for 4 min, followed by 33 cycles at 94 °C for 30 s, 65 °C (tHsc70) or 60 °C (β-actin) for 10 s, and 72 °C for 15 s.
2.9. Turtle tHsc70 mRNA expression analysis under different heat shock treatments by quantitative PCR
Quantitative PCR was performed to detect relative tHsc70 mRNA expression levels (β-actin RNA as internal control) under different heat shock treatments using cDNA and the SYBR® Premix Ex Taq™ kit (TaKaRa Biotechnology Co., Ltd.). The 7.5 μl of PCR master mix, 0.4 μl of 10 μmol/L primers (β-actin-F/β-actin-R or Hsc70-qF/Hsc70-qR, Table 1), and 1 μl of cDNA were mixed for quantification using the iQ5™ real-time multicolor PCR detector (Bio-Rad). The cycling profile was 95 °C pre-denaturation for 1 min, followed by 40 cycles at 95 °C for 10 s, 60 °C (β-actin) or 65 °C (tHsc70) for 10 s, and 72 °C for 15 s. Fold changes in gene expression were calculated as 2−ΔΔCt (Livak and Schmittgen, 2001).
2.10. Data analysis
All reactions were conducted in triplicate and relative mRNA expression was shown as mean±standard deviation (SD) (turtle n=3). SPSS software (Version 13.0, IBM Corporation, NY, USA) was used to evaluate the quantitative PCR data by one-way analysis of variance (ANOVA). Significance was declared if P<0.05.
3. Results
3.1. Cloning and sequence analysis of turtle tHsc70
The 1 157-bp length fragment was amplified by PCR from the liver cDNA using the degenerate primer pairs and sequenced. Sequence analysis shows that the gene fragment has 88% nucleotide and 99.5% amino acid identity to the Hsc70 of Alligator mississippiensis (accession No. AB_306280.1 and BAF_94143.1, respectively). Next, we designed pairs of gene-specific overlapping primers according to 1 157-bp sequence for 3′- and 5′-end RACE PCRs. A 933-bp sequence containing the stop codon (TAA), the polyadenylation signal sequence (AATAAA), and a poly(A) tail were obtained by 3′-RACE PCR (Fig. 1). Subsequently, a 664-bp fragment containing the start codon was obtained by 5′-RACE PCR. The above fragments were assembled to get a 2 272-bp sequence containing the full-length cDNA of turtle tHsc70. The ORF was further amplified with specific primers and cloned into pET30a(+).
Fig. 1.
Complete nucleotide and deduced amino acid sequences of tHsc70 in P. sinensis
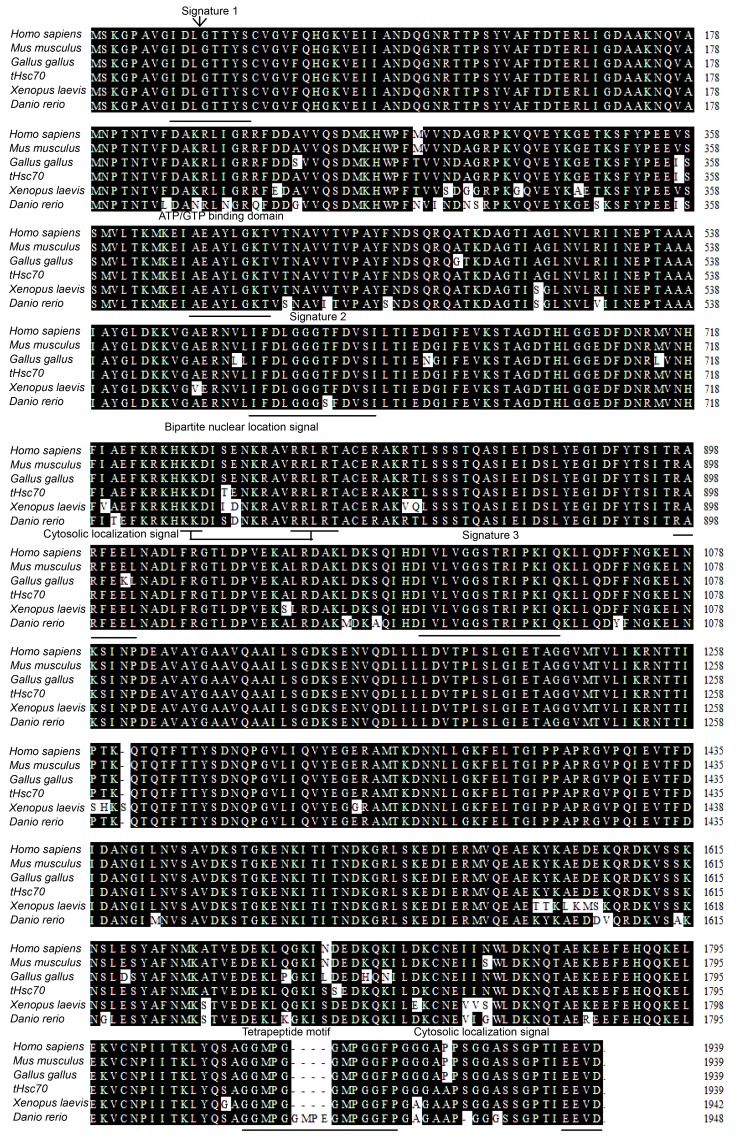
* indicates the start codon (ATG) and the stop codon (TAA). The polyadenylation signal sequence (AATAAA) is dash-underlined. Characteristic Hsc70 signatures are underlined. The putative ATP-GTP binding site is in bold, the putative bipartite nuclear location signal (KK and RRLRT) in grey, and the potential non-organelle eukaryotic consensus motif is boxed. Double underlines represent two consecutive repeats of the tetrapeptide motif GGMP. The ATPase domain (left) and the peptide-binding domain (right) are separated by a double vertical line
