Abstract
Now that the human genome has been sequenced, the challenge of assigning function to human genes has become acute. Existing approaches using microarrays or proteomics frequently generate very large volumes of data not directly related to biological function, making interpretation difficult. Here, we describe a technique for integrative systems biology in which: (i) primary cells are cultured under biologically meaningful conditions; (ii) a limited number of biologically meaningful readouts are measured; and (iii) the results obtained under several different conditions are combined for analysis. Studies of human endothelial cells overexpressing different signaling molecules under multiple inflammatory conditions show that this system can capture a remarkable range of functions by a relatively small number of simple measurements. In particular, measurement of seven different protein levels by ELISA under four different conditions is capable of reconstructing pathway associations of 25 different proteins representing four known signaling pathways, implicating additional participants in the NF-κBorRAS/mitogen-activated protein kinase pathways and defining additional interactions between these pathways.
The completion of the human genome has made the full complement of human genes available, offering unparalleled opportunities for biology and medicine (1). Realizing this potential, however, will require practical and rapid methods for discovering the functions of novel genes in complex biological systems (2). Existing attempts to meet this challenge have generally used whole-cell measurements of gene expression levels (3), protein–protein interactions (4), or cellular metabolism (5) to model system responses. In yeast, genes have been successfully assigned to particular functional or signaling pathways on the basis of their coordinate regulation in genomewide expression analyses using DNA microarrays (3). But despite extensive efforts to develop improved statistical techniques for predicting functional networks from large data sets (6) the transition from whole-cell molecular measurements to useful models of cellular responses in higher eukaryotes remains daunting.
In seeking to address this problem, we have developed an alternative technique with three main features. First, to reduce artifacts induced by the use of cell lines, we have concentrated on measuring the responses of primary human cells cultured in biologically relevant contexts. Second, rather than measuring the expression levels of every gene or the physical interactions of every protein, we have focused on a small set of biologically relevant parameters. And third, instead of performing these measurements under only one set of culture conditions, we have studied the same cell type in multiple different contexts (culture conditions differing in cell activation stimuli), so as to allow functional characterization of a wide range of protein activities from a manageably small number of measurements.
To implement this approach, which we have termed biologically multiplexed activity profiling (BioMAP), it is important to assemble a set of measurements and cell systems (cells in different defined contexts) broad enough to encompass most or all of the signaling pathways relevant to a particular biological process. The responses of these systems to genetic or other experimental perturbation is then registered by changes in the selected parameters. Here, we show by using vascular endothelium in four contexts defined by stimulation with different proinflammatory cytokines that BioMAP analysis can predict functional relationships of proteins within pathways and reveal interactions between different pathways that could not have been deduced from analysis of cells in any single context. BioMAP analyses will be a useful tool for modeling the signaling networks operating in human cells.
Methods
Cytokines, Antibodies, and Cell Culture. Recombinant human IFN-γ, tumor necrosis factor α (TNF-α), and IL-1β were from R & D Systems. Murine IgG was from Sigma, mouse anti-human intercellular adhesion molecule 1 (ICAM-1) (clone B4H10) was from Beckman Coulter, and mouse anti-human E-selectin (clone ENA1) was from HyCult Biotechnology (Uden, The Netherlands). Un-conjugated mouse antibodies against human vascular cell adhesion molecule 1 (VCAM-1) (clone 51–10C9), CD31 (clone WM-59), HLA-DR (clone G46–6), monokine induced by IFN-γ (MIG) (clone B8–77), and monocyte chemoattractant protein 1 (MCP-1) (clone 5D3-F7) were from BD Biosciences (San Diego). Mouse anti-human IL-8 (clone 6217.111) was from R & D Systems. PD098059 was from Calbiochem. EGM-2 medium and required supplements were from Clonetics (San Diego). Human umbilical vein endothelial cells were from Clonetics and cultured in microtiter plates in EGM-2 medium containing manufacturer's supplements plus 2% heat-inactivated FBS. Confluent cell were stimulated with cytokines (1 ng/ml IL-1β, 5 ng/ml TNF-α, or 100 ng/ml IFN-γ) for 24 h. PD098059 (3.7 μM final concentration) was added 1 h before stimulation and was present during the whole 24-h stimulation period.