The CSST tHsc70 cDNA sequence (GenBank No. HQ_219723.1) contains a 1 941-bp ORF encoding a polypeptide of 646 amino acid (aa) residues with a predicted molecular mass of 70.8 kDa and an isoelectric point (pI) of 5.38 by DNAstar program (Madison, WUI, USA). There are three characteristic motifs of the Hsp70s from the deduced amino acid sequence: signature 1 (9–16 aa, IDLGTTYS), signature 2 (197–210 aa, DLGGGTFD), and signature 3 (334–348 aa, IVLVGGSTRIPKIQK) by website search (http://www.expasy.org) (Tabeta and Yamazaki, 2010). The other conserved motifs, such as two consecutive repeats of the tetrapeptide motif GGMP (Luan et al., 2010; Yang et al., 2010), the putative ATP-GTP binding domain, the putative bipartite nuclear location signal, and two cytosolic localization signals (a potential non-organelle eukaryotic consensus motif and the extreme C-terminal domain EEVD) (Shim et al., 2006; Tabeta and Yamazaki, 2010; Xie et al., 2010) were also identified (Fig. 2). The complete tHsc70 amino acid sequence can be separated into two parts—the ATPase domain (1–385 aa) and the peptide-binding domain (386–646 aa) (Fig. 2) (Luan et al., 2010).
Fig. 2.
Alignment of deduced amino acid sequences of Hsc70s from P. sinensis and other vertebrates
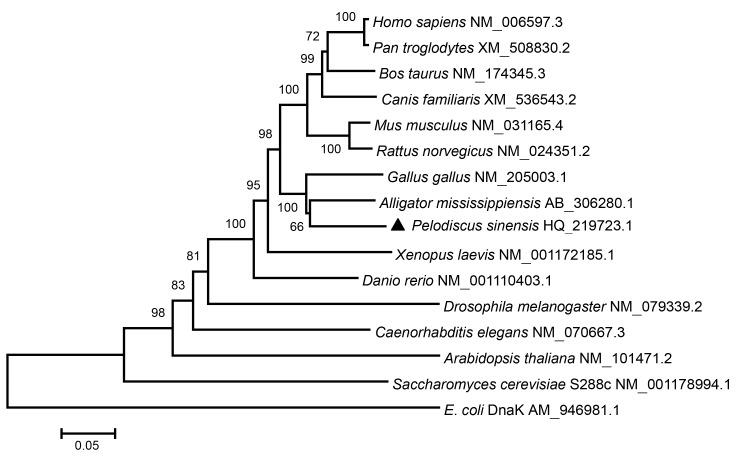
The nucleotide and amino acid sequences of tHsc70 closely match those of Hsc70s in other species and the identity scores range from 53.9% to 87.7% at nucleotide levels, and 49.1% to 99.5% at amino acid levels (Table 2). tHsc70 has the highest similarity to Hsc70 of the other reptile species A. mississippiensis and also shares high identity to those from other vertebrates (Fig. 2). Phylogenetic analysis reveals that tHsc70 forms a sister subgroup with reptile and avian species (Fig. 3).
Table 2.
Sequence identities of Hsc70 from P. sinensis with those of other animal species
| Species | Nucleotide |
Amino acid |
||
| GenBank accession No. | Hsc70 identity (%) | GenBank accession No. | Hsc70 similarity (%) | |
| Homo sapiens | NM_006597.3 | 84.2 | NP_006588.1 | 99.1 |
| Mus musculus | NM_031165.4 | 83.5 | NP_112442.2 | 98.9 |
| Pan troglodytes | XM_508830.2-1 | 84.4 | XP_508830.2 | 99.1 |
| Rattus norvegicus | NM_024351.2 | 83.6 | NP_077327.1 | 98.8 |
| Canis familiaris | XM_536543.2 | 83.3 | XP_536543.1 | 99.1 |
| Bos taurus | NM_174345.3 | 84.2 | NP_776770.2 | 98.9 |
| Gallus gallus | NM_205003.1 | 86.5 | NP_990334.1 | 97.8 |
| Alligator mississippiensis | AB_306280.1 | 87.7 | BAF_94143.1 | 99.5 |
| Xenopus laevis | NM_001172185.1 | 81.6 | NP_001165656.1 | 95.2 |
| Danio rerio | NM_001110403.1 | 80.0 | NP_001103873.1 | 95.5 |
| Drosophila melanogaster | NM_079339.2 | 70.6 | NP_524063.1 | 80.8 |
| Caenorhabditis elegans | NM_070667.3 | 71.8 | NP_503068.1 | 85.9 |
| Saccharomyces cerevisiae S288c | NM_001178994.1 | 66.0 | NP_011029.1 | 75.4 |
| Arabidopsis thaliana | NM_101471.2 | 66.4 | NP_173055.1 | 76.0 |
| E. coli DnaK | AM_946981.1 | 53.9 | CAQ_30531.1 | 49.1 |
* Bold means the highest identity and similarity (%)
Fig. 3.