Cell-Based ELISAs. Cell-based ELISAs were carried out as described (7). Briefly, microtiter plates containing treated and stimulated human umbilical vein endothelial cells were blocked, and then incubated with primary antibodies or isotype control antibodies (0.01–0.5 μg/ml) for 1 h. After washing, plates were then incubated with a peroxidase-conjugated anti-mouse IgG secondary antibody (Promega) for 1 h. Plates were washed and developed with TMB substrate (Clinical Science Products, Mansfield, MA), and the OD was read at 450 nm (subtracting the background absorbance at 650 nm) on a SpectraMAX 190 plate reader (Molecular Devices).
Retroviral Gene Transduction. Test genes were cloned into a vector derived from the Moloney murine leukemia virus (MoMLV)-based vector pFB (Stratagene) downstream of the MoMLV LTR. A truncated form of the human nerve growth factor receptor preceded by an internal ribosomal entry site (8) was used as a marker gene. Retroviral vector plasmid DNA was transfected into AmphoPack-293 cells (Clontech) by a modified calcium phosphate method according to the manufacturer's protocol (MBS transfection kit, Stratagene). Cell supernatants were harvested 48 h posttransfection, filtered to remove cell debris (0.45 μm), and transferred onto exponentially growing human umbilical vein endothelial cells. DEAE dextran (10 μg/ml) was added to facilitate transduction. After 5–8 h, the viral supernatant was removed, and cells were cultured for an additional 40 h. Gene transfer efficiency was determined by flow cytometric analysis using a nerve growth factor receptor-specific mAb and was typically ≥70%.
Statistical Analysis. The value of each parameter was measured three times per experiment, and two to four experiments were carried out for each overexpressed gene. Within each experiment, the mean value obtained for each parameter was then divided by the mean value from a sample transduced with empty vector to generate a ratio. All ratios were then log10-transformed, the transformed ratios were averaged from repeat experiments, and nonparametric analysis was used to compare the profile of these ratios to the envelope of control profiles. Those profiles containing ratio values that exceeded the 95% prediction level envelope for control profiles were used to calculate pairwise Pearson correlation coefficients (partek pro, version 5.1). To select statistically significant correlation coefficients, 100 randomized data sets were created by permuting the original expression data, and the pairwise correlation coefficients were calculated for each randomized set. Correlation limits were then selected so as to exclude all but a defined minimal number of correlations from the randomized data sets. For the four cellular environments combined, limits of [–0.5035, 0.546] excluded all but 2.5% of the “correlations” derived from the randomized data sets (in other words, at these limits 2.5% of the correlations observed are potentially false positives). Limits used to filter correlations obtained in individual cellular environments were: IL-1β-treated cells [–0.87, 0.88], TNF-α-treated cells [–0.87, 0.90], IFN-γ-treated cells [–0.86, 0.88], and control cells [–0.84, 0.89] (see also Tables 2–4, which are published as supporting information on the PNAS web site). Significantly anticorrelated profiles were observed as well, although too few examples of negative correlations occurred to permit biological interpretation. The arrangement of genes in two dimensions was automatically determined from the entire set of correlation values by a multidimensional scaling method using AT&T graphviz software (www.research.att.com/sw/tools/graphviz); the statistically significant correlations are highlighted by connecting lines seen in Fig. 2 c–g.
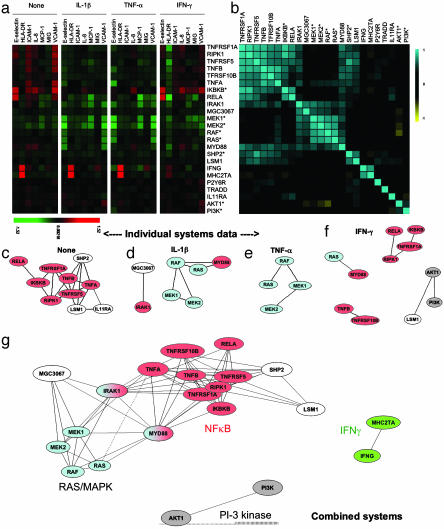
Fig. 2.