Phylogenetic analysis of Hsc70 genes from 13 species using the neighbor-joining method
The number in each branch shows the bootstrap confidence value (percentage of 1 000 replicates). The tHsc70 of P. sinensis identified in this study is labeled with ▲. The GenBank accession numbers of all the sequences are given after the latin names
3.2. Identification of tHsp70 by SDS-PAGE and Western blotting
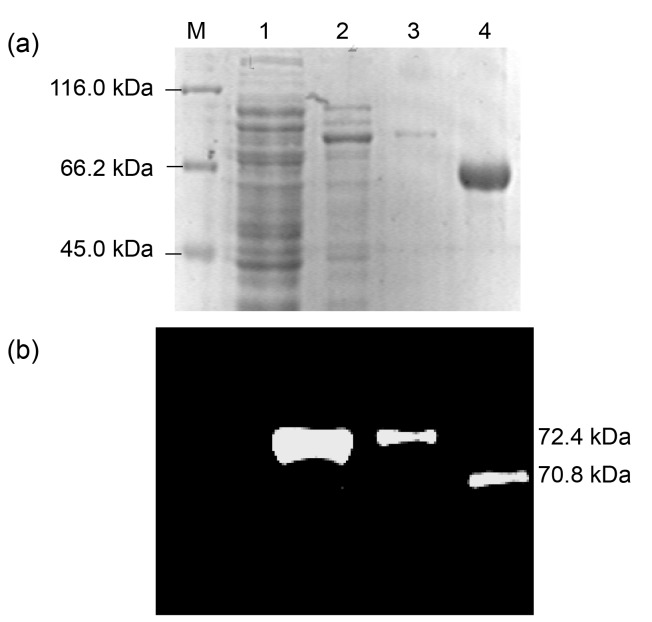
SDS-PAGE and Western blotting analyses further revealed that the recombinant fusion protein tHsc70 reacted with a specific monoclonal Hsc70 antibody (Fig. 4), indicating that the tHsc70 gene we obtained is correct. The slower motility of tHsc70 than that of Hsc70 (shown as higher molecular mass) could be due to presence of the His-tag of recombinant tHsc70 as well as possible conformational difference.
Fig. 4.
SDS-PAGE (a) and Western blotting (b) analyses of the recombinant protein tHsc70 expressed in E. coli using anti-Hsc70 monoclonal antibody
Lane 1: pET30a vector control; Lane 2: pET30-tHsc70 expression product; Lane 3: purified protein tHsc70; Lane 4: human Hsc70 protein
3.3. Expression profiles of turtle tHsc70 mRNA in different tissues under normal temperature
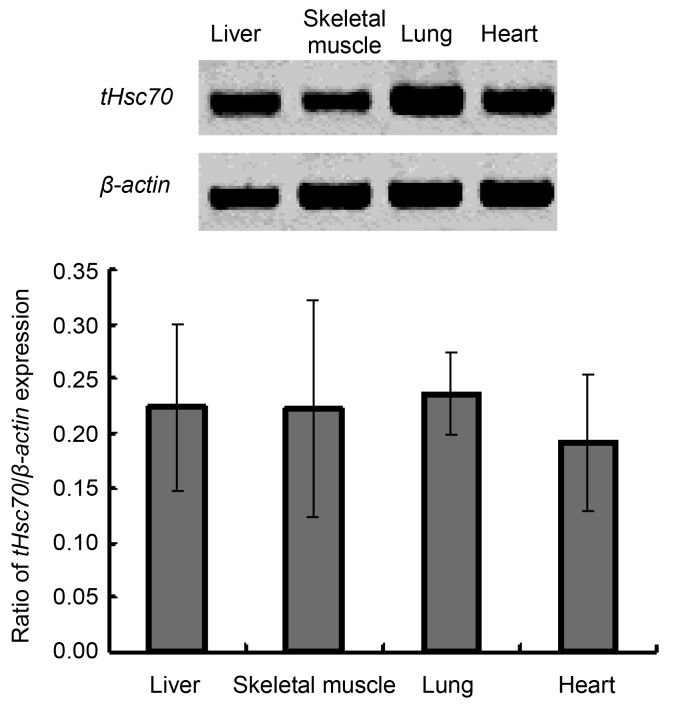
The turtle tHsc70 mRNA expression levels in the liver, skeletal muscle, lung and heart were examined by semi-quantitative RT-PCR. The results show that tHsc70 was expressed in these four tissues. However, there was no significant difference in expression levels among the tissues (Fig. 5).