Functional classification of genes in multiple cellular contexts. (a) Endothelial cells transduced with retroviral vectors expressing the genes listed to the right were treated with IL-1β, TNF-α, IFN-γ, or media alone (Control). Shown are relative increase (red), decrease (green), or lack of change (black) in the mean log expression ratio of each parameter relative to nontransduced cells in two to four experiments. (b) Pairwise Pearson correlation analysis of gene-specific profiles using the combined 28 parameter profile comprising all seven readouts from each of the four cellular systems (cells plus cytokine-defined contexts) combined into a single data string for calculations. Positive correlation is shown in blue, and negative correlation is shown in yellow. The order of genes was automatically determined by multidimensional scaling of the Pearson correlation metric (see Methods). (c and d) 2D representations of the functional similarity of gene profiles revealed in each individual system [cells in medium alone (c), IL-1β-treated cells (d), TNF-α-treated cells (e), and IFN-γ-treated cells (f)]. Pearson correlation analysis was performed as before, using the seven readouts within a given system, and multidimensional scaling was used to represent the extent of similarity of gene activities in the systems indicated. Only genes whose responses showed significant similarity to other genes in the indicated system are shown. (g) The relationships revealed by combined systems analysis are shown. In this case, the 28 parameter combined systems profiles (encompassing the seven readouts from each of the four cell systems) was used for correlation analysis and 2D representation. The arrangement of genes in two dimensions was automatically determined by multidimensional scaling (see Methods), and statistically significant correlations are shown by the connecting lines. Genes are color-coded to indicate participation in common pathways (red, NF-κB; blue, RAS/MAPK; green, IFN-γ; gray, PI3K/Akt; white, unique genes). SHP2 is a dominant negative and may stimulate NF-κB by suppressing RAS/MAPK pathway.
Results
Analysis of Endothelial Cells Overexpressing Signaling Proteins. Endothelial cells control vascular inflammation by regulating leukocyte traffic and express immunomodulatory cytokines and chemokines. To analyze this range of activity, we overexpressed genes encoding key elements of the NF-κB signaling pathway, the phosphatidylinositol 3-kinase (PI3K)/Akt pathway, and the RAS/mitogen-activated protein kinase (MAPK) pathway in cultures of primary endothelial cells and stimulated individual proinflammatory pathways (genes overexpressed are listed in Table 1). Some genes (denoted by an asterisk in Table 1) were overexpressed in a constitutively active form (with the exception of SHP2, dominant negative) to maximize their activity. The effects were then assessed by measuring the levels of surface proteins known to be regulated by inflammation and/or to reflect the functional state of the cells, including VCAM-1, ICAM-1, and E-selectin (vascular adhesion molecules for leukocytes), HLA-DR (MHC class II; the protein responsible for antigen presentation), MIG/CXCL9 and IL-8/CXCL8 (chemokines that mediate selective leukocyte recruitment from the blood), and platelet-endothelial cell adhesion molecule 1/CD31 (a protein controlling leukocyte transmigration).
Table 1. Genes overexpressed.
| Gene | Gene description | GenBank accession no. |
|---|---|---|
| TNFRSF1A | TNF-α receptor type I | BC010140 |
| RIPK1 | Receptor-interacting serine threonine kinase 1 (RIP) | NM_003804 |
| TNFRSF5 | CD40 | BC012419 |
| TNFB | TNF-β (lymphotoxin A) | D12614 |
| TNFRSF10B | TRAIL receptor 2 | BC001281 |
| TNFA | TNF-α | NM_000594 |
| IKBKB* | I-κB kinase β (IKKB), constitutively active (15) | AF031416 |
| RELA | NF-κB subunit 3 (p65) | NM_021975 |
| IRAK1 | IL-1 receptor-associated kinase 1 | BC014963 |
| MGC3067 | Hypothetical protein MGC3067 | BC002457 |
| MEK1* | MAP2K1, constitutively active R4F (16) | NM_002755 |
| MEK2* | MAP2K2, constitutively active K71W (16) | L11285 |
| RAF* | Raf1, constitutively active (17) | L00212 |
| RAS* | H-Ras, constitutively active V12 (18) | NM_005343 |
| MYD88 | Myeloid differentiation primary response gene 88 | NM_002468 |
| SHP2** | Phosphotyrosyl-protein phosphatase (SH-PTP2), dominant negative (19) | L03535 |
| LSM1 | Sm-like protein 1 (CASM) | BC001767 |
| IFNG | IFN-γ | NM_000619 |
| MHC2TA | MHC class II transactivator (C2TA) | NM_000246 |
| P2Y6R | Pyrimidinergic receptor P2Y | BC000571 |
| TRADD | TNF receptor type 1-associated death domain protein | BC004491 |
| IL11RA | IL-11 receptor α | BC003110 |
| AKT1* | AKT1-estrogen receptor fusion, constitutively active upon tamoxifen treatment (20) | BC000479 |
| PI3K* | p110 subunit of PI3K, constitutively active (21) | M93252 |
Genes to be overexpressed were introduced into endothelial cells by retroviral transduction. After waiting 48 h to ensure that the encoded proteins were expressed, the cells were incubated for an additional 24 h in the presence of proinflammatory cytokines (IL-1β, TNF-α, or IFN-γ) or medium alone, and levels of readout proteins were measured by ELISA (7). Figs. 1 and 2a show that the levels of readout proteins were a function of the gene being overexpressed and the cell context (presence of proinflammatory cytokines). For example, TNFRSF1A (the gene encoding TNF receptor I) elicited strong responses in IFN-γ-treated and medium-treated cells, whereas RAS* (encoding a constitutively active form of RAS) was most active in the context of IL-1β and TNF-α treatment (Fig. 1). Fig. 2a summarizes the effect of each gene on the level of each readout protein in the four different cell systems (cells plus contexts) used.