Fig. 5.
Semi-quantitative RT-PCR analysis of tHsc70 expression in the liver, skeletal muscle, lung, and heart of P. sinensis
3.4. Expression profiles of turtle tHsc70 mRNA in different tissues under heat stress treatments
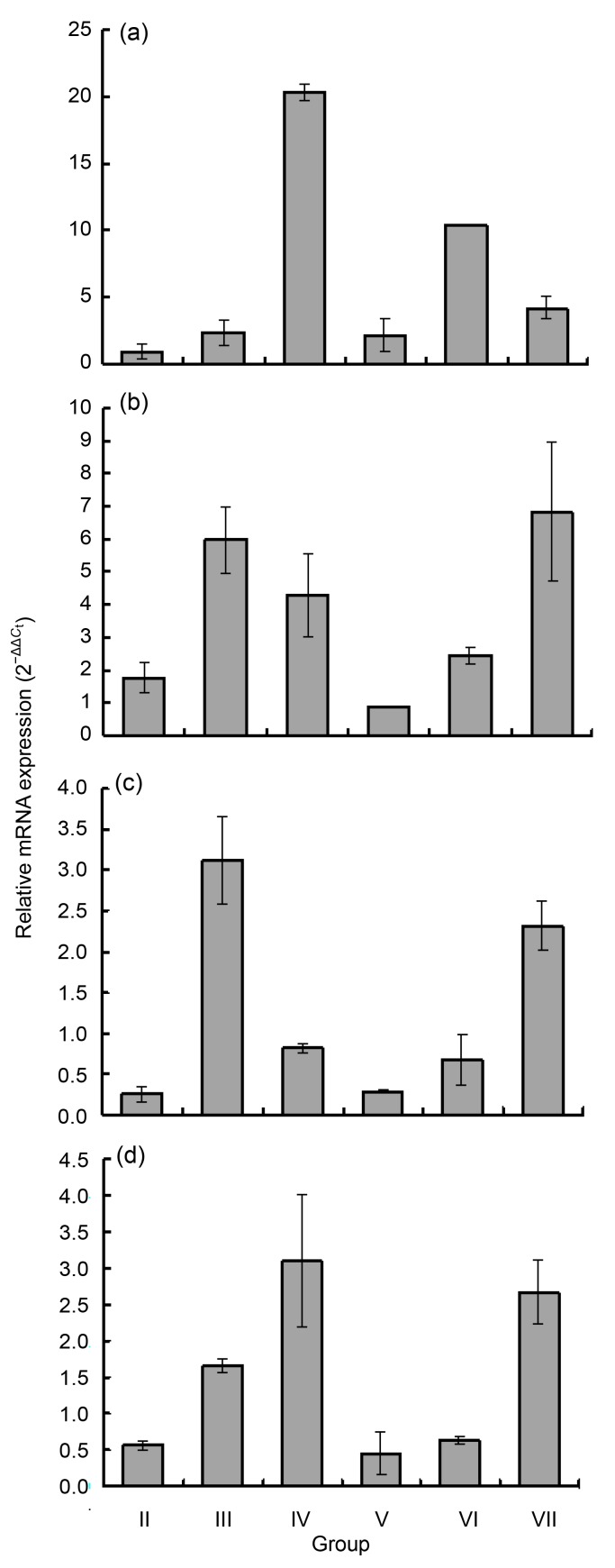
To determine whether tHsc70 responds to heat stress treatments, its expression was compared between turtles subjected to heat treatments and those of control Group I. In general, 1-h heat shock did not induce significant expression of tHsc70 except in the skeletal muscle. The expression in liver was generally similar to that in the heart (Figs. 6a and 6d). The expression profiles were also similar between skeletal muscle and lung (Figs. 6b and 6c). Expression of tHsc70 increased in the liver and heart with duration of treatment (Figs. 6a and 6d), while its transcripts in skeletal muscle and lung were highest in Group III and increased, but to a moderate degree, in Group IV (Figs. 6b and 6c). The highest expression was seen in the liver at 4 h of exposure to heat (approximately a 20-fold increase). Significant changes of relative mRNA expression could be found between different treatment groups of the same tissue as follows: Group II (0.88±0.56) vs. Group IV (20.35±0.64) (P<0.01) and Group III (2.29±0.97) vs. Group IV (20.35±0.64) (P<0.01) in the liver, Group II (1.74±0.47) vs. Group III (5.97±1.03) (P<0.05) in the skeletal muscle, Group II (0.25±0.09) vs. Group III (3.12±0.54) (P<0.05) and Group II (0.25±0.09) vs. Group IV (0.86±0.06) (P<0.05) in the lung, and Group II (0.56±0.05) vs. Group III (1.65±0.08) (P<0.05) in the heart (Fig. 6).
Fig. 6.