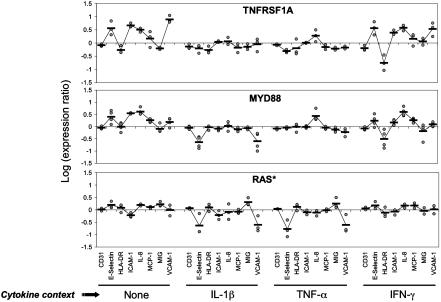
Fig. 1.
Response profiles induced in endothelial cells overexpressing selected genes and stimulated with proinflammatory cytokines. Endothelial cells transduced with retroviral vectors expressing the genes TNFRSF1A, MYD88, and RAS* were treated with IL-1β, TNF-α, IFN-γ, or media alone (Control). The relative levels of readout parameters (CD31, E-selectin, etc.) were measured by ELISA. Data presented are log expression ratios (see Methods) from three (TNFRSF1A, RAS*) or four (MYD88) repeat experiments. The black line representing the overall shape of each profile connects the mean values of the data points.
Analysis of Gene Function by Correlating Responses. We next asked whether the readout profiles could be used to identify functional relationships between the overexpressed genes. We initially performed pairwise comparisons of the readout profiles induced by all overexpressed genes in each individual cell system, measuring the similarity between profiles by using Pearson correlation coefficients (r). The relationships implied by these correlations were visualized by using multidimensional scaling to represent them in two dimensions (Fig. 2 c–f), drawing lines between pairs of genes whose profiles were significantly correlated (details of the statistical techniques used are given in Methods; the individual value of each correlation appears in Table 4).
Strikingly, the readout profiles of genes with closely related functions were indeed strongly correlated, but the strength of the correlation highly depended on the cell context. For example, the profiles produced by MEK1* and MEK2* were strongly correlated in IL-1β- and TNF-α-treated cells (r = 0.95 and 0.98, respectively), but the correlation between the two did not survive significance filtering in IFN-γ-treated or control cells (r = 0.69 and 0.68, respectively). Similarly, the profiles produced by TNFA and TNFB were highly correlated in control cells (r = 0.98), but the correlations in IFN-γ-, IL-1β- and TNF-α-treated cells were not statistically significant (r = 0.77, 0.68, and 0.74, respectively).
Context-dependent correlations were also seen between members of the same signaling pathway. For example, genes encoding members of the NF-κB pathway (9) (including TNF-α, TNF-β, their receptor TNFRSF1A, and the intracellular signaling molecules RIPK1, IKBKB*, and RELA) all produced correlated profiles in control cells and to a lesser extent in IFN-γ-treated cells, but not in cells treated with IL-1β or TNF-α. By contrast, genes encoding members of the RAS/MAPK pathway (10) (including RAS*, RAF*, MEK1*, and MEK2*) produced correlated profiles in IL-1β- and TNF-α-treated cells, but not in cells treated with IFN-γ or control cells. Thus, only some of the possible functional relationships can be mapped in any one cellular context. Conversely, some genes whose products are known to belong to the same signaling pathway (such as IRAK1 and MYD88, which both encode key components of the IL-1 signaling pathway, or IFNG, which induces the transcription of MHC2TA) did not produce significantly correlated responses in any of the individual cell systems tested.
Enhanced Resolution of Biological Activity in Correlations of Combined Profiles. Because the functional relationships observed depended so strongly on the cellular context, we hypothesized that an analysis that simultaneously encompasses the data from multiple context-defined systems should increase the sensitivity of our approach. We therefore concatenated the gene-induced readout profiles from the four cellular systems, yielding for each gene a combined profile comprising 28 normalized parameter readouts (the seven measured parameters from each of the four systems: no cytokine, and IL-1-, TNF-, and IFN-γ-treated endothelial cells). (As examples, the 28 parameter readouts illustrated inside the rectangles in Fig. 1 comprise the multisystem profiles for the TNF receptor, MYD88, or RAS*.) We performed pairwise comparisons of these 28-parameter profiles, measuring the similarity between profiles with Pearson correlations (summarized in Fig. 2b) and representing the implied relationships in two dimensions as before (Fig. 2g).
Virtually all of the relationships observed in individual systems were still apparent, but many other relationships also could be detected, including those between IRAK1 and MYD88 and between IFNG and MHC2TA. The only relationships that were no longer evident were those previously detected between AKT1 and LSM1 in cells treated with IFN-γ and between IL11RA and TNFA or LSM1 in control cells. In fact, AKT1, LSM1, and IL11RA induced very different responses in other cellular contexts, indicating their distinct biological functions: the responses to AKT1 and LSM1 were generally related to those induced by PI3K and members of the NF-κB pathway, respectively, whereas IL11RA induced responses, especially robust in IL-1β- and TNF-α-treated cells, that were not significantly correlated to those produced by any other genes tested. Combining data obtained in multiple cell contexts thus improved the specificity and the sensitivity of the analysis.
Interactions Between Signaling Pathways. One benefit of the greater detail revealed by multisystem BioMAP analysis was a much clearer separation of the genes whose products participate in different pathways. Genes encoding members of the NF-κB and RAS/MAPK pathways, for instance, define separate highly interconnected clusters in Fig. 2g. Even more strikingly, however, additional routes by which pathways can interact could also be detected. As shown in Fig. 2g, MYD88 and IRAK1 were functionally related to genes encoding members of both the NF-κB and RAS/MAPK pathways, suggesting that MYD88 and IRAK1 can interact with both of these pathways.
To explore this observation further, we re-examined the response to MYD88 and genes encoding representative members of the RAS/MAPK and NF-κB pathways (RAS* and TNFRSF1A, respectively) in all four cell systems. As shown in Fig. 1, overexpression of MYD88 and TNFRSF1A increased E-selectin, ICAM-1, IL-8, and VCAM-1 levels in IFN-γ-treated and control endothelial cells, consistent with the known ability of MYD88 and TNFRSF1A to activate the NF-κB pathway (9). By contrast, the response induced by MYD88 in IL-1β-treated cells was similar to that induced by RAS*, the main effect being to inhibit expression of the adhesion molecules VCAM-1 and E-selectin. Overexpression of MYD88 thus appears to stimulate the RAS/MAPK pathway under these conditions. Blocking the RAS/MAPK pathway by treatment with the MEK inhibitor PD098059 reversed the effect of MYD88 or RAS* overexpression (Fig. 3a), confirming that the effects induced by both genes were mediated by the RAS/MAPK pathway. MYD88 (and IRAK1) are known to be involved in IL-1-induced but not TNF-induced signaling (9), and PD098059 indeed had no effect on VCAM-1 expression in TNF-α-treated cells (Fig. 3a). On the other hand, treating TNF-α-treated cells with low doses of IL-1β did reduce the level of VCAM-1 expression (Fig. 3b), as predicted from the effect of MYD88 in IL-1β-treated cells. The inhibitory effect of RAS* could be overcome by overexpressing RELA or IKBKB* (Fig. 3c), indicating that the interaction between the two pathways occurs upstream of IKBKB kinase. A schematic summary is presented in Fig. 3d. Multisystem analysis can thus detect novel functional interelationships between different signaling pathways.
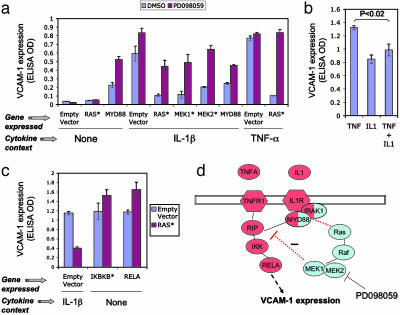
Fig. 3.