Real-time RT-PCR analyses of tHsc70 expression in the liver (a), skeletal muscle (b), lung (c), and heart (d) of P. sinensis after heat stress for 1, 2, and 4 h with 1-h adaptive recovery at 25 °C (Groups V, VI, and VII) or without adaptation (Groups II, III, and IV)
Expression levels of the control samples were set at 100% to normalize over the corresponding tissues from three normal turtles. Error bars represent the mean±SD (n=3)
We further examined relative tHsc70 mRNA expression in different tissues of CSST adapted for 1 h at 25 °C after varying periods of heat shock. The 1-h adaptation of turtles in Groups V and VI increased the expression in the liver, as compared to that in Groups II and III (Fig. 6a). However, adaptation suppressed the expression of tHsc70 for Group V turtles (Fig. 6a). Adaptation seemed to suppress the expression in the skeletal muscle, lung and heart from turtles heat-shocked for 2 h, but did not affect the expression for those heat-shocked for 4 h (Figs. 6b–6d).
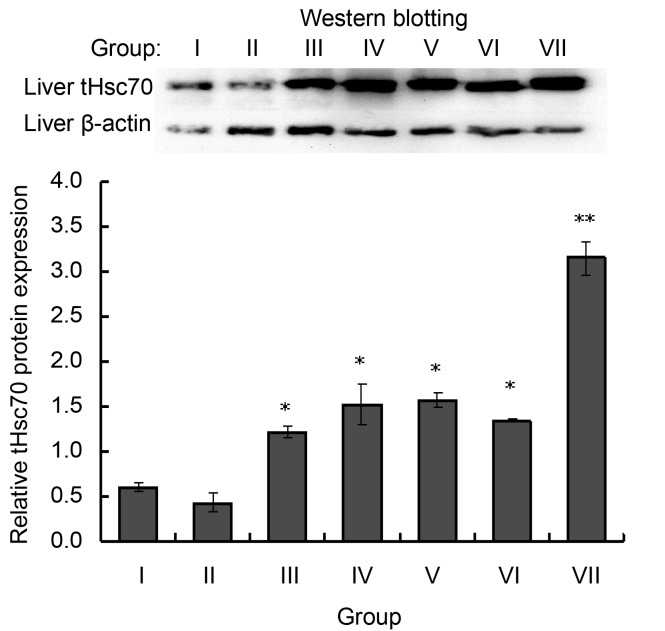
Western blotting shows that heat shock for 1 h did not affect tHsc70 in the liver. However, 2- to 4-h heat treatment increased its expression significantly (Group III (1.57±0.04) vs. Group I (1.57±0.04) (P<0.05), Group IV (1.11±0.07) vs. Group I (1.57±0.04) (P<0.05)). Expression of tHsc70 in the liver was even higher in Groups V, VI, and VII compared with Groups II, III, and IV (Fig. 7).
Fig. 7.
Western blotting analysis of tHsc70 expression in the liver of P. sinensis after heat stress for 1, 2, or 4 h with 1-h adaptive recovery at 25 °C (Groups V, VI, and VII) or without adaptation (Groups II, III, and IV)
Error bars represent the mean±SD (n=3). * P<0.05 and ** P<0.01 indicate statistical difference as compared with the untreated control
4. Discussion
In this study, a novel tHsc70 molecule from a Chinese soft-shelled turtle was cloned and identified. The full-length cDNA and its predicted protein sizes are similar to those of other Hsc70s. The tetrapeptide motif GGMP repeat, which is thought to be involved in co-chaperone-binding activities of Hsc70, was found near the C-terminal of tHsc70 (Demand et al., 1998), similar to FcHsc70 in Chinese shrimp Fenneropenaeus chinensis (Luan et al., 2010). Also present in the tHsc70 is the motif EEVD, which is found in nearly all eukaryotic cytosolic/nuclear Hsc70s (Demand et al., 1998; Saxena et al., 2008) and plays an important role in ATPase and peptide-binding activity (Pockley et al., 2008). For example, the EEVD motif may facilitate Hsp70 to bind with co-chaperones such as Hop, Hsp40, and Hip, through its interaction with Hsp90 (Demand et al., 1998). The EEVD motif was also thought to be involved in Listeria monocytogenes uptake by trophoblast giant cells and virus propagation (Watanabe et al., 2006; 2009). The presence of Hsc70 family signatures and the ATP-GTP binding domain suggests that tHsc70 is a member of the Hsp70 family.
Hsp70s are conserved proteins among living organisms during evolution. In this study, the phylogenetic tree reveals eight major evolutionarily related groups from fungi to mammalian species (Fig. 3). We have found that Hsc70 molecules of mammalian and avian species as well as the reptiles form two distinct subgroups (the upper part of Fig. 3 above P. sinensis). Similar results were seen with non-classical class I MHC and MyD88 genes (Glaberman et al., 2008; Li et al., 2011). Chicken and turtle are phylogenetically close, as with the case of their close evolutionary relationship (Kawai et al., 2007; Pokorná et al., 2011). Matsuda et al. (2005) suggested that there were six large chromosomes being almost equivalent to each other between turtles and chickens, and homology to chicken chromosomes was higher in turtle than in snake.