IL-1 activates the RAS/MAPK pathway through MYD88, stimulating a MAPK-dependent negative feedback loop modulating endothelial VCAM-1 expression. (a) Endothelial cells overexpressing MYD88, RAS*, MEK1*, or MEK2* were stimulated with IL-1β, TNF-α, or media alone (None), and VCAM-1 expression was measured by ELISA. MEK inhibitor PD098059 (3.7 μM) or DMSO (0.1%) as buffer control was added to cells 1 h before cytokine stimulation. Note that blockade of the RAS/MAPK pathway with PD098059 increases VCAM-1 expression when the pathway is activated through RAS*, MEK1*, MEK2*, or IL-1/MYD88, but not in cells treated with TNF. Error bars indicate SD from triplicate samples. (b) Endothelial cells were stimulated with TNF-α (10 ng/ml), IL-1β (1 ng/ml), or a mixture of TNF and IL-1 (10 ng/ml TNF plus 1 ng/ml IL-1), and VCAM-1 expression was measured by ELISA. Note that IL-1 modulates the VCAM-1 expression induced by TNF. (c) Endothelial cells were cotransduced with RAS* plus empty vector, RAS* plus IKBKB*, or RAS* plus RELA. Expression of individual genes in cotransduced cells was confirmed by quantitative RT-PCR. Cells transduced with RAS* plus empty vector were treated with IL-1β to stimulate the NF-κB pathway. In cells transduced with RAS* plus IKBKB*or RAS* plus RELA cells the NF-κB pathway is stimulated by overexpression of IKBKB* and RELA themselves. Note that RAS* has no effect on VCAM expression in cells expressing IKBKB* or RELA. (d) Schematic diagram of the interactions between the NF-κB and RAS/MAPK pathways in endothelial cells. Genes are color-coded according to the pathways to which they belong (red, NF-κB; blue, RAS/MAPK). The split coloration of MYD88 and IRAK1 genes indicates that they participate in both pathways. Red dotted lines represent pathway interactions revealed by the present study.
Pathway Participants and Mechanisms. BioMAP analysis is also capable of identifying additional participants in signaling pathways and defining their network interactions. For example, the intracellular phosphatase SHP2 is known to have a role in growth factor-induced signaling (11). In our experiments, however, SHP2** showed clear functional similarity to members of the NF-κB pathway (Fig. 2g), reflecting, for example, a similar up-regulation of ICAM-1 and VCAM-1 in control cells and down-regulation of HLA-DR in IFN-γ-treated cells, and suggesting that this protein can regulate NF-κB signaling in endothelial cells. In fibroblasts, SHP2 has indeed been shown to interact physically with the NF-κB complex and is required for the NF-κB-dependent production of IL-6 (11). Similarly, our studies reveal similarity of function of the hypothetical protein MGC3067 to IRAK1, MEK1, and MEK, leading to the testable hypothesis that it plays a role in the RAS/MAPK pathway.
Multisystem BioMAP analysis also revealed additional effects of known genes. TRADD, IL11RA, and P2Y6R, for example, all induced unique profiles that were not significantly related to any known pathway. P2Y6R is a G protein-coupled receptor that binds UDP (12). The precise relationship between this activity and the vascular responses to inflammation remain to be determined, but it is intriguing that P2Y6R also plays a role in monocyte responses to cytokine stimulation (12).
Discussion
The BioMAP technique we describe represents a simplification of existing approaches to systems biology. A very wide range of biological behavior can be examined by overexpressing signaling proteins in primary cells and evaluating the cells' responses in a range of biologically relevant environments. Surprisingly, only a small number of measurements from each perturbed cell state is required to reveal a great deal of information about the function of the perturbing gene product. Using this approach with endothelial cells in several contexts in which inflammatory signaling pathways are activated, we have rapidly reconstructed key pathway relationships of gene products, correctly identifying genes involved in several known inflammatory signaling pathways, and also revealing additional mediators of pathway interactions in endothelial cells. In addition, we have identified genes with unique activities in endothelial responses (e.g., P2Y6R, IL11RA) and others with activities similar to members of the NF-κB or RAS pathways (SHP2 dominant negative and MGC3067), leading to testable hypotheses about their pathway interactions. Thus BioMAP analysis is useful for discovery and characterization of pathways and pathway interactions and for defining key nodal and regulatory points in cell signaling networks.
The BioMAP approach should allow analysis of signaling networks in other endothelial processes (e.g., angiogenesis) and other cells types as well. Application to a given biology will, of course, depend on the empirical selection of systems (cell types and contexts) and parameters that provide a sufficient sensitivity and diversity of responses to perturbations of the physiologic processes being studied. In practice, these are selected iteratively by evaluating different test sets of cell contexts and parameters for their ability to detect and discriminate benchmarking agents (e.g., select genes or functional proteins representing diverse relevant pathways). In the endothelial system we used here, the readout parameters were chosen to detect and discriminate signaling driven by three key cytokine drivers of the inflammatory process, IL-1β, TNF-α, and IFN-γ, that were also used to define three of the cell contexts studied. Nevertheless, this set of parameters also revealed the activity of other known signaling pathways (for example, the RAS/MAPK and PI3K/Akt pathways) and that of additional pathways (such as signaling through the UDP receptor P2Y6R or the IL-11 receptor).
This broad sensitivity may be an innate property of complex cellular systems, in which the level and state of each protein are actually an indirect reflection of the interactions between tens or hundreds of proteins. If we assume that we can experimentally identify both an appropriate set of readout parameters and a sufficient number of distinct contexts to capture the responses induced by overexpressing each gene, as few as 10 independent parameters would be sufficient to generate unique profiles for all human genes. (Assuming that there are 40,000 genes and that a readout parameter can have three states, up, down, or unchanged, allows 310 = 59,049 profiles.) In practice, the breadth of pathway coverage and functional discrimination will depend on the cellular contexts and readout parameters selected.
Our data clearly show that parallel interrogation of cells in multiple contexts allows classification of gene function by using only a small set of readout parameters. From a theoretical perspective, it is clear that each gene product, and the network in which it participates, has evolved not to carry out a function in one particular cell context or environment, rather it has evolved to provide appropriate integration of inputs and outputs from any context the cell may encounter. Thus, the physiologic function of a gene product can only be defined by its effects within multiple cell contexts. The ability of BioMAP analyses to efficiently classify gene function by using only a few readouts shows that multisystem analyses contribute enormously to the biological information content. Indeed, multisystem analyses may be essential for modeling signaling networks from measurements of cell states no matter how many parameters are used.
Our results also suggest why it has been hard to extend earlier successful systems biology studies of yeast to higher eukaryotes. Studies based on whole-cell measurements of gene expression levels under a single set of conditions might well link ICAM, VCAM, E-selectin, IL-8, and MCP-1, because these proteins are coordinately up-regulated in IL-1β- or TNF-treated cells. If an additional input (such as overexpression of IL11RA) is provided, however, the coordinated up-regulation is lost. Overexpression of IL11RA blocks IL-1β-driven VCAM expression and further increases E-selectin levels, but has no effect on the increased expression of ICAM, IL-8, and MCP-1 (Fig. 2a). Thus the observed correlation between output parameters depends on the gene being overexpressed, disrupting pathway classification. In the BioMAP technique, by contrast, such alterations actually increase the discriminatory power of the analysis.
In this study we used specific proteins as readouts, both because these proteins are directly relevant to the biology of vascular inflammation and their levels can be readily measured in high-throughput assays, but other readouts such as transcript levels could certainly be used. Similarly, although we used gene overexpression to perturb selected pathways, it should also be possible to carry out a complementary analysis in which gene activity is suppressed by using small interfering RNA or chemical compounds. Indeed, compound profiling with the BioMAP technique is a powerful tool for characterizing potential drug candidates (E. Kunkel, E.L.B., E.C.B., and I.P., unpublished work).
One of the findings in this study is the inhibition of the NF-κB pathway by IL-1, MYD88, RAS, and MEK in primary endothelial cells (Fig. 3d). Our results suggest that the RAS/MAPK pathway may help to prevent overstimulation of the NF-κB pathway and expression of adhesion molecules in endothelial cells, so moderating immune responses and leukocyte recruitment. By contrast, RAS has been shown to activate the NF-κB pathway in transformed fibroblast and epithelial cell lines (13, 14), suggesting that the same signaling molecule may have different biological roles in different cell types (or in transformed as opposed to primary cells).
The BioMAP technique we describe provides an independent system for classifying gene function, which is complementary to methods relying on sequence homology, protein–protein interactions, or expression profiling. It is well suited to large-throughput analyses, and as such will allow a “discovery science” approach to defining signaling networks in human cells, as shown by the reconstruction of functional relationships in the NF-κB, Ras, and PI3K pathways. By providing critical insights into functional relationships and networks, BioMAP analyses should accelerate the systematic reconstruction of signaling pathways in mammalian cells.