Demand et al. (1998) showed that the 10-kDa C-terminal domain of Hsc70 of rat consists of a tightly folded α-helical subdomain, repeats of the tetrapeptide GGMP, and the EEVD motif at the extreme C-terminus that provides binding sites for a distinct set of cofactors, such as Hsp40 (Hdj-1) and Hop (p60/Sti1). In this study, alignment analysis indicates that there was partial deletion of amino acids EGMP between the two GGMP motif repeats in Homo sapiens, Mus musculus, Gallus gallus, P. sinensis, and Xenopus laevis species, as compared to Danio rerio (Fig. 2). If EGMP acts as the isoform of the GGMP motif involved in co-chaperone binding, this deletion might affect binding affinity because of reduction of repeats. This has been seen with cortactin; its F-actin binding ability is mostly correlated with the number of cortactin repeats (Katsube et al., 2004).
The tHsc70 was expressed in four tissues at 25 °C (Fig. 5). There was no significant difference among these tissues. However, transcription was up-regulated after heat stress with different expression profiles. Relative expression of tHsc70 increased in all tissues from animals receiving heat stress (0.25-fold to 20.35-fold), as compared to control animals (Fig. 6). Expression was highest in the liver with 4-h heat treatment, over 20-fold higher compared to the control animals. This phenomenon was also observed in the diamondback moth (Plutella xylostella), tiger shrimp (Penaeus monodon), common killifish (Fundulus heteroclitus), and endoparasitoid (Pteromalus puparum) (Luan et al., 2010; Xiang et al., 2010; Zimmerman et al., 2010). There are similar reports showing that transcription of tHsc70 in the rat cortex and cerebellum was up-regulated after heat shock (Schroderus et al., 2010). Its increased expression may represent response to heat in order to stabilize proteins during folding in conjunction with other chaperones (Wang et al., 2008).
The turtle tHsc70 transcripts in the skeletal muscle and lung were highest upon 2-h heat treatment but had moderate increases at 4-h treatment (Figs. 6b and 6c). Similar scenarios were seen in earlier studies in which the increase in Hsc70 transcription in P. puparum and Drosophila melanogaster was higher with short-time heat exposure than with extended exposure (Wang et al., 2008). This may suggest that tHsc70 is only important for fitness in the beginning of heat stress, and other HSPs are induced then to play their parts in thermo-tolerance.
Adaptation for 1 h after heat stress exhibited different patterns of tHsc70 expression among different tissues. With tissues like skeletal muscle and lung having more direct exposure to heat shock, adaptation after 2-h heat shock could have helped the tissues recover from stress as shown by reduced tHsc70 expression. However, 4-h heat stress might have a stronger effect on heat shock response such that adaptation for 1 h was not enough to help the recovery in the muscle and lung in Group VII turtles (Figs. 6b and 6c), shown as elevated tHsc70 transcription. However, the level of tHsc70 transcript in the liver from the same group was considerably reduced as compared with other tissues. This may be due to shorter half-life (less stable) of mRNA in the liver than in other tissues such as heart and muscle (Connor et al., 1996). The increased tHsc70 protein expression in the liver in Group VII was in contrast to lower mRNA transcription (Fig. 6a vs. Fig. 7). Quick decay of Hsc70 mRNA by post-transcriptional regulation in the liver during post-stress adaptation could be one possibility. Significant decay of Hsp70 mRNA was seen in a Drosophila cell line Schneider 2 cells during the post-stress adaptation period (Bönish et al., 2007).
In summary, the tHsc70 molecule that we characterized is important in protecting turtles from heat stress. Further research is under way to examine the roles of tHsc70 in response to microbial infections at elevated temperatures that may occur in intensified turtle farming.
Acknowledgments
The authors thank Mr. Guo-qiang XIONG, Director of the Hangzhou Zhongde Aquatic Culture Co., Ltd., Zhejiang, China, for providing the turtles.