Supplementary Material
Acknowledgments
We thank S. R. Watson, C. Laudanna, L. J. Picker, N. Short, and N. Sigal for critical reading of the manuscript and R. Tibshirani and T. Hastie for help with statistical analysis. This study was supported in part by Small Business Innovation Research Grants R43 AI048255 (to E.L.B.) and R43 AI049048 (to I.P.).
Abbreviations: BioMAP, biologically multiplexed activity profiling; TNF-α, tumor necrosis factor α; ICAM, intercellular adhesion molecule; VCAM, vascular cell adhesion molecule; MIG, monokine induced by IFN-γ; MCP-1, monocyte chemoattractant protein 1; PI3K, phosphatidylinositol 3-kinase; MAPK, mitogen-activated protein kinase.
References
- 1.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Ideker, T., Galitski, T. & Hood, L. (2001) Annu. Rev. Genomics Hum. Genet. 2, 343–372. [DOI] [PubMed] [Google Scholar]
- 3.Roberts, C. J., Nelson, B., Marton, M. J., Stoughton, R., Meyer, M. R., Bennett, H. A., He, Y. D., Dai, H., Walker, W. L., Hughes, T. R., et al. (2000) Science 287, 873–880. [DOI] [PubMed] [Google Scholar]
- 4.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson, J. K. & Wilsin, I. D. (2003) Nat. Rev. Drug Discov. 2, 668–676. [DOI] [PubMed] [Google Scholar]
- 6.Steffen, M., Petti, A., Aach, J., D'haeseleer, P. & Church, G. (2002). BMC Bioinformatics 3, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melrose, J., Tsurushita, N., Liu, G. & Berg, E. L. (1998) J. Immunol. 161, 2457–2464. [PubMed] [Google Scholar]
- 8.Gan, W., LaCelle, M. & Rhoads, R. E. (1998) J. Biol. Chem. 273, 5006–5012. [DOI] [PubMed] [Google Scholar]
- 9.Li, Q. & Verma, I. M. (1998) Nat. Rev. Immunol. 2, 725–734. [DOI] [PubMed] [Google Scholar]
- 10.Kolch, W. (2000) Biochem. J. 351, 289–305. [PMC free article] [PubMed] [Google Scholar]
- 11.You, M., Flick, L. M., Yu, D. & Feng, G. S. (2001) J. Exp. Med. 193, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warny, M., Aboudola, S., Robson, S. C., Sevigny, J., Communi, D., Soltoff, S. P. & Kelly, C. P. (2001) J. Biol. Chem. 276, 26051–26056. [DOI] [PubMed] [Google Scholar]
- 13.Kim, B.-Y., Gaynor, R. B., Song, K., Dritschilo, A. & Jung, M. (2002) Oncogene 21, 4490–4497. [DOI] [PubMed] [Google Scholar]
- 14.Millan, O., Ballester, A., Castrillo, A., Oliva, J. L., Traves, P. G., Rojas, J. M. & Bosca, L. (2003) Oncogene 22, 477–483. [DOI] [PubMed] [Google Scholar]
- 15.Mercurio, F., Zhu, H., Murray, B. W., Shevchenko, A., Bennett, B. L., Li, J., Young, D. B., Barbosa, M., Mann, M., Manning, A. & Rao, A. (1997) Science 278, 860–866. [DOI] [PubMed] [Google Scholar]
- 16.Mansour, S. J., Matten, W. T., Hermann, A. S., Candia, J. M., Rong, S., Fukasawa, K., Vande Woude, G. F. & Ahn, N. G. (1994) Science 265, 966–970. [DOI] [PubMed] [Google Scholar]
- 17.Bosch, E., Cherwinski, H., Peterson, D. & McMahon, M. (1997) Oncogene 15, 1021–1033. [DOI] [PubMed] [Google Scholar]
- 18.Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. (1997) Cell 88, 593–602. [DOI] [PubMed] [Google Scholar]
- 19.Aoki, Y., Huang, Z., Thomas, S. S., Bhide, P. G., Huang, I., Moskowitz, M. A. & Reeves, S. A. (2000) FASEB J. 14, 1965–1973. [DOI] [PubMed] [Google Scholar]
- 20.Luo, Z., Fujio, Y., Kureishi, Y., Rudic, R. D., Daumerie, G., Fulton, D., Sessa, W. C. & Walsh, K. (2000) J. Clin. Invest. 106, 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seasholtz, T. M., Zhang, T., Morissette, M. R., Howes, A. L., Yang, A. H. & Brown, J. H. (2001) Circ. Res. 89, 488–495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.