Footnotes
Project (No. 20082332Y03) supported by the Science and Technology Department of Hangzhou City, China
References
- 1.Bönish C, Temme C, Moritz B, Wahle E. Degradation of hsp70 and other mRNAs in Drosophila via the 5′-3′ pathway and its regulation by heat shock. J Biol Chem. 2007;282(30):21818–21828. doi: 10.1074/jbc.M702998200. [DOI] [PubMed] [Google Scholar]
- 2.Brocchieri L, de Macario EC, Macario AJL. hsp70 genes in the human genome: conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol Biol. 2008;8:19. doi: 10.1186/1471-2148-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chuang KH, Ho SH, Song YL. Cloning and expression analysis of heat shock cognate 70 gene promoter in tiger shrimp (Penaeus monodon) Gene. 2007;405(1-2):10–18. doi: 10.1016/j.gene.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Connor MK, Takahashi M, Hood DA. Tissue-specific stability of nuclear- and mitochondrially encoded mRNAs. Arch Biochem Biophys. 1996;333(1):103–108. doi: 10.1006/abbi.1996.0369. [DOI] [PubMed] [Google Scholar]
- 5.de Maio A. Extracellular heat shock proteins, cellular export vesicles, and the Stress Observation System: a form of communication during injury, infection, and cell damage. It is never known how far a controversial finding will go! Dedicated to Ferruccio Ritossa. Cell Stress Chaperones. 2011;16(3):235–249. doi: 10.1007/s12192-010-0236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demand J, Lüders J, Höhfeld J. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol. 1998;18(4):2023–2028. doi: 10.1128/mcb.18.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glaberman S, du Pasquier L, Caccone A. Characterization of a nonclassical class I MHC gene in a reptile, the Galápagos marine iguana (Amblyrhynchus cristatus) PLoS One. 2008;3(8):e2859. doi: 10.1371/journal.pone.0002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunt CR, Parsian AJ, Goswami PC, Kozak CA. Characterization and expression of the mouse Hsc70 gene. Biochim Biophys Acta. 1999;1444(3):315–325. doi: 10.1016/S0167-4781(98)00285-1. [DOI] [PubMed] [Google Scholar]
- 9.Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14(1):105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karouna-Renier NK, Yang WJ, Rao KR. Cloning and characterization of a 70 kDa heat shock cognate gene (Hsc70) from two species of Chironomus . Insect Mol Biol. 2003;12(1):19–26. doi: 10.1046/j.1365-2583.2003.00383.x. [DOI] [PubMed] [Google Scholar]
- 11.Katsube T, Togashi S, Hashimoto N, Ogiu T, Tsuji H. Filamentous actin binding ability of cortactin isoforms is responsible for their cell-cell junctional localization in epithelial cells. Arch Biochem Biophys. 2004;427(1):79–90. doi: 10.1016/j.abb.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Kawai A, Nishida-Umehara C, Ishijima J, Tsuda Y, Ota H, Matsuda Y. Different origins of bird and reptile sex chromosomes inferred from comparative mapping of chicken Z-linked genes. Cytogenet Genome Res. 2007;117(1-4):92–102. doi: 10.1159/000103169. [DOI] [PubMed] [Google Scholar]
- 13.Li XL, Zhang CL, Fang WH, Lin FC. White-spot disease of Chinese soft-shelled turtles (Trionyx sinens) caused by Paecilomyces lilacinus . J Zhejiang Univ Sci B. 2008;9(7):578–581. doi: 10.1631/jzus.B0720009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li XL, Zhu BL, Chen N, Hu HX, Chen JS, Zhang XY, Li J, Fang WH. Molecular characterization and functional analysis of MyD88 in Chinese soft-shelled turtle Trionyx sinensis . Fish Shellfish Immunol. 2011;30(1):33–38. doi: 10.1016/j.fsi.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Lindquist S, Craig EA. The heat shock protein. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Lo WY, Liu KF, Liao IC, Song YL. Cloning and molecular characterization of heat shock cognate 70 from tiger shrimp (Penaeus monodon) Cell Stress Chaperones. 2004;9(4):332–343. doi: 10.1379/CSC-47R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luan W, Li F, Zhang J, Wen R, Li Y, Xiang J. Identification of a novel inducible cytosolic Hsp70 gene in Chinese shrimp Fenneropenaeus chinensis and comparison of its expression with the cognate Hsc70 under different stresses. Cell Stress Chaperones. 2010;15(1):83–93. doi: 10.1007/s12192-009-0124-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuda Y, Nishida-Umehara C, Tarui H, Kuroiwa A, Yamada K, Isobe T, Ando J, Fujiwara A, Hirao Y, Nishimura O, et al. Highly conserved linkage homology between birds and turtles: bird and turtle chromosomes are precise counterparts of each other. Chromosome Res. 2005;13(6):601–615. doi: 10.1007/s10577-005-0986-5. [DOI] [PubMed] [Google Scholar]
- 20.Parent R, Qu X, Petit MA, Beretta L. The heat shock cognate protein 70 is associated with hepatitis C virus particles and modulates virus infectivity. Hepatology. 2009;49(6):1798–1809. doi: 10.1002/hep.22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pockley AG. Heat shock proteins as regulators of the immune response. Lancet. 2003;362(9382):469–476. doi: 10.1016/S0140-6736(03)14075-5. [DOI] [PubMed] [Google Scholar]
- 22.Pockley AG, Muthana M, Calderwood SK. The dual immunoregulatory roles of stress proteins. Trends Biochem Sci. 2008;33(2):71–79. doi: 10.1016/j.tibs.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Pokorná M, Giovannotti M, Kratochvíl L, Kasai F, Trifonov VA, O′Brien PCM, Caputo V, Olmo E, Ferguson-Smith MA, Rens W. Strong conservation of the bird Z chromosome in reptilian genomes is revealed by comparative painting despite 275 million years divergence. Chromosoma. 2011;120(5):455–468. doi: 10.1007/s00412-011-0322-0. [DOI] [PubMed] [Google Scholar]
- 24.Santacruz H, Vriz S, Angelier N. Molecular characterization of a heat shock cognate cDNA of zebrafish, hsc70, and developmental expression of the corresponding transcripts. Dev Genet. 1997;21(3):223–233. doi: 10.1002/(SICI)1520-6408(1997). [DOI] [PubMed] [Google Scholar]
- 25.Saxena V, Lienesch DW, Zhou M, Bommireddy R, Azhar M, Doetschman T, Singh RR. Dual roles of immunoregulatory cytokine TGF-beta in the pathogenesis of autoimmunity-mediated organ damage. J Immunol. 2008;180(3):1903–1912. doi: 10.4049/jimmunol.180.3.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroderus E, Jokinen I, Koivula M, Koskela E, Mappes T, Mills SC, Oksanen TA, Poikonen T. Intra- and intersexual trade-offs between testosterone and immune system: implications for sexual and sexually antagonistic selection. Am Nat. 2010;176(4):e90–e97. doi: 10.1086/656264. [DOI] [PubMed] [Google Scholar]
- 27.Scotto-Lavino E, Du G, Frohman MA. 3′ end cDNA amplification using classic RACE. Nat Protoc. 2006;1(6):2742–2745. doi: 10.1038/nprot.2006.481. [DOI] [PubMed] [Google Scholar]
- 28.Shim JK, Jung DO, Park JW, Kim DW, Ha DM, Lee KY. Molecular cloning of the heat-shock cognate 70 (Hsc70) gene from the two-spotted spider mite, Tetranychus urticae, and its expression in response to heat shock and starvation. Comp Biochem Physiol B Biochem Mol Biol. 2006;145(3-4):288–295. doi: 10.1016/j.cbpb.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Su X, Sykes JB, Ao L, Raeburn CD, Fullerton DA, Meng X. Extracellular heat shock cognate protein 70 induces cardiac functional tolerance to endotoxin: differential effect on TNF-alpha and ICAM-1 levels in heart tissue. Cytokine. 2010;51(1):60–66. doi: 10.1016/j.cyto.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabeta K, Yamazaki K. Analysis of immune responses to purified recombinant antigens of periodontal pathogens. Methods Mol Biol. 2010;666:345–357. doi: 10.1007/978-1-60761-820-121. [DOI] [PubMed] [Google Scholar]
- 31.Tsan MF, Gao B. Heat shock proteins and immune system. J Leukoc Biol. 2009;85(6):905–910. doi: 10.1189/jlb.0109005. [DOI] [PubMed] [Google Scholar]
- 32.Uenishi R, Gong P, Suzuki K, Koizumi S. Cross talk of heat shock and heavy metal regulatory pathways. Biochem Biophys Res Commun. 2006;341(4):1072–1077. doi: 10.1016/j.bbrc.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Dong SZ, Li K, Hu C, Ye GY. A heat shock cognate 70 gene in the endoparasitoid, Pteromalus puparum, and its expression in relation to thermal stress. BMB Rep. 2008;41(5):388–393. doi: 10.5483/BMBRep.2008.41.5.388. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe K, Fuse T, Asano I, Tsukahara F, Maru Y, Nagata K, Kitazato K, Kobayashi N. Identification of Hsc70 as an influenza virus matrix protein (M1) binding factor involved in the virus life cycle. FEBS Lett. 2006;580(24):5785–5790. doi: 10.1016/j.febslet.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe K, Tachibana M, Kim S, Watarai M. EEVD motif of heat shock cognate protein 70 contributes to bacterial uptake by trophoblast giant cells. J Biomed Sci. 2009;16(1):113. doi: 10.1186/1423-0127-16-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang LX, He D, Dong WR, Zhang YW, Shao JZ. Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genomics. 2010;11:472. doi: 10.1186/1471-2164-11-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie JH, Zhu HY, Guo L, Ruan YY, Wang L, Sun LL, Zhou L, Wu WB, Yun XJ, Shen AG, et al. Lectin-like oxidized low-density lipoprotein receptor-1 delivers heat shock protein 60-fused antigen into the MHC class I presentation pathway. J Immunol. 2010;185(4):2306–2313. doi: 10.4049/jimmunol.0903214. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita M, Hirayoshi K, Nagata K. Characterization of multiple members of the HSP70 family in platyfish culture cells: molecular evolution of stress protein HSP70 in vertebrates. Gene. 2004;336(2):207–218. doi: 10.1016/j.gene.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 39.Yang AF, Zhou ZC, Dong Y, Jiang B, Wang XY, Chen Z, Guan XY, Wang B, Sun DP. Expression of immune-related genes in embryos and larvae of sea cucumber Apostichopus japonicus. Fish Shellfish Immunol. 2010;29(5):839–845. doi: 10.1016/j.fsi.2010.07.023. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerman LM, Vogel LA, Bowden RM. Understanding the vertebrate immune system: insights from the reptilian perspective. J Exp Biol. 2010;213(5):661–671. doi: 10.1242/jeb.038315. [DOI] [PubMed] [Google Scholar